Abstract
Serratia marcescens has long been recognized as an important opportunistic pathogen, but the underlying pathogenesis mechanism is not completely clear. Here, we report a key pathogenesis pathway in S. marcescens comprising the RssAB two-component system and its downstream elements, FlhDC and the dominant virulence factor hemolysin ShlBA. Expression of shlBA is under the positive control of FlhDC, which is repressed by RssAB signaling. At 37°C, functional RssAB inhibits swarming, represses hemolysin production, and promotes S. marcescens biofilm formation. In comparison, when rssBA is deleted, S. marcescens displays aberrant multicellularity favoring motile swarming with unbridled hemolysin production. Cellular and animal infection models further demonstrate that loss of rssBA transforms this opportunistic pathogen into hypervirulent phenotypes, leading to extensive inflammatory responses coupled with destructive and systemic infection. Hemolysin production is essential in this context. Collectively, a major virulence regulatory pathway is identified in S. marcescens.
The Gram-negative bacterium Serratia marcescens is an important opportunistic pathogen that causes a wide range of diseases and clinical presentations with high morbidity (25). S. marcescens frequently causes outbreaks in intensive and neonatal care units, and the occurrence of multiple-antibiotic-resistant strains has further exacerbated clinical treatment difficulties (3, 39). Despite years of study, the mechanism of pathogenesis of S. marcescens and why it behaves as an opportunistic pathogen remain poorly understood. Unraveling the underlying mechanism of pathogenesis is thus very important for developing strategies to prevent and treat S. marcescens infection.
The hemolysin ShlA was shown to be a dominant virulence factor in S. marcescens pathogenesis using a murine lung infection model (35). ShlA is responsible for the hemolytic and cytotoxic effects on erythrocytes and cultured cells, with the aid of an outer membrane protein, ShlB (28, 29, 47, 53, 54). ShlA also contributes to the release of inflammatory mediators, increases uropathogenicity, and triggers microtubule-dependent invasion of S. marcescens into epithelial cells (27, 30, 34, 40). However, the mechanism by which the expression of shlA is regulated, especially in response to any bacterial signaling system control, remains uncharacterized. Only one reported study has indicated that iron is involved in the regulation of shlBA expression in S. marcescens (46).
S. marcescens exhibits swarming, which is recognized as a highly coordinated multicellular surface migration behavior (24, 51, 62) that is correlated with virulence capability, antibiotic resistance, and hemolysin production in other bacteria (1, 17, 44). S. marcescens swarms on Luria-Bertani (LB) agar surfaces at 30°C, but not at 37°C (36). Our previous studies showed that activation of a bacterial two-component system, RssAB, comprising a sensor kinase, RssA, and a response regulator, RssB, inhibited swarming and reduced hemolysin production in S. marcescens CH-1 at 37°C (36, 57, 63). In contrast, loss of rssBA resulted in a precocious swarming phenotype and a significant increase in hemolytic activity (36). These findings strongly indicate the presence of an important relationship between RssAB signaling, swarming regulation, and hemolysin production. However, subsequent experiments failed to show evidence of direct binding to the shlBA promoter region and repression of shlBA expression by phosphorylated RssB (63), indicating that another factor is involved in RssAB-regulated hemolysin production.
Recently, we showed that activated RssAB signaling directly represses expression of flhDC and swarming in S. marcescens CH-1 at 37°C (57). FlhDC is known to be a major checkpoint of swarmer cell differentiation in S. marcescens MG1 (formerly Serratia liquefaciens MG1) (13) and is also involved in early stages of biofilm formation in Escherichia coli (48). In addition, studies in Proteus mirabilis and Xenorhabdus nematophila indicated that FlhDC positively regulates hemolysin production (10, 17). Notably, swarming motility and biofilm formation are inversely related (4, 41), and both of them participate in modulation of virulence capability and pathogenesis of bacterial infections (32, 45). Since hemolysin ShlA is critical for S. marcescens pathogenesis (35) and its overexpression is correlated with precocious swarming (36) and defective biofilm formation (58), we hypothesized that RssAB might regulate S. marcescens pathogenesis through coordination of multicellularity of swarming, biofilm formation, and hemolysin production in conjunction with FlhDC.
This study investigated the in vitro and in vivo pathogenesis mechanism of an S. marcescens clinical isolate. Special attention was given to the role of hemolysin in pathogenesis and the underlying regulatory mechanism of shlBA expression by RssAB at 37°C. Repression of shlBA expression and hemolysin production by RssAB signaling was found to depend on FlhDC. Loss of rssBA in S. marcescens resulted in multiple aberrant multicellular behaviors coupled with elevated hemolysin production, leading to increased destruction of bronchial epithelium cells, high mortality in a rat acute-pneumonia model, and even systemic infection in a rat sublethal-pneumonia model. In contrast, when shlBA was deleted, S. marcescens showed little virulence. Together, these findings demonstrate that a hierarchical regulatory relationship exists between RssAB, FlhDC, and ShlBA in S. marcescens. The RssAB-FlhDC-ShlBA pathway plays a major role in controlling S. marcescens multicellularity and pathogenesis.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains and plasmids used in this study are summarized in Table 1. Bacteria were routinely cultured with agitation in LB broth (Difco) at 37°C or 30°C with adequate antibiotics when necessary. A clinical isolate, S. marcescens CH-1, was used as the wild-type strain. All mutants in which the specific gene locus was replaced with a streptomycin resistance (Smr) or gentamicin resistance (Gmr) cassette were generated through homologous recombination.
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Genotype or relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Invitrogen |
| CC118 | λ-pir lysogen [Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1]; permissive host for suicide plasmids requiring Pir protein | 26 |
| S17-1 | λ-pir lysogen [thi pro hsdR hsdM+recA RP4 2-Tc::Mu-Km::Tn7 (Tpr Smr)]; permissive host able to transfer suicide plasmid pDM4 to recipient cells via conjugation | 26 |
| S. marcescens strains | ||
| CH-1 | Wild-type strain; clinical isolate | 36 |
| ΔrssBA | rssBA knockout mutant; Gmr | This study |
| ΔshlBA | shlBA knockout mutant; Smr | This study |
| ΔrssBA-shlBA | rssBA and shlBA knockout mutant; Gmr Smr | This study |
| ΔflhDC | flhDC knockout mutant; Smr | This study |
| ΔrssBA-flhDC | rssBA and flhDC knockout mutant; Gmr Smr | This study |
| Plasmids | ||
| pGEM-T Easy | TA cloning vector; Ampr | Promega |
| pBluescript II SK(+/−) | Cloning vector; Ampr | Stratagene |
| pUT-Sm | Suicide plasmid containing mini-Tn5 (Smr); requires Pir protein for replication | 11 |
| pUT-Km1 | Suicide plasmid containing mini-Tn5 (Kmr); requires Pir protein for replication | 11 |
| pACYC184 | Cloning vector; Tcr Cmr | 5 |
| pEGFP-C3 | GFPmut1 variant; cytomegalovirus (CMV) promoter; f1 origin; simian virus 40 (SV40) origin; pUC origin; kanamycin resistance gene | BD Bioscience Clontech |
| pBAD18-Cm | pBAD18; arabinose regulation; Cmr | 23 |
| pBAD24-Amp | pBAD24; arabinose regulation; Ampr | 23 |
| pBAD24EGFP::Sm | pBAD24 expressing egfp | This study |
| pRepoEGFP | pACYC184 containing promoterless egfp | This study |
| pRepoEGFP-shlB | pRepoGFP with shlB promoter | This study |
| pRssAB | pACYC184 containing full-length rssBA with promoter | This study |
| pFlhDC | pBAD18 expressing flhDC | This study |
| pshlBAKO | Derived from pUT; contains shlB and shlA homologous region; Smr Kmr | This study |
| prssBAKO | Derived from pUT; contains rssB and rssA homologous region; Gmr Smr | This study |
| pflhDCKO | Derived from pUT; contains flhD and flhC homologous region; Smr Kmr | This study |
Amp, ampicillin; Sm, streptomycin; Cm, chloramphenicol; Km, kanamycin; Gm, gentamicin; Tc, tetracycline.
Enzymes, chemicals, reagents, and primers.
DNA restriction endonucleases and modifying enzymes were purchased from Roche (Germany) and New England Biolabs. DNA polymerase and PCR-related products were purchased from Stratagene, Takara (Japan), and Perkin Elmer. F-12K medium and RPMI 1640 medium without phenol red and culture-related products were purchased from Invitrogen. Other laboratory grade chemicals were purchased from Sigma, Merck (Germany), and BDH (United Kingdom). The primers used in this study are summarized in Table 2.
TABLE 2.
PCR primers used in the study
| Primer | Sequence (5′-3′)a |
|---|---|
| shlBF | AGATCTAATCACCGCCTTGACGCT |
| shlBR | AAGCTTCTGATGCACTGTTCCGGC |
| shlAF2 | AAGCTTGAGTGAACAAAGTGGCCGAC |
| shlAR | GAATTCTCAACGCCCTGTTTCGCC |
| SalFlhDCKOF1 | GTCGACTAATGGTTCGGGGGTAGAGTT |
| HindFlhDCKOR1 | AAGCTTCATATTCCCCATATTCCCCA |
| HindFlhDCKOF2 | AAGCTTGGCCGATATTATTCCTCAACTGC |
| EcoRFlhDCKOR2 | GAATTCCGCCGATAATCAGAAACTCC |
| FlhDCCF1 | GGAGCTCTCTGTCGGGATGGGGAATAT |
| FlhDCCR1 | GTCTAGATCACACGTCGGTATTCATTGCC |
| FlhDCRTF | GCCAAAAGGAATGTTACCGT |
| FlhDCRTR | CAGTTGCGGCGAAAGTTTAC |
| 16SrDNAF | AACTGGAGGAAGGTGGGGAT |
| 16SrDNAR | AGGAGGTGATCCAACCGCA |
| SmaEGFPF | TCCCCCGGGATGGTGAGCAAGGGCGAGGAGCTG |
| XbaEGFPRstop | GCTCTAGAGGTTACTTGTACAGCTCGTCCATGCC |
| HindpBAD24SDF | CCCAAGCTTAGGAGGAATTCACCATGGTAGCCG |
| PstEGFPRstop | AAACTGCAGTTACTTGTACAGCTCGTCCATGCC |
| PstrrnBTF | AAACTGCAGGCTGTTTTGGCGGATGAGAGAAG |
| XbarrnBTR | GCTCTAGAGTTTGTAGAAACGCAAAAAGG |
| SalrrnBTF | CGCGTCGACGGCTGTTTTGGCGGATGAGAGAAG |
| LM13R | GGAAACAGCTATGACCATGATTACGCCAAG |
| ShlBPF | CGGATCCTCGTATTTCCCACTGGTCGG |
| ShlBPR | CCAAGCTTACTCGCCCATCGACATATCCC |
The underlined sequences represent cutting sites of restriction endonucleases.
RT-PCR assay.
Total bacterial RNA was purified by using a Trizol kit (Invitrogen). To analyze the transcript amounts of flhDC and shlBA genes, S. marcescens CH-1, S. marcescens ΔrssBA, S. marcescens CH-1 harboring pFlhDC (CH-1/pFlhDC), S. marcescens ΔflhDC, and S. marcescens ΔrssBA-flhDC were cultivated in LB broth with agitation at 37°C for 3 and 4 h, respectively. To induce flhDC, 0.1% arabinose was added to the LB broth medium of CH-1/pFlhDC at the second hour. The relative amounts of transcripts were evaluated by reverse transcription (RT)-PCR according to the instructions for the SuperScript III First-Strand Synthesis System (Invitrogen). The targets of flhDC, shlBA, and 16S rRNA (internal control) were amplified using the primer pairs FlhDCRTF and FlhDCRTR, shlBF and shlBR, and 16SrDNAF and 16SrDNAR, respectively. Electrophoresis images were edited with Image J (NIH). The transcript amounts of shlBA and flhDC for each strain were semiquantified by Image J and are shown as relative mRNA expression, which was normalized to that of 16S rRNA and compared to that of S. marcescens CH-1.
Swarming assay.
The swarming motility of each S. marcescens strain was analyzed as previously described (36). Swarmer cells were defined as the bacteria around the outer layer after swarming at 37°C.
Biofilm attachment assay.
A biofilm attachment assay was performed as described previously (58). Briefly, an overnight culture of each strain was diluted 1:100 in 150 μl fresh LB broth containing 0.5% glucose onto a 96-well polyvinylchloride (PVC) microtiter plate (BD), followed by incubation at 37°C without agitation for 24 h. After the supernatant was removed, the well was washed twice with 200 μl phosphate-buffered saline (PBS) and stained with 200 μl of crystal violet (1%) for 20 min. After the crystal violet was removed, the stained biofilm was imaged and extracted with 95% ethanol. The quantification of attachment was estimated as the absorbance at 630 nm (A630). Biofilm cells were defined as the bacteria attached to the PVC microtiter plate after it was washed twice with PBS. Each experiment was performed in triplicate independently. Data were expressed as means with standard deviations.
Hemolysis assay.
Hemolytic activity was determined as described elsewhere (29). Briefly, an overnight culture of each S. marcescens strain was diluted 1:100 in fresh LB broth and cultivated for 4 h with agitation at 37°C. For the induction of flhDC expression, arabinose was added to a final concentration of 0.1% at the second hour. Equal numbers of bacteria were collected and diluted in PBS, followed by incubation with 8% sheep erythrocytes for 15 min at 22°C. After centrifugation, the released hemoglobin was determined as the absorbance at 405 nm. The absorbance of each sample was normalized to the total released hemoglobin from the sheep erythrocytes incubated in distilled water and expressed as percent hemolysis. To determine the hemolytic activity of swarmer or biofilm cells, a swarming assay as described previously (36) or a biofilm attachment assay followed by the collection of swarmer or biofilm cells, respectively, was performed for each strain. After quantification of the same number of bacteria, the hemolytic activity of each strain was determined. Each experiment was performed in triplicate independently. Data were expressed as means with standard deviations.
Construction of enhanced green fluorescent protein (EGFP)-expressing plasmids.
To construct the vector pRepoEGFP, which was used to detect the promoter activity of shlB, the egfp gene was first amplified from the plasmid pEGFP-C3 (BD Biosciences Clontech) using the primer pair SmaEGFPF-XbaEGFPRstop, digested with SmaI and XbaI, and ligated into the SmaI and XbaI sites of pBAD24 to form pBAD24EGFP. The primer pair HindpBAD24SDF-PstEGFPRstop was used to amplify SD-EGFP containing a high-efficiency Shine-Dalgarno box with pBAD24EGFP. The PCR product was cloned into the HindIII-PstI site of plasmid pBAD24 to form pBAD24SD-EGFP(R). The primer pair PstrrnBTF-XbarrnBTR was used to amplify the terminator, rrnBt, with pBAD24 and cloned into the PstI-XbaI sites of pBAD24SD-EGFP(R) to form pBAD24SD-EGFP-rrnBT(R) and into the SmaI site of pBluescript II SK (Stratagene). pBAD24SD-EGFP-rrnBT(R) was then digested by HindIII and XbaI to obtain SD-EGFP-rrnBT and was cloned into the HindIII and XbaI sites of pACYC184 to generate pSD-EGFP-rrnBT. The primer pair SalrrnBTF-LM13R was used to amplify another rrnBt with pBluescript II SK containing rrnBt and cloned into the SalI-BamHI sites of pSD-EGFP-rrnBT to obtain pRepoEGFP. pRepoEGFP-shlB was constructed through the ligation of the shlB promoter region, which was amplified using Pfu polymerase via the primer pair ShlBPF and ShlBPR, into the BamHI and HindIII sites of pRepoEGFP. pBAD24EGFP::Sm was derived from pBAD24EGFP with the insertion of an Smr cassette into the HindIII site
Cell culture.
BEAS-2B human bronchial epithelial cells (ATCC CRL-9609) were obtained from the Bioresource Collection and Research Center (BCRC), Taiwan. They were maintained in F-12K medium (Gibco) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, and penicillin-streptomycin (complete F-12K medium) at 37°C under a 5% CO2 atmosphere.
Cellular cytotoxicity assay.
BEAS-2B cells were seeded at 5 × 104 cells/well in complete F-12K medium in a 96-well microplate and incubated for 24 h at 37°C under a 5% CO2 atmosphere. The medium was replaced with basal RPMI 1640 medium (Gibco) before infection. The bacteria were cultivated to mid-log phase, pelleted by centrifugation, and washed twice with PBS. The bacteria were then diluted in basal RPMI 1640 medium to infect the cells at a multiplicity of infection (MOI) of 10 for 4 h. The cytotoxicity was determined by measuring the release of lactate dehydrogenase (LDH) using the CytoTox 96 NonRadioactive Cytotoxicity Assay (Promega) and was expressed as percent cytotoxicity, calculated as the ratio of the average experimental values obtained from each bacterial strain to those obtained from cells treated with the lysis solution provided in the kit. The assay was performed in triplicate and repeated three times independently. The percent cytotoxicity was expressed as the average of the experiments with the standard deviation.
Cellular invasion assay.
BEAS-2B cells were seeded at 1 × 105 cells/well in complete F-12K medium in a 24-well plate and incubated for 24 h at 37°C under a 5% CO2 atmosphere. Before infection, the medium was replaced with basal RPMI 1640 medium. Each strain prepared for the cytotoxicity assay was inoculated onto a cell monolayer at an MOI of 5 to infect the cells for 2 h. The antibiotic protection assay was performed as described previously (31) with some modifications. The use of gentamicin, streptomycin, or chloramphenicol at 100 μg/ml in the basal RPMI 1640 medium killed extracellular S. marcescens cells during an additional 2-hour incubation. The cells were then washed with PBS three times and lysed with 1% Triton X-100 to release viable intracellular bacteria. The supernatants were serially diluted and plated on LB plates to determine the CFU. The data were expressed as means with standard deviations. Fluorescence microscopy was applied to visualize the invasion of S. marcescens into cells. pBAD24EGFP::Sm was introduced into S. marcescens CH-1, S. marcescens ΔrssBA, and S. marcescens ΔrssBA harboring pRssAB (ΔrssBA/pRssAB). All bacterial strains were cultivated to mid-log phase in LB broth supplemented with 0.1% arabinose to induce egfp expression in pBAD24EGFP::Sm. BEAS-2B cells were cultivated on sterile round coverslips (12 mm) in 24-well plates in complete F-12K medium for 24 h at 37°C under a 5% CO2 atmosphere, followed by replacement of the medium with RPMI 1640 medium before infection. The cells were infected by bacteria at an MOI of 5 for 2 h and washed twice with PBS. To locate the cell membrane and nucleus, the cells were stained with 4 μM FM 4-64 (Molecular Probes) and 10 μM Hoechst 33342 (Molecular Probes) for 5 min at 37°C under a 5% CO2 atmosphere. The stained cells and bacteria expressing EGFP were imaged on a Leica DM2500 fluorescence microscope (Germany) equipped with a 63× oil immersion objective. The images were captured with a Spot RT3 Slider charge-coupled device (CCD) camera (Diagnostic Instruments Inc.), edited, and merged with Spot (Diagnostic Instruments Inc.). To quantify bacteria invading cells, the bacteria in 200 cells were counted. Each result was calculated as an average of triplicate tests with the standard deviation.
Promoter assay.
S. marcescens CH-1 and S. marcescens ΔrssBA harboring pRepoEGFP (as a background control) or pRepoEGFP-shlB, respectively, were used to infect BEAS-2B cells at an MOI of 5 for 1 h. The supernatants with bacteria were collected for fluorescence analysis by Victor III (Perkin Elmer). Promoter activity was determined with a fluorometer as relative florescence units (RFU), with excitement at 485 nm and detection at 535 nm. Each result was calculated as an average of triplicate tests with the standard deviation.
Rat acute-pneumonia model.
All experiments for animal studies were approved by the National Laboratory Animal Center in Taiwan and followed the guidelines of the Institutional Animal Care and Use Committee in accordance with the Taiwan law for animal protection.
Male Sprague-Dawley (SD) rats (6 to 8 weeks old; 150 to 200 g) were purchased from the National Laboratory Animal Center, Taiwan, and allowed to acclimatize for at least 7 days before use. Food and water were supplied ad libitum. The rat acute-pneumonia model basically followed the protocols described previously (38) with minor modifications. Briefly, bacteria were cultivated in LB broth at 37°C with agitation to mid-log phase. The bacterial suspensions were washed with normal saline and diluted to 1 × 109 CFU in 1 ml of normal saline for rat inoculation. The rats were lightly anesthetized with halothane. The trachea was exposed in a sterile atmosphere, and 100 μl of S. marcescens inoculum was administered intratracheally (i.t.) using a sterile 26-gauge needle. To perform the survival study, 12 rats per group were infected i.t. with S. marcescens CH-1, S. marcescens ΔrssBA, S. marcescens ΔrssBA-shlBA, or normal saline (vehicle) and closely monitored for 1 day. The survival rate was calculated from three independent experiments. After the rats were sacrificed, the whole lungs from each group were dissected and imaged. Lung tissues were fixed in 10% formalin for 24 h and embedded in paraffin; 4-μm sections were cut, stained with hematoxylin and eosin (H&E), and processed for light microscopy. Images were captured and analyzed with a Leica DM 2500 and a Spot RT3 Slider CCD camera.
Quantification of the bacterial burden in lungs and BAL fluid.
The experimental protocols described by Wang et al. (61) were basically followed. Twenty-four hours after i.t. inoculation, rats were sacrificed and the thoracic cavity was exposed. The left lung was removed intact with the endotracheal tube in place and lavaged twice with 3 ml of normal saline. The lungs were perfused with normal saline, and the right lung was excised; special care was taken to exclude hilar tissues or proximal bronchi. The right lung was minced, suspended in 10 ml of normal saline, and homogenized; 10-μl of aliquots from each specimen were serially diluted and plated on LB agar plates containing tetracycline (12.5 μg/ml), followed by incubation at 37°C for 24 h. Bacteria were counted as the average number of CFU per gram of lung and CFU per ml of bronchoalveolar lavage (BAL) fluid from three independent experiments with the standard deviation.
BAL fluid cellularity analysis.
Twenty-four hours postinfection, cells harvested from 100 μl of BAL fluid were subjected to hemocytometer (Hausser Scientific) analysis for total leukocytes, and the differential cell populations of neutrophils, eosinophils, basophils, monocytes, and lymphocytes were expressed as percentages. Data were expressed as averages of three independent experiments with the standard deviations.
Cytokine measurement.
Concentrations of tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β), and IL-6 in BAL fluid and sera were determined by enzyme-linked immunosorbent assay (ELISA) (Biosource, United Kingdom) according to the manufacturer's instructions.
Rat sublethal-pneumonia model.
The procedures of infection in the rat sublethal-pneumonia model were basically similar to those used in the rat acute-pneumonia model with some modifications. Six rats from each group were i.t. inoculated with 1 × 105 CFU of each strain (S. marcescens CH-1, S. marcescens ΔrssBA, S. marcescens ΔshlBA, and S. marcescens ΔrssBA-shlBA) in 100 μl normal saline. The normal saline was used as a vehicle. Body weight was monitored daily, and the rats were sacrificed 3 days postinfection. Sera collected from the abdominal aorta and BAL fluids were used for subsequent determination of the bacterial loads and cytokines. The lungs, livers, and spleens of the rats were dissected and imaged. To analyze the bacterial load, the organs were weighed and homogenized in 500 μl of PBS. Homogenates of each organ, serum, or BAL fluid was plated on LB agar plates containing tetracycline (12.5 μg/ml) and incubated at 37°C for 24 h. The number of bacteria was expressed as the average number of CFU per gram of organ or CFU per ml of serum or BAL fluid from three independent experiments with the standard deviations.
RESULTS
RssAB represses hemolytic activity through downregulation of flhDC expression.
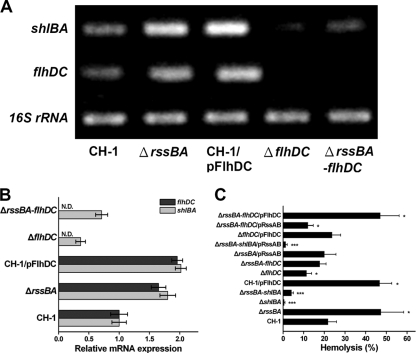
To evaluate whether RssAB regulates shlBA expression through control of flhDC expression at 37°C, we constructed rssBA (ΔrssBA), flhDC (ΔflhDC), and rssBA-flhDC (ΔrssBA-flhDC) knockout mutants derived from the parent strain, S. marcescens CH-1, to analyze the transcription levels of flhDC and shlBA by RT-PCR. Elevated mRNA levels of flhDC and shlBA were found in S. marcescens ΔrssBA compared with S. marcescens CH-1 cells (Fig. 1 A and B). When flhDC was further deleted in S. marcescens ΔrssBA to form S. marcescens ΔrssBA-flhDC, the shlBA expression was significantly reduced to a level lower than that of S. marcescens CH-1 (Fig. 1A and B). In contrast, S. marcescens ΔflhDC had the lowest shlBA mRNA level (Fig. 1A and B). Complementation of wild-type rssBA in S. marcescens ΔrssBA or flhDC in S. marcescens ΔflhDC restored the mutant shlBA expression level to that of S. marcescens CH-1 (C. S. Lin and H. C. Lai, unpublished data). Notably, overexpression of flhDC in S. marcescens CH-1 (CH-1/pFlhDC) also increased the shlBA mRNA level, similar to that observed in S. marcescens ΔrssBA (Fig. 1A and B). Together, these results indicate that RssAB represses shlBA expression through the inhibition of flhDC expression.
FIG. 1.
RssAB represses hemolysin synthesis and hemolytic activity through downregulation of flhDC expression. (A) The mRNA expression levels of shlBA and flhDC in different S. marcescens strains grown to mid-log phase in LB broth at 37°C were determined by RT-PCR as shown on the agarose gel. (B) The transcript amount was semiquantified by Image J and is shown as relative mRNA expression, which is normalized to the 16S rRNA expression level and compared to that of S. marcescens CH-1. N.D., not detectable. (C) The percentage of relative hemolytic activity of each S. marcescens strain grown in LB broth at 37°C was determined by hemolysis assay. All results are shown as the average of three independent experiments, with the standard deviations indicated by error bars. *, P < 0.05, and ***, P < 0.001 in comparison to CH-1 (Student's t test).
The role of the RssAB-FlhDC-ShlBA pathway in hemolytic activity was evaluated. As expected, deletion of shlBA or rssBA to form S. marcescens ΔshlBA and S. marcescens ΔrssBA, respectively, resulted in the lowest and highest hemolytic activities among all S. marcescens cells tested (Fig. 1C). The mutant phenotypes were restored to the level of S. marcescens CH-1 through complementation by rssBA with its own promoter (Fig. 1C) and shlBA (Lin and Lai, unpublished). Further deletion of shlBA in S. marcescens ΔrssBA (ΔrssBA-slhBA) significantly reduced hemolytic activity to a level close to that of S. marcescens ΔshlBA (Fig. 1C), and reintroduction of shlBA into S. marcescens ΔrssBA-shlBA restored the hemolytic activity to the level of S. marcescens ΔrssBA (Lin and Lai, unpublished), indicating that shlBA is required for the hyperhemolytic activity of S. marcescens ΔrssBA. Furthermore, deletion of flhDC in S. marcescens ΔrssBA to form S. marcescens ΔrssBA-flhDC significantly reduced hemolytic activity (Fig. 1C). In comparison, rssBA complementation (ΔrssBA-flhDC/pRssAB) further inhibited hemolytic activity and overexpression of flhDC (ΔrssBA-flhDC/pFlhDC) significantly increased hemolytic activity (Fig. 1C), indicating that the hyperhemolytic activity observed in S. marcescens ΔrssBA relies on the presence of flhDC. When flhDC was overexpressed in S. marcescens CH-1 (CH-1/pFlhDC), the elevated hemolytic activity was similar to that of S. marcescens ΔrssBA, while loss of flhDC in S. marcescens CH-1 resulted in decreased hemolytic activity that could be rescued by flhDC (Fig. 1C). Thus, RssAB represses hemolytic activity through downregulation of flhDC expression.
Roles of RssAB, FlhDC, and ShlBA in biofilm formation and swarming.
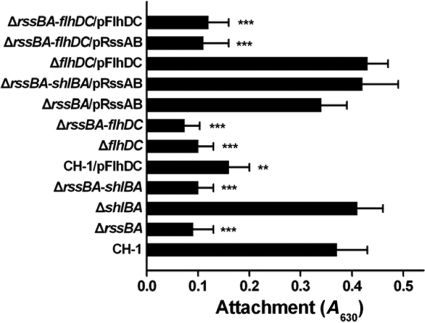
Surface-associated multicellular behaviors, such as swarming and biofilm formation, display advantages that protect bacteria against harsh environmental conditions and are correlated with each other and accompanied by virulence factor production and the ability to infect the host (60). Based on the correlation of rssBA disruption with precocious swarming and derepression of flhDC expression in S. marcescens CH-1 (36, 57), we further examined whether RssAB and FlhDC influence biofilm formation in S. marcescens CH-1. S. marcescens ΔrssBA displayed reduced biofilm formation on PVC microtiter plates (Fig. 2), which could be rescued to the wild-type level by restoration of rssBA with its own promoter, suggesting RssAB is important for biofilm formation in S. marcescens CH-1. Interestingly, overexpression of flhDC in S. marcescens CH-1 also resulted in defective biofilm attachment similar to that of S. marcescens ΔrssBA (Fig. 2). These findings led us to consider whether the reduced biofilm attachment observed in S. marcescens ΔrssBA is, at least in part, attributable to derepression of flhDC, which increases flagellar motility and reduces biofilm attachment (50). In contrast, S. marcescens ΔrssBA-flhDC also showed reduced biofilm attachment (Fig. 2), indicating that proper expression of flhDC is important for early stages of biofilm formation in S. marcescens. By comparison, deletion of shlBA in S. marcescens CH-1 and S. marcescens ΔrssBA, respectively, did not significantly affect their parent strains' biofilm formation abilities (Fig. 2). Taken together, these results indicate that both RssAB and FlhDC, but not ShlBA, play vital roles in biofilm formation in S. marcescens CH-1. We further evaluated swarming motility in these mutants. In contrast to the precocious swarming observed in S. marcescens ΔrssBA and S. marcescens ΔrssBA-shlBA at 37°C, S. marcescens CH-1, S. marcescens ΔshlBA, S. marcescens ΔflhDC, and S. marcescens ΔrssBA-flhDC did not swarm, while artificial induction of flhDC expression in S. marcescens CH-1(pFlhDC) resulted in elevated swarming motility (Lin and Lai, unpublished), indicating an important role of flhDC expression in S. marcescens swarming, which is consistent with other studies (59).
FIG. 2.
RssAB and FlhDC are required for efficient biofilm attachment. Shown is the quantification of bacterial attachment efficacy by microtiter plate assay. The results were expressed as the A630. Expression of flhDC in S. marcescens CH-1/pFlhDC was induced by supplementation with 0.1% arabinose. All results are shown as the average of three independent experiments, with the standard deviation indicated by error bars. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 in comparison to CH-1 (Student's t test).
RssAB-FlhDC-ShlBA controls virulence capability against human bronchial epithelial cells.
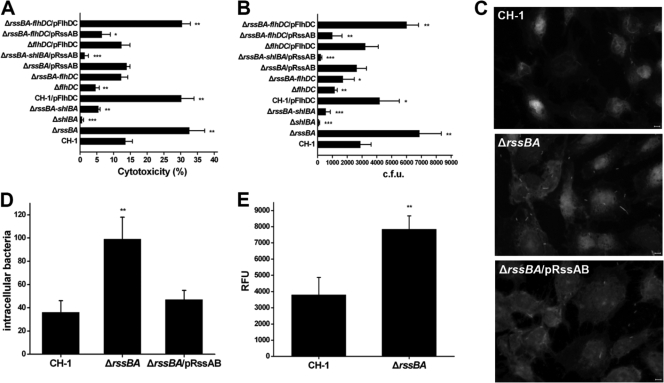
As S. marcescens shows cytotoxic effects on many cultured cells exerted by the dominant virulence factor ShlA (29, 35), which is under RssAB-FlhDC control (Fig. 1), we further examined whether the RssAB-FlhDC pathway modulates the cytotoxic capability of S. marcescens CH-1 through the regulation of hemolysin ShlA. The human bronchial epithelial cell line BEAS-2B was used. Consistent with its high hemolytic potency, S. marcescens ΔrssBA displayed the highest cytotoxic effect against BEAS-2B cells (Fig. 3 A). Nevertheless, further deletion of shlBA in S. marcescens ΔrssBA significantly reduced the cytotoxic effect to a level almost as low as that of S. marcescens ΔshlBA (Fig. 3A). Furthermore, S. marcescens ΔflhDC showed about half the cytotoxic activity of S. marcescens CH-1, while overexpression of flhDC in S. marcescens CH-1 cells resulted in a cytotoxic effect similar to that of S. marcescens ΔrssBA (Fig. 3A). S. marcescens ΔrssBA-flhDC also possessed a cytotoxic effect similar to that of S. marcescens CH-1, and the cytotoxic effects of S. marcescens ΔrssBA and S. marcescens ΔrssBA-flhDC could be restored through complementation with rssBA and flhDC, respectively (Fig. 3A). Together, these results indicate that the RssAB-FlhDC-ShlBA pathway manipulates the cytotoxic potency of S. marcescens CH-1 against BEAS-2B cells.
FIG. 3.
Deletion of rssBA increases S. marcescens cellular cytotoxicity, invasive activity, and shlBA promoter activity. (A) Each S. marcescens strain was cultivated in LB broth to mid-log phase at 37°C before infection of the human bronchial epithelial cell line BEAS-2B at an MOI of 10 for 4 h. The percent cytotoxicity was determined by release of lactate dehydrogenase. (B) Invasion of BEAS-2B cells by different S. marcescens strains was determined by an antibiotic protection assay. The bacterial growth conditions were the same as those used in the cytotoxicity assay. The results are expressed as the average number of CFU from three independent experiments. (C) Each bacterial strain harboring pBAD24EGFP::Sm was cultivated in LB broth containing 0.1% arabinose to mid-log phase and harvested. Bacteria at an MOI of 5 were used to infect BEAS-2B cells cultured on a coverslip at 37°C for 2 h. The cells were washed twice with PBS and stained with 5 μM FM 4-64 and 10 μM Hoechst. Under fluorescence microscopy, bacteria were localized and imaged. Scale bars, 5 μm. (D) The bacteria invading cells were counted in 200 BEAS-2B cells. (E) The shlBA promoter activities of S. marcescens CH-1 and S. marcescens ΔrssBA that harbored pRepoEGFP or pRepoEGFP-shlB were measured by fluorometer while they were infecting BEAS-2B cells. The results are shown as RFU. The data are shown as the mean from three independent experiments, with the standard deviations indicated by error bars. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 in comparison to CH-1 (Student's t test).
Since hemolysin and hemolytic activity are required for invasion, virulence, and spreading within hosts (30, 35, 40), we further analyzed the correlation of the RssAB-FlhDC-ShlBA pathway in S. marcescens CH-1 with potency of invasion into BEAS-2B cells. Antibiotic protection assay (31) results showed that S. marcescens ΔrssBA possessed the highest invasiveness, and this was reduced to the level of S. marcescens CH-1 by complementation with rssBA (Fig. 3B). Strains with deletion of shlBA did not exhibit an invasive phenotype (Fig. 3B). In contrast, overexpression of flhDC in S. marcescens CH-1 resulted in a more invasive phenotype, whereas deletion of flhDC in S. marcescens CH-1 or S. marcescens ΔrssBA decreased their invasive capacities (Fig. 3B).
To directly observe the invasiveness of S. marcescens in BEAS-2B cells, we infected the cells with the EGFP-expressing S. marcescens CH-1, S. marcescens ΔrssBA, and S. marcescens ΔrssBA/pRssAB strains at an MOI of 5 for 2 h. The invasiveness of S. marcescens ΔrssBA was 2.5-fold higher than that of S. marcescens CH-1 but could be restored to the CH-1 level via complementation with rssBA (Fig. 3C and D). To determine whether RssAB plays a role in the regulation of hemolysin expression during infection of BEAS-2B cells, the recombinant plasmid pRepoEGFP-shlB, in which egfp expression was under the control of the shlBA promoter, was constructed and transformed into S. marcescens CH-1 and S. marcescens ΔrssBA. After infection for 1 h, the media containing bacteria were collected for fluorescence intensity analysis. A 2-fold increase in the promoter activity of the shlBA operon was observed in S. marcescens ΔrssBA compared to that in S. marcescens CH-1 (Fig. 3E).
RssAB and ShlBA dominantly control pathogenesis in a rat acute-pneumonia model.
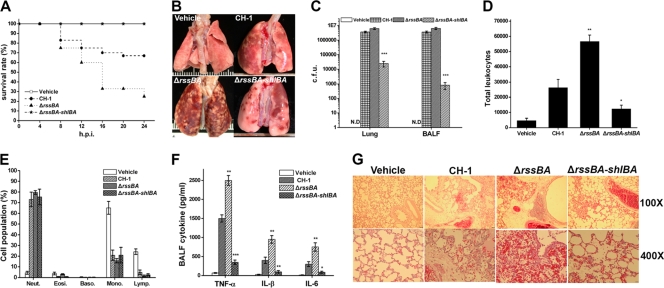
The role of RssAB in S. marcescens in vivo animal infection was further examined. Since S. marcescens is an important nosocomial pathogen frequently causing pneumonia (25), the acute-pneumonia model established in Sprague-Dawley rats (38) was used for evaluation. Rats were infected i.t. with 1 × 108 CFU of S. marcescens CH-1, S. marcescens ΔrssBA, S. marcescens ΔrssBA-shlBA or normal saline (vehicle), followed by monitoring for 24 h. While all rats from the vehicle group survived, S. marcescens ΔrssBA caused a higher mortality rate (75%) than the CH-1 strain (33.3%) (Fig. 4 A). Rats infected by S. marcescens ΔrssBA-shlBA all survived (Fig. 4A). This suggested that deletion of rssAB leads to high morbidity in acute lung infection, in which hemolysin SlhA plays a dominant role in S. marcescens pathogenesis.
FIG. 4.
ShlBA under RssAB control plays an important role in S. marcescens acute-pneumonia pathogenesis. (A) Rats (n = 12 in each group for independent experiments) were i.t. inoculated with 1 × 108 CFU of S. marcescens CH-1, S. marcescens ΔrssBA, and S. marcescens ΔrssBA-shlBA and vehicle. The survival rate was monitored every 4 h for 24 h postinfection (h.p.i.). (B) The whole lung of a rat was dissected for a gross view and imaged 24 h p.i. (C) Bacterial burdens from the lung and BAL fluid (BALF) at 24 h p.i. were analyzed by plate counting. The results are expressed as the average number of CFU per gram of lung and CFU per ml of BALF. N.D., not detectable. (D) Total leukocyte counts in BALF from each group were determined by hemocytometer analysis. (E) The cell differential count was determined and expressed as an average ratio of total leukocyte counts. (F) The supernatant of BALF from each group 1 day postinfection was collected for measurement of TNF-α, IL-1β, and IL-6, determined as pg/ml, by ELISA. (G) Dissected lungs fixed in 10% formalin for 24 h and paraffin embedded were cut as 4-μm sections for H&E staining. The images were analyzed and captured with a Leica DM 2500 under ×100 and ×400 magnification. Scale bars, 40 μm and 10 μm for ×100 and ×400, respectively. All results are shown as the average of three independent experiments, with the standard deviations indicated by error bars. *, P < 0.05; **, P < 0.01; and ***, P < 0.001 in comparison to CH-1 (Student's t test).
Gross examination of whole lungs from the sacrificed animals revealed more severe disseminated pulmonary edema and consolidation in S. marcescens ΔrssBA-challenged lungs than in those challenged with S. marcescens CH-1 (Fig. 4B). In comparison, restricted and mild signs of pneumonia were observed in the S. marcescens ΔrssBA-shlBA-infected group (Fig. 4B). While no bacteria were detected in the vehicle group, no significant difference in bacterial load was observed between S. marcescens ΔrssBA- and S. marcescens CH-1-infected lungs (Fig. 4C). In contrast, a significant reduction in the bacterial load occurred in S. marcescens ΔrssBA-shlBA-infected rats (Fig. 4C), and a similar trend of bacterial load in BAL fluids was also observed (Fig. 4C).
To further characterize the inflammatory states associated with infection, BAL fluids from rats of each group were collected for cell population analysis and cytokine measurement. Significant leukocyte infiltration was observed in the bacterium-infected group compared to the vehicle group (Fig. 4D). Intensive leukocyte infiltration, among which neutrophils constituted the major population, was more significant in infections caused by S. marcescens ΔrssBA, with a 2-fold increase of total leukocytes in comparison to S. marcescens CH-1 cells, whereas significantly less infiltration was found in the S. marcescens ΔrssBA-shlBA-infected group (Fig. 4D and E). Additionally, compared to S. marcescens CH-1, S. marcescens ΔrssBA induced more extensive production of proinflammatory cytokines, whereas concomitant deletion of shlBA diminished this phenomenon (Fig. 4F). Further histological examination results were consistent with these findings, with the S. marcescens ΔrssBA-infected group showing more severe edema in the alveolar septa, disruption of the alveolar-capillary barrier, damage to alveolar cells, hemorrhage, and cell sloughing associated with infiltration of neutrophils (Fig. 4G). Only partial damage associated with neutrophil aggregation, but no obvious barrier disruption and hemorrhage, were observed in the S. marcescens ΔrssBA-shlBA-infected group (Fig. 4G). Cumulatively, production of hemolysin SlhA under RssAB control is important for S. marcescens pathogenesis in vivo.
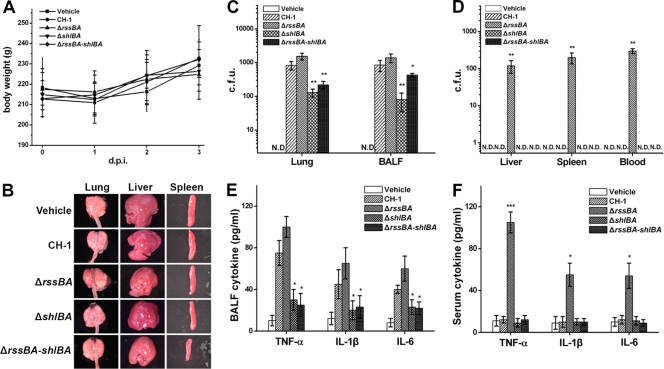
Deletion of rssBA in S. marcescens leads to systemic infection in immunocompetent rats.
As few S. marcescens systemic infections have been reported in immunocompetent patients (55), a rat sublethal-pneumonia model was used to evaluate the roles of RssAB and ShlBA in infections of immunocompetent animals. Rats were i.t. inoculated with 1 × 105 CFU each of S. marcescens CH-1, S. marcescens ΔrssBA, S. marcescens ΔshlBA, and S. marcescens ΔrssBA-shlBA and with vehicle, followed by monitoring for 3 days. Although all bacterium-infected rats appeared weak on the first day, none of the rats exhibited significant symptoms of pneumonia. On the day of sacrifice, rats in the S. marcescens ΔrssBA-infected group all appeared weak and stayed aggregated. However, body weights were not significantly different among the rats in each group (Fig. 5 A). After sacrifice, the lungs, livers, and spleens were dissected and examined. No significant pathological change in the gross morphology of organs was observed (Fig. 5B). These results indicate that the sublethal S. marcescens pneumonia did not result in severe pathological changes in the local lungs or other organs in each group of rats. Notably, bacterial growth in the local area of the lung, BAL fluid, liver, spleen, and serum did show different results. Higher bacterial loads in both lung and BAL fluid samples were detected in S. marcescens ΔrssBA-infected rats than in rats infected by S. marcescens CH-1, with increased proinflammatory cytokines, although this difference was not significant (Fig. 5C and E). In contrast, S. marcescens ΔshlBA and S. marcescens ΔrssBA-shlBA were efficiently eliminated by the rats (Fig. 5C) and were incapable of inducing proinflammatory cytokine production at the level of S. marcescens CH-1 or S. marcescens ΔrssBA (Fig. 5E). Remarkably, bacteria were detected in the sera and even in the livers and spleens of S. marcescens ΔrssBA-infected animals, along with higher levels of proinflammatory cytokines in sera, whereas no bacteria or significantly elevated cytokines could be detected in other experimental groups (Fig. 5D and F). Importantly, no signs of disseminated infection and inflammatory responses were found in rats inoculated with S. marcescens ΔrssBA with concomitant deletion of shlBA (Fig. 5D and F). Consistent with the in vitro results, RssAB reduced the virulence of S. marcescens in animal infection.
FIG. 5.
The rssBA deletion leads to systemic infection of S. marcescens in a sublethal-pneumonia model of immunocompetent rats. Rats (n = 6 in each group for independent experiments) were i.t. infected with 1 × 105 CFU of S. marcescens CH-1, S. marcescens ΔrssBA, S. marcescens ΔshlBA, and S. marcescens ΔrssBA-shlBA cells for 3 days postinfection (d.p.i.). Normal saline was used as a vehicle control. (A) Body weight was monitored daily. (B) On the third day p.i., the rats were sacrificed, and organs, including lungs, livers, and spleens, were removed for gross morphology observation accompanied by collection of BALF and sera. (C and D) The bacterial loads of organ homogenates, sera, and BALF were determined as the average number of CFU per gram of organ and per ml of serum or BALF. N.D., not detectable. (E and F) BALF and sera from each group 3 days p.i. were utilized to measure TNF-α, IL-1β, and IL-6 as determined by ELISA. All results are shown as the average of three independent experiments, with the standard deviations indicated by error bars. *, P < 0.05, and **, P < 0.01 in comparison to CH-1 (Student's t test).
DISCUSSION
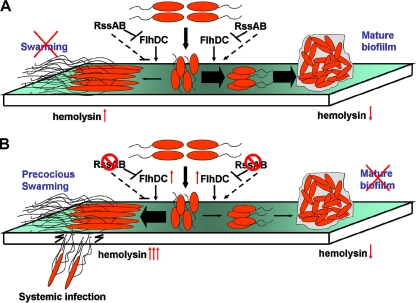
S. marcescens produces a variety of virulence proteins, including hemolysin, which was reported to be the dominant virulence factor in the bacterium (35). Even so, S. marcescens has long been considered only an opportunistic human pathogen, and the underlying mechanism of pathogenesis is still not clearly understood. This study provides strong evidence that RssAB-FlhDC-ShlBA works as an important multicellularity and pathogenesis regulation pathway in S. marcescens. Although our present evidence could not exclude the possibility that other, intermediate regulators might be involved in this pathway, RssAB-mediated repression of the ability for cellular infection substantially depends on modulation of flhDC. In response to the environmental temperature of 37°C and other uncharacterized factors, RssAB signaling negatively modulates expression of the important regulator FlhDC, which positively regulates hemolysin SlhA synthesis and coordinates bacterial multicellularity. Our results using an in vitro cell line model and in vivo animal studies indicated hemolysin as the dominant virulence factor of S. marcescens, in agreement with previous studies (35). Based on our findings, a model for the control by RssAB of flhDC expression and subsequent hemolysin production and bacterial multicellularity in S. marcescens is presented (Fig. 6). During transformation from planktonic cells to multicellularity, RssAB plays an important role and determines whether S. marcescens should live in the biofilm or swarming stage. In parallel with these processes, hemolysin production was coordinately regulated, although it did not affect bacterial multicellularity. At the human body temperature of 37°C, activation of RssAB signaling reduces expression of flhDC, resulting in reduction of swarming ability and favoring the transition of S. marcescens cells toward biofilm formation and concomitant downregulation of hemolysin expression (Fig. 6A). In contrast, instead of sessile biofilm formation, loss of RssAB engenders a hypervirulent capability in S. marcescens that is associated with precocious swarming and elevated hemolysin production, further leading to disseminated infection (Fig. 6B). This phenomenon has never been reported in wild-type S. marcescens cells. The temperature shift also plays an important role in the regulation of S. marcescens virulence. Downregulation of swarming motility and hemolysin production was observed in response to a temperature upshift from 30°C to 37°C, and RssAB conducts this change through repression of flhDC (Fig. 1) (36, 57). Of note, we could not exclude the possibility that other S. marcescens virulence factors might also be repressed by RssAB, because the hemolytic and cytotoxic activities of S. marcescens ΔrssBA-shlBA were higher than those of S. marcescens ΔshlBA (Fig. 1C and 3A). Indeed, Shimuta et al. have indicated that S. marcescens utilizes phospholipase A to exploit hemolytic and cytotoxic effects and potentially to regulate hemolysin ShlA activity (56). Intriguingly, synthesis of phospholipase is positively regulated by FlhDC in S. marcescens (21, 37), indicating that the higher hemolytic and cytotoxic activities of S. marcescens ΔrssBA-shlBA might be due to upregulation of flhDC expression, which further induces phospholipase production. Although other virulence factors might also influence the virulence capability of S. marcescens, hemolysin was shown to play a dominant role in our established S. marcescens-mediated pulmonary infection model in rats. These results indicate that by nature, S. marcescens is relatively less virulent at 37°C and that RssAB plays an important role in the regulation of these processes.
FIG. 6.
Proposed mechanisms by which RssAB controls multicellularity and pathogenesis in S. marcescens. S. marcescens utilizes RssAB to coordinate multicellular behaviors, such as biofilm formation and swarming motility, with the concomitant modulation of virulence and hemolysin production, by regulating flhDC expression at 37°C. (A) Activation of RssAB signaling reduces flhDC expression and hemolysin production and favors biofilm formation over swarming. (B) In the absence of RssAB, flhDC expression is increased. S. marcescens becomes hypervirulent, with increased hemolysin production accompanied by precocious swarming and defective biofilm formation. This gives S. marcescens the capability to cause disseminated infection. The dashed arrows represent unidentified determinants probably involved in the proposed model.
Coordinate regulation of hemolysin production and multicellularity is vital for bacterial pathogenesis.
In this study, we indicated that S. marcescens CH-1 utilizes hemolysin as a dominant virulence factor that is required for induction of multiple proinflammatory cytokines, including TNF-α, IL-1β, and IL-6, which are highly correlated with bacterium-induced pathogenesis (49, 52). Hemolysin also plays important roles in infection by other bacteria. Staphylococcus aureus α-hemolysin (Hla) is essential for the pathogenesis of S. aureus-mediated pneumonia (2). In addition, group B streptococcal β-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8 (12). The pore-forming toxin HlyA can enhance uropathogenic E. coli (UPEC) virulence in mouse urinary tract infection (UTI) models and modulate the host immune response, and HlyA expression correlates closely with increased clinical severity in UTI patients (64). These findings highlight the importance of hemolysin as a common and important virulence factor during bacterial infections. As a result, strict regulation of hemolysin production is a substantial issue for bacterial pathogenesis and immune responses to avoid excessive host injury, which is consistent with our finding that RssAB-repressed hemolysin production controls S. marcescens virulence capability.
Besides hemolysin production, bacterial motility is influential in bacterial pathogen-host interactions (43). In E. coli and Salmonella enterica serovar Typhimurium, the flagellar system comprises around 50 genes organized into three hierarchical transcriptional classes, at the top of which is the class I master operon flhDC (6). FlhDC has been proposed as a global regulator controlling flagellum biogenesis, cell septation, and expression of virulence factors during swarming (6, 18, 22). In P. mirabilis, FlhDC-conducted swarming coupled with increased hemolysin (HpmA) expression promotes the ability to invade host cells (17, 51). In X. nematophila, FlhDC coordinately controls motility and extracellular hemolysis (20). Both of these studies indicated the involvement of FlhDC in coregulation of swarming and hemolysin production. Besides swarming, FlhDC also regulates biofilm formation. In E. coli, FlhDC is required for biofilm formation, especially that involved in the first stage of reversible attachment (48, 50). Interestingly, we found that loss or overexpression of flhDC both resulted in impaired biofilm attachment in S. marcescens (Fig. 2), indicating that optimal expression of flhDC is required for biofilm formation.
Role of antivirulence genes in pathogen fitness.
In agreement with our findings, a two-component system, CovRS, from another opportunistic pathogen, Streptococcus pyogenes, also directly represses the expression of multiple virulence factors (8). Deletion of covRS also leads to a hypervirulence phenotype in S. pyogenes (15). Furthermore, a sampling of clinical isolates from invasive human diseases showed that 7 of 16 isolates lacked functional CovS or CovR (14). These observations, together with our results, indicated that conservation of a system that negatively regulates virulence implies a counteradaptive consequence for loss-of-function mutations (16). Besides two-component systems, defects in some enzymes or secretion systems may also lead to a significant increase in virulence and pathogenesis. In S. enterica, the pcgL gene encoding a periplasmic d-alanyl-d-alanine (d-Ala-d-Ala) dipeptidase allows bacteria to metabolize d-Ala-d-Ala released during cell wall synthesis (42). Pcgl is required for survival in the nonhost environment under nutrient-limiting conditions (42), and mutation of pcgL resulted in a hypervirulence phenotype of S. enterica in C3H/HeN and CBA/J mouse models (42). Recently, a novel secretion system, ZirTS, of S. enterica was reported to act as an antivirulence modulator, and during salmonellosis, zirTS showed a unique expression pattern throughout the gastrointestinal tract rather than at systemic sites (19). In this study, we showed that the virulence capability of S. marcescens might rely on the RssAB signaling control, the environmental conditions, and the host immunity situation. Consistent with our findings, reduced biofilm attachment accompanied by increased virulence capability has also been reported in some other bacterial infections, although the underlying mechanism remains unknown (7, 9). A possible explanation for the hypervirulent capability of the rssBA mutant may be increased swarming motility coupled with elevated hemolysin production, which efficiently disrupts the tissue barrier, resulting in inflammatory injury and easier dissemination in the host, as evidenced by our in vivo animal studies (Fig. 4 and 5).
Pathophysiological role of RssAB in S. marcescens.
While RssAB signaling plays an important role in the regulation of S. marcescens pathogenesis, the results of rssBA DNA sequence analysis of 38 S. marcescens clinical isolates failed to detect any significant amino acid changes in conserved domains compared with those from S. marcescens CH-1 (see Table S1 in the supplemental material). Also, the hemolytic activities of these strains were similar to that of S. marcescens CH-1. Furthermore, when we introduced pFlhDC into three other clinical strains, increased hemolytic activity was observed compared to those harboring vector (Lin and Lai, unpublished), suggesting FlhDC-dependent regulation of hemolytic activity was not an artifact in this study. The next question raised is why these S. marcescens strains retain a functionally complete RssAB, at least with respect to signaling transfer. In prokaryotic cells, the critical role of the two-component signal transduction system is the integration of stimuli from either subtle extracellular or intracellular changes to adapt to diverse environments. Basically, the role of a functional RssAB may be in fine tuning the pathophysiological behaviors of S. marcescens to cope with bacterium-host interaction and to achieve longer-term survival during infection. From an evolutionary perspective, premature death of the host is not advantageous for bacteria to evolve and spread. As an important opportunistic pathogen with increasing incidences of nosocomial outbreak of multidrug-resistant strains (33), RssAB-directed repression of hemolysin expression might explain why S. marcescens is not as virulent at 37°C as the other major enterobacterial pathogens, Salmonella and Shigella. The conservation of functional RssAB in this clinical survey probably supports this perspective. However, even though S. marcescens becomes hypervirulent when rssBA is deleted, S. marcescens clinical strains, which might contain functionally complete RssAB signaling, are still pathogenic. Thus, infection by S. marcescens in these patients may be due to either transient inactivation of RssAB by unknown factors or inadequate innate immunity for prevention of infection by S. marcescens with functional RssAB signaling. These characteristics remain to be further addressed.
Supplementary Material
Acknowledgments
This work was supported by grants NSC98-2320-B-182-007-MY3 from the National Science Council, Taiwan, and CMRPD170243 from Chang Gung Memorial hospital, which are much appreciated.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 16 August 2010.
REFERENCES
- 1.Allison, C., N. Coleman, P. L. Jones, and C. Hughes. 1992. Ability of Proteus mirabilis to invade human urothelial cells is coupled to motility and swarming differentiation. Infect. Immun. 60:4740-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bubeck Wardenburg, J., and O. Schneewind. 2008. Vaccine protection against Staphylococcus aureus pneumonia. J. Exp. Med. 205:287-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buffet-Bataillon, S., V. Rabier, P. Betremieux, A. Beuchee, M. Bauer, P. Pladys, E. Le Gall, M. Cormier, and A. Jolivet-Gougeon. 2009. Outbreak of Serratia marcescens in a neonatal intensive care unit: contaminated unmedicated liquid soap and risk factors. J. Hosp. Infect. 72:17-22. [DOI] [PubMed] [Google Scholar]
- 4.Caiazza, N. C., J. H. Merritt, K. M. Brothers, and G. A. O'Toole. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:3603-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, K. H., and M. G. Caparon. 2005. Patterns of virulence gene expression differ between biofilm and tissue communities of Streptococcus pyogenes. Mol. Microbiol. 57:1545-1556. [DOI] [PubMed] [Google Scholar]
- 8.Churchward, G. 2007. The two faces of Janus: virulence gene regulation by CovR/S in group A streptococci. Mol. Microbiol. 64:34-41. [DOI] [PubMed] [Google Scholar]
- 9.Coulthurst, S. J., S. Clare, T. J. Evans, I. J. Foulds, K. J. Roberts, M. Welch, G. Dougan, and G. P. Salmond. 2007. Quorum sensing has an unexpected role in virulence in the model pathogen Citrobacter rodentium. EMBO Rep. 8:698-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cowles, K. N., and H. Goodrich-Blair. 2005. Expression and activity of a Xenorhabdus nematophila haemolysin required for full virulence towards Manduca sexta insects. Cell Microbiol. 7:209-219. [DOI] [PubMed] [Google Scholar]
- 11.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 12.Doran, K. S., J. C. Chang, V. M. Benoit, L. Eckmann, and V. Nizet. 2002. Group B streptococcal beta-hemolysin/cytolysin promotes invasion of human lung epithelial cells and the release of interleukin-8. J. Infect. Dis. 185:196-203. [DOI] [PubMed] [Google Scholar]
- 13.Eberl, L., M. K. Winson, C. Sternberg, G. S. Stewart, G. Christiansen, S. R. Chhabra, B. Bycroft, P. Williams, S. Molin, and M. Givskov. 1996. Involvement of N-acyl-L-hormoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20:127-136. [DOI] [PubMed] [Google Scholar]
- 14.Engleberg, N. C., A. Heath, A. Miller, C. Rivera, and V. J. DiRita. 2001. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J. Infect. Dis. 183:1043-1054. [DOI] [PubMed] [Google Scholar]
- 15.Engleberg, N. C., A. Heath, K. Vardaman, and V. J. DiRita. 2004. Contribution of CsrR-regulated virulence factors to the progress and outcome of murine skin infections by Streptococcus pyogenes. Infect. Immun. 72:623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foreman-Wykert, A. K., and J. F. Miller. 2003. Hypervirulence and pathogen fitness. Trends Microbiol. 11:105-108. [DOI] [PubMed] [Google Scholar]
- 17.Fraser, G. M., L. Claret, R. Furness, S. Gupta, and C. Hughes. 2002. Swarming-coupled expression of the Proteus mirabilis hpmBA haemolysin operon. Microbiology 148:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser, G. M., and C. Hughes. 1999. Swarming motility. Curr. Opin. Microbiol. 2:630-635. [DOI] [PubMed] [Google Scholar]
- 19.Gal-Mor, O., D. L. Gibson, D. Baluta, B. A. Vallance, and B. B. Finlay. 2008. A novel secretion pathway of Salmonella enterica acts as an antivirulence modulator during salmonellosis. PLoS Pathog. 4:e1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Givaudan, A., and A. Lanois. 2000. flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J. Bacteriol. 182:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Givskov, M., L. Eberl, G. Christiansen, M. J. Benedik, and S. Molin. 1995. Induction of phospholipase- and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol. Microbiol. 15:445-454. [DOI] [PubMed] [Google Scholar]
- 22.Givskov, M., J. Ostling, L. Eberl, P. W. Lindum, A. B. Christensen, G. Christiansen, S. Molin, and S. Kjelleberg. 1998. Two separate regulatory systems participate in control of swarming motility of Serratia liquefaciens MG1. J. Bacteriol. 180:742-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harshey, R. M. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57:249-273. [DOI] [PubMed] [Google Scholar]
- 25.Hejazi, A., and F. R. Falkiner. 1997. Serratia marcescens. J. Med. Microbiol. 46:903-912. [DOI] [PubMed] [Google Scholar]
- 26.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hertle, R. 2000. Serratia type pore forming toxins. Curr. Protein Pept. Sci. 1:75-89. [DOI] [PubMed] [Google Scholar]
- 28.Hertle, R., S. Brutsche, W. Groeger, S. Hobbie, W. Koch, U. Konninger, and V. Braun. 1997. Specific phosphatidylethanolamine dependence of Serratia marcescens cytotoxin activity. Mol. Microbiol. 26:853-865. [DOI] [PubMed] [Google Scholar]
- 29.Hertle, R., M. Hilger, S. Weingardt-Kocher, and I. Walev. 1999. Cytotoxic action of Serratia marcescens hemolysin on human epithelial cells. Infect. Immun. 67:817-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hertle, R., and H. Schwarz. 2004. Serratia marcescens internalization and replication in human bladder epithelial cells. BMC Infect. Dis. 4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isberg, R. R., and S. Falkow. 1985. A single genetic locus encoded by Yersinia pseudotuberculosis permits invasion of cultured animal cells by Escherichia coli K-12. Nature 317:262-264. [DOI] [PubMed] [Google Scholar]
- 32.Justice, S. S., D. A. Hunstad, L. Cegelski, and S. J. Hultgren. 2008. Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6:162-168. [DOI] [PubMed] [Google Scholar]
- 33.Knowles, S., C. Herra, E. Devitt, A. O'Brien, E. Mulvihill, S. R. McCann, P. Browne, M. J. Kennedy, and C. T. Keane. 2000. An outbreak of multiply resistant Serratia marcescens: the importance of persistent carriage. Bone Marrow Transplant. 25:873-877. [DOI] [PubMed] [Google Scholar]
- 34.König, W., Y. Faltin, J. Scheffer, H. Schoffler, and V. Braun. 1987. Role of cell-bound hemolysin as a pathogenicity factor for Serratia infections. Infect. Immun. 55:2554-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kurz, C. L., S. Chauvet, E. Andres, M. Aurouze, I. Vallet, G. P. Michel, M. Uh, J. Celli, A. Filloux, S. De Bentzmann, I. Steinmetz, J. A. Hoffmann, B. B. Finlay, J. P. Gorvel, D. Ferrandon, and J. J. Ewbank. 2003. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 22:1451-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lai, H. C., P. C. Soo, J. R. Wei, W. C. Yi, S. J. Liaw, Y. T. Horng, S. M. Lin, S. W. Ho, S. Swift, and P. Williams. 2005. The RssAB two-component signal transduction system in Serratia marcescens regulates swarming motility and cell envelope architecture in response to exogenous saturated fatty acids. J. Bacteriol. 187:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, J. H., M. J. Lai, S. Ang, J. C. Shu, P. C. Soo, Y. T. Horng, W. C. Yi, H. C. Lai, K. T. Luh, S. W. Ho, and S. Swift. 2000. Role of flhDC in the expression of the nuclease gene nucA, cell division and flagellar synthesis in Serratia marcescens. J. Biomed. Sci. 7:475-483. [DOI] [PubMed] [Google Scholar]
- 38.Lu, C. C., H. C. Lai, S. C. Hsieh, and J. K. Chen. 2008. Resveratrol ameliorates Serratia marcescens-induced acute pneumonia in rats. J. Leukoc. Biol. 83:1028-1037. [DOI] [PubMed] [Google Scholar]
- 39.Maragakis, L. L., A. Winkler, M. G. Tucker, S. E. Cosgrove, T. Ross, E. Lawson, K. C. Carroll, and T. M. Perl. 2008. Outbreak of multidrug-resistant Serratia marcescens infection in a neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 29:418-423. [DOI] [PubMed] [Google Scholar]
- 40.Marre, R., J. Hacker, and V. Braun. 1989. The cell-bound hemolysin of Serratia marcescens contributes to uropathogenicity. Microb. Pathog. 7:153-156. [DOI] [PubMed] [Google Scholar]
- 41.Merritt, J. H., K. M. Brothers, S. L. Kuchma, and G. A. O'Toole. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189:8154-8164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mouslim, C., F. Hilbert, H. Huang, and E. A. Groisman. 2002. Conflicting needs for a Salmonella hypervirulence gene in host and non-host environments. Mol. Microbiol. 45:1019-1027. [DOI] [PubMed] [Google Scholar]
- 43.Ottemann, K. M., and J. F. Miller. 1997. Roles for motility in bacterial-host interactions. Mol. Microbiol. 24:1109-1117. [DOI] [PubMed] [Google Scholar]
- 44.Overhage, J., M. Bains, M. D. Brazas, and R. E. Hancock. 2008. Swarming of Pseudomonas aeruginosa is a complex adaptation leading to increased production of virulence factors and antibiotic resistance. J. Bacteriol. 190:2671-2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 46.Poole, K., and V. Braun. 1988. Iron regulation of Serratia marcescens hemolysin gene expression. Infect. Immun. 56:2967-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poole, K., E. Schiebel, and V. Braun. 1988. Molecular characterization of the hemolysin determinant of Serratia marcescens. J. Bacteriol. 170:3177-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 49.Prince, A. S., J. P. Mizgerd, J. Wiener-Kronish, and J. Bhattacharya. 2006. Cell signaling underlying the pathophysiology of pneumonia. Am. J. Physiol. Lung Cell. Mol. Physiol. 291:L297-L300. [DOI] [PubMed] [Google Scholar]
- 50.Prüss, B. M., C. Besemann, A. Denton, and A. J. Wolfe. 2006. A complex transcription network controls the early stages of biofilm development by Escherichia coli. J. Bacteriol. 188:3731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rather, P. N. 2005. Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 7:1065-1073. [DOI] [PubMed] [Google Scholar]
- 52.Sadikot, R. T., T. S. Blackwell, J. W. Christman, and A. S. Prince. 2005. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am. J. Respir. Crit. Care Med. 171:1209-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schiebel, E., and V. Braun. 1989. Integration of the Serratia marcescens haemolysin into human erythrocyte membranes. Mol. Microbiol. 3:445-453. [DOI] [PubMed] [Google Scholar]
- 54.Schiebel, E., H. Schwarz, and V. Braun. 1989. Subcellular location and unique secretion of the hemolysin of Serratia marcescens. J. Biol. Chem. 264:16311-16320. [PubMed] [Google Scholar]
- 55.Shimizu, S., H. Kojima, C. Yoshida, K. Suzukawa, H. Y. Mukai, Y. Hasegawa, S. Hitomi, and T. Nagasawa. 2003. Chorioamnionitis caused by Serratia marcescens in a non-immunocompromised host. J. Clin. Pathol. 56:871-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimuta, K., M. Ohnishi, S. Iyoda, N. Gotoh, N. Koizumi, and H. Watanabe. 2009. The hemolytic and cytolytic activities of Serratia marcescens phospholipase A (PhlA) depend on lysophospholipid production by PhlA. BMC Microbiol. 9:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soo, P. C., Y. T. Horng, J. R. Wei, J. C. Shu, C. C. Lu, and H. C. Lai. 2008. Regulation of swarming motility and flhDCSm expression by RssAB signaling in Serratia marcescens. J. Bacteriol. 190:2496-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soo, P. C., J. R. Wei, Y. T. Horng, S. C. Hsieh, S. W. Ho, and H. C. Lai. 2005. Characterization of the dapA-nlpB genetic locus involved in regulation of swarming motility, cell envelope architecture, hemolysin production, and cell attachment ability in Serratia marcescens. Infect. Immun. 73:6075-6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stella, N. A., E. J. Kalivoda, D. M. O'Dee, G. J. Nau, and R. M. Shanks. 2008. Catabolite repression control of flagellum production by Serratia marcescens. Res. Microbiol. 159:562-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verstraeten, N., K. Braeken, B. Debkumari, M. Fauvart, J. Fransaer, J. Vermant, and J. Michiels. 2008. Living on a surface: swarming and biofilm formation. Trends Microbiol. 16:496-506. [DOI] [PubMed] [Google Scholar]
- 61.Wang, J., R. A. Barke, R. Charboneau, and S. Roy. 2005. Morphine impairs host innate immune response and increases susceptibility to Streptococcus pneumoniae lung infection. J. Immunol. 174:426-434. [DOI] [PubMed] [Google Scholar]
- 62.Wei, J. R., and H. C. Lai. 2006. N-acylhomoserine lactone-dependent cell-to-cell communication and social behavior in the genus Serratia. Int. J. Med. Microbiol. 296:117-124. [DOI] [PubMed] [Google Scholar]
- 63.Wei, J. R., Y. H. Tsai, P. C. Soo, Y. T. Horng, S. C. Hsieh, S. W. Ho, and H. C. Lai. 2005. Biochemical characterization of RssA-RssB, a two-component signal transduction system regulating swarming behavior in Serratia marcescens. J. Bacteriol. 187:5683-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiles, T. J., B. K. Dhakal, D. S. Eto, and M. A. Mulvey. 2008. Inactivation of host Akt/protein kinase B signaling by bacterial pore-forming toxins. Mol. Biol. Cell 19:1427-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.