Abstract
In the course of establishing its replication niche within the cytosol of infected host cells, the facultative intracellular bacterial pathogen Listeria monocytogenes must efficiently regulate the secretion and activity of multiple virulence factors. L. monocytogenes encodes two predicted posttranslocation secretion chaperones, PrsA1 and PrsA2, and evidence suggests that PrsA2 has been specifically adapted for bacterial pathogenesis. PrsA-like chaperones have been identified in a number of Gram-positive bacteria, where they are reported to function at the bacterial membrane-cell wall interface to assist in the folding of proteins translocated across the membrane; in some cases, these proteins have been found to be essential for bacterial viability. In this study, the contributions of PrsA2 and PrsA1 to L. monocytogenes growth and protein secretion were investigated in vitro and in vivo. Neither PrsA2 nor PrsA1 was found to be essential for L. monocytogenes growth in broth culture; however, optimal bacterial viability was found to be dependent upon PrsA2 for L. monocytogenes located within the cytosol of host cells. Proteomic analyses of prsA2 mutant strains in the presence of a mutationally activated allele of the virulence regulator PrfA revealed a critical requirement for PrsA2 activity under conditions of PrfA activation, an event which normally takes place within the host cell cytosol. Despite a high degree of amino acid similarity, no detectable degree of functional overlap was observed between PrsA2 and PrsA1. Our results indicate a critical requirement for PrsA2 under conditions relevant to host cell infection.
During the course of infection, bacterial pathogens are dependent upon the secretion of multiple protein products that modulate host cell physiology and facilitate bacterial growth. A number of protein secretion systems have been identified and functionally characterized for Gram-negative bacteria for which the existence of both an inner and outer membrane presents a significant barrier to protein translocation (12, 17, 20, 35, 62, 79, 120). In Gram-positive bacteria, secreted proteins are translocated across the single bacterial cell membrane in an unfolded state and delivered to the compartment existing between the membrane and the cell wall (80). The cell walls of Gram-positive bacteria consist of a thick matrix of peptidoglycan layers and glycopolymers, including teichoic acids and lipoteichoic acids (111), and these abundant anionic polymers have a high capacity to bind divalent metal ions and cationic molecules (5, 70, 108). Proteins that are translocated across the bacterial membrane therefore enter a challenging environment for protein folding based on the high density of negative charge, high concentrations of cations, and low pH (80, 108). Within this environment, secreted proteins may additionally require further posttranslational modification, proteolytic activation, or sequestration prior to release for interaction with host cell targets. It should be noted that not all secreted proteins are found in the extracellular milieu, as many are specifically localized at the membrane or within the cell wall. Proteins present in bacterial culture supernatants thus constitute a group of exoproteins to which numerous pathogenic traits can be attributed (17).
For the facultative intracellular pathogen Listeria monocytogenes, protein secretion has been reported to occur primarily via the Sec-mediated secretion pathway (16). Proteins secreted via Sec-dependent secretion include well-characterized virulence factors, such as the internalins InlA and InlB, which mediate host cell invasion (6, 32, 50-52, 60, 72, 87), listeriolysin-O (LLO) and the broad-range phosphatidyl-choline phospholipase (PC-PLC), which mediate vacuole membrane lysis (21, 36, 37, 41, 47, 65, 82-84, 101, 118), and the surface protein ActA, which mediates actin polymerization and cell-to-cell spread within the host (3, 9, 18, 54, 90, 91, 93, 115). These proteins are critical for the establishment of the L. monocytogenes replication niche within the cytosol of infected host cells (28, 40, 86, 102).
L. monocytogenes PrsA1 and PrsA2 are secreted proteins that are predicted to function as parvulin-type peptidyl-prolyl isomerase (PPIase) chaperones at the bacterial membrane-cell wall interface to assist in the folding and stability of secreted proteins (1). PrsA2 appears to be primarily adapted for L. monocytogenes pathogenesis, based on the regulation of prsA2 expression by the central virulence transcriptional activator PrfA and on the essential requirement for PrsA2 for bacterial virulence in mice (1, 74, 121). The loss of PrsA2 dramatically reduces bacterial cell-to-cell spread in monolayers of mouse fibroblast cells and also reduces LLO stability and impedes the processing of PC-PLC to its enzymatically active form (1, 13, 121). Zemansky et al. have additionally demonstrated that prsA2 deletion mutants are defective for bacterial flagellum-mediated swimming motility, an observation that suggests multiple roles for PrsA2 both inside and outside infected host cells (121). In contrast to its homologue in Bacillus subtilis, PrsA2 is not required for L. monocytogenes viability, and ΔprsA2 mutants replicate very similarly to wild-type strains in broth culture and on agar medium (1, 80).
Unlike L. monocytogenes ΔprsA2 mutants, strains lacking prsA1 are fully virulent in mouse models of infection (1). prsA1 is not required for bacterial growth in broth culture, and its potential contributions to other aspects of L. monocytogenes physiology are as yet undefined. PrsA2 and PrsA1 are highly similar at the amino acid sequence level; thus, it is possible that PrsA2 and PrsA1 share some degree of functional overlap (1). In B. subtilis, the depletion of PrsA leads to the induction of the CssR/S two-component system and increased expression of the HtrA chaperone/protease in response to the accumulation of misfolded proteins at the bacterial membrane-cell wall interface (48, 80). The loss or depletion of both PrsA2 and PrsA1 in L. monocytogenes could potentially elicit a similar membrane stress response if one or both are required for the folding of a large number of secreted proteins.
In this study, we investigated the potential functional overlap of PrsA2 and PrsA1 through the construction of an L. monocytogenes ΔprsA1 ΔprsA2 double mutant. In addition, exoproteomic analyses were used to identify proteins whose secretion or localization was altered due to the lack of either PrsA2 or PrsA1. Our findings indicate a critical PrsA1-independent role for PrsA2 in the maintenance of full bacterial viability in the host cell cytosol, further strengthening the link between PrsA2 function and L. monocytogenes virulence.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
All bacterial strains used in this study are listed in Table 1. L. monocytogenes 10403S (NF-L100) and the 10403S prfA(L140F) strain (NF-L1167) were used as the parent strains for the construction of gene deletion mutants (67, 74, 116). All strains were grown overnight at 37°C with agitation in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, MI) or Luria broth (LB) (Invitrogen, Carlsbad, CA) unless otherwise specified. Antibiotics were used at the following concentrations: streptomycin (200 μg/ml), chloramphenicol (7.5 and 5 μg/ml), erythromycin (1 μg/ml), and neomycin (10 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain | Description | Designation | Reference |
|---|---|---|---|
| TOP10 | E. coli host strain used for recombinant pPL2 plasmids | ||
| SM10 | E. coli host strain for conjugation of pPL2 plasmids | ||
| NF-L100 | L. monocytogenes 10403S parent strain | ||
| NF-L1167 | 10403S actA-gus-neo prfA(L140F) | L140F strain | 116 |
| NF-L1651 | 10403S with ΔprsA2::erm | ΔprsA2 mutant | 1 |
| NF-L1656 | NF-L1651 (ΔprsA2::erm) with integrated pPL2-prsA2 (pNF1255) | ΔprsA2 + pPL2-prsA2 mutant | 1 |
| NF-L1483 | HEL 402-10403S with a prsA1 in-frame deletion | ΔprsA1 mutant | 1 |
| NF-L1637 | NF-L1167 [prfA(L140F)] with ΔprsA2::erm | prfA(L140F) ΔprsA2 mutant | 1 |
| NF-L1631 | NF-L1438 (ΔprsA1) transduced with ΔprsA2::erm | ΔprsA1 ΔprsA2 mutant | This work |
| NF-L1605 | 10403S with a ΔhtrA in-frame deletion | ΔhtrA mutant | 113 |
| NF-L1633 | NF-L1605 (ΔhtrA) transduced with ΔprsA2::erm | ΔhtrA ΔprsA2 mutant | This work |
| NF-L1665 | NF-L1633 (ΔhtrA ΔprsA2) with integrated pPL2-prsA2 (pNF1255) | ΔhtrA ΔprsA2 + pPL2-prsA2 mutant | This work |
| NF-L1670 | NF-L1651 (ΔprsA2::erm) with integrated pPL2-PprsA2-prsA1 (pNF1611) | ΔprsA2 + pPL2-PprsA2-prsA1 mutant | This work |
| pNF1255 | pPL2 containing the prsA2 open reading frame for complementation | pPL2-prsA2 mutant | 1 |
| pNF1611 | pPL2 containing PprsA2-prsA1 SOE product | pPL2-PprsA2-prsA1 mutant | This work |
Construction of L. monocytogenes ΔprsA1 ΔprsA2 and ΔhtrA ΔprsA2 mutant strains.
Strains containing multiple in-frame deletion mutations were generated by phage-mediated transduction of L. monocytogenes ΔprsA1 (NF-L1483) and ΔhtrA (NF-L1605) mutant strains (1, 113). U153 bacteriophage-mediated transduction was performed as previously described (1, 116). Briefly, phage lysates were prepared from the L. monocytogenes ΔprsA2::erm mutant (NF-L1651) (1). Lysates were mixed at a 1:1 ratio with bacteria (108 phage to 108 CFU of each strain) and incubated at room temperature for 40 min in the presence of CaCl2 and MgCl2 (final concentration, 10 mM). Mixtures of bacteria and bacteriophage lysates were then spread onto BHI plates containing 1 μg/ml erythromycin to select for transductants. For each transductant, the replacement of the wild-type prsA2 allele with the prsA2::erm mutation was confirmed by PCR amplification of the appropriate chromosomal region. Confirmed mutant strains were designated as follows: ΔprsA1 ΔprsA2 (NF-L1631) and ΔhtrA ΔprsA2 (NF-L1633) double mutants.
Construction of an L. monocytogenes ΔprsA2 strain expressing prsA1 under the regulation of the prsA2 promoter.
A PprsA2-prsA1 (prsA2 promoter to prsA1 open reading frame [ORF]) fusion was generated as follows: two DNA fragments were generated by PCR using primer pairs P2A (5′-GGC-GAGCTC-CCTAAAATCAATCAAC-3′)/P2B (5′-CATCACTTTTTTTAATTTTGTCAATAAATAAAACACACTCCTTAG-3′) and P1A (5′-CTAAGGAGTGTGTTTTATTTATTGACAAAATTAAAAAAAGTGATG-3′)/P1B (5′-GGC-GGTACC-TTAGTTAGATGTAGTCGTTGA-3′). The fragments were purified and used in a splicing-by-overlap extension (SOE) PCR along with primer pair P2A and P1B to generate a 1,520-bp fragment. The fragment was digested with KpnI and SacI and subcloned into the plasmid vector pPL2 to generate pNF1611. pNF1611 was introduced into the L. monocytogenes ΔprsA2 mutant strain (NF-L1651) via conjugation as previously described (29, 57). The resultant strain was designated NF-L1670.
Growth curves and bacterial cell viability assays.
Bacterial growth was measured in BHI broth beginning with a 1:20 dilution of overnight culture into fresh BHI. Growth was measured each hour by determining the absorbance at an optical density at 600 nm (OD600) in a spectrophotometer. For measurement of growth in terms of the number of CFU, 1-ml culture aliquots were removed at each hour and serially diluted into phosphate-buffered saline (PBS) (0.144 g/ml KH2PO4, 9 g/ml NaCl, 0.795 g/ml Na2HPO4 [anhydrous]) and CFU were enumerated after overnight incubation on BHI plates at 37°C. Cell viability was measured using the Live/Dead BacLight bacterial viability kit (Molecular Probes, Invitrogen, Carlsbad, CA). Briefly, bacterial strains were diluted 1:20 from an overnight culture into 19 ml of BHI. Cultures were grown for 3 h to an approximate OD600 of ∼0.6, at which point 10 ml of each culture was removed and centrifuged at 8,500 rpm for 15 min. The bacterial pellets were washed with 1 ml of PBS and resuspended in 1 ml of PBS prior to staining. A 2× propidium iodide-Cyto9 solution was prepared according to the manufacturer's instructions in 5 ml of water and mixed 1:1 with the bacterial cell suspension. After incubation for 15 min at room temperature, 8 μl of cell suspension was placed onto a glass microscope slide and immediately examined using a DeltaVision fluorescent microscope (Applied Precision, Issaquah, WA). Images were acquired using Softworx Image Acquisition software (Applied Precision, Issaquah, WA). A minimum of 10 fields were viewed for each strain and total bacteria per field (live and dead) from at least three independent experiments were enumerated.
Bacterial intracellular growth in tissue culture cells.
J774 macrophage-like cells were maintained as previously described (10, 68, 96). Macrophages (2 × 106) were seeded onto glass coverslips in tissue culture dishes the night prior to infection. Overnight cultures of L. monocytogenes were used to infect cells at a multiplicity of infection of 0.1 bacterium to 1 macrophage. The infection was allowed to proceed for 30 min, followed by three washes with PBS and the addition of fresh medium containing gentamicin (30 μg/ml) to kill extracellular bacteria. At the indicated time points, coverslips were removed and lysed in 5 ml H2O with vigorous vortexing. Lysates were spread onto LB agar plates and incubated at 37°C overnight. Bacterial CFU were enumerated the following day. The data shown (see Fig. 2) are representative of results from three independent experiments.
Plaque assays.
Plaque assays were conducted as previously described (95). Briefly, L2 fibroblasts were infected with L. monocytogenes at a multiplicity of infection (MOI) of 30 to 1. After 1 h, gentamicin was added in a Dulbecco's modified Eagle's medium (DMEM) agarose (0.7%) overlay. Plaque formation was monitored at 72 h after staining with Neutral Red solution (Sigma, St. Louis, MO) in at least three independent experiments.
Hemolysin assays.
Hemolytic activity assays were conducted as previously described with some modifications (1, 11, 53). Overnight cultures of L. monocytogenes in LB broth were diluted 1:10 in fresh LB and grown for 5 h at 37°C with shaking. OD600 readings were taken, and 1.2 ml of culture was centrifuged at maximum speed for 5 min in a tabletop centrifuge. Bacterial supernatants were normalized based on the OD600 of the original cultures to account for any differences in culture density, such that supernatants from cultures with greater optical densities were diluted into a suitable volume of LB to match the culture with the lowest OD600. Serial dilutions of normalized supernatants were made in PBS (pH 5.5) containing 1 mM dithiothreitol (DTT) and 5% washed sheep's red blood cells (RBCs) (Cocalico Biologicals, Reamstown, PA), and samples were incubated for 30 min at 37°C. Hemolytic units are described as the reciprocal of the dilution required for 50% lysis of RBCs and are derived from the results of at least five independent experiments.
Detection of PC-PLC activity.
Egg yolk agar was used to measure phospholipase activity (68). Chicken egg yolks were separated and mixed 1:1 (vol/vol) with PBS by vortexing, and 5 ml of the egg yolk suspension was then mixed with molten LB agar at 42°C containing 0.2% activated charcoal (Fisher Scientific, Pittsburgh, PA) and 25 mM glucose-6-phosphate (Sigma, St. Louis, MO) to enhance plcB expression (119). The degree of phospholipase activity was detected as a zone of opacity surrounding the bacterial streak after overnight growth at 37°C.
Isolation of bacterial exoproteins for two-dimensional gel electrophoresis.
Bacterial exoproteins were prepared from culture supernatants as previously described (1, 74). Bacterial strains [10403S and the ΔprsA2, ΔprsA1, ΔprsA1 ΔprsA2, prfA(L140F), and ΔprsA2 prfA(L140F) mutants] were grown in 20 ml BHI overnight at 37°C with shaking. Bacteria were diluted 1:20 into 200 ml fresh BHI, and growth was monitored for 5 h to an approximate OD600 of 1.2. Bacterial cells were centrifuged at 9,000 rpm for 20 min to recover the supernatant to which trichloroacetic acid was added to a final volume of 10%. The exoproteins were precipitated on ice for 30 min, and the protein pellets were recovered following centrifugation at 9,000 rpm for 15 min followed by a wash in 12 ml of ice-cold acetone and an additional centrifugation at 9,000 rpm for 15 min. The pellets were air dried and resuspended in 400 μl 1× SDS boiling buffer without β-mercaptoethanol (5% SDS, 10% glycerol, 60 mM Tris, pH 6.8). Total protein was determined by a bicinchoninic acid (BCA) assay (Fisher Scientific, Pittsburgh, PA) and was used to normalize loading onto two-dimensional (2-D) polyacrylamide gels. Prior to 2-D SDS-PAGE, all samples were run on 1-D SDS polyacrylamide gels to verify the consistency and quality of the sample preparation. All samples were prepared on three independent occasions and used for 2-D SDS-PAGE, resulting in a minimum of two biological replicates for all gels. Samples from biological replicates were each analyzed a minimum of three times, resulting in three technical replicates to validate consistency between samples.
Two-dimensional polyacrylamide gel electrophoresis.
Two-dimensional electrophoresis was performed according to the carrier Ampholine method of isoelectric focusing (IEF) (69) by Kendrick Labs, Inc. (Madison, WI), as follows: isoelectric focusing was carried out in a glass tube with an inner diameter of 2.0 mm by using 2% Ampholine mix (pH 4 to 8) (GE Healthcare, Piscataway, NJ, and Serva, Heidelberg, Germany) for 9,600 V·h. One microgram of an IEF internal standard, tropomyosin, was added to the sample. This protein migrated as a doublet with a lower polypeptide spot that had a molecular weight (MW) of 33,000 and a pI of 5.2. A tube gel pH gradient plot was determined with a surface pH electrode. After isoelectric focusing and equilibration for 10 min in buffer O (10% glycerol, 50 mM dithiothreitol, 2.3% SDS, and 0.0625 M Tris, pH 6.8), each tube gel was sealed to the top of a stacking gel that overlaid a 10% acrylamide slab gel (0.75 mm thick). SDS slab gel electrophoresis was carried out for about 4 h at 15 mA/gel. The following proteins were used as molecular-weight standards: myosin (220,000), phosphorylase A (94,000), catalase (60,000), actin (43,000), carbonic anhydrase (29,000), and lysozyme (14,000) (Sigma-Aldrich, St. Louis, MO). These standards appear along the basic edge of the Coomassie blue-stained 10% acrylamide slab gel. The Coomassie blue-stained gels were dried between sheets of cellophane with the acid edge to the left.
Protein identification using liquid chromatography-tandem mass spectrometry (LC-MS/MS).
All proteomics data were acquired from two independent biological replicates and two or more technical replicates per strain. Gel spots were considered unique if present in one gel but completely absent in another as determined by visual inspection of gels on a light box. Modest differences in protein abundances were thus excluded for these profiles. All unique protein spots were cut out of gels using a fresh scalpel blade and placed into Eppendorf tubes containing 200 μl of sterile water. The remainder of the protein extraction and digestion was performed by the proteomics core facility at the University of Illinois at Chicago Research Resources Center. Briefly, identified spots were cut into 1-mm cubes with a scalpel followed by being washed with 100 mM ammonium bicarbonate, reduction with dithiothreitol in ammonium bicarbonate, and alkylation with iodoacetamide in the dark. Samples were digested overnight with Promega modified sequencing-grade trypsin in ammonium bicarbonate (Promega, Madison, WI). Peptides were liberated using three consecutive extractions with ammonium bicarbonate at 37°C, followed by sample concentration using a SpeedVac.
LC-MS/MS was carried out using a Thermo Scientific LTQ FT instrument equipped with a Dionex UltiMate 3000 two-dimensional microcapillary high-performance liquid chromatography (HPLC) system (Thermo Fisher Scientific, Waltham, MA). Peptides were separated on a C18 column after being eluted with a gradient. Generated peak lists were extracted from the resulting chromatograms as mascot generic format (MGF) files by using ReAdW (Institute for Systems Biology, Seattle, WA) and in-house software and then searched using a Mascot 2.2 search engine (Matrix Science, Boston, MA) against the List_monocyt Listeria monocytogenes NCBI database (53,458 sequences; 14,892,194 residues) by using a peptide tolerance of 10 ppm and carbamidomethylation of cysteine and oxidation of methionine as variable modifications. Scaffold 2.4 (Proteome Software, Portland, OR) software was used to merge and display only the results with a 95% confidence and 2 or more unique peptide matches. The average false discovery rate was between 3% and 5%, as estimated by Mascot and automated decoy database searching.
Mouse intravenous infections.
Animal procedures were IACUC approved and performed in the Biological Resources Laboratory at the University of Illinois at Chicago. Bacterial cells were prepared as previously described (1). Female Swiss Webster mice (6 to 8 weeks old) were inoculated via the tail vein with 200 μl PBS containing 2 × 104 CFU of each bacterial strain tested. After 72 h, mice were sacrificed and both the liver and spleen were isolated. Each organ was homogenized in 5 ml of H2O using a Tissue Master 125 homogenizer (Omni, Marietta, GA), and dilutions were spread onto BHI agar plates to quantify the bacterial burden in each organ.
Statistics.
Statisical analysis was performed using Prism Software (GraphPad version 2.0) and is described in the figure legends of experiments for which statistics were necessary. Where appropriate, a Student t test or one-way analysis of variance (ANOVA) with Tukey's multiple comparison test was used to identify statistically significant differences. In all cases, a P value of <0.05 was considered significant.
RESULTS
In vitro characterization of an L. monocytogenes ΔprsA1 ΔprsA2 double mutant.
L. monocytogenes ΔprsA1 and ΔprsA2 mutants exhibit apparently normal growth characteristics in broth culture (1). Given that PrsA homologues fulfill essential functions in some Gram-positive bacteria (such as B. subtilis), we sought to determine if L. monocytogenes PrsA1 and PrsA2 shared any functional redundancy that contributed to bacterial growth in broth culture. In contrast to PrsA's essential function in B. subtilis, an L. monocytogenes mutant lacking both prsA1 and prsA2 was fully viable (Fig. 1A). In tissue culture cells, the growth of the ΔprsA1 ΔprsA2 mutant resembled that of the single ΔprsA2 mutant based on the slightly lower levels of bacterial uptake into J774 macrophages and a leveling off of bacterial growth at late time points (Fig. 1B). The successful construction of the ΔprsA1 ΔprsA2 double mutant demonstrated that neither gene product serves an essential function for L. monocytogenes growth in vitro.
FIG. 1.
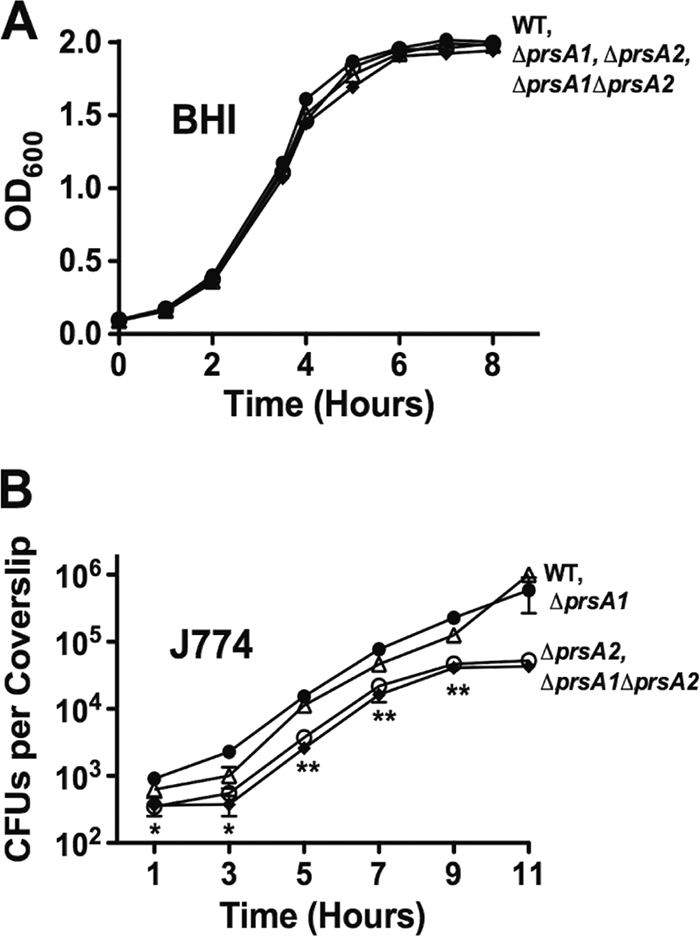
L. monocytogenes ΔprsA1 ΔprsA2 mutants exhibit normal growth patterns in broth culture and resemble single ΔprsA2 mutants in infected tissue culture cells. (A) Bacterial growth in BHI broth as determined by optical density measurements at 600 nm. The data shown are representative of results from three independent experiments, each performed in duplicate. (B) J774 macrophage-like cells were infected at an MOI of 0.1 bacterium to 1 macrophage, and intracellular bacterial replication was measured at the indicated time points following the addition of gentamicin (30 μg/ml) at 1 h postinfection. Data shown are the averages of results from three independent experiments. •, wild type; ○, ΔprsA2 mutant; ▵, ΔprsA1 mutant; ♦, ΔprsA1 ΔprsA2 mutant. Statistical significance was determined using one-way analysis of variance with Tukey's multiple comparison test (*, P < 0.01; **, P < 0.001).
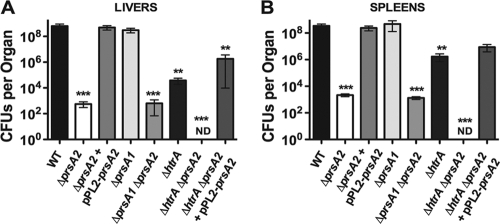
Loss of prsA1 does not exacerbate the virulence-associated defects of a ΔprsA2 mutant.
The loss of prsA2 reduces LLO-associated hemolytic activity as well as PC-PLC-associated phospholipase activity and severely diminishes the capacity of L. monocytogenes to spread to adjacent cells (1, 121). ΔprsA2 mutants are highly attenuated in a mouse infection model, with a greater than 100,000-fold reduction in bacterial burdens recovered from the livers and spleens of infected animals (1, 121). In contrast, a prsA1 deletion mutant exhibits none of the prsA2-associated defects either in vitro or in vivo (1). To determine whether PrsA1 shared any detectable degree of functional overlap with respect to PrsA2-associated activities in L. monocytogenes, the ΔprsA1 ΔprsA2 double mutant was assessed for hemolytic activity, phospholipase activity, cell-to-cell spread in tissue culture cells, and bacterial virulence in infected mice. In all cases, the phenotypes observed for the double-deletion mutant resembled those of the single prsA2 deletion mutant, with a 2-fold reduction in hemolytic activity and a similar reduction in phospholipase activity on egg yolk agar plates (Fig. 2A and B). The ΔprsA1 ΔprsA2 mutant also resembled the ΔprsA2 mutant with respect to its ability to spread cell-to-cell in monolayers of L2 fibroblast cells (plaque formation) (Fig. 2C) and in the magnitude of its virulence defect in infected mice (Fig. 3). Additionally, expression of prsA1 under the control of the prsA2 promoter in a ΔprsA2 background was unable to complement any in vitro defect associated with a prsA2 mutant (Fig. 2). Thus, although PrsA2 and PrsA1 share a high degree of amino acid similarity, PrsA1 does not compensate to any measurable degree for any PrsA2-associated phenotype.
FIG. 2.
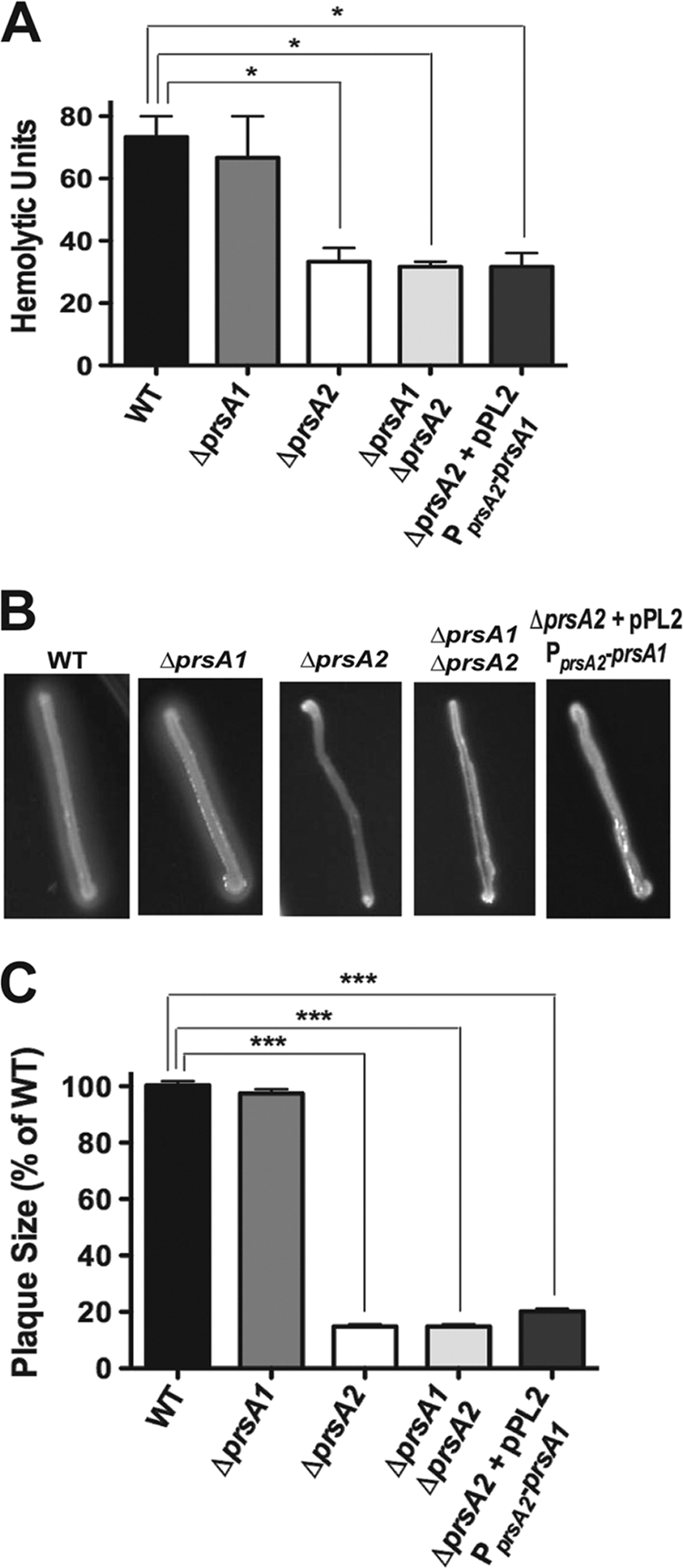
PrsA1 shares no apparent functional overlap with PrsA2 in in vitro assays. All assays were conducted using the wild-type strain or ΔprsA2, ΔprsA1, ΔprsA1 ΔprsA2, or ΔprsA2 plus pPL2-PprsA2-prsA1 mutant strains. (A) Measurement of LLO-associated bacterial hemolytic activity. Dilutions of bacterial culture supernatants were assessed for their ability to lyse sheep's red blood cells (RBCs) in vitro. The reciprocal of the supernatant dilution that resulted in 50% lysis of RBCs (hemolytic units) was determined in a minimum of five independent experiments, each conducted in duplicate, and the average of results is shown. (B) Phospholipase activity as determined by the incubation of bacterial strains on egg yolk agar plates followed by the observation of a zone of opacity surrounding the bacterial streak. A representative image from one of five plates is shown. (C) Plaque formation in L2 fibroblast monolayers in the presence of gentamicin. At 72 h postinfection, plaques were measured using a micrometer and the plaque size for the wild type was set to 100%. Plaque assays were repeated at least three times, with results from a minimum of 20 plaques averaged per experiment. Statistical significance was determined using one-way analysis of variance with Tukey's multiple comparison test (*, P < 0.01; ***, P < 0.0001).
FIG. 3.
Loss of htrA significantly impairs the survival of ΔprsA2 mutants in mice. Mice were injected via the tail vein with 2 × 104 CFU of either the wild-type strain or ΔprsA2, ΔprsA2 plus pPL2-prsA2, ΔprsA1, ΔprsA1 ΔprsA2, ΔhtrA, ΔhtrA ΔprsA2, or ΔhtrA ΔprsA2 plus pPL2-prsA2 mutant strains. The liver and spleen of infected mice were recovered at 72 h postinoculation, and the bacterial burden in each organ was determined. A minimum of 5 mice were inoculated per strain tested, and the means and standard deviations are shown. Statistical significance was determined using one-way analysis of variance with Tukey's multiple comparison test (**, P < 0.001; ***, P < 0.0001).
Proteomic analyses reveal altered exoprotein patterns for strains lacking prsA2 but not those lacking prsA1.
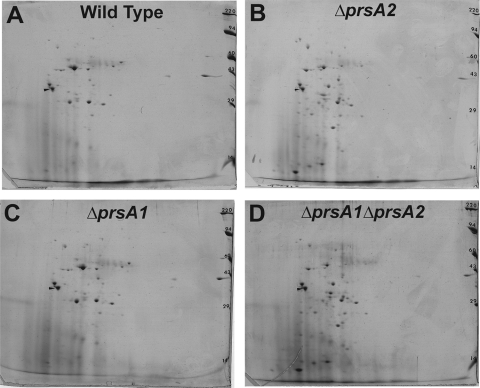
It has been previously reported that both LLO stability and activity as well as the processing of the phospholipase PC-PLC to its mature form are reduced in L. monocytogenes ΔprsA2 strains (1, 121). Altered SDS-PAGE polypeptide profiles have also been observed for supernatant proteins derived from ΔprsA2 strains versus those derived from the wild type (121). We sought to better define how the loss of prsA2 or prsA1 affects the L. monocytogenes exoproteome by identifying changes in the supernatant protein profiles for the ΔprsA1, ΔprsA2, and ΔprsA1 ΔprsA2 mutant strains in comparison to the wild type by using two-dimensional polyacrylamide gel electrophoresis.
An examination of the exoprotein profiles for supernatants derived from the wild-type and ΔprsA2, ΔprsA1, and ΔprsA1 ΔprsA2 mutant strains resulted in a number of interesting observations (Fig. 4). First, consistent with the absence of a detectable phenotype for a ΔprsA1 mutant in comparison to the wild-type strain (Fig. 1 to 3), no notable differences in secreted polypeptides were observed for this mutant compared to those of the wild type (Fig. 4A and 4C). In contrast, the supernatant protein profiles derived from a ΔprsA2 mutant exhibited striking differences compared to the wild-type and ΔprsA1 mutant profiles, while the ΔprsA1 ΔprsA2 double mutant profile resembled that of the ΔprsA2 mutant (Fig. 4). Table 2 lists the identities of the proteins isolated based on differences in exoprotein profiles. Twenty-three proteins were identified based on their presence in the supernatants derived from wild-type strains (and the ΔprsA1 mutant) in comparison to strains lacking prsA2. The majority of the identified proteins (16/23) could be classified into four functional categories: virulence factors, cell surface/cell wall metabolism, transport/binding proteins, and proteins involved in stress responses/detoxification/adaptation to atypical conditions. Included among the virulence proteins was LLO, whose stability has already been linked to PrsA2 activity (1, 121). Additional proteins were identified with functions associated with motility, metabolism, and membrane bioenergetics, as well as a hypothetical protein and amino-acyl tRNA synthetase.
FIG. 4.
Two-dimensional electrophoresis of bacterial exoproteins reveals significantly altered protein profiles for ΔprsA2 mutants. Supernatants from the wild-type strain or ΔprsA1, ΔprsA2, or ΔprsA1 ΔprsA2 mutant strains were trichloroacetic acid (TCA) precipitated and subjected to two-dimensional SDS-PAGE, revealing a significantly altered protein profile for ΔprsA2 mutant supernatants (B) in comparison to wild-type supernatants (A). A ΔprsA1 mutant supernatant profile (C) closely resembled that of the wild type, while a ΔprsA1 ΔprsA2 mutant profile (D) was remarkably similar to the ΔprsA2 single mutant.
TABLE 2.
Proteins present in wild-type gels but absent from ΔprsA2 mutant gels
| Function or functional class | Gene no. | Protein name | Protein description | No. of peptide matches | % coverage | Protein score | Reference(s) |
|---|---|---|---|---|---|---|---|
| Virulence factors | lmo0202 | LLO | Listeriolysin-O | 17 | 47 | 586 | 83 and 86 |
| Cell surface and cell wall metabolism | lmo2467 | Chitin binding protein/carbohydrate binding protein | 10 | 35 | 979 | 59 | |
| lmo1521 | N-Acetylmuramoyl-l-alanine amidase | 8 | 30 | 1298 | |||
| lmo1883 | Chitinase | 4 | 16 | 477 | 59 | ||
| lmo2522 | LysM domain penicillin binding protein | 2 | 12 | 443 | 74 | ||
| lmo2039 | PbpB | Penicillin binding protein | 5 | 8.9 | 414 | 43 | |
| lmo1892 | PbpA | Penicillin binding protein 2A | 6 | 11 | 257 | 43 | |
| lmo0540 | Penicillin binding protein, putative | 5 | 20 | 118 | 43 | ||
| lmo2591 | GW repeat surface protein | 3 | 24 | 210 | |||
| lmo2504 | Peptidase M48 family | 5 | 21 | 309 | |||
| Transport/binding proteins and lipoproteins | lmo2196 | OppA | Oligopeptide ABC transporter, oligopeptide binding protein | 9 | 26 | 489 | 8 |
| lmo0135 | CtaP | Cysteine transport-associated protein (lipoprotein) | 14 | 38 | 1668 | 74 and 117 | |
| Detoxification | lmo1439 | Sod | Superoxide dismutase | 3 | 16 | 670 | 2 and 100 |
| lmo0927 | IspB | Sulfatase family protein | 4 | 9.8 | 456 | 110 | |
| lmo2079 | Putative lipoprotein | 6 | 30 | 1099 | |||
| Mobility and chemotaxis | lmo0690 | FlaA | Flagellin | 2 | 11 | 81 | 109 |
| Hypotheticals | lmo1752 | Hypothetical protein | 4 | 22 | 249 | ||
| Metabolism | lmo2459 | Gap | Glyceraldehyde 3-phosphate dehydrogenase | 6 | 20 | 1323 | 81 |
| lmo1620 | Similar to dipeptidase PepV | 7 | 18 | 420 | |||
| lmo2455 | Eno | Enolase | 2 | 20 | 74 | 81 | |
| Specific pathways | lmo0429 | Glycosyl hydrolase | 2 | 2 | 20 | ||
| Membrane bioenergetics | lmo0013 | QoxA | AA3-600 quinol oxidase subunit II | 5 | 17 | 464 | |
| Amino-acyl tRNA synthetases | lmo1755 | GatA | Glutamyl-tRNA (Gln) amidotransferase subunit A | 2 | 6 | 110 |
Table 3 lists exoproteins identified in ΔprsA2 mutant supernatants but absent in supernatants derived from the wild-type and ΔprsA1 strains. In contrast to the majority of the proteins identified in wild-type supernatants, none of the 24 differing proteins identified within ΔprsA2 mutant supernatants were predicted to be bona fide secreted proteins based on the presence of a recognizable signal peptide sequence. All were predicted to be located within the bacterial cytosol with roles in protein folding, protein synthesis, amino acid synthesis, sugar and lipid metabolism, and glycolysis, as well as other physiological pathways. Despite the lack of a secretion signal sequence, nine of the proteins predicted to reside within the bacterial cytosol have been previously identified in L. monocytogenes exoproteome analyses (23, 97). It thus appears that a number of additional bacterial proteins with cytosolic functions have altered localization in the absence of functional PrsA2.
TABLE 3.
Proteins present in ΔprsA2 mutant gels but absent from wild-type gels
| Function or functional class | Gene no. | Protein name | Protein description | No. of peptide matches | % coverage | Protein score | Reference(s) |
|---|---|---|---|---|---|---|---|
| Detoxification/adaptation to atypical conditions | lmo2785 | Kat | Catalase | 13 | 41 | 791 | 26 and 58 |
| lmo1439 | Sod | Superoxide dismutase | 8 | 51 | 536 | 2 and 100 | |
| lmo2468 | ClpP | ATP-dependent Clp-protease | 3 | 27 | 146 | 33 and 34 | |
| lmo0943 | Fri | Nonheme iron-containing ferritin | 5 | 50 | 970 | 25 | |
| Hypotheticals | lmo0995 | Hypothetical, similar to B. subtilis YkrP protein | 2 | 13 | 100 | ||
| lmo1597 | Hypothetical protein | 2 | 9 | 126 | |||
| Protein folding | lmo1267 | Tig | Trigger factor (prolyl isomerase) | 7 | 27 | 395 | 7 |
| Protein synthesis | lmo0239 | CysS | Cysteinyl tRNA synthetase | 5 | 17 | 263 | 61 |
| lmo1657 | Tsf | Translation elongation factor Ts | 11 | 51 | 1091 | ||
| lmo1314 | Frr | Ribosome recycling factor | 2 | 14 | 102 | ||
| lmo0250 | RplJ | 50S ribosomal protein L10 | 5 | 36 | 362 | 27 | |
| Termination | lmo2543 | Prf1 | Peptide chain release factor 1 | 2 | 8 | 63 | |
| Amino acid synthesis | lmo0223 | CysK | Cysteine synthase A | 11 | 59 | 977 | 22 |
| Metabolism | lmo2459 | Gap | Glyceraldehyde-3-phosphate dehydrogenase | 5 | 19 | 228 | 81 |
| Metabolism of lipids | lmo1372 | 2-Oxoisovalerate dehydrogenase E1 component | 3 | 17 | 187 | 27 | |
| Main glycolytic pathways | lmo2456 | Pgm | Phosphoglycerate mutase | 12 | 36 | 723 | |
| lmo2457 | Tpi | Triose-phosphate isomerase | 2 | 16 | 169 | ||
| Specific pathways | lmo2556 | FbaA | Fructose 1,6-bisphosphate aldolase | 4 | 28 | 149 | |
| lmo2103 | Pta | Phosphotransacetylase | 5 | 29 | 251 | 42 | |
| lmo1571 | Pfk | 6-Phosphofructokinase | 6 | 29 | 651 | 112 | |
| 1-Phosphofructokinase | 3 | 17 | 131 | ||||
| lmo0210 | Ldh | l-Lactate dehydrogenase | 2 | 4 | 106 | ||
| lmo0191 | Phospho-beta-glucosidase | 2 | 16 | 85 | |||
| lmo1376 | 6-Phosphogluconate dehydrogenase | 13 | 34 | 749 |
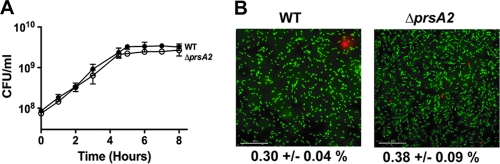
The presence of 15 proteins of predicted cytosolic functions within ΔprsA2 mutant supernatants could potentially reflect compromised bacterial membrane permeability and/or bacterial lysis. While differences in bacterial growth rates and cell densities were not apparent based on optical density measurements (Fig. 1A), limited bacterial cell lysis could have occurred in late-log- or stationary-phase cultures, resulting in the release of cytosolic proteins into the supernatant. Bacterial viability was therefore directly measured for ΔprsA2 mutants by the assessment of the number of bacterial CFU during growth in broth culture. Wild-type and ΔprsA2 mutant strains exhibited no detectable differences either in growth rate or in the numbers of viable cells, as the total number of CFU of the ΔprsA2 mutant remained nearly identical to that of the wild type (Fig. 5A). Similarly, an assessment of membrane integrity and cell viability via Live/Dead staining revealed no differences between the wild-type and ΔprsA2 strains (Fig. 5B). Thus, the presence of bacterial cytosolic proteins in ΔprsA2 mutant supernatants does not appear to be due to increased levels of cell lysis or to gross changes in cell membrane permeability.
FIG. 5.
ΔprsA2 mutants are fully viable in broth culture despite an increased abundance of bacterial cytosol-associated proteins in culture supernatants. (A) Broth culture growth curves of the wild type and the ΔprsA2 mutant as measured by plating dilutions of bacterial culture onto solid medium at the indicated time points. Data represent results from one of three experiments performed in duplicate. •, wild type; ○, ΔprsA2 mutant. (B) Bacterial Live/Dead staining as performed on log-phase cultures of the wild type or a ΔprsA2 mutant. Live bacterial cells are stained with Cyto9 (green), while dead or membrane-compromised bacterial cells are stained with propidium iodide (red). The percentage of cells positive for propidium iodide (red) was determined by counting the number of total cells in a minimum of 10 randomly chosen fields from two independent experiments. Statistical significance was determined using a Student t test (P > 0.05).
PrfA activation dramatically alters the exoproteome of a ΔprsA2 mutant.
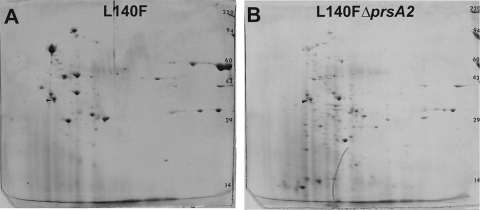
PrfA is the major transcriptional regulator of L. monocytogenes-secreted virulence factors (28, 30). PrfA exists in a low-activity state during bacterial growth in broth culture but becomes activated upon contact with host cells and bacterial entry into the cytosol, resulting in the expression and secretion of a number of gene products required for host cell invasion, bacterial intracellular growth, and cell-to-cell spread (86). The signal that leads to PrfA activation is not known, but mutations have been identified within prfA that result in constitutive PrfA activation in broth culture (prfA* mutations) (66, 67, 77, 88, 116). Given that prsA2 expression is PrfA regulated (74) and that L. monocytogenes prfA* strains constitutively express significant amounts of secreted virulence gene products (68, 74, 88), we performed additional exoproteome analyses in ΔprsA2 strains in the presence of prfA* (Fig. 6A and B). As previously observed, the presence of the prfA* allele prfA(L140F) dramatically increased the number and abundance of L. monocytogenes exoproteins (74) (compare Fig. 4A with Fig. 6A). In the absence of prsA2, the prfA(L140F) exoprotein profile was dramatically altered (Fig. 6B). Numerous polypeptides that were abundant in prfA(L140F) strain supernatants were completely absent in ΔprsA2 prfA(L140F) strains, and an increased number of lower-molecular-weight spots were observed, suggestive of increased levels of protein degradation.
FIG. 6.
Two-dimensional electrophoresis of bacterial exoproteins in the presence of PrfA activation reveals significantly altered protein profiles for ΔprsA2 mutants. Supernatant fractions derived from prfA(L140F) or ΔprsA2 prfA(L140F) mutant strains were TCA precipitated and subjected to two-dimensional SDS-PAGE, thereby revealing a significantly altered protein profile for the ΔprsA2 prfA(L140F) strain (B) in comparison to the prfA(L140F) strain with the wild-type prsA2 strain (A). As previously noted, the prfA(L140F) strain supernatant protein profile (A) is distinct from that of wild-type supernatants in which PrfA is not mutationally activated (Fig. 3A).
Ten proteins were identified in the supernatants derived from prfA(L140F) strains that were not present in ΔprsA2 prfA(L140F) mutant supernatants (Table 4). Of note were four well-characterized virulence gene products (ActA, LLO, PC-PLC, and Mpl), two of which (LLO and PC-PLC) have been functionally associated with PrsA2 activity (1, 121). Additional proteins involved in stress responses/detoxification/adaptation to atypical conditions, cell surface/wall metabolism, and transport/binding were identified. In contrast to the 10 proteins identified as present in prfA(L140F) strain supernatants, a large number of polypeptides were identified as present in ΔprsA2 prfA(L140F) mutant supernatants but absent in prfA(L140F) strain supernatants (Table 5). Fifteen of the 44 identified gene products were predicted to be secreted proteins and may potentially represent PrsA2 substrates whose abundance and/or localization is altered in the absence of prsA2. Virulence factors, cell surface/cell wall proteins, and transport/binding proteins were included within this group. The virulence factors identified were identical to those found in prfA(L140F) strain supernatants (Table 5); however, they were excised from portions of the gel that reflected smaller-than-expected molecular masses and thus likely represent degradation products or proteins with altered mobility. Similar to a number of the proteins associated with ΔprsA2 mutant supernatants, 29 proteins were predicted to be cytosolic and to serve as intermediates in metabolic pathways (Table 5). The increased abundance of bacterial cytosolic proteins present in the supernatant fraction of ΔprsA2 prfA(L140F) strains suggests that PrfA activation enhances and/or exacerbates the aberrant transit of these proteins across the bacterial membrane.
TABLE 4.
Proteins present in L140F strain gels but absent from ΔprsA2(L140F) mutant gels
| Function or functional class | Gene no. | Protein name | Protein description | No. of peptide matches | % coverage | Protein score | Reference(s) |
|---|---|---|---|---|---|---|---|
| Virulence factors | lmo0204 | ActA | Actin assembly-inducing protein | 14 | 28 | 1239 | 18 and 54 |
| lmo0202 | LLO | Listeriolysin-O | 22 | 58 | 3398 | 83 and 86 | |
| lmo0205 | PlcB | Phospholipase-C | 13 | 32 | 745 | 92 and 101 | |
| lmo0203 | Mpl | Zinc metalloproteinase precursor | 1 | 2.7 | 62 | 63, 64, 75, and 76 | |
| Detoxification | lmo0927 | IspB | Sulfatase family protein | 13 | 32 | 874 | 110 |
| lmo0644 | Sulfatase family protein | 9 | 21 | 526 | 110 | ||
| Cell surface and cell wall metabolism | lmo1540 | N-Acetyl muramoyl l-alanine amidase | 11 | 25 | 522 | ||
| lmo2505 | P45 | Peptidoglycan lytic protein | 3 | 11 | 280 | 85 | |
| lmo2504 | Peptidase M48 family | 2 | 4.4 | 273 | |||
| Transport/binding proteins and lipoproteins | lmo2416 | Conserved hypothetical lipoprotein | 4 | 20 | 174 |
TABLE 5.
Proteins present in ΔprsA2 prfA(L140F) mutant gels but absent from prfA(L140F) gels
| Function or functional class | Gene no. | Protein name | Protein description | No. of peptide matches | % coverage | Protein score | Reference(s) |
|---|---|---|---|---|---|---|---|
| Virulence factors | lmo1786 | InlC | Internalin C | 8 | 29 | 568 | 19 and 24 |
| lmo0204 | ActA | Actin assembly-inducing protein | 7 | 33 | 1819 | 18 and 54 | |
| lmo0202 | LLO | Listeriolysin-O | 12 | 44 | 1444 | 83 and 86 | |
| lmo0205 | PlcB | Phospholipase-C | 3 | 17 | 36 | 92 and 101 | |
| lmo0203 | Mpl | Zinc metalloproteinase precursor | 2 | 9 | 179 | 63, 64, 75, and 76 | |
| Cell surface and cell wall metabolism | lmo2505 | P45 | Peptidoglycan lytic protein | 9 | 23 | 4374 | 85 |
| lmo2522 | LysM domain-containing protein | 5 | 34 | 736 | 74 | ||
| lmo1547 | MreC | Rod shape-determining protein | 4 | 11 | 468 | 98 | |
| Transport/binding proteins and lipoproteins | lmo2416 | ABC transporter, substrate binding protein | 2 | 11 | 204 | ||
| lmo0369 | Conserved hypothetical protein | 2 | 14 | 109 | |||
| lmo1388 | TcsA | CD4+ T cell-stimulating antigen (lipoprotein) | 2 | 6.9 | 227 | 74 | |
| lmo1757 | Putative lipoprotein | 2 | 8.4 | 395 | |||
| lmo2637 | Hypothetical pheromone-like protein | 3 | 13 | 687 | 74 | ||
| lmo1847 | ABC transporter, manganese binding protein | 4 | 15 | 642 | |||
| lmo1002 | PtsH | PTS phosphocarrier protein (Hpr) | 4 | 41 | 782 | 14 | |
| Protein folding | lmo1474 | GrpE | Heat shock protein/cochaperone | 4 | 36 | 246 | |
| lmo1267 | Tig | Trigger factor (prolyl isomerase) | 8 | 27 | 1379 | 7 | |
| Metabolism | |||||||
| Carbohydrates | lmo2455 | Eno | Enolase | 16 | 50 | 3741 | 81 |
| lmo2367 | Pgi | Glucose-6-phosphate isomerase | 2 | 6.2 | 225 | ||
| lmo2456 | Pgm | Phosphoglycerate mutase, 2,3-biphosphoglycerate independent | 10 | 31 | 1668 | ||
| lmo2459 | Gap | Glyceraldehyde 3-phosphate dehydrogenase | 2 | 8.6 | 416 | 81 | |
| Lipids | lmo1808 | FabD | Malonyl coenzyme A acyl carrier protein transacylase | 3 | 13 | 296 | |
| Coenzymes | lmo2101 | Pyridoxine biosynthesis protein | 4 | 22 | 744 | ||
| Amino acids | lmo2414 | SufD | Hypothetical FeS assembly protein | 3 | 11 | 246 | |
| Nucleic acids | lmo2611 | Adk | Adenylate kinase | 5 | 26 | 875 | |
| Detoxification/adaptation to atypical conditions | lmo0927 | IspB | Sulfatase family protein | 6 | 13 | 820 | 110 |
| lmo2785 | Kat | Catalase | 2 | 5.1 | 102 | 26 and 58 | |
| lmo1439 | Sod | Superoxide dismutase | 6 | 37 | 2726 | 2 and 100 | |
| lmo2468 | ClpP | ATP-dependent Clp protease proteolytic subunit | 3 | 20 | 602 | 33 and 34 | |
| lmo0943 | Fri | Nonheme iron-binding ferritin | 4 | 36 | 181 | 25 | |
| Regulation and transcription | lmo0443 | Transcriptional regulator (putative) | 8 | 28 | 1212 | ||
| lmo1280 | CodY | Transcriptional repressor | 7 | 37 | 1050 | 4 | |
| lmo1496 | GreA | Transcription elongation factor | 5 | 44 | 606 | ||
| Protein synthesis | lmo0239 | CysS | Cysteinyl-tRNA synthetase | 2 | 7.6 | 284 | 61 |
| lmo1657 | Tsf | Translation elongation factor Ts | 7 | 26 | 853 | ||
| lmo2654 | Fus | Elongation factor G | 2 | 4.6 | 479 | ||
| lmo1314 | Frr | Ribosome recycling factor | 5 | 31 | 676 | ||
| lmo0250 | RplJ | 50S ribosomal protein L10 | 4 | 24 | 617 | 27 | |
| lmo0251 | RplL | 50S ribosomal protein L7/L12 | 2 | 20 | 305 | ||
| Amino acid synthesis | lmo0223 | CysK | Cysteine synthase A | 8 | 43 | 631 | 22 |
| Specific pathways | lmo2556 | FbaA | Fructose bisphosphate aldolase | 5 | 18 | 1045 | |
| lmo1339 | Glucokinase | 2 | 8.4 | 363 | |||
| lmo0429 | Glycosyl hydrolase | 2 | 8 | 205 | |||
| lmo0811 | Carbonic anhydrase | 2 | 11 | 399 |
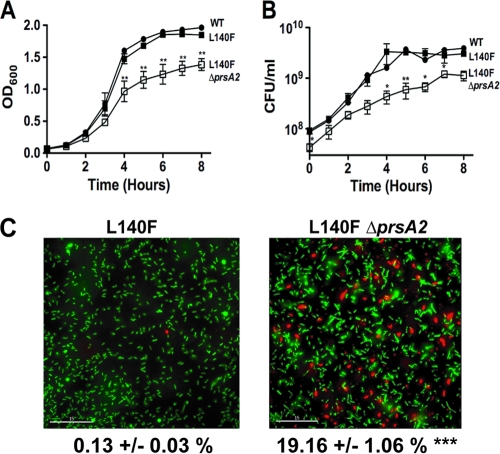
Mutants lacking prsA2 exhibit decreased viability and altered membrane integrity in the presence of mutationally activated prfA*.
As the supernatants derived from ΔprsA2 prfA(L140F) strains contained increased numbers of cytosolic proteins in comparison to ΔprsA2 strains containing the wild-type prfA allele (29 versus 15 bacterial cytosolic proteins, respectively), we examined whether prfA activation exacerbated the phenotype of the ΔprsA2 mutant and resulted in compromised membrane integrity or reduced bacterial viability. In the presence of prfA*, the ΔprsA2 mutant was dramatically impaired for growth in broth culture, as indicated both by optical density measurements at 600 nm and by enumeration of bacterial CFU (Fig. 7A and B). The assessment of the membrane integrity of the ΔprsA2 prfA(L140F) mutant by using Live/Dead staining reagents indicated a substantial number of cells that stained positive for propidium iodide uptake [19.16% ± 1.06% (mean ± standard deviation) for the ΔprsA2 prfA(L140F) mutant compared to 0.38% ± 0.09% for the ΔprsA2 mutant and 0.13% ± 0.03% for the prfA(L140F) mutant]. These results strongly suggest that PrsA2 plays a critical role in maintaining bacterial cell viability and membrane integrity under the conditions of increased protein secretion resulting from constitutive PrfA activation.
FIG. 7.
PrfA activation reduces the viability of ΔprsA2 mutants. (A and B) Broth culture growth of the wild-type strain and prfA(L140F) and ΔprsA2 prfA(L140F) mutants as measured by optical density at 600 nm or by plating dilutions of bacterial culture onto solid medium at the indicated time points. Data represent results from one of three experiments performed in duplicate. •, wild type; ▪, prfA(L140F) strain; □, ΔprsA2 prfA(L140F) mutant. (C) Bacterial Live/Dead staining as performed on log-phase cultures of the prfA(L140F) or ΔprsA2 prfA(L140F) mutant. Live bacterial cells are stained with Cyto9 (green), while dead cells or cells with a compromised membrane are stained with propidium iodide (red). The percentage of cells positive for propidium iodide (red) was determined by counting the number of total cells in a minimum of 10 randomly chosen fields from two independent experiments. Statistical significance for panels A and B was determined using a one-way analysis of variance with Tukey's multiple comparison test (*, P < 0.01; **, P < 0.001) and for panel C using a Student t test (***, P < 0.0001).
The loss of the htrA-encoded chaperone/protease further exacerbates the viability defects associated with ΔprsA2 in cytosolic bacteria.
In B. subtilis, PrsA and the secreted chaperone/protease HtrA appear to be functionally linked (48, 80). Hyyryläinen et al. have demonstrated that depletion of B. subtilis PrsA results in an increase in HtrA expression, and it has been hypothesized that HtrA functions to reduce the accumulation of misfolded proteins at the bacterial membrane-cell wall interface that occurs in the absence of PrsA (48). Given that the viability of a ΔprsA2 mutant in L. monocytogenes is severely compromised in the presence of constitutively activated PrfA (Fig. 7) and that the loss of PrsA2 is likely to result in an increase of misfolded proteins at the bacterial membrane, we examined whether the loss of L. monocytogenes HtrA might further exacerbate the ΔprsA2 defects associated with protein secretion under more natural conditions of PrfA activation. A ΔprsA2 ΔhtrA double-deletion mutant exhibited a modest defect in bacterial growth, as measured in rich broth culture and as evidenced by its slightly increased doubling time (67.4 ± 0.58 min for the ΔprsA2 ΔhtrA mutant compared to an average of approximately 57.5 ± 1.4 min for the wild type and single-deletion mutants) (Fig. 8A). In contrast, the growth of the ΔprsA2 ΔhtrA mutant within the cytosol of J774 macrophages was dramatically reduced in comparison to the wild-type and single-mutant strains (Fig. 8B). The number of ΔprsA2 ΔhtrA mutant CFU was observed to increase modestly during the first 5 h postinfection; however, after this time point, bacterial numbers declined, indicative of a loss of bacterial viability. Consistent with the dramatic reduction in intracellular bacterial viability, no bacteria were detected in the livers or spleens of mice infected with ΔprsA2 ΔhtrA mutants at 72 h postinfection (Fig. 3A and B). These results indicate that the combined loss of prsA2 and htrA dramatically impacts L. monocytogenes viability under conditions of PrfA activation.
FIG. 8.
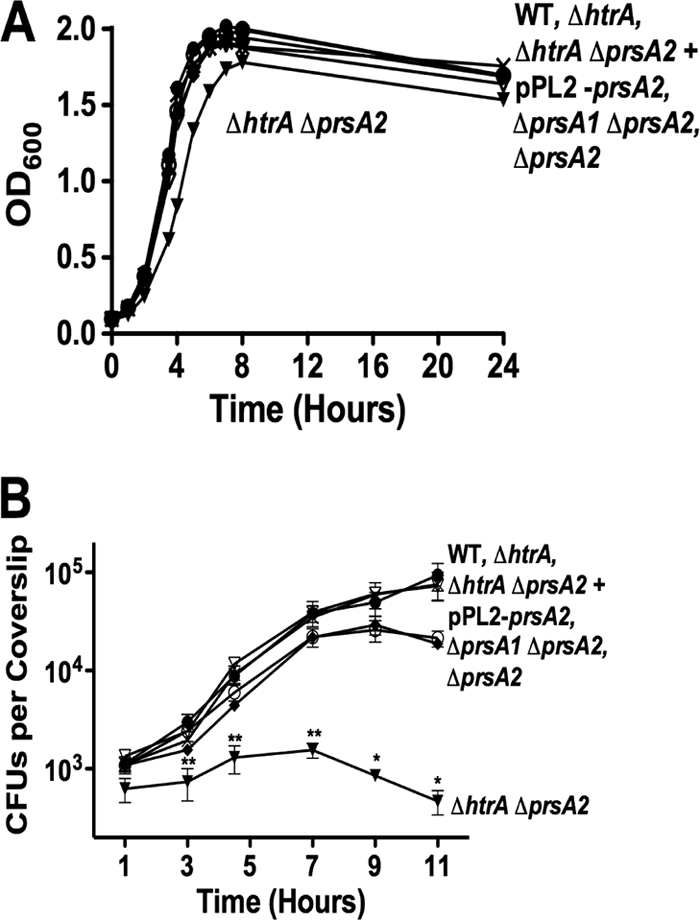
The loss of htrA exacerbates ΔprsA2-associated bacterial viability defects within infected host cells. (A) Bacterial growth in BHI broth was determined via optical density measurements at 600 nm. The data shown represent results from one of three independent experiments, each performed in duplicate. (B) J774 macrophage-like cells were infected at an MOI of 0.1 bacterium to 1 macrophage, and bacterial intracellular growth was measured in the presence of gentamicin at the indicated time points. Data shown are the averages of results from three independent experiments. •, wild type; ○, ΔprsA2 mutant; X, ΔhtrA mutant; ♦, ΔprsA1 ΔprsA2 mutant; ▾, ΔhtrA ΔprsA2 mutant; ▿, ΔhtrA ΔprsA2 plus pPL2-prsA2 mutant. Statistical significance was determined using one-way analysis of variance with Tukey's multiple comparison test (*, P < 0.01; **, P < 0.001).
DISCUSSION
As a facultative intracellular bacterial pathogen, L. monocytogenes relies on the secretion and activity of a number of factors that enable the bacterium to establish its replicative niche within host cells (28, 40, 86, 102). Proteins essential for bacterial virulence must be rapidly translocated across the bacterial membrane and properly folded to carry out target functions. PrsA1 and PrsA2 were recently identified as posttranslocation chaperones with predicted roles in protein folding and stability at the L. monocytogenes membrane-cell wall interface (1). Experimental evidence indicated that PrsA2 was distinct from PrsA1 in its requirement for L. monocytogenes virulence; however, it remained possible that PrsA1 and PrsA2 shared a limited degree of functional overlap. Here, we demonstrate that PrsA2 alone has been functionally adapted for a role in L. monocytogenes virulence and for maintaining bacterial viability within infected host cells.
The successful construction of the ΔprsA1 ΔprsA2 double mutant indicates that neither gene product is required for bacterial viability in broth culture, a result which contrasts with the essential requirement for the single prsA gene in B. subtilis (107). The ΔprsA1 ΔprsA2 double mutant was indistinguishable from a ΔprsA2 single mutant for all in vitro and in vivo phenotypes examined, further substantiating the absence of functional redundancy between the two proteins. It remains possible that PrsA1 contributes to L. monocytogenes protein secretion under as-yet-undefined environmental conditions that may be encountered outside infected hosts. PrsA1 is present in L. monocytogenes broth culture supernatants (F. Alonzo and N. Freitag, unpublished data), but little information exists thus far regarding its expression patterns or functional activity. It is interesting that despite the high degree of amino acid sequence similarity shared between PrsA1 and PrsA2 (58% identify and 75% similarity), PrsA1 was not capable of compensating for the loss of PrsA2 even when the gene was expressed from the prsA2 promoter (Fig. 2). This result suggests that unique residues and/or regions of PrsA2 are critical for its functional differentiation from PrsA1.
PrsA2 plays an important role in L. monocytogenes pathogenesis, as indicated by the greater than 100,000-fold reduction in bacterial burdens recovered from the livers and spleens of mice infected with the ΔprsA2 mutant in comparison to mice infected with wild-type L. monocytogenes (1, 121). Based on the mutant's severely attenuated phenotype, it was somewhat surprising that ΔprsA2 mutants exhibited only relatively modest reductions in secreted LLO and PC-PLC activity. Previous work has indicated that L. monocytogenes mutants that produce as little as one-tenth of the LLO normally secreted in broth culture are still capable of efficient escape from host cell vacuoles (31). Consistent with this observation, the ΔprsA2 mutant gained access to the host cytosol with kinetics that resembled those observed for cells infected with the wild-type strain (1). PC-PLC has been demonstrated to facilitate bacterial escape from the secondary vacuoles formed during cell-to-cell spread; however, a complete-loss-of-function ΔplcB mutant is attenuated for virulence in mice by only approximately 10-fold (92, 101). While modest reductions in LLO and PC-PLC activity seem unlikely to account for the full magnitude of the virulence defect observed for ΔprsA2 mutants, it remains possible that the reduction in LLO and PC-PLC activity combined with a loss in activity of other PrsA2-dependent substrate proteins might be sufficient to account for the severe attenuation. Proteomic analyses of bacterial supernatants have identified a number of proteins whose localization and/or stability was altered in the absence of PrsA2 (Fig. 4). While these proteins may not all be bona fide PrsA2 substrates, it seems feasible that a subset of these proteins represent PrsA2-interacting partners with roles in bacterial virulence. At least 3 of the proteins identified have been associated with PrsA2 activity and/or PrsA2-dependent phenotypes (LLO, PC-PLC, and FlaA) (1, 121). Additionally, similar proteomics-based approaches have been used successfully to identify substrates for PrsA of B. subtilis and for the SurA PPIase chaperone of Escherichia coli (103, 106).
Proteins identified based on differential expression patterns in the presence or absence of PrsA2 included recognized virulence factors as well as a substantial number of proteins with reported roles in cell surface and cell wall metabolism. Several gene products were identified as potential penicillin binding proteins (PBPs), including Lmo2522, Lmo2039, Lmo1892, and Lmo0540 (43, 74). PBPs are associated with the synthesis and structural integrity of the bacterial cell wall via their transglycosylase and transpeptidase activities and have been most studied in L. monocytogenes as targets of β-lactam antibiotics (38, 39, 44-46, 55, 56, 71, 73, 99, 104, 105). It is possible that in the absence of PrsA2, altered PBP activity negatively impacts the integrity of the cell wall and results in the aberrant secretion of a number of proteins. PrsA-dependent maintenance of PBP activity/stability was recently demonstrated to be critical for the maintenance of cell wall integrity in B. subtilis (49). In addition to PBPs, a number of transport/binding proteins were identified, including OppA and CtaP. OppA and CtaP are oligopeptide- and cysteine-associated transport proteins (respectively) that contribute to bacterial survival within the host (8, 74, 117). CtaP secretion is increased following PrfA activation, and the loss of CtaP has been associated with alterations in bacterial membrane integrity; thus, CtaP could potentially contribute to the integrity defect observed for ΔprsA2 prfA(L140F) mutants (74, 117). Lastly, proteins involved in the protection of the bacterium from reactive oxygen species (catalase and superoxide dismutase) were found to be altered in localization in prsA2 mutant strains. However, neither ΔprsA2 nor ΔprsA2 prfA(L140F) mutants demonstrated any increase in sensitivity to reactive oxygen intermediates in in vitro assays (F. Alonzo and N. Freitag, unpublished).
One informative finding resulting from the proteomic analyses of both ΔprsA2 and ΔprsA2 prfA(L140F) mutant-derived supernatants was the identification of multiple proteins associated with functional roles in the bacterial cytosol. For ΔprsA2 mutants, at least 9 of these cytosol-associated proteins have previously been detected in L. monocytogenes supernatants, and no obvious alteration in membrane permeability or evidence of increased bacterial cell lysis/death was observed for ΔprsA2 mutants in comparison to wild-type strains (Fig. 5) (23, 97). An increase in the number of proteins associated with the bacterial cytosol was especially evident in the supernatants derived from ΔprsA2 prfA(L140F) strains, and these mutants were confirmed to exhibit significant alterations in membrane integrity as well as increased cell lysis. Based on these observations, we speculate that PrsA2 plays a critical role not only in facilitating normal protein secretion but also in maintaining bacterial viability following PrfA activation and the associated increased secretion of PrfA-dependent virulence gene products.
For the examination of the effects of PrfA activation on PrsA2-dependent functions, the mutationally activated prfA* allele was a useful tool for rapidly comparing differences in exoprotein profiles in culture (74). PrsA2-associated defects in bacterial viability were not obvious for strains containing wild-type prfA in broth culture (conditions under which PrfA is not activated) but were detectable for bacteria located within the cytosol after several hours of growth, as shown by a plateau in bacterial numbers beginning at approximately 9 h postinfection (Fig. 1 and 8). The bacterial growth defect observed for intracellular ΔprsA2 mutants was exacerbated by the deletion of a second posttranslocation chaperone with protease activity, HtrA (94, 113, 114). Interestingly, work with E. coli has demonstrated that the loss of the PPIase chaperone SurA in combination with HtrA is lethal, inducing cell death in response to the accumulation of misfolded proteins at the membrane (15, 78, 89). For L. monocytogenes, the most significant decrease in bacterial viability was evident only under conditions of PrfA activation (within the host cell cytosol or within infected mice) (Fig. 3 and 8), indicating that neither PrsA2 nor HtrA is required for normal L. monocytogenes physiology in broth culture.
PrsA2 thus appears to have been adapted to facilitate L. monocytogenes survival under conditions of increased protein secretion, such as occurs during PrfA activation within host cells. We speculate that the dramatic attenuation observed for ΔprsA2 mutants in animal infection models reflects not only the diminished activity of PrsA2 substrates (such as LLO) but also the decrease in bacterial viability that results from the accumulation of misfolded proteins at the membrane-cell wall interface. It will be interesting to determine if the combined loss of PrsA2 and HtrA under conditions of PrfA activation serves to trigger the induction of a membrane stress response that ultimately leads to bacterial cell death, similar to the loss of SurA and HtrA in E. coli (15, 78, 89).
Acknowledgments
We thank Rebecca Wilson for providing the ΔhtrA deletion mutant in 10403S, Bobbi Xayarath for assistance with animal studies, and members of the Freitag lab for helpful discussions. Proteomics and informatic services were provided by the CBC-UIC Research Resources Center Proteomics and Informatics Services Facility, which was established by a grant from the Searle Funds at the Chicago Community Trust to the Chicago Biomedical Consortium.
This work was supported by Public Health Service grant AI41816 (N.E.F.) from NIAID.
The contents of the article are solely the responsibility of the authors and do not necessarily represent the official views of the funding sources.
Editor: A. Camilli
Footnotes
Published ahead of print on 7 September 2010.
REFERENCES
- 1.Alonzo, F., III, G. C. Port, M. Cao, and N. E. Freitag. 2009. The posttranslocation chaperone PrsA2 contributes to multiple facets of Listeria monocytogenes pathogenesis. Infect. Immun. 77:2612-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Archambaud, C., M. A. Nahori, J. Pizarro-Cerda, P. Cossart, and O. Dussurget. 2006. Control of Listeria superoxide dismutase by phosphorylation. J. Biol. Chem. 281:31812-31822. [DOI] [PubMed] [Google Scholar]
- 3.Auerbuch, V., L. L. Lenz, and D. A. Portnoy. 2001. Development of a competitive index assay to evaluate the virulence of Listeria monocytogenes actA mutants during primary and secondary infection of mice. Infect. Immun. 69:5953-5957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett, H. J., D. M. Pearce, S. Glenn, C. M. Taylor, M. Kuhn, A. L. Sonenshein, P. W. Andrew, and I. S. Roberts. 2007. Characterization of relA and codY mutants of Listeria monocytogenes: identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63:1453-1467. [DOI] [PubMed] [Google Scholar]
- 5.Beveridge, T. J., and R. G. Murray. 1980. Sites of metal deposition in the cell wall of Bacillus subtilis. J. Bacteriol. 141:876-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bierne, H., C. Sabet, N. Personnic, and P. Cossart. 2007. Internalins: a complex family of leucine-rich repeat-containing proteins in Listeria monocytogenes. Microbes Infect. 9:1156-1166. [DOI] [PubMed] [Google Scholar]
- 7.Bigot, A., E. Botton, I. Dubail, and A. Charbit. 2006. A homolog of Bacillus subtilis trigger factor in Listeria monocytogenes is involved in stress tolerance and bacterial virulence. Appl. Environ. Microbiol. 72:6623-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boujemaa-Paterski, R., E. Gouin, G. Hansen, S. Samarin, C. Le Clainche, D. Didry, P. Dehoux, P. Cossart, C. Kocks, M. F. Carlier, and D. Pantaloni. 2001. Listeria protein ActA mimics WASp family proteins: it activates filament barbed end branching by Arp2/3 complex. Biochemistry 40:11390-11404. [DOI] [PubMed] [Google Scholar]
- 10.Brundage, R. A., G. A. Smith, A. Camilli, J. A. Theriot, and D. A. Portnoy. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. U. S. A. 90:11890-11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camilli, A., C. R. Paynton, and D. A. Portnoy. 1989. Intracellular methicillin selection of Listeria monocytogenes mutants unable to replicate in a macrophage cell line. Proc. Natl. Acad. Sci. U. S. A. 86:5522-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cascales, E. 2008. The type VI secretion toolkit. EMBO Rep. 9:735-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatterjee, S. S., H. Hossain, S. Otten, C. Kuenne, K. Kuchmina, S. Machata, E. Domann, T. Chakraborty, and T. Hain. 2006. Intracellular gene expression profile of Listeria monocytogenes. Infect. Immun. 74:1323-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christensen, D. P., A. K. Benson, and R. W. Hutkins. 1998. Cloning and expression of the Listeria monocytogenes Scott A ptsH and ptsI genes, coding for HPr and enzyme I, respectively, of the phosphotransferase system. Appl. Environ. Microbiol. 64:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443-455. [DOI] [PubMed] [Google Scholar]
- 16.Desvaux, M., and M. Hebraud. 2006. The protein secretion systems in Listeria: inside out bacterial virulence. FEMS Microbiol. Rev. 30:774-805. [DOI] [PubMed] [Google Scholar]
- 17.Desvaux, M., M. Hebraud, R. Talon, and I. R. Henderson. 2009. Secretion and subcellular localizations of bacterial proteins: a semantic awareness issue. Trends Microbiol. 17:139-145. [DOI] [PubMed] [Google Scholar]
- 18.Domann, E., J. Wehland, M. Rohde, S. Pistor, M. Hartl, W. Goebel, M. Leimeister-Wachter, M. Wuenscher, and T. Chakraborty. 1992. A novel bacterial gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 11:1981-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Domann, E., S. Zechel, A. Lingnau, T. Hain, A. Darji, T. Nichterlein, J. Wehland, and T. Chakraborty. 1997. Identification and characterization of a novel PrfA-regulated gene in Listeria monocytogenes whose product, IrpA, is highly homologous to internalin proteins, which contain leucine-rich repeats. Infect. Immun. 65:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donnenberg, M. S. 2000. Pathogenic strategies of enteric bacteria. Nature 406:768-774. [DOI] [PubMed] [Google Scholar]
- 21.Dramsi, S., and P. Cossart. 2003. Listeriolysin O-mediated calcium influx potentiates entry of Listeria monocytogenes into the human Hep-2 epithelial cell line. Infect. Immun. 71:3614-3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duche, O., F. Tremoulet, P. Glaser, and J. Labadie. 2002. Salt stress proteins induced in Listeria monocytogenes. Appl. Environ. Microbiol. 68:1491-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dumas, E., M. Desvaux, C. Chambon, and M. Hebraud. 2009. Insight into the core and variant exoproteomes of Listeria monocytogenes species by comparative subproteomic analysis. Proteomics 9:3136-3155. [DOI] [PubMed] [Google Scholar]
- 24.Engelbrecht, F., S. K. Chun, C. Ochs, J. Hess, F. Lottspeich, W. Goebel, and Z. Sokolovic. 1996. A new PrfA-regulated gene of Listeria monocytogenes encoding a small, secreted protein which belongs to the family of internalins. Mol. Microbiol. 21:823-837. [DOI] [PubMed] [Google Scholar]
- 25.Fiorini, F., S. Stefanini, P. Valenti, E. Chiancone, and D. De Biase. 2008. Transcription of the Listeria monocytogenes fri gene is growth-phase dependent and is repressed directly by Fur, the ferric uptake regulator. Gene 410:113-121. [DOI] [PubMed] [Google Scholar]
- 26.Fisher, C. W., D. Lee, B. A. Dodge, K. M. Hamman, J. B. Robbins, and S. E. Martin. 2000. Influence of catalase and superoxide dismutase on ozone inactivation of Listeria monocytogenes. Appl. Environ. Microbiol. 66:1405-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Folio, P., P. Chavant, I. Chafsey, A. Belkorchia, C. Chambon, and M. Hebraud. 2004. Two-dimensional electrophoresis database of Listeria monocytogenes EGDe proteome and proteomic analysis of mid-log and stationary growth phase cells. Proteomics 4:3187-3201. [DOI] [PubMed] [Google Scholar]
- 28.Freitag, N. E. 2006. From hot dogs to host cells: how the bacterial pathogen Listeria monocytogenes regulates virulence gene expression. Future Microbiol. 1:89-101. [DOI] [PubMed] [Google Scholar]
- 29.Freitag, N. E. 2000. Genetic tools for use with Listeria monocytogenes, p. 488-498. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-postive pathogens. ASM Press, Washington, DC.
- 30.Freitag, N. E., G. C. Port, and M. D. Miner. 2009. Listeria monocytogenes—from saprophyte to intracellular pathogen. Nat. Rev. Microbiol. 7:623-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freitag, N. E., and D. A. Portnoy. 1994. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol. Microbiol. 12:845-853. [DOI] [PubMed] [Google Scholar]
- 32.Gaillard, J. L., P. Berche, C. Frehel, E. Gouin, and P. Cossart. 1991. Entry of L. monocytogenes into cells is mediated by internalin, a repeat protein reminiscent of surface antigens from gram-positive cocci. Cell 65:1127-1141. [DOI] [PubMed] [Google Scholar]
- 33.Gaillot, O., S. Bregenholt, F. Jaubert, J. P. Di Santo, and P. Berche. 2001. Stress-induced ClpP serine protease of Listeria monocytogenes is essential for induction of listeriolysin O-dependent protective immunity. Infect. Immun. 69:4938-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 35.Gerlach, R. G., and M. Hensel. 2007. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int. J. Med. Microbiol. 297:401-415. [DOI] [PubMed] [Google Scholar]
- 36.Glomski, I. J., A. L. Decatur, and D. A. Portnoy. 2003. Listeria monocytogenes mutants that fail to compartmentalize listerolysin O activity are cytotoxic, avirulent, and unable to evade host extracellular defenses. Infect. Immun. 71:6754-6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glomski, I. J., M. M. Gedde, A. W. Tsang, J. A. Swanson, and D. A. Portnoy. 2002. The Listeria monocytogenes hemolysin has an acidic pH optimum to compartmentalize activity and prevent damage to infected host cells. J. Cell Biol. 156:1029-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gravesen, A., B. Kallipolitis, K. Holmstrom, P. E. Hoiby, M. Ramnath, and S. Knochel. 2004. pbp2229-mediated nisin resistance mechanism in Listeria monocytogenes confers cross-protection to class IIa bacteriocins and affects virulence gene expression. Appl. Environ. Microbiol. 70:1669-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gravesen, A., K. Sorensen, F. M. Aarestrup, and S. Knochel. 2001. Spontaneous nisin-resistant Listeria monocytogenes mutants with increased expression of a putative penicillin-binding protein and their sensitivity to various antibiotics. Microb. Drug Resist. 7:127-135. [DOI] [PubMed] [Google Scholar]
- 40.Gray, M. J., N. E. Freitag, and K. J. Boor. 2006. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect. Immun. 74:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grundling, A., M. D. Gonzalez, and D. E. Higgins. 2003. Requirement of the Listeria monocytogenes broad-range phospholipase PC-PLC during infection of human epithelial cells. J. Bacteriol. 185:6295-6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gueriri, I., S. Bay, S. Dubrac, C. Cyncynatus, and T. Msadek. 2008. The Pta-AckA pathway controlling acetyl phosphate levels and the phosphorylation state of the DegU orphan response regulator both play a role in regulating Listeria monocytogenes motility and chemotaxis. Mol. Microbiol. 70:1342-1357. [DOI] [PubMed] [Google Scholar]
- 43.Guinane, C. M., P. D. Cotter, R. P. Ross, and C. Hill. 2006. Contribution of penicillin-binding protein homologs to antibiotic resistance, cell morphology, and virulence of Listeria monocytogenes EGDe. Antimicrob. Agents Chemother. 50:2824-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gutkind, G. O., M. E. Mollerach, and R. A. De Torres. 1989. Penicillin-binding proteins in Listeria monocytogenes. APMIS 97:1013-1017. [DOI] [PubMed] [Google Scholar]
- 45.Gutkind, G. O., S. B. Ogueta, A. C. de Urtiaga, M. E. Mollerach, and R. A. de Torres. 1990. Participation of PBP 3 in the acquisition of dicloxacillin resistance in Listeria monocytogenes. J. Antimicrob. Chemother. 25:751-758. [DOI] [PubMed] [Google Scholar]
- 46.Hakenbeck, R., and H. Hof. 1991. Relatedness of penicillin-binding proteins from various Listeria species. FEMS Microbiol. Lett. 68:191-195. [DOI] [PubMed] [Google Scholar]
- 47.Henry, R., L. Shaughnessy, M. J. Loessner, C. Alberti-Segui, D. E. Higgins, and J. A. Swanson. 2006. Cytolysin-dependent delay of vacuole maturation in macrophages infected with Listeria monocytogenes. Cell. Microbiol. 8:107-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hyyrylainen, H. L., A. Bolhuis, E. Darmon, L. Muukkonen, P. Koski, M. Vitikainen, M. Sarvas, Z. Pragai, S. Bron, J. M. van Dijl, and V. P. Kontinen. 2001. A novel two-component regulatory system in Bacillus subtilis for the survival of severe secretion stress. Mol. Microbiol. 41:1159-1172. [DOI] [PubMed] [Google Scholar]
- 49.Hyyrylainen, H. L., B. C. Marciniak, K. Dahncke, M. Pietiainen, P. Courtin, M. Vitikainen, R. Seppala, A. Otto, D. Becher, M. P. Chapot-Chartier, O. P. Kuipers, and V. P. Kontinen. 2010. Penicillin-binding protein folding is dependent on the PrsA peptidyl-prolyl cis-trans isomerase in Bacillus subtilis. Mol. Microbiol. 77:108-127. [DOI] [PubMed]
- 50.Ireton, K. 2007. Entry of the bacterial pathogen Listeria monocytogenes into mammalian cells. Cell. Microbiol. 9:1365-1375. [DOI] [PubMed] [Google Scholar]
- 51.Kim, H., K. J. Boor, and H. Marquis. 2004. Listeria monocytogenes sigmaB contributes to invasion of human intestinal epithelial cells. Infect. Immun. 72:7374-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim, H., H. Marquis, and K. J. Boor. 2005. SigmaB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology 151:3215-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kingdon, G. C., and C. P. Sword. 1970. Biochemical and immunological effects of Listeria monocytogenes hemolysin. Infect. Immun. 1:363-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kocks, C., E. Gouin, M. Tabouret, P. Berche, H. Ohayon, and P. Cossart. 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521-531. [DOI] [PubMed] [Google Scholar]
- 55.Korsak, D., W. Vollmer, and Z. Markiewicz. 2005. Listeria monocytogenes EGD lacking penicillin-binding protein 5 (PBP5) produces a thicker cell wall. FEMS Microbiol. Lett. 251:281-288. [DOI] [PubMed] [Google Scholar]
- 56.Korsak, D., J. J. Zawadzka, M. E. Siwinska, and Z. Markiewicz. 2002. Penicillin-binding proteins of Listeria monocytogenes—a re-evaluation. Acta Microbiol. Pol. 51:5-12. [PubMed] [Google Scholar]
- 57.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leblond-Francillard, M., J. L. Gaillard, and P. Berche. 1989. Loss of catalase activity in Tn1545-induced mutants does not reduce growth of Listeria monocytogenes in vivo. Infect. Immun. 57:2569-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leisner, J. J., M. H. Larsen, R. L. Jorgensen, L. Brondsted, L. E. Thomsen, and H. Ingmer. 2008. Chitin hydrolysis by Listeria spp., including L. monocytogenes. Appl. Environ. Microbiol. 74:3823-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lingnau, A., E. Domann, M. Hudel, M. Bock, T. Nichterlein, J. Wehland, and T. Chakraborty. 1995. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect. Immun. 63:3896-3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu, S., J. E. Graham, L. Bigelow, P. D. Morse II, and B. J. Wilkinson. 2002. Identification of Listeria monocytogenes genes expressed in response to growth at low temperature. Appl. Environ. Microbiol. 68:1697-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marlovits, T. C., and C. E. Stebbins. 2009. Type III secretion systems shape up as they ship out. Curr. Opin. Microbiol. 13:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marquis, H., V. Doshi, and D. A. Portnoy. 1995. The broad-range phospholipase C and a metalloprotease mediate listeriolysin O-independent escape of Listeria monocytogenes from a primary vacuole in human epithelial cells. Infect. Immun. 63:4531-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marquis, H., H. Goldfine, and D. A. Portnoy. 1997. Proteolytic pathways of activation and degradation of a bacterial phospholipase C during intracellular infection by Listeria monocytogenes. J. Cell Biol. 137:1381-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marquis, H., and E. J. Hager. 2000. pH-regulated activation and release of a bacteria-associated phospholipase C during intracellular infection by Listeria monocytogenes. Mol. Microbiol. 35:289-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miner, M. D., G. C. Port, H. G. Bouwer, J. C. Chang, and N. E. Freitag. 2008. A novel prfA mutation that promotes Listeria monocytogenes cytosol entry but reduces bacterial spread and cytotoxicity. Microb. Pathog. 45:273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miner, M. D., G. C. Port, and N. E. Freitag. 2008. Functional impact of mutational activation on the Listeria monocytogenes central virulence regulator PrfA. Microbiology 154:3579-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mueller, K. J., and N. E. Freitag. 2005. Pleiotropic enhancement of bacterial pathogenesis resulting from the constitutive activation of the Listeria monocytogenes regulatory factor PrfA. Infect. Immun. 73:1917-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 70.Petit-Glatron, M. F., L. Grajcar, A. Munz, and R. Chambert. 1993. The contribution of the cell wall to a transmembrane calcium gradient could play a key role in Bacillus subtilis protein secretion. Mol. Microbiol. 9:1097-1106. [DOI] [PubMed] [Google Scholar]
- 71.Pierre, J., A. Boisivon, and L. Gutmann. 1990. Alteration of PBP 3 entails resistance to imipenem in Listeria monocytogenes. Antimicrob. Agents Chemother. 34:1695-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pizarro-Cerda, J., and P. Cossart. 2006. Subversion of cellular functions by Listeria monocytogenes. J. Pathol. 208:215-223. [DOI] [PubMed] [Google Scholar]
- 73.Poros-Gluchowska, J., M. Kloszewska, and Z. Markiewicz. 2003. Ampicillin resistance in Listeria monocytogenes acquired as a result of transposon mutagenesis. Acta Microbiol. Pol. 52:131-142. [PubMed] [Google Scholar]
- 74.Port, G. C., and N. E. Freitag. 2007. Identification of novel Listeria monocytogenes secreted virulence factors following mutational activation of the central virulence regulator, PrfA. Infect. Immun. 75:5886-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Poyart, C., E. Abachin, I. Razafimanantsoa, and P. Berche. 1993. The zinc metalloprotease of Listeria monocytogenes is required for maturation of phosphatidylcholine phospholipase C: direct evidence obtained by gene complementation. Infect. Immun. 61:1576-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Raveneau, J., C. Geoffroy, J. Beretti, J. Gaillard, J. E. Alouf, and P. Berche. 1992. Reduced virulence of a Listeria monocytogenes phospholipase-deficient mutant obtained by transposon insertion into the zinc metalloprotease gene. Infect. Immun. 60:916-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ripio, M.-T., G. Dominguez-Bernal, M. Lara, M. Suarez, and J.-A. Vazquez-Boland. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rizzitello, A. E., J. R. Harper, and T. J. Silhavy. 2001. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J. Bacteriol. 183:6794-6800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Russel, M. 1998. Macromolecular assembly and secretion across the bacterial cell envelope: type II protein secretion systems. J. Mol. Biol. 279:485-499. [DOI] [PubMed] [Google Scholar]
- 80.Sarvas, M., C. R. Harwood, S. Bron, and J. M. van Dijl. 2004. Post-translocational folding of secretory proteins in Gram-positive bacteria. Biochim. Biophys. Acta 1694:311-327. [DOI] [PubMed] [Google Scholar]
- 81.Schaumburg, J., O. Diekmann, P. Hagendorff, S. Bergmann, M. Rohde, S. Hammerschmidt, L. Jansch, J. Wehland, and U. Karst. 2004. The cell wall subproteome of Listeria monocytogenes. Proteomics 4:2991-3006. [DOI] [PubMed] [Google Scholar]
- 82.Schnupf, P., J. Hofmann, J. Norseen, I. J. Glomski, H. Schwartzstein, and A. L. Decatur. 2006. Regulated translation of listeriolysin O controls virulence of Listeria monocytogenes. Mol. Microbiol. 61:999-1012. [DOI] [PubMed] [Google Scholar]
- 83.Schnupf, P., and D. A. Portnoy. 2007. Listeriolysin O: a phagosome-specific lysin. Microbes Infect. 9:1176-1187. [DOI] [PubMed] [Google Scholar]
- 84.Schnupf, P., J. Zhou, A. Varshavsky, and D. A. Portnoy. 2007. Listeriolysin O secreted by Listeria monocytogenes into the host cell cytosol is degraded by the N-end rule pathway. Infect. Immun. 75:5135-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schubert, K., A. M. Bichlmaier, E. Mager, K. Wolff, G. Ruhland, and F. Fiedler. 2000. P45, an extracellular 45 kDa protein of Listeria monocytogenes with similarity to protein p60 and exhibiting peptidoglycan lytic activity. Arch. Microbiol. 173:21-28. [DOI] [PubMed] [Google Scholar]
- 86.Scortti, M., H. J. Monzo, L. Lacharme-Lora, D. A. Lewis, and J. A. Vazquez-Boland. 2007. The PrfA virulence regulon. Microbes Infect. 9:1196-1207. [DOI] [PubMed] [Google Scholar]
- 87.Seveau, S., J. Pizarro-Cerda, and P. Cossart. 2007. Molecular mechanisms exploited by Listeria monocytogenes during host cell invasion. Microbes Infect. 9:1167-1175. [DOI] [PubMed] [Google Scholar]
- 88.Shetron-Rama, L. M., K. Mueller, J. M. Bravo, H. G. Bouwer, S. S. Way, and N. E. Freitag. 2003. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol. Microbiol. 48:1537-1551. [DOI] [PubMed] [Google Scholar]
- 89.Sklar, J. G., T. Wu, D. Kahne, and T. J. Silhavy. 2007. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 21:2473-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Skoble, J., V. Auerbuch, E. D. Goley, M. D. Welch, and D. A. Portnoy. 2001. Pivotal role of VASP in Arp2/3 complex-mediated actin nucleation, actin branch-formation, and Listeria monocytogenes motility. J. Cell Biol. 155:89-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Skoble, J., D. A. Portnoy, and M. D. Welch. 2000. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J. Cell Biol. 150:527-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Smith, G. A., H. Marquis, S. Jones, N. C. Johnston, D. A. Portnoy, and H. Goldfine. 1995. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 63:4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith, G. A., and D. A. Portnoy. 1997. How the Listeria monocytogenes ActA protein converts actin polymerization into a motile force. Trends Microbiol. 5:272-276. [DOI] [PubMed] [Google Scholar]
- 94.Stack, H. M., R. D. Sleator, M. Bowers, C. Hill, and C. G. Gahan. 2005. Role for HtrA in stress induction and virulence potential in Listeria monocytogenes. Appl. Environ. Microbiol. 71:4241-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sun, A. N., A. Camilli, and D. A. Portnoy. 1990. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 58:3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Theriot, J. A., T. J. Mitchison, L. G. Tilney, and D. A. Portnoy. 1992. The rate of actin-based motility of intracellular Listeria monocytogenes equals the rate of actin polymerization. Nature 357:257-260. [DOI] [PubMed] [Google Scholar]
- 97.Trost, M., D. Wehmhoner, U. Karst, G. Dieterich, J. Wehland, and L. Jansch. 2005. Comparative proteome analysis of secretory proteins from pathogenic and nonpathogenic Listeria species. Proteomics 5:1544-1557. [DOI] [PubMed] [Google Scholar]
- 98.van den Ent, F., M. Leaver, F. Bendezu, J. Errington, P. de Boer, and J. Lowe. 2006. Dimeric structure of the cell shape protein MreC and its functional implications. Mol. Microbiol. 62:1631-1642. [DOI] [PubMed] [Google Scholar]
- 99.Van de Velde, S., S. Carryn, F. Van Bambeke, C. Hill, P. M. Tulkens, and R. D. Sleator. 2009. Penicillin-binding proteins (PBP) and Lmo0441 (a PBP-like protein) play a role in beta-lactam sensitivity of Listeria monocytogenes. Gut Pathog. 1:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Vasconcelos, J. A., and H. G. Deneer. 1994. Expression of superoxide dismutase in Listeria monocytogenes. Appl. Environ. Microbiol. 60:2360-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vazquez-Boland, J., C. Kocks, S. Dramsi, H. Ohayon, C. Geoffroy, J. Mengaud, and P. Cossart. 1992. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect. Immun. 60:219-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vertommen, D., N. Ruiz, P. Leverrier, T. J. Silhavy, and J. F. Collet. 2009. Characterization of the role of the Escherichia coli periplasmic chaperone SurA using differential proteomics. Proteomics 9:2432-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vicente, M. F., J. Berenguer, M. A. de Pedro, J. C. Perez-Diaz, and F. Baquero. 1990. Penicillin binding proteins in Listeria monocytogenes. Acta Microbiol. Hung. 37:227-231. [PubMed] [Google Scholar]
- 105.Vicente, M. F., J. C. Perez-Daz, F. Baquero, M. Angel de Pedro, and J. Berenguer. 1990. Penicillin-binding protein 3 of Listeria monocytogenes as the primary lethal target for beta-lactams. Antimicrob. Agents Chemother. 34:539-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vitikainen, M., I. Lappalainen, R. Seppala, H. Antelmann, H. Boer, S. Taira, H. Savilahti, M. Hecker, M. Vihinen, M. Sarvas, and V. P. Kontinen. 2004. Structure-function analysis of PrsA reveals roles for the parvulin-like and flanking N- and C-terminal domains in protein folding and secretion in Bacillus subtilis. J. Biol. Chem. 279:19302-19314. [DOI] [PubMed] [Google Scholar]
- 107.Vitikainen, M., T. Pummi, U. Airaksinen, E. Wahlstrom, H. Wu, M. Sarvas, and V. P. Kontinen. 2001. Quantitation of the capacity of the secretion apparatus and requirement for PrsA in growth and secretion of alpha-amylase in Bacillus subtilis. J. Bacteriol. 183:1881-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wahlstrom, E., M. Vitikainen, V. P. Kontinen, and M. Sarvas. 2003. The extracytoplasmic folding factor PrsA is required for protein secretion only in the presence of the cell wall in Bacillus subtilis. Microbiology 149:569-577. [DOI] [PubMed] [Google Scholar]
- 109.Way, S. S., L. J. Thompson, J. E. Lopes, A. M. Hajjar, T. R. Kollmann, N. E. Freitag, and C. B. Wilson. 2004. Characterization of flagellin expression and its role in Listeria monocytogenes infection and immunity. Cell. Microbiol. 6:235-242. [DOI] [PubMed] [Google Scholar]
- 110.Webb, A. J., M. Karatsa-Dodgson, and A. Grundling. 2009. Two-enzyme systems for glycolipid and polyglycerolphosphate lipoteichoic acid synthesis in Listeria monocytogenes. Mol. Microbiol. 74:299-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Weidenmaier, C., and A. Peschel. 2008. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat. Rev. Microbiol. 6:276-287. [DOI] [PubMed] [Google Scholar]
- 112.Wemekamp-Kamphuis, H. H., J. A. Wouters, P. P. de Leeuw, T. Hain, T. Chakraborty, and T. Abee. 2004. Identification of sigma factor sigma B-controlled genes and their impact on acid stress, high hydrostatic pressure, and freeze survival in Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 70:3457-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wilson, R. L., L. L. Brown, D. Kirkwood-Watts, T. K. Warren, S. A. Lund, D. S. King, K. F. Jones, and D. E. Hruby. 2006. Listeria monocytogenes 10403S HtrA is necessary for resistance to cellular stress and virulence. Infect. Immun. 74:765-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wonderling, L. D., B. J. Wilkinson, and D. O. Bayles. 2004. The htrA (degP) gene of Listeria monocytogenes 10403S is essential for optimal growth under stress conditions. Appl. Environ. Microbiol. 70:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wong, K. K., H. G. Bouwer, and N. E. Freitag. 2004. Evidence implicating the 5′ untranslated region of Listeria monocytogenes actA in the regulation of bacterial actin-based motility. Cell. Microbiol. 6:155-166. [DOI] [PubMed] [Google Scholar]
- 116.Wong, K. K., and N. E. Freitag. 2004. A novel mutation within the central Listeria monocytogenes regulator PrfA that results in constitutive expression of virulence gene products. J. Bacteriol. 186:6265-6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xayarath, B., H. Marquis, G. C. Port, and N. E. Freitag. 2009. Listeria monocytogenes CtaP is a multifunctional cysteine transport-associated protein required for bacterial pathogenesis. Mol. Microbiol. 74:956-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yeung, P. S., Y. Na, A. J. Kreuder, and H. Marquis. 2007. Compartmentalization of the broad-range phospholipase C activity to the spreading vacuole is critical for Listeria monocytogenes virulence. Infect. Immun. 75:44-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yeung, P. S., N. Zagorski, and H. Marquis. 2005. The metalloprotease of Listeria monocytogenes controls cell wall translocation of the broad-range phospholipase C. J. Bacteriol. 187:2601-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Young, G. M., D. H. Schmiel, and V. L. Miller. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. U. S. A. 96:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zemansky, J., B. C. Kline, J. J. Woodward, J. H. Leber, H. Marquis, and D. A. Portnoy. 2009. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J. Bacteriol. 191:3950-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]