Abstract
Trachoma is the leading infectious cause of blindness and is endemic in 52 countries. There is a critical need to further our understanding of the host response during disease and infection, as millions of individuals are still at risk of developing blinding sequelae. Infection of the conjunctival epithelial cells by the causative bacterium, Chlamydia trachomatis, stimulates an acute host response. The main clinical feature is a follicular conjunctivitis that is incompletely defined at the tissue-specific gene expression and molecular levels. To explore the features of disease and the response to infection, we measured host gene expression in conjunctival samples from Gambian children with active trachoma and healthy controls. Genome-wide expression and transcription network analysis identified signatures characteristic of the expected infiltrating immune cell populations, such as neutrophils and T/B lymphocytes. The expression signatures were also significantly enriched for genes in pathways which regulate NK cell activation and cytotoxicity, antigen processing and presentation, chemokines, cytokines, and cytokine receptors. The data suggest that in addition to polymorph and adaptive cellular responses, NK cells may contribute to a significant component of the conjunctival inflammatory response to chlamydial infection.
Chlamydia trachomatis is an obligate intracellular bacterium that infects millions of people worldwide. Infection of conjunctival epithelial cells causes trachoma, which is the world's leading infectious cause of blindness, affecting over 40 million people in the developing world (50). C. trachomatis is also the world's most common bacterial sexually transmitted infection (STI), with an estimated 92 million new cases of C. trachomatis urogenital infections occurring annually (74). Asymptomatic infection is common, and untreated cases are at risk of developing complications related to fertility and pregnancy (6). A vaccine to prevent C. trachomatis infection or disease progression would be of great value, but the development of such a vaccine is handicapped by the fact that the immunological correlates of protective immunity and pathogenesis are not well understood.
The immune and inflammatory responses initiated by C. trachomatis infection, although important for successful control and resolution of infection, are thought to be at least partly responsible for tissue damage and its sequelae (10). Some progress has been made in dissecting correlates of protective immunity and immunopathology in humans (14), but reports are dominated by studies using the mouse as a model system (reviewed in reference 54). The extrapolation of results obtained from murine experimental models needs cautious interpretation. Several papers have demonstrated the exquisite and often subtle adaptations of chlamydial parasites to their natural host tissues and the specificity of the molecular pathways that control intracellular replication (53, 58, 67).
Ocular C. trachomatis infection is readily accessible to examination and investigation. As a result, the clinical and epidemiological features of trachoma and the phases of disease are well documented in many populations (49). Trachomatous inflammation (follicular) (TF) and/or trachomatous inflammation (intense) (TI) is characterized by C. trachomatis-driven conjunctival inflammation and is most prevalent during childhood (<10 years). Conversely, the sequelae of these earlier stages, i.e., trachomatous conjunctival scarring (TS), trachomatous trichiasis (TT), and corneal opacity (CO), are most prevalent in adults (>45 years) and require repeated or persistent episodes of infection and inflammation.
Gene expression profiling studies have used a variety of array platforms to study the host response to chlamydial infection (2, 18, 32, 43, 44, 47, 64, 66, 70, 86, 89, 93). With the exception of two recent studies (43, 64), each has described the in vitro response in experimentally infected cell lines or cells isolated from tissue. Numerous genes and pathways have been implicated as being important in the innate response to infection. Thus far, there have been no transcriptome-defining studies of human tissues that are infected or diseased as a result of natural chlamydial infection. To gain a better understanding of the immune and inflammatory responses to ocular C. trachomatis infection in humans, we collected conjunctival swabs from the upper tarsal conjunctiva of Gambian children with active trachoma and examined their transcriptomes using genome-wide expression arrays.
The extraction of biological meaning from microarray data is challenging and complex. This has led to the development of many new computational tools and methods for their analysis. We used statistical methods to define differential gene expression and a graph theoretic method to define networks of coexpressed transcripts (27). The latter method analyzes the degree of correlation (coexpression) between transcripts (79) and can help define the transcriptional networks which are characteristic of the cell types present in conjunctival samples.
MATERIALS AND METHODS
Ethical approval.
The study was approved by the Gambia Government/Medical Research Council Joint Ethics Committee (reference L2006.47) and by the Ethics Committee of the London School of Hygiene and Tropical Medicine (LSHTM). Written informed consent was obtained from all study subjects or their guardians. Children diagnosed with clinical signs of active trachoma were treated according to National Eye Care Programme guidelines with topical tetracycline or a single oral dose of azithromycin (20 mg/kg).
Study subjects.
Infants and children from the North Bank, Western, and Central River regions in the Gambia were screened for clinical signs of trachoma. Sterile polyester-tipped swab (Hardwood Products, ME) samples were collected from the upper tarsal conjunctiva from 200 children with clinical signs of inflammatory trachoma and from control subjects without clinical signs of active trachoma. Clinical diagnosis was made according to the WHO detailed trachoma grading system (30); each subject was graded based on follicular score (F0 to F3), papillary score (P0 to P3), and conjunctival scarring score (C0 to C3), with the presence of clinically active trachoma indicated by a follicular score of 2 or 3 (F2/F3) or a papillary score of 3 (P3). These are equivalent to trachomatous inflammation (follicular) (TF) and trachomatous inflammation (intense) (TI) of the WHO simplified trachoma grading system, respectively (81).
Anesthetic eye drops (Minims Proxymetacaine 0.5%; Chauvin Pharmaceuticals, Romford, United Kingdom) were administered prior to swab collection. Swab collection was performed in a standardized manner (12). Two swabs per eye were collected; the first swab (swab A) was collected into RNAlater (Ambion, Huntingdon, United Kingdom), and the second swab (swab B) was collected in a dry polypropylene tube. Both samples were stored on ice until frozen (−20°C) in the laboratory on the same day. Samples were then shipped on dry ice to the London School of Hygiene and Tropical Medicine (LSHTM).
Processing of conjunctival swab samples for human gene expression studies.
Total RNA was extracted from swab A samples using the RNeasy micro kit (Qiagen, Crawley, United Kingdom) according to the manufacturer's instructions with modifications previously described (56). Total RNA was immediately stored at −80°C until processed for gene expression (within 1 month). For each total RNA sample, a 2-μl aliquot was used to estimate the concentration of RNA using a NanoDrop ND-1000 spectrophotometer (ThermoScientific, Waltham, MA). RNA quality was assessed with an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA).
Processing of conjunctival swab samples for diagnosis and quantification of C. trachomatis.
DNA was extracted from dry conjunctival swabs (swab B) for diagnosis and quantification of C. trachomatis as described by Solomon et al. (72). The Amplicor CT/NG diagnostic kit (Roche Molecular Systems, Branchburg, NJ) was used as a qualitative PCR for C. trachomatis. Both Amplicor-positive and -negative samples were then purified using DNA extraction spin columns (QIAmp DNA mini kit; Qiagen, Crawley, United Kingdom) and tested by a C. trachomatis quantitative PCR (qPCR) which amplifies a 123-bp fragment within constant domain 3 of the C. trachomatis ompA gene using sets of primers which amplify each of the reference ocular serovars. The samples were then stratified into high-load infections and low-load infections, defined as >390 and <34 copies of C. trachomatis ompA per swab, respectively (12).
Microarray analysis.
Samples for analysis were selected from subjects based on clinical diagnosis, C. trachomatis infection status (determined by a positive result in the CT/NG Amplicor diagnostic test), and adequate quantity and quality of extracted total RNA. A total of 60 samples were selected for microarray analysis: 20 samples were from subjects with active trachoma (all had ≥F2 with or without ≥P0) and C. trachomatis infection (group DI), 20 were from subjects with clinical signs of active trachoma but without detectable C. trachomatis ocular infection (group D), and 20 were from subjects without clinical signs of active trachoma in both eyes and absence of C. trachomatis infection (group N).
Total RNA (100 ng) from each subject was subjected to two rounds of in vitro transcription with biotin-labeled nucleotides (two-cycle cDNA synthesis and labeling kit; Affymetrix, Santa Clara, CA). A hybridization cocktail was made according to the Affymetrix protocol using 15 μg of biotinylated and fragmented cRNA and hybridized to Affymetrix Human Genome U133 Plus 2.0 arrays (GeneChips) for 16 h at 45°C according to the manufacturer's standard protocols. GeneChips were washed and stained following the standard Affymetrix protocol and scanned using the Affymetrix confocal laser scanner 3000. These GeneChips contained 54,675 probe sets representing ∼47,400 human transcripts. The analyzed data are available at http://www.macrophages.com/index_GE_microarray.html.
Microarray data processing and statistical analysis.
Array data files (.cel) were run through Bioconductor R (http://www.bioconductor.org/) in order to perform quality assessment and normalization. The data set was normalized using the Robust Multichip Average (RMA) algorithm (40). Normalized data were then subjected to an interquartile range (IQR) filtering, in which array features with an IQR of <0.5 log2 intensity unit were discarded, removing outliers and weakly expressed probe sets. Group mean differences in average expression for each transcript were tested through empirical Bayes moderated t tests (Bioconductor package limma). The simultaneous testing of a large number of variables was accounted for by controlling the false-discovery rate (FDR) at the 1% level (8). Large-scale differential gene expression was estimated in two ways: first by calculation of B values (log odds of p [transcript is differentially expressed]/p [transcript is not differentially expressed]), where a B value of ≥0 indicates that transcripts are more likely than not to be differentially expressed, and second by fold change gene expression, defined as transcripts with an adjusted P value of <0.01 and log2 fold change (FC) of <−0.7 (FC < 0.6) and >0.7 (FC > 1.6) or <−1.0 (FC < 0.5) and >1.0 (FC > 2.0).
Network analysis.
A sample-to-sample Pearson correlation matrix of normalized total gene expression was calculated using the cor function within R. A graph was then generated using the matrix file in BioLayout Express3D (www.biolayout.org/) (27). An overall gene expression graph was then generated using the normalized IQR-filtered gene expression data and was then arranged according to patient clinical and infection classification (i.e., N, D, and DI). Briefly, Pearson correlation coefficients (r) were calculated for all pairs of probe sets on the array, and r values of ≥0.7 were stored. These were used to construct networks represented in 3D by BioLayout Express3D in which nodes represent the probe sets (transcript) and are linked by an edge if the Pearson correlation r between the probe sets exceeds a cutoff value. After selecting a cutoff r value of ≥0.85, the graph was then partitioned using the Markov Clustering (MCL) algorithm (85) at an MCL inflation value of 2.2 (27). The inflation value controls the granularity of clustering (the higher the value, the more granular the clustering), and this value has been determined empirically as being optimal for most large expression graphs (27). The partitioned clusters contain transcripts that exhibit a high degree of coexpression across the samples that are statistically differentially expressed between subject groupings, as well as those that show no differential expression between groups (39, 48).
Gene set enrichment analysis.
Gene lists consisting of statistical and differentially expressed genes or MCL defined gene transcription clusters from network analysis were tested for enrichment or overrepresentation of content in biological pathways and cell types using the NIH database for annotation, visualization, and integrated discovery (DAVID) (http://david.abcc.ncifcrf.gov/) (37, 38). Default value settings were used, and fold enrichments with unadjusted P values (EASE scores) were calculated using a modification of the Fisher's exact test. To complement DAVID v6 analysis, two additional enrichment tools were used Onto-tools Pathway Express (http:vortex.cs.wayne.edu/projects.htm) (42) and the Molecular Signatures Database (MSigDB) (78) using the C2 “curated gene sets” and canonical pathways (www.broadinstitute.org/gsea/msigdb). Analogous to DAVID, these analysis tools calculate a hypergeometric P value that indicates the likelihood that the content of the gene list is overrepresented (or enriched) in particular terms, functions, or membership of canonical pathways.
Selection of transcripts for classification of disease and infection.
For classification we selected uniquely expressed transcripts from each of the clinical and infection groups. For the DI group, 23 transcripts that had the largest changes in fold change expression were selected. Probe sets that targeted the same transcript were not included. This set of probe sets was then used to classify the samples using a standard k nearest-neighbor (kNN) algorithm and validated within the data set using leave-one-out cross-validation (LOOCV).
Validation of differentially expressed transcripts.
Independent confirmation of differential gene expression by quantitative reverse transcription-PCR (qRT-PCR) with the same samples was not possible, due to the limited amount of RNA obtained in each sample. The data obtained from 60 samples tested on U133 plus 2.0 arrays were compared with data from an independent set of samples from the same population in which gene expression was measured using Affymetrix HG-focused target arrays. The series of HG-focused target arrays consisted of 29 arrays in total. Twenty-five of these were from clinical and infection categories equivalent to those described for the U133 plus 2.0 arrays. RNA amplification, labeling, and hybridization were carried out with the same methods described above. The samples were from 8 participants with no clinical signs of active trachoma (F0 and <P2) and absence of C. trachomatis infection, 10 participants with active disease (≥F2) but negative for C. trachomatis infection, and 7 participants with active trachoma (≥F2) with a positive test for C. trachomatis infection. The remaining four arrays in this series were from participants without the clinical signs of active trachoma but positive by Amplicor PCR. In addition, data were available from previous studies of conjunctival gene expression, measured by qRT-PCR, in populations in areas where trachoma is endemic (24).
Microarray data accession numbers.
The raw data obtained for the 60 U133 plus 2.0 arrays were deposited in the NCBI archive (www.ncbi.nlm.nih.gov/geo/) Gene Expression Omnibus under accession number GSE20436. The raw data for the 29 HG-focused target arrays were deposited in the Gene Expression Omnibus under accession number GSE20430.
RESULTS
C. trachomatis infection.
Age and gender distributions were comparable among the clinical groups (see Table S1 in the supplemental material). The median age of participants was 6 years (range, 1 to 13 years). The C. trachomatis infection loads ranged from <19 to 185,270 C. trachomatis ompA copies per swab. A positive result by Amplicor and increasing conjunctival load were significantly associated with clinical severity (F score) (see Table S2 in the supplemental material) (36). High-load infection was detected in the majority (11/20) of Amplicor-positive participants. The high-load infections accounted for all but one of the participants with the highest clinical severity scores. The quantified load in a minority of Amplicor-positive participants amplified with a reverse primer which binds genovar B ompA strain types more efficiently than genovar A ompA strain types, and these samples are indicated in Table S1 in the supplemental material (35).
Clustering of study subjects by global conjunctival gene expression correlation.
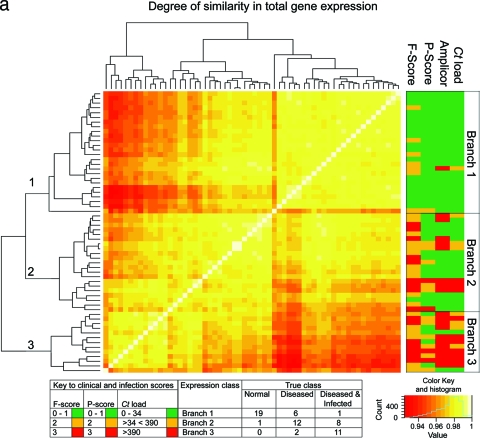
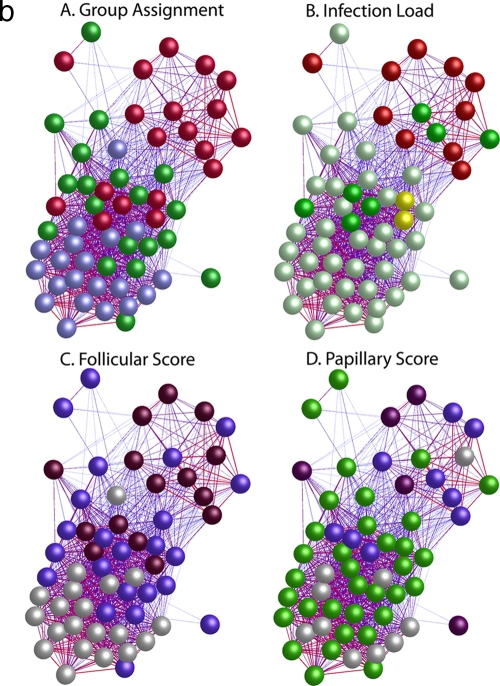
Quality control of Affymetrix gene expression data demonstrated that all data were of a consistent and high quality. The pairwise Pearson correlation coefficient of global gene expression was used as the distance metric to construct a matrix from which the individual samples were hierarchically clustered using a agglomerative clustering algorithm with complete linkage (Fig. 1a). This clustered the samples into three main branches. Branch 1 (n = 26) contains 95% of the samples from healthy subjects. Of the remaining samples in this branch, seven had F scores of ≥2 and a single sample had evidence of current intermediate-level infection (134 copies per swab). One participant also had papillary inflammation (P2). Branch 2 (n = 21) contained the single remaining sample from a subject with a normal conjunctiva. The remaining 20 samples in this branch were collected from subjects with follicular scores of ≥F2 or papillary scores of ≥P3. Fifteen of these had no or low-level infection (<34). Three samples had intermediate infection loads, and three had high-load infection (>390 copies per swab). Branch 3 (n = 13) contained subjects with the highest infection loads and clinical grades of disease (≥F2 or ≥P2). Eighty-four percent of the participants in branch three were infected, of whom over half (8/13) had high (>390) infection loads. It is not surprising that the overall range of correlation between these 60 arrays is narrow (between 0.9 and 1.0), since the overwhelming majority of genes do not have altered expression between the clinical and infection states. Pairwise correlation of global expression stratified by disease severity or infection load was also explored using BioLayout Express3D (Fig. 1b), and this confirms that the expression profiles cluster with clinical and infection phenotypes.
FIG. 1.
(a) Similarity of global gene expression clusters samples into three distinct groups. The axes of the matrix are clustered with a simple tree, which represents sample distance based on the global similarity of gene expression (sample-to-sample Pearson correlation coefficient). The trachoma clinical grading and infection scores are shown on the right hand side (F, follicular score; P, papillary hypertrophy score; Amplicor, C. trachomatis PCR positive or negative; C. trachomatis load, qPCR estimate of the number of C. trachomatis ompA copies per swab). The genome-wide expression profiles of conjunctival tissue from 60 Gambian subjects resident in communities where trachoma is endemic were used to construct the 60-by-60 matrix. The expression across all 54,675 probe sets on the HG-U133 plus 2.0 GeneChip were used. Three main branches (1, 2, and 3) cluster the samples into groups based on the biological signature of their overall gene expression profile. Samples defined by a branch share the greatest similarity with each other. Overall, the separation achieved by the unsupervised clustering of the samples has a true class accuracy value of 70% and implies that the three groups have a different expression signature. (b) Sample-to-sample correlation network represented in BioLayout Express3D. A matrix file was generated by comparing the overall expression pattern of all samples (as in panel a) and used to generate a graph of the data. In this context, nodes represent samples and the edges correlation values between them greater than the selected threshold (r = 0.95). The thicker/redder is the edge, the higher is the sample-to-sample correlation that it represents. Panel A, nodes colored light blue represent samples derived from healthy participants showing no clinical signs of disease or infection, those colored green represent participants who had clinical signs of disease but no detectable infection, and those colored red represent participants with clinical signs of disease and C. trachomatis infection. Panel B, nodes colored gray represent samples derived from participants showing no detectable levels of infection, those in yellow represent those with C. trachomatis loads of <34 copies per swab, those in green represent C. trachomatis loads of 34 to 389, and those in red represent loads f >390. Panel C, nodes colored gray represent participants with an F score of 0, those colored blue represent participants with an F score of 2, and those colored maroon represent participants with an F score of 3. Panel D, nodes colored gray represent participants with a P score of 0, those colored green represent participants with a P score of 1, those colored blue represent participants with a P score of 2, and those colored maroon represent participants with a P score of 3.
Identification of genes differentially expressed during infection and disease.
Large numbers of transcripts were both up- and downregulated when healthy and disease groups (with and without current infection) were compared (Table 1). There was a >2-fold change in the level of expression in 2,316 transcripts in samples derived from individuals with clinical disease and infection compared to healthy, infection-free individuals. However, the presence of disease signs alone induced differential expression of only 421 transcripts compared to healthy individuals. Among individuals with disease, the additional presence of C. trachomatis infection induced differential expression in 341 transcripts (the full gene list can be found in Table S3 in the supplemental material).
TABLE 1.
Summary of the total number of transcripts with altered expression associated with each comparison of clinical and ocular infection status
| Comparison | No. of transcripts |
||||||
|---|---|---|---|---|---|---|---|
| B ≥ 0 | Fold change | n | Adjusted P < 0.01 | Expected false positives | Direction of gene expression change | ||
| N vs DI | 18,792 | <0.6 and >1.6 | 4,993 | 4,845 | 48 | Up | 2,024 |
| Down | 2,821 | ||||||
| <0.5 and >2.0 | 2,365 | 2,316 | 23 | Up | 1,104 | ||
| Down | 1,212 | ||||||
| N vs D | 8,325 | <0.6 and >1.6 | 1,457 | 1,172 | 12 | Up | 491 |
| Down | 681 | ||||||
| <0.5 and >2.0 | 468 | 421 | 4 | Up | 195 | ||
| Down | 226 | ||||||
| D vs DI | 4,100 | <0.6 and >1.6 | 1,418 | 894 | 9 | Up | 445 |
| Down | 459 | ||||||
| <0.5 and >2.0 | 482 | 341 | 3 | Up | 151 | ||
| Down | 190 | ||||||
The major and fine biologies of the differentially regulated genes were explored using the DAVID v6 database. Each of the differentially regulated gene lists showed significant gene enrichment for numerous annotation terms. The top-ranked gene ontology (GO) terms were typical of immune system activation, epithelial cell integrity, apoptosis, cell death, leukocyte migration, and interleukin receptor activity (Table 2). In particular, genes involved in pathways that control cell adhesion, chemokine signaling, antigen (Ag) processing and presentation, and regulation of the actin cytoskeleton were strongly represented.
TABLE 2.
Principal biological annotation and functional terms identified by gene set enrichment analysis of gene lists made up only of genes with a high fold change in expression (adjusted P > 0.01)
| Comparison | No. of functional annotation clusters (enrichment score, ≥1.3) | No. of functional annotation terms (enrichment of ≥ 1.5 and P < 0.05) | Principal GO terms (cellular component, biological process, molecular function) | Fold enrichment | P value | Principal static KEGG pathway maps | Principal canonical pathways (MSigDB) |
|---|---|---|---|---|---|---|---|
| N vs D | 27 (14.09) | 97 | GO:0009897, external side of plasma membrane | 9.20 | 5.73 × 10−8 | hsa04060, cytokine-cytokine receptor interaction; hsa04640, hematopoietic cell lineage; hsa04630, Jak-STAT signaling pathway; hsa04510, focal adhesion | VERHAAK_AML_NPM1_MUT_VS_WT_UP, ZHAN_MMPC_PC, BOQUEST_CD31PLUS_VS_CD31MINUS_UP |
| GO:0006935, chemotaxis | 5.68 | 1.30 × 10−7 | |||||
| GO:0042330, taxis | 5.68 | 1.30 × 10−7 | |||||
| GO:0051240, positive regulation of multicellular organism process | 4.83 | 2.20 × 10−4 | |||||
| GO:0006954, inflammatory response | 4.79 | 2.84 × 10−11 | |||||
| N vs DI | 66 (46.62) | 166 | GO:0030595, leukocyte chemotaxis | 6.12 | 3.61 × 10−6 | hsa04060, cytokine-cytokine receptor interaction; hsa04650, natural killer cell-mediated cytotoxicity; hsa04640, hematopoietic cell lineage; hsa04514, cell adhesion molecules | CARIES_PULP_UP, ZHAN_MMPC_PC, LEE_TCELLS2_UP |
| GO:0019965, interleukin binding | 5.72 | 8.91 × 10−9 | |||||
| GO:0004907, interleukin receptor activity | 5.57 | 3.69 × 10−7 | |||||
| GO:0002274, myeloid leukocyte activation | 5.55 | 1.11 × 10−6 | |||||
| GO:0050900, leukocyte migration | 5.04 | 3.67 × 10−6 | |||||
| D vs DI | 12 (15.43) | 77 | GO:0003823, antigen binding | 27.17 | 2.44 × 10−21 | hsa04060, cytokine-cytokine receptor interaction | CARIES_PULP_UP, EEA1PATHWAY, LEE_TCELLS2_UP |
| GO:0050896, response to stimulus | 7.16 | 7.23 × 10−8 | |||||
| GO:0042330, taxis | 7.16 | 7.23 × 10−8 | |||||
| GO:0007626, locomotory behavior | 5.39 | 1.89 × 10−8 | |||||
| GO:0006955, immune response | 5.16 | 1.1 × 10−18 |
Identification of differentially regulated transcripts for classification.
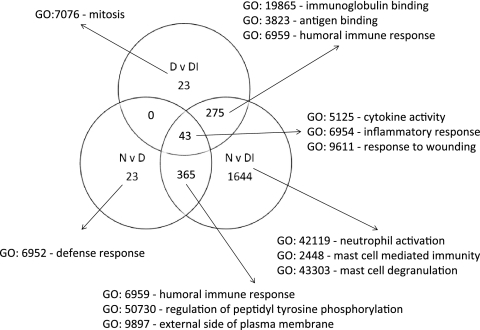
The probe sets that were differentially expressed (>2-fold change and adjusted P value of <0.01) were separated in a Venn diagram that identified probe sets with significant differential expression unique to each comparison (Fig. 2). The samples from participants who were diseased and infected contained the vast majority of the transcripts with greatest changes in expression compared to normal conjunctiva. Other comparisons had relatively few unique transcripts (N versus D = 23 and D versus DI = 23). The principal newly identified GO terms uncovered indicated enrichment of genes characteristic of neutrophils and mast cell biology during disease and infection episodes. The use of a set of 63 probe sets defined as uniquely expressed in each comparison classified the samples with 75% accuracy across the three clinical groups (Fig. 3; see Table S4 in the supplemental material). The true class and the kNN class are shown in Fig. 3 with a heat map of expression intensity. Samples are ordered by clinical category and within-group similarity of expression.
FIG. 2.
Identification of genes with high-degree changes in expression unique to each clinical and infection category, based on >2-fold changes. The biology of each of the gene lists was investigated by testing for enrichment or membership of pathways using DAVID v6, and the major GO terms are listed. The main features reflect those identified based only on high-degree differential expression (Table 2). Influx and activation of cells important in innate immune responses, such as neutrophils and mast cells, are prominent during disease and infection. Humoral and cytokine/chemokine responses typical of adaptive responses appear as expected and are shared between groups. Genes unique to each category were used to generate a small list of genes that could be used to classify participants based on expression profiles and to identify either biomarkers or important expression pathways.
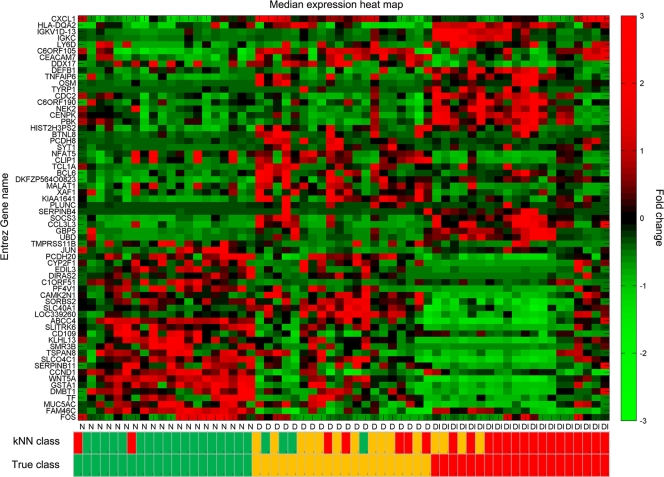
FIG. 3.
Classification of individuals based on differentially expressed transcripts. Transformed expression levels are indicated by the color scale, with red representing relatively high expression and green indicating relatively low expression compared to the median expression for each gene across all participants. Hierarchical clustering of 63 gene classifiers obtained from selecting genes with >2-fold changes in expression which are unique to each participant category is shown. Selection of a small subset from the largest group of uniquely expressed genes (1,644 in the disease and infection class) was based on the fold change and absence of correlation of expression with the genes identified for the other unique classes (23 genes in each). Selection of a relatively small list of genes as potential biomarkers is a requirement for the application of computationally efficient methods. A supervised learning algorithm (kNN) was used to test the capacity of the classifier subset of genes to discriminate three classes of participants. Leave-one-out cross-validation of the training set with 63 genes classified the samples with 75% accuracy. The predicted class is indicated by light-color solid rectangles (green for healthy participants, amber/yellow for participants with clinical disease signs only, and red for participants with disease signs with current ocular C. trachomatis infection). Participants from all categories are misclassified; however, misclassified participants are those with disease signs only (9/20). Two patients with clinical disease and infection were classified as healthy uninfected participants, and no healthy participants were classified as diseased and infected.
Comparison of Affymetrix array gene expression results with previous independent studies of gene expression in trachoma.
We compared the data obtained from this set of samples and arrays with data from previous studies in which participants were of equivalent age range and in which the diagnosis of current infection was made using the same PCR test (CT/NG Amplicor). Data were available from an independent set of samples from the same population in which gene expression was measured using Affymetrix HG-focused target arrays. The comparative results are shown in Tables S5a to S5d in the supplemental material. Overall there was strong correlation of fold change for all probe sets common to each array platform. This became very strong when genes with high levels of fold change were considered. Some probe sets, such as CXCL13 and S100A7, had high levels of fold change on one array platform (U133 plus 2.0) and low levels on the second platform; however, in each case the fold changes were statistically significant. Previous studies using qRT-PCR to estimate gene expression with conjunctival samples from children with active trachoma were also correlated with gene expression by microarray analysis. The highest degree of correlation was always obtained from the comparison of group N with group DI in all studies and platforms.
Transcription networks in the conjunctiva.
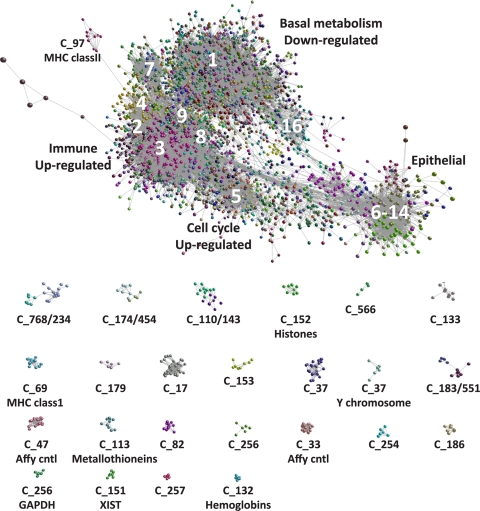
The undirected network graph based on a Pearson correlation (r) threshold of ≥0.85 contained 9,993 nodes (probe sets) representing approximately 8,359 genes connected by 245,457 edges. MCL clustering partitioned the network into 577 clusters of coexpressed genes. These clusters ranged in size from 1,148 to 4 transcripts and accounted for 7,719 (77.3%) of the probe sets in the original network. Probe sets that formed clusters containing <4 members that were part of the network were not assigned a cluster number (no class) and accounted for remaining 2,274 probe sets. Clusters established by MCL were assigned a number according to the number of probe sets they contained; MCL1 contained the largest number of probe sets (1,148), while MCL577 contained four probe sets. Each MCL cluster was assigned an arbitrary color, and the graph (Fig. 4) was explored for evidence of functional enrichment of gene clusters and the relationships between genes and clusters (the full list of transcripts and cluster assignment is available in Table S6 in the supplemental material). The graph (Fig. 4) shows the interrelationships and overlapping nature of the main large clusters and the discrete separation of other, smaller clusters. A number of small but interesting clusters confirm the power of this approach in identifying and grouping coexpressed genes. For example, MCL33 and -47 are derived from the probes for the Affymetrix labeling controls (24 probe sets) and the Affymetrix hybridization controls (19 probe sets), respectively. MCL37 was made up entirely of transcripts derived from the Y chromosome, which are expressed only in males. The clusters MCL12, -13, and -22 were all highly enriched with ribosomal genes.
FIG. 4.
Pairwise gene expression correlation network analysis of microarray data derived from 60 trachoma cases and healthy controls. Each node (sphere) represents a single probe set, and the edges (fine gray lines) connecting them represent a Pearson correlation (r) of ≥0.85. The color of the nodes represents their membership in a cluster (MCL). The major clusters described in the text are labeled. The smaller isolated clusters are distinct from the main component and are shown below the main component. The main component shows the overlapping nature and interrelationship of the coexpression clusters described in the text.
The partitioned graph of 577 clusters contained three basic classes of clusters: genes in which expression was unchanged across all samples, genes whose expression was increased during infection and disease, and genes whose expression was downregulated during infection and disease. The gene/transcript content of each cluster and their associated biological function are provided in Table S6 in the supplemental material. The major and fine biologies of the members of each of the major transcriptional networks with the numbers of differentially regulated genes are summarized in Tables 3 and 4.
TABLE 3.
Summary of the principal biological annotation and functional terms identified by gene set enrichment analysis of gene lists selected by membership of a coexpression (Biolayout Express) network
| Coexpression cluster | No. of functional annotation clusters (enrichment score, ≥1.3) | No. of functional annotation terms (enrichment of ≥ 1.5 and P < 0.05) | Principal GO terms (cellular component, biological process, molecular function) | Fold enrichment | P value | Principal static KEGG pathway identified or principal canonical pathway identified |
|---|---|---|---|---|---|---|
| MCL1 (tissue physiology and homeostasis) | 23 (5.91) | 36 | GO:0007423, sensory organ development | 4.04 | 1.78 × 10−5 | hsa04310, Wnt signaling pathway; hsa04530, tight junction; |
| GO:0009790, embryonic development | 2.51 | 1.39 × 10−5 | hsa04916, melanogenesis | |||
| GO:0008134, transcription factor binding | 2.2 | 1.99 × 10−5 | ||||
| MCL2 (cellular immune responses) | 56 (23.78) | 134 | GO:0042101, T cell receptor complex | 22.48 | 6.31 ×10−10 | hsa04650, natural killer cell mediated cytotoxicity; |
| GO:0015026, coreceptor activity | 15.07 | 4.08 × 10−7 | hsa04660, T cell receptor signaling pathway; hsa04612, | |||
| GO:0030101, natural killer cell activation | 13.96 | 9.69 × 10−8 | antigen processing and presentation | |||
| MCL3 (innate immunity) | 17 (16.95) | 50 | GO:0019955, cytokine binding | 12.03 | 1.37 × 10−7 | hsa04060, cytokine-cytokine receptor interaction |
| GO:0006935, chemotaxis | 10.78 | 1.99 × 10−11 | ||||
| GO:0001816, cytokine production | 9.96 | 9.96 × 10−8 | ||||
| MCL4 (cell cycle) | 31 (50.7) | 76 | GO:0007051, spindle organization and biogenesis | 73.46 | 5.85 × 10−17 | hsa04110, cell cycle |
| GO:0000070, mitotic sister chromatid segregation | 43.08 | 8.72 × 10−13 | ||||
| GO:0000775, chromosome, pericentric region | 43.05 | 4.92 × 10−29 | ||||
| MCL5 (basic cell processes) | 8 (2.97) | 19 | GO:0042578, phosphoric ester hydrolase activity | 4.07 | 7.45 × 10−4 | No enriched genes in KEGG pathways identified; gene |
| GO:0009057, macromolecule catabolic process | 3.16 | 0.004 | content is typical of basic metabolic processes, zinc | |||
| GO:0016788, hydrolase activity, acting on ester bonds | 2.80 | 0.001 | finger and metal ion binding and splice variants; MSigDB gene sets identified were TAKEDA_NUP8_HOXA9_16DN, PPARG pathway, and ALZHEIMERS_DISEASE_UP | |||
| MCL6 (keratinocyte cell biology) | 8 (4.55) | 57 | GO:0001533, cornified envelope | 53.36 | 2.33 × 10−9 | hsa04120, ubiquitin-mediated proteolysis; hsa01430, cell |
| GO:0030216, keratinocyte differentiation | 26.87 | 2.53 × 10−6 | communication | |||
| GO:0031424, keratinization | 20.62 | 9.36 × 10−5 | ||||
| MCL7 (plasma cells) | 6 (13.84) | 21 | GO:0003823, antigen binding | 36.20 | 1.03 × 10−14 | No enriched genes in KEGG pathways identified; gene |
| GO:0006955, immune response | 7.73 | 4.46 × 10−11 | content is typical of immunoglobulin response | |||
| GO:0002376, immune system process | 6.62 | 1.11 × 10−9 | signature: MSigDB gene sets identified were TARTE_PC, HBX_NL_DN, and PENG_GLUTAMINE_UP | |||
| MCL8 (type II interferon response) | 8 (7.8) | 73 | GO:0007260, tyrosine phosphorylation of STAT protein | 50.1 | 2.5 × 10−6 | hsa04060, cytokine-cytokine receptor interaction; |
| GO:0007259, JAK-STAT cascade | 24.64 | 4.77 × 10−5 | hsa04630, Jak-STAT signaling pathway; hsa04620, Toll- | |||
| GO:0018108, peptidyl-tyrosine phosphorylation | 22.1 | 6.81 × 10−6 | like receptor signaling pathway | |||
| MCL9 (B cells) | 5 (3.63) | 24 | GO:0006959, humoral immune response | 25.66 | 3.77 × 10−5 | No enriched genes in KEGG pathways identified; gene |
| GO:0009986, cell surface | 11.59 | 8 × 10−4 | content is typical of a immunoglobulin response | |||
| GO:0002376, immune system process | 4.82 | 6.87 × 10−7 | signature; MSigDB gene sets identified were KLEIN_PEL_DN, KUMAR_HOXA_DIFF, and ZHAN_MMPC_PC | |||
| MCL10 (type I interferon response) | 8 (8.87) | 63 | GO:0009615, response to virus | 39.61 | 5.02 × 10−15 | No enriched genes in KEGG pathways identified; gene |
| GO:0051707, response to other organism | 19.51 | 1.31 × 10−11 | content is typical of type I (INTERFERONα/β) | |||
| GO:0009607, response to biotic stimulus | 12.53 | 1.49 × 10−9 | signaling response signature; MSigDB gene sets identified were TAKEDA_NUP8_HOXA9_16DN, BECKER_IFN_INDUCIBLE_SUBSET_1, and IFNA_HCMV_6HRS_UP |
TABLE 4.
Summary of the number of transcripts with differential expression within the major coexpression networks based on a stringent level of fold change in expression (>2-fold) or likelihood that the transcript is differentially expressed irrespective of the fold change (B value)
| Criterion | Comparison | No. of transcripts with altered expression in clustera: |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MCL1 | MCL2 | MCL3 | MCL4 | MCL5 | MCL6 | MCL7 | MCL8 | MCL9 | MCL10 | MCL11-259 | MCL260-577 | ||
| Tissue physiology and homeostasis (1,148) | Cellular immune responses (958) | Innate immunity (271) | Cell cycle (175) | Basic cell processes (174) | Keratinocyte cell biology (162) | Immunoglobulins (130) | Type II interferon signature (117) | Immunoglobulins (89) | Type I interferon response (88) | Epithelial cell barrier integrity (3,036) | Regulation of epithelial and conjunctiva-associated lymphoid tissue response with an inflammatory stimulus (1,371) | ||
| B ≥ 0 | N vs DI | 1,132 | 861 | 221 | 133 | 4 | 24 | 120 | 82 | 78 | 31 | 2,068 | 1,031 |
| N vs D | 874 | 207 | 146 | 1 | 5 | 1 | 45 | 2 | 1,163 | 521 | |||
| D vs DI | 277 | 359 | 4 | 136 | 2 | 106 | 34 | 9 | 464 | 243 | |||
| FC of <0.5 or >2.0; adjusted P < 0.01 | N vs DI | 368 | 402 | 112 | 20 | 19 | 74 | 42 | 54 | 9 | 455 | 440 | |
| N vs D | 49 | 25 | 55 | 1 | 1 | 1 | 24 | 1 | 97 | 99 | |||
| D vs DI | 6 | 2 | 65 | 17 | 2 | 81 | 75 | ||||||
Numbers in parentheses indicate the total number of transcripts in the cluster.
Individuals with active disease and infection exhibit altered expression profiles that are enriched with genes indicative of T cell and NK cell activation.
The largest cluster of upregulated genes was MCL2. Manual inspection of this cluster and DAVID v6 analysis indicated that this cluster contained genes typical of NK cells, T cells, and macrophages. Of particular interest was the upregulation of genes associated with NK and T cell ligand-receptor complexes (PTPN6, NCR3-CD3Z, CD244-CD48, NKG2D-DAP10, CD86, CD8, CD28, CD2, CD45, CD4, CD3D, and CD3E), intracellular signal-transduction mediators of the T-cell receptor (TCR) cascade, and nuclear proteins mediating expression of genes controlling the functions of Th1 lymphocytes (TXK and NFATC) and activation of major histocompatibility complex (MHC) class II transcription (RFX5). This cluster also contains members of the killer cell immunoglobulin-like receptors (KIRs), some of which regulate the cytotoxic function of NK cells via MHC class I ligands. These include inhibitors of NK-mediated activation (KIR2DL5A), cell triggering complexes associated with NK mobilization and cytotoxicity (FCER1G/KIR2DL4, KIR3DL2, KIR2DS2, and KIR3DL1), and genes of the C-type lectin superfamily involved in the regulation of NK cell function and costimulation of CD8 T cells (KLRD1and KLRK1). The activating natural killer (NK) cell receptor NCR3, perforin (PRF1), and granzyme B (GZMB) are also represented in the cluster. Other prominently expressed genes present in this cluster encode secreted proteins regulating NK and Th1 cell recruitment and activation at sites of inflammation. These include the adhesion molecules ADAM19, ITGB7, ICAM3, and VCAM1; chemokines and receptors (CCR5, CCL5, and CXCR6); and cytokines and receptors (IFNG, IL2RG, interleukin-16 [IL-16], IL12RB1, IL12RB2, IL-32, LTA, IL10RA, and CSF1R). Evident within MCL2 are genes controlling homeostatic regulation of inflammatory mediators (e.g., anaphylatoxins [C3AR1]) via increased expression of PDE4B and PDE3B, GPR132, PTGDR, prostaglandins PLA2G2D and, the tumor necrosis factor (TNF) receptor superfamily (TNFRSF25 and TNFSF8).
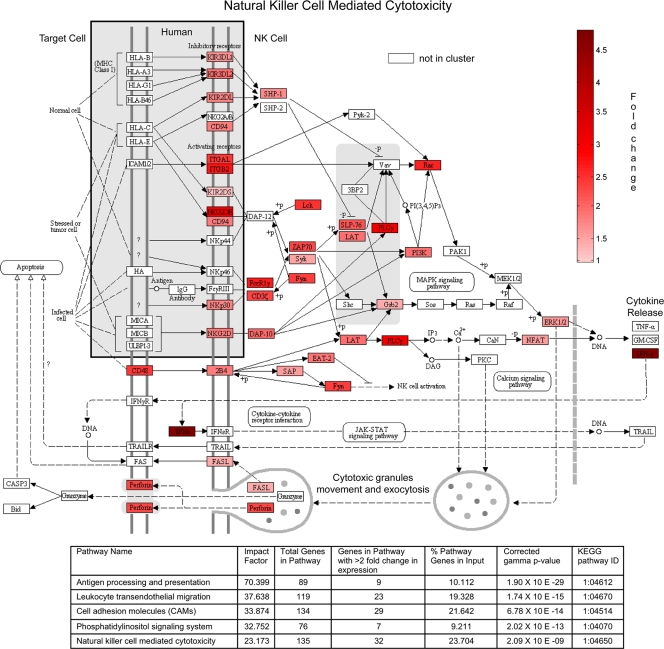
Using network analysis and gene set enrichment, we consistently identified overrepresentation of transcripts with a role in NK cell biology. Therefore, the fold change expression values for each probe set from MCL2 were imported into the KEGG database defined pathway map for NK cell-mediated cytotoxicity (Fig. 5). This provides a visual representation of the canonical pathway for the activation of NK cells. The greatest changes in expression intensity are evident in the subjects with infection and disease signs. Since analysis based purely on differential expression (2-fold changes) (Tables 1 and 2) also identified enrichment of genes in this pathway, we submitted the list of differentially regulated genes to an expression perturbation analysis tool (Onto-tools Pathway Express). The results indicated that the largest predicted effects based on enrichment and fold changes were on the same MCL2-enriched pathways and provided further supporting evidence for stimulation of NK cell-mediated cytotoxicity.
FIG. 5.
Gene expression within cluster MCL2 is enriched in genes affecting NK cell-mediated cytotoxicity. Fold change in gene expression from MCL2 is shown on the KEGG pathway map (modified from map04650, natural killer cell-mediated cytotoxicity, at the Kyoto Encyclopedia of Genes and Genomes [KEGG] website http://www.genome.jp/kegg/kegg1.html, with permission). KIR2DL, phosphatidylinositol 3-kinase (PI3K), and phospholipase Cγ (PLCγ) depicted in this pathway are represented by multiple gene transcripts in MCL2; in these cases genes are colored according to the transcript with the largest fold change in expression within a multigene or transcript family (KIR2DL4, PI3KCD, and PLCG2). Participants with disease and infection had the greatest changes in gene expression (shown on the map by increasing intensities of red). Onto-tools Pathway Express expression perturbation analysis based only on differentially regulated genes (<0.5 and >2.0 at an adjusted P value of <0.01) comparing healthy subjects with diseased and infected subjects also identifies NK cell-mediated cytotoxicity as substantially affected. An overlapping effect on antigen processing and presentation related primarily to NK cell activity was also observed. The impact factor is the estimated effect on each pathway as a result of changes in transcript expression in addition to enrichment.
Transcripts typical of neutrophils and interferon signaling form overlapping clusters.
MCL3 consists of 271 transcripts, and the average profile of expression is similar to that of MCL2; this can be visualized in Fig. 4, which shows the proximity of these two clusters in the coexpression network. The cluster is dominated by genes typical of leukocyte biology and innate host defense (cytokines, chemokines, and Toll-like receptors [TLRs]). Several members of the Toll-like receptor family (TLR1, TLR2, TLR4, and TLR8) were present in this cluster. The expression signature of cytokine receptors involved in cell-cell signaling is characteristic of the innate immune response that induces transcription of inflammation-related genes (IL8RA, IL8RB, CSF3R, CSF2RB, IL1B, IL1A, IL1R2, and IL6R) as well as those involved in humoral and adaptive immunity. MCL3 also contains receptors for granulocyte-macrophage colony-stimulating factors (CSF3R and CSF2RB) and mediators of neutrophil migration to sites of inflammation (CXCL5 and MMP25). Consequently, genes involved in many of the processes of phagocytosis, cell-mediated cytotoxicity, chemotaxis, or cellular activation of NK cells, monocytes, and neutrophils are represented (CR1, C5AR1, and CFP), including those genes modulating iron metabolism (SLC11A1) and intracellular superoxide production (FPR1, NCF2, and NCF4). MCL3 also has upregulation of genes that balance inflammatory responses, such as SLA, LILRB3, SOCS3, and protease inhibitors (SERPINA1). Manual annotation and enrichment analysis therefore suggests that this cluster is comprised of genes expressed predominately in neutrophils.
The expression profiles of MCL8 and -10 are highly similar, are upregulated during disease, and are further upregulated with high C. trachomatis loads (Fig. 6). These clusters have a signature of type I and II interferon (IFN)-regulated genes. These include assembly of MHC class I molecules and the presentation of endogenous peptides to CD8+ T cells (TAP1 and TAP2), induction of apoptosis in infected cells presenting antigen-MHC-I complexes by cytotoxic T lymphocytes (CTL) (PRF1, GZMA, and GZMB), and expression of CTL cell adhesion molecules (ITGAL and CD226). There is a chemokine and cytokine pattern indicative of cell-mediated adaptive immune responses (IL2RA, IL15RA, JAK2, STAT1, STAT4, and IL-22) and IFN-inducible genes (IFI1/2/3/5/7, STAT1/2, and IRF1/7). Other well-established markers of the IFN-γ response, such as GBP1/2/5, were upregulated in this cluster, as, importantly, was INDOL1. INDOL1 is an IFN-γ-induced enzyme that is structurally and functionally similar to the immunomodulatory and antimicrobial indoleamine 2,3-dioxygenase (IDO) (IDO, although not part of the MCL8 coexpression network, was strongly upregulated in those with disease and infection). Further evidence of activation of other genes in known IFN-γ-induced anti-C. trachomatis pathways was ATF5 and WARS.
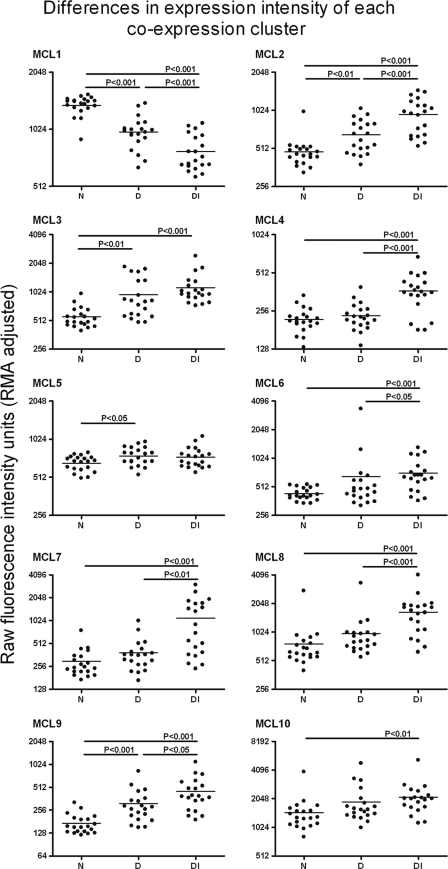
FIG. 6.
Mean expression levels of all the transcripts in a coexpression cluster for each array are summarized as a single point for each array. Differences in the expression level for each transcript cluster between each clinical and infection category was then tested by analysis of variance. Where differences between the groups are significant, they are indicated with the appropriate P value, and this indicates where the changes of expression are attributable.
MCL8 was also enriched with genes which regulate or oppose the activation of Th1 cells and IFN-γ-induced genes, such as SOCS1, TFEC, PTGER2, HAVCR2, and TRAFD1 (FLN29). Negative regulators of CTL and NK cell effector mechanisms such as the serine protease inhibitor SERPINB9 and cell surface receptors (CTLA4 and KLRD1) were also induced. Although similar in profile, MCL10 is enriched with genes associated with type I IFN signaling. These include interferon-induced genes (IFIH1, IFIT2, IFIT3, IFI15, and ISG20), the interferon regulatory transcription factor IRF7, and genes associated with host defense against intracellular infections (MX1, MX2, OAS1, OAS2, OAS3 OASL, DDX58, RIG-1, and PML).
MCL7 and -9 were comprised of 219 transcripts that were strongly upregulated during disease with infection. MCL9 contained genes characteristic of the B cell lineage. Genes coding for members of the B cell antigen receptor complex (FCRL1, FCRL2, FCRL4, FCRL5, and FCRLA), immunoglobulins, and the CD79A/CD79B heterodimer were present in this cluster, as well as the classical B cell marker CD19. Other immunoglobulin-related genes included those involved in signal transduction pathways (BLK, SPIB, BACH2, TECs, and PTKs) triggered by the B cell antigen receptor (BCR and BRDG1). On the other hand MCL7 was highly enriched with immunoglobulin genes predominately expressed in plasma cells, e.g., IGH@, IGAH1, IGHD, IGHG1, IGHM, IGHV1, IGKC, IGKL@, and IGJ3.
To illustrate the effect of clinical status on the levels of expression in healthy, diseased, and diseased and infected individuals, the geometric mean fluorescence intensities of all the transcripts which belong to a particular MCL or cluster were calculated. The raw intensity levels and the differences in these levels between the clinical groups show the effect of either disease or disease with infection on the level of expression (Fig. 6). The largest changes in expression across a cluster of genes are seen in the diseased-with-infection group. Only rarely did current infection have little effect on expression (e.g., MCL5).
Other clusters significant in the biology of trachoma.
MCL4 was made up almost entirely of genes encoding proteins involved in the S phase of the cell cycle, which were upregulated with high infection loads. MCL42 is a cluster of upregulated transcripts which function in lipid metabolism and/or trafficking (APOE, APOC1, SCARB1, and SLC16A1) and innate intracellular defense. Genes in MCL29 were upregulated during disease and infection and included 24 genes coding for extracellular matrix proteins of connective tissue collagens (COL1A1, COL1A2, COL3A1, COL4A1, COL4A2, COL5A2, COL6A2, and COL6A3) and extracellular matrix (ECM) glycoproteins (TNC and ASPN). MCL133 was composed of nine transcripts covering eight genes that are characteristic of conjunctival goblet cells, such as the trefoil factors (TFF1 and TFF3) and mucins (MUC5AC and CAPN8). All the genes in this cluster were downregulated in disease and infection.
Downregulated clusters.
Genes that were downregulated in disease and infection clustered together in the network (MCL1). MCL1 is the largest single cluster in the network and was enriched for transcripts in the Wnt signaling pathway. Other clusters in this downregulated category include MCL11, -16, -19, -21, -23, and -24.
DISCUSSION
In trachoma, unresolved or repeated ocular C. trachomatis infection and inflammation causes a progressive fibrotic response that can ultimately result in blindness. The conjunctival scarring observed is more common in those with a severe inflammatory response to infection. This study describes tissue-specific transcriptional networks associated with the response to ocular C. trachomatis infection and inflammation. Previous data suggest that in communities with endemic trachoma, individuals who are at different positions in the disease and infection cycle can be distinguished based on their host response profile (11, 24, 25). We now show that global transcription profiles can also cluster individuals into the same types of disease and infection categories. Furthermore, a reduced subset of genes that were statistically differentially regulated was also able to separate individuals with increased accuracy into the same categories. This suggests that discrete expression patterns are associated with these different biological states and illustrates the potential to identify an expression signature for those at most risk of severe inflammation and the likely development of scarring sequelae.
An initial wave of polymorphonuclear leukocyte influx is a well-recognized feature of ocular and genital chlamydial infection that is documented in animal models and in human infection (69, 92). The prominent neutrophil gene signature identified in the conjunctiva (MCL3) supports this observation and suggests that the network and enrichment analyses are robust. Indeed, signatures typical of granulopoeisis and neutrophil activation have frequently been identified using genome expression profiling in response to other classes of infection (9, 30, 63). In some of these infections, in which the focus of research has been adaptive “protective” T cells, transcriptome signatures have identified previously hidden type I interferon signaling pathways present in neutrophils and have suggested that they are crucial in the control of infection (9). In some chlamydial infections, depletion of neutrophils delayed the clearance of infection and increased chlamydial shedding in the murine genital tract (5). Neutrophils were found to be a requirement for the recruitment of T cells, particularly CD8+ T cells, to the site of infection (19), and in the lung, an increased influx of neutrophils was associated with a greater chlamydial burden of infection in a susceptible mouse strain relative to a resistant strain (41). The influx of neutrophils in the conjunctiva could be attributed to chemokines such as IL-8 and CXCL1, -2, -5, and -6, all of which were upregulated. Extensive work on the mechanism of neutrophil influx into the cornea following infection with Onchocerca volvulus has demonstrated the dependence of this influx on multiple factors originating from cells that are resident in and infiltrate the tissue (28, 46). The resulting neutrophil influx and activation was then responsible for keratitis and corneal haze (29). It therefore seems likely that the control of neutrophil recruitment and activation either by adaptive CD4+ Th cells (e.g., Th17 cells) or by chemokines secreted by infected epithelial cells will be crucial, and this requires further investigation.
We found strong induction of gene expression for IFN-γ (MCL2) and IDO in active trachoma and C. trachomatis infection. The production of IFN-γ has a pivotal role in chlamydial disease via control of pathogen growth and replication (68). IFN-γ-induced IDO can inhibit proliferation of C. trachomatis in vivo through consumption of the essential amino acid Trp (65, 80). Previously we have also shown upregulation of IDO expression in the conjunctiva of subjects with increasing loads of ocular C. trachomatis infection. IDO also has recognized immunoregulatory properties in both human and murine cells (61). Therefore, IDO could control the balance between T cell subset differentiation and local DC priming, suggesting that C. trachomatis might exploit IDO expression to induce immunoregulation (5). The identification of inflammatory cells (neutrophils: MCL3) and the upregulation of IFN-γ with T cell receptor signaling pathways (MCL2) together provide evidence that the arrays reveal elements of the expected and previously known cellular and gene expression patterns observed in C. trachomatis disease and infection. We suggest that this expected result supports the novel observation that transcripts associated with NK cells and NK cell cytotoxicity, which are found in MCL2, are over represented and suggests an important contribution of NK cells in the response to C. trachomatis infection and disease.
There are a limited number of studies in which NK cells have been reported to have a demonstrable impact on chlamydial disease or infection (33, 34), yet depletion of NK cells exacerbated the course of disease and infection in mice (83). Using different gene enrichment approaches, we consistently found evidence for the contribution of NK cell activation and cytotoxicity in the conjunctiva of participants with trachoma. In addition to cytotoxic effects, NK cells could also be a major source of crucial cytokines such as IFN-γ and IL-22. Thus, while NK cells may not be essential for the resolution of infection, they may be critical in the inflammatory process and in the bridge between the adaptive and innate responses. Work with murine models suggests that adaptive CD4+ Th1 cells which produce IFN-γ are required for the resolution of primary infections and that in secondary responses other immune cells can contribute but are not an absolute requirement (87, 90, 91). NK cells can polarize the CD4+ T cell response via dendritic cells (DC), which results in an amplification of IFN-γ production by T cells. NK cells can be helped to produce IFN-γ by other innate cells, such as neutrophils, or by chemokines derived from infected epithelial cells, such as IL-12 and IL-18. It is also well recognized that T cell-derived IL-2 can activate NK cells, and recent evidence suggests that Ag-specific IL-2 from effector memory T cells can in effect immediately “recall” NK cells which degranulate and secrete IFN-γ (35, 36). Thus, in the presence of IL-2, IL-12, and IL-18, the local inflammatory responses are directed toward strong type I and IFN-γ responses (1, 26, 73). Therefore, the strong expression signature of NK cells observed in these conjunctival samples, which reflects those seen after several natural ocular challenge infections or episodes of disease, could be explained by the boosting effect of antigen-specific effector memory T cells. We suggest that this interaction warrants further in vitro study and investigation.
The regulation of NK cell activity is complex; epistatic effects between HLA ligands and KIRs control the activity of NK cells. The level of diversity in the KIR gene system reflects its coevolution with MHC class I (71), which encodes the ligands of some KIRs (15-17, 31). This diversity is generated by haplotypic, allelic (84, 88), and transcriptional (3) variation that results in a unique KIR expression repertoire. The unusual nature of KIR polymorphism and expression can confound the interpretation of microarray expression studies with respect to individual KIR alleles or genes. Nevertheless, KIR2DL4, which is found on all NK cells, can be regarded as a KIR “framework” locus present (with only rare exceptions) in all KIR haplotypes (62). Since we find this marker and several other markers of NK cells to be highly expressed in the conjunctiva, we suggest that their activity in inflammatory trachoma is significant.
The majority of cells from conjunctival swabs are epithelial cells, and it is well established that infected host epithelial cells are the source of many of the initiating factors that drive inflammation (20, 21). This led Stephens to suggest an alternative paradigm for the pathogenesis of chlamydial diseases (75). We found strong induction of many chemokines, pattern recognition genes (TLRs and NOD [inflammasome] [NOD2, NLRP3, and NLRP7]), and mediators of inflammation (IL1B). Clustering of coexpressed genes and annotation of the gene content of the clusters suggests that infiltrating cells, largely neutrophils, are a major cellular source of many of these factors. The largest fold changes in expression were seen for CXCL5 (epithelial cell-derived neutrophil-activating protein [ENA-78]), -11, and -13. Strong induction of Cxcl13 has been described in the development of murine salpingitis, and this has been suggested as the primary chemokine required for the development of organized lymphoid tissue in the genital tract (43). Fractalkine (CX3CL1), a δ-chemokine expressed by epithelial cells, DC, and some T cells, was upregulated, and its expression in response to chlamydial infection has not been described before. Induction of CXCR3, -4, and -6 was also observed and is consistent with the recruitment of T cells, NK cells, monocytes, macrophages, and neutrophils (7, 47, 66). The increased expression of CXCR6 in chlamydial infection has not been previously identified. Its cellular distribution overlaps with that of CCR6, but it is also found on neutrophils and NKT cells.
Of note among the chemokines and receptors expressed by the cells entering the conjunctiva were CCL18 and -19. CCL18 is selectively chemotactic for T lymphocytes and has been shown to be important in pulmonary fibrosis and inflammation (59, 60). CCL18 can be produced by macrophages, alternatively activated macrophages, dendritic cells (DC-CK1), and in some circumstances neutrophils (4). The receptor for CCL18 remains to be identified, but it is expressed on T cells that infiltrate epithelial surfaces. CCL19 is known to mediate the entry of naive lymphocytes into secondary lymphoid tissue and, similar to CXCL13, is important in the organization of lymphoid tissue. Uniquely, we identify CCR10 and the orphan receptor CCRL2 as upregulated in active trachoma. CCRL2 has the unusual property of focusing responses, enhancing chemotaxis of leukocytes by binding and presenting nonchemokine chemoattractants to cells with the appropriate chemokine-like receptors. The roles of many CC and CXC ligands and receptors in chlamydial diseases have been investigated, mostly on a candidate gene basis or by a selective targeted approach (7, 64). Our results are consistent for the most part with the majority of these studies (51, 52). Differences between our results and those for other tissues likely reflect tissue compartmentalization or the differing repertoire of chemokines shared between Homo sapiens and Mus musculus.
The control of innate responses appears at the center of several networks, and expression of cytokines such as IL-12 and IL-23 is at the fulcrum of protective acquired immune responses. Recent work with mice, which differ in susceptibility to chlamydial pulmonary infection, has shown that the IL-12/IL-23 balance is altered in DC isolated from susceptible BALB/c strains. The excessive IL-23 production observed was suggested to favor the later development of Th17 cells, which were associated with a larger burden of infection in the lungs (41). In our study, the expression pattern of IL23A in the conjunctiva is contained in a transcription network that is characteristic of human epidermal keratinocytes (MCL6). IL-23 promotes inflammatory responses that include upregulation of MMP9 (45), polymorphism of which is associated with scarring trachoma. The overall effect of this polymorphism is complex, since several genes involved in the immune (IFNG and IL-10) and inflammatory (IL-8 and CSF2) responses alter its protective or risk-associated effects (55, 57). Although IL-23 is not involved in Th17 differentiation, it is thought to play an important role in maintaining Th17 effector function (76, 77) and therefore in local tissue inflammation. IL-22 expressed by Th17 or Th22 (22, 82) or NK (13) cells was upregulated in active disease with C. trachomatis infection, whereas its decoy receptor IL22RA2 was upregulated during active disease episodes free of infection. IL-22 allows cross talk between the immune system and epithelial cells, and it has been suggested to have an important role both in host defense and in the pathogenesis of inflammatory skin diseases such as psoriasis. Th22 cells, which are a CD4+ subset that home to the skin and are important in the repair of the epithelial barrier (23), therefore would appear to be important in C. trachomatis infection and immunity. However, the means to identify a clear and separate CD4+ T helper subtype signature within the background of the transcriptome expression profiles are not yet available.
Interpretation of expression studies of disease versus control tissue is often confounded by the very dramatic differences in the cell populations present. An immune response is orchestrated by the activity of numerous leukocyte populations, with each cell expressing genes specific to that population, and in principle the level of these genes in each sample contributes to the overall transcriptional signature. Identification of clusters of coexpressed transcripts that are indicative of these cell types offers the opportunity to attribute some of the differences in expression to the cellular content of the tissue. Furthermore, unlike experimental models of infection where genetically homogeneous animals or cell lines are challenged with equivalent levels of infectious organisms of the same strain, naturally observed infection in the human population results in considerable variation, and this must be taken into account as we interpret the data.
Several observations suggest that transcription network-based analysis provides a powerful approach and an added advantage that allows the identification of genes expressed by particular cell types or those under the influence of the same transcriptional activators. Combining this with pairwise differential gene expression, we show that the major networks of coexpressed and highly regulated genes in the conjunctiva of participants with active trachoma and C. trachomatis infection are dominated by genes involved in innate immune responses and IFN-γ-mediated signaling. We have demonstrated the prominence of innate responses (NK cells and neutrophils), underpinned by the balance of IL-12/IL-23 and expression characteristic of several CD4+ helper phenotypes (Th1,Th17, and Th22 cells).
Supplementary Material
Acknowledgments
We thank the study participants, field workers, and laboratory staff at the Medical Research Council Laboratories in the Gambia for their assistance.
This work was supported by the Wellcome Trust (079246/Z/06/Z) and MRC (United Kingdom) core budget, the Gambia.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 7 September 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Agaugue, S., E. Marcenaro, B. Ferranti, L. Moretta, and A. Moretta. 2008. Human natural killer cells exposed to IL-2, IL-12, IL-18, or IL-4 differently modulate priming of naive T cells by monocyte-derived dendritic cells. Blood 112:1776-1783. [DOI] [PubMed] [Google Scholar]
- 2.Alvesalo, J., D. Greco, M. Leinonen, T. Raitila, P. Vuorela, and P. Auvinen. 2008. Microarray analysis of a Chlamydia pneumoniae-infected human epithelial cell line by use of gene ontology hierarchy. J. Infect. Dis. 197:156-162. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, S. K. 2006. Transcriptional regulation of NK cell receptors. Curr. Top. Microbiol. Immunol. 298:59-75. [DOI] [PubMed] [Google Scholar]
- 4.Auer, J., M. Blass, H. Schulze-Koops, S. Russwurm, T. Nagel, J. R. Kalden, M. Rollinghoff, and H. U. Beuscher. 2007. Expression and regulation of CCL18 in synovial fluid neutrophils of patients with rheumatoid arthritis. Arthritis Res. Ther. 9:R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barteneva, N., I. Theodor, E. M. Peterson, and L. M. de la Maza. 1996. Role of neutrophils in controlling early stages of a Chlamydia trachomatis infection. Infect. Immun. 64:4830-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baud, D., L. Regan, and G. Greub. 2008. Emerging role of Chlamydia and Chlamydia-like organisms in adverse pregnancy outcomes. Curr. Opin. Infect. Dis. 21:70-76. [DOI] [PubMed] [Google Scholar]
- 7.Belay, T., F. O. Eko, G. A. Ananaba, S. Bowers, T. Moore, D. Lyn, and J. U. Igietseme. 2002. Chemokine and chemokine receptor dynamics during genital chlamydial infection. Infect. Immun. 70:844-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289-300. [Google Scholar]
- 9.Berry, M. P. R., C. M. Graham, F. W. McNab, Z. Xu, S. A. A. Bloch, T. Oni, K. A. Wilkinson, R. Banchereau, J. Skinner, R. J. Wilkinson, C. Quinn, D. Blankenship, R. Dhawan, J. J. Cush, A. Mejias, O. Ramilo, O. M. Kon, V. Pascual, J. Banchereau, D. Chaussabel, and A. O'Garra. 2010. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466:973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brunham, R. C., and J. Rey-Ladino. 2005. Immunology of Chlamydia infection: implications for a Chlamydia trachomatis vaccine. Nat. Rev. Immunol. 5:149-161. [DOI] [PubMed] [Google Scholar]
- 11.Burton, M. J., R. L. Bailey, D. Jeffries, D. C. Mabey, and M. J. Holland. 2004. Cytokine and fibrogenic gene expression in the conjunctivas of subjects from a Gambian community where trachoma is endemic. Infect. Immun. 72:7352-7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burton, M. J., M. J. Holland, N. Faal, E. A. Aryee, N. D. Alexander, M. Bah, H. Faal, S. K. West, A. Foster, G. J. Johnson, D. C. Mabey, and R. L. Bailey. 2003. Which members of a community need antibiotics to control trachoma? Conjunctival Chlamydia trachomatis infection load in Gambian villages. Invest. Ophthalmol. Vis. Sci. 44:4215-4222. [DOI] [PubMed] [Google Scholar]
- 13.Cella, M., A. Fuchs, W. Vermi, F. Facchetti, K. Otero, J. K. Lennerz, J. M. Doherty, J. C. Mills, and M. Colonna. 2009. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature 457:722-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen, C. R., K. M. Koochesfahani, A. S. Meier, C. Shen, K. Karunakaran, B. Ondondo, T. Kinyari, N. R. Mugo, R. Nguti, and R. C. Brunham. 2005. Immunoepidemiologic profile of Chlamydia trachomatis infection: importance of heat-shock protein 60 and interferon-gamma. J. Infect. Dis. 192:591-599. [DOI] [PubMed] [Google Scholar]
- 15.Colonna, M., G. Borsellino, M. Falco, G. B. Ferrara, and J. L. Strominger. 1993. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc. Natl. Acad. Sci. U. S. A. 90:12000-12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colonna, M., E. G. Brooks, M. Falco, G. B. Ferrara, and J. L. Strominger. 1993. Generation of allospecific natural killer cells by stimulation across a polymorphism of HLA-C. Science 260:1121-1124. [DOI] [PubMed] [Google Scholar]
- 17.Colonna, M., and J. Samaridis. 1995. Cloning of immunoglobulin-superfamily members associated with HLA-C and HLA-B recognition by human natural killer cells. Science 268:405-408. [DOI] [PubMed] [Google Scholar]
- 18.Coombes, B. K., and J. B. Mahony. 2001. cDNA array analysis of altered gene expression in human endothelial cells in response to Chlamydia pneumoniae infection. Infect. Immun. 69:1420-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Oca, R. M., A. J. Buendia, L. Del Rio, J. Sanchez, J. Salinas, and J. A. Navarro. 2000. Polymorphonuclear neutrophils are necessary for the recruitment of CD8(+) T cells in the liver in a pregnant mouse model of Chlamydophila abortus (Chlamydia psittaci serotype 1) infection. Infect. Immun. 68:1746-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dessus-Babus, S., T. L. Darville, F. P. Cuozzo, K. Ferguson, and P. B. Wyrick. 2002. Differences in innate immune responses (in vitro) to HeLa cells infected with nondisseminating serovar E and disseminating serovar L2 of Chlamydia trachomatis. Infect. Immun. 70:3234-3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dessus-Babus, S., S. T. Knight, and P. B. Wyrick. 2000. Chlamydial infection of polarized HeLa cells induces PMN chemotaxis but the cytokine profile varies between disseminating and non-disseminating strains. Cell. Microbiol. 2:317-327. [DOI] [PubMed] [Google Scholar]
- 22.Duhen, T., R. Geiger, D. Jarrossay, A. Lanzavecchia, and F. Sallusto. 2009. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 10:857-863. [DOI] [PubMed] [Google Scholar]
- 23.Eyerich, S., K. Eyerich, D. Pennino, T. Carbone, F. Nasorri, S. Pallotta, F. Cianfarani, T. Odorisio, C. Traidl-Hoffmann, H. Behrendt, S. R. Durham, C. B. Schmidt-Weber, and A. Cavani. 2009. Th22 cells represent a distinct human T cell subset involved in epidermal immunity and remodeling. J. Clin. Invest. 119:3573-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faal, N., R. L. Bailey, D. Jeffries, H. Joof, I. Sarr, M. Laye, D. C. Mabey, and M. J. Holland. 2006. Conjunctival FOXP3 expression in trachoma: do regulatory T cells have a role in human ocular Chlamydia trachomatis infection? PLoS Med. 3:e266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faal, N., R. L. Bailey, I. Sarr, H. Joof, D. C. Mabey, and M. J. Holland. 2005. Temporal cytokine gene expression patterns in subjects with trachoma identify distinct conjunctival responses associated with infection. Clin. Exp. Immunol. 142:347-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortin, C. F., T. Ear, and P. P. McDonald. 2009. Autocrine role of endogenous interleukin-18 on inflammatory cytokine generation by human neutrophils. FASEB J. 23:194-203. [DOI] [PubMed] [Google Scholar]
- 27.Freeman, T. C., L. Goldovsky, M. Brosch, S. van Dongen, P. Maziere, R. J. Grocock, S. Freilich, J. Thornton, and A. J. Enright. 2007. Construction, visualisation, and clustering of transcription networks from microarray expression data. PLoS Comput. Biol. 3:2032-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gentil, K., and E. Pearlman. 2009. Gamma interferon and interleukin-1 receptor 1 regulate neutrophil recruitment to the corneal stroma in a murine model of Onchocerca volvulus keratitis. Infect. Immun. 77:1606-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillette-Ferguson, I., K. Daehnel, A. G. Hise, Y. Sun, E. Carlson, E. Diaconu, H. F. McGarry, M. J. Taylor, and E. Pearlman. 2007. Toll-like receptor 2 regulates CXC chemokine production and neutrophil recruitment to the cornea in Onchocerca volvulus/Wolbachia-induced keratitis. Infect. Immun. 75:5908-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffiths, M. J., M. J. Shafi, S. J. Popper, C. A. Hemingway, M. M. Kortok, A. Wathen, K. A. Rockett, R. Mott, M. Levin, C. R. Newton, K. Marsh, D. A. Relman, and D. P. Kwiatkowski. 2005. Genomewide analysis of the host response to malaria in Kenyan children. J. Infect. Dis. 191:1599-1611. [DOI] [PubMed] [Google Scholar]
- 31.Gumperz, J. E., V. Litwin, J. H. Phillips, L. L. Lanier, and P. Parham. 1995. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J. Exp. Med. 181:1133-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hess, S., J. Peters, G. Bartling, C. Rheinheimer, P. Hegde, M. Magid-Slav, R. Tal-Singer, and A. Klos. 2003. More than just innate immunity: comparative analysis of Chlamydophila pneumoniae and Chlamydia trachomatis effects on host-cell gene regulation. Cell. Microbiol. 5:785-795. [DOI] [PubMed] [Google Scholar]
- 33.Hook, C. E., M. K. Matyszak, and J. S. Gaston. 2005. Infection of epithelial and dendritic cells by Chlamydia trachomatis results in IL-18 and IL-12 production, leading to interferon-gamma production by human natural killer cells. FEMS Immunol. Med. Microbiol. 45:113-120. [DOI] [PubMed] [Google Scholar]
- 34.Hook, C. E., N. Telyatnikova, J. C. Goodall, V. M. Braud, A. J. Carmichael, M. R. Wills, and J. S. Gaston. 2004. Effects of Chlamydia trachomatis infection on the expression of natural killer (NK) cell ligands and susceptibility to NK cell lysis. Clin. Exp. Immunol. 138:54-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horowitz, A., R. H. Behrens, L. Okell, A. R. Fooks, and E. M. Riley. 2010. NK cells as effectors of acquired immune responses: effector CD4+ T cell-dependent activation of NK cells following vaccination. J. Immunol. [DOI] [PubMed]
- 36.Horowitz, A., K. C. Newman, J. H. Evans, D. S. Korbel, D. M. Davis, and E. M. Riley. 2010. Cross-talk between T cells and NK cells generates rapid effector responses to Plasmodium falciparum-infected erythrocytes. J. Immunol. 184:6043-6052. [DOI] [PubMed] [Google Scholar]
- 37.Huang, D. W., B. T. Sherman, and R. A. Lempicki. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44-57. [DOI] [PubMed] [Google Scholar]
- 38.Huang, D. W., B. T. Sherman, X. Zheng, J. Yang, T. Imamichi, R. Stephens, and R. A. Lempicki. 2009. Extracting biological meaning from large gene lists with DAVID. Curr. Protoc. Bioinformatics, Chapter 13, unit 13-11. [DOI] [PubMed]
- 39.Hume, D. A., K. M. Summers, S. Raza, J. K. Baillie, and T. C. Freeman. 2010. Functional clustering and lineage markers: insights into cellular differentiation and gene function from large-scale microarray studies of purified primary cell populations. Genomics 95:328-338. [DOI] [PubMed] [Google Scholar]
- 40.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249-264. [DOI] [PubMed] [Google Scholar]
- 41.Jiang, X., C. Shen, H. Yu, K. P. Karunakaran, and R. C. Brunham. 2010. Differences in innate immune responses correlate with differences in murine susceptibility to Chlamydia muridarum pulmonary infection. Immunology 129:556-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khatri, P., C. Voichita, K. Kattan, N. Ansari, A. Khatri, C. Georgescu, A. L. Tarca, and S. Draghici. 2007. Onto-Tools: new additions and improvements in 2006. Nucleic Acids Res. 35:W206-W211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King, M., H. Poya, J. Rao, S. Natarajan, A. W. Butch, N. Aziz, S. Kok, M. H. Chang, J. M. Lyons, K. Ault, and K. A. Kelly. 2009. CXCL13 expression in Chlamydia trachomatis infection of the female reproductive tract. Drugs Today (Barc.) 45(Suppl. B):125-134. [PMC free article] [PubMed] [Google Scholar]
- 44.Lad, S. P., E. Y. Fukuda, J. Li, L. M. de la Maza, and E. Li. 2005. Up-regulation of the JAK/STAT1 signal pathway during Chlamydia trachomatis infection. J. Immunol. 174:7186-7193. [DOI] [PubMed] [Google Scholar]
- 45.Langowski, J. L., X. Zhang, L. Wu, J. D. Mattson, T. Chen, K. Smith, B. Basham, T. McClanahan, R. A. Kastelein, and M. Oft. 2006. IL-23 promotes tumour incidence and growth. Nature 442:461-465. [DOI] [PubMed] [Google Scholar]
- 46.Lin, M., P. Jackson, A. M. Tester, E. Diaconu, C. M. Overall, J. E. Blalock, and E. Pearlman. 2008. Matrix metalloproteinase-8 facilitates neutrophil migration through the corneal stromal matrix by collagen degradation and production of the chemotactic peptide Pro-Gly-Pro. Am. J. Pathol. 173:144-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu, W., and K. A. Kelly. 2008. Prostaglandin E2 modulates dendritic cell function during chlamydial genital infection. Immunology 123:290-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mabbott, N. A., J. Kenneth Baillie, D. A. Hume, and T. C. Freeman. 2010. Meta-analysis of lineage-specific gene expression signatures in mouse leukocyte populations. Immunobiology 215:724-736. [DOI] [PubMed] [Google Scholar]
- 49.Mabey, D. C., R. L. Bailey, and Y. J. F. Hutin. 1992. The epidemiology and pathogenesis of trachoma. Rev. Med. Microbiol. 3:112-119. [Google Scholar]
- 50.Mariotti, S. P., D. Pascolini, and J. Rose-Nussbaumer. 2009. Trachoma: global magnitude of a preventable cause of blindness. Br. J. Ophthalmol. 93:563-568. [DOI] [PubMed] [Google Scholar]
- 51.Maxion, H. K., and K. A. Kelly. 2002. Chemokine expression patterns differ within anatomically distinct regions of the genital tract during Chlamydia trachomatis infection. Infect. Immun. 70:1538-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maxion, H. K., W. Liu, M. H. Chang, and K. A. Kelly. 2004. The infecting dose of Chlamydia muridarum modulates the innate immune response and ascending infection. Infect. Immun. 72:6330-6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McClarty, G., H. D. Caldwell, and D. E. Nelson. 2007. Chlamydial interferon gamma immune evasion influences infection tropism. Curr. Opin. Microbiol. 10:47-51. [DOI] [PubMed] [Google Scholar]
- 54.Morrison, R. P., and H. D. Caldwell. 2002. Immunity to murine chlamydial genital infection. Infect. Immun. 70:2741-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Natividad, A., G. Cooke, M. J. Holland, M. J. Burton, H. M. Joof, K. Rockett, D. P. Kwiatkowski, D. C. Mabey, and R. L. Bailey. 2006. A coding polymorphism in matrix metalloproteinase 9 reduces risk of scarring sequelae of ocular Chlamydia trachomatis infection. BMC Med. Genet. 7:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Natividad, A., M. J. Holland, K. A. Rockett, J. Forton, N. Faal, H. M. Joof, D. C. Mabey, R. L. Bailey, and D. P. Kwiatkowski. 2008. Susceptibility to sequelae of human ocular chlamydial infection associated with allelic variation in IL10 cis-regulation. Hum. Mol. Genet. 17:323-329. [DOI] [PubMed] [Google Scholar]
- 57.Natividad, A., J. Hull, G. Luoni, M. Holland, K. Rockett, H. Joof, M. Burton, D. Mabey, D. Kwiatkowski, and R. Bailey. 2009. Innate immunity in ocular Chlamydia trachomatis infection: contribution of IL8 and CSF2 gene variants to risk of trachomatous scarring in Gambians. BMC Med. Genet. 10:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nelson, D. E., D. P. Virok, H. Wood, C. Roshick, R. M. Johnson, W. M. Whitmire, D. D. Crane, O. Steele-Mortimer, L. Kari, G. McClarty, and H. D. Caldwell. 2005. Chlamydial IFN-gamma immune evasion is linked to host infection tropism. Proc. Natl. Acad. Sci. U. S. A. 102:10658-10663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pochetuhen, K., I. G. Luzina, V. Lockatell, J. Choi, N. W. Todd, and S. P. Atamas. 2007. Complex regulation of pulmonary inflammation and fibrosis by CCL18. Am. J. Pathol. 171:428-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prasse, A., D. V. Pechkovsky, G. B. Toews, W. Jungraithmayr, F. Kollert, T. Goldmann, E. Vollmer, J. Muller-Quernheim, and G. Zissel. 2006. A vicious circle of alveolar macrophages and fibroblasts perpetuates pulmonary fibrosis via CCL18. Am. J. Respir. Crit. Care Med. 173:781-792. [DOI] [PubMed] [Google Scholar]
- 61.Puccetti, P., and U. Grohmann. 2007. IDO and regulatory T cells: a role for reverse signalling and non-canonical NF-kappaB activation. Nat. Rev. Immunol. 7:817-823. [DOI] [PubMed] [Google Scholar]
- 62.Rajagopalan, S., and E. O. Long. 1999. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J. Exp. Med. 189:1093-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ramilo, O., W. Allman, W. Chung, A. Mejias, M. Ardura, C. Glaser, K. M. Wittkowski, B. Piqueras, J. Banchereau, A. K. Palucka, and D. Chaussabel. 2007. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 109:2066-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rank, R. G., H. M. Lacy, A. Goodwin, J. Sikes, J. Whittimore, P. B. Wyrick, and U. M. Nagarajan. 2010. Host chemokine and cytokine response in the endocervix within the first developmental cycle of Chlamydia muridarum. Infect. Immun. 78:536-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rapoza, P. A., S. G. Tahija, J. P. Carlin, S. L. Miller, M. L. Padilla, and G. I. Byrne. 1991. Effect of interferon on a primary conjunctival epithelial cell model of trachoma. Invest. Ophthalmol. Vis. Sci. 32:2919-2923. [PubMed] [Google Scholar]
- 66.Ren, Q., S. J. Robertson, D. Howe, L. F. Barrows, and R. A. Heinzen. 2003. Comparative DNA microarray analysis of host cell transcriptional responses to infection by Coxiella burnetii or Chlamydia trachomatis. Ann. N. Y. Acad. Sci. 990:701-713. [DOI] [PubMed] [Google Scholar]
- 67.Roshick, C., H. Wood, H. D. Caldwell, and G. McClarty. 2006. Comparison of gamma interferon-mediated antichlamydial defense mechanisms in human and mouse cells. Infect. Immun. 74:225-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rottenberg, M. E., A. Gigliotti-Rothfuchs, and H. Wigzell. 2002. The role of IFN-gamma in the outcome of chlamydial infection. Curr. Opin. Immunol. 14:444-451. [DOI] [PubMed] [Google Scholar]
- 69.Schachter, J. 1999. Infection and disease epidemiology, p. 139. In R. S. Stephens (ed.), Chlamydia: intracellualr biology, pathogenesis, and immunity. ASM Press, Washington, DC.
- 70.Schrader, S., A. Klos, S. Hess, H. Zeidler, J. G. Kuipers, and M. Rihl. 2007. Expression of inflammatory host genes in Chlamydia trachomatis-infected human monocytes. Arthritis Res. Ther. 9:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Single, R. M., M. P. Martin, X. Gao, D. Meyer, M. Yeager, J. R. Kidd, K. K. Kidd, and M. Carrington. 2007. Global diversity and evidence for coevolution of KIR and HLA. Nat. Genet. 39:1114-1119. [DOI] [PubMed] [Google Scholar]
- 72.Solomon, A. W., M. J. Holland, M. J. Burton, S. K. West, N. D. Alexander, A. Aguirre, P. A. Massae, H. Mkocha, B. Munoz, G. J. Johnson, R. W. Peeling, R. L. Bailey, A. Foster, and D. C. Mabey. 2003. Strategies for control of trachoma: observational study with quantitative PCR. Lancet 362:198-204. [DOI] [PubMed] [Google Scholar]
- 73.Sporri, R., N. Joller, H. Hilbi, and A. Oxenius. 2008. A novel role for neutrophils as critical activators of NK cells. J. Immunol. 181:7121-7130. [DOI] [PubMed] [Google Scholar]
- 74.Starnbach, M. N., and N. R. Roan. 2008. Conquering sexually transmitted diseases. Nat. Rev. Immunol. 8:313-317. [DOI] [PubMed] [Google Scholar]
- 75.Stephens, R. S. 2003. The cellular paradigm of chlamydial pathogenesis. Trends Microbiol. 11:44-51. [DOI] [PubMed] [Google Scholar]
- 76.Stockinger, B., and M. Veldhoen. 2007. Differentiation and function of Th17 T cells. Curr. Opin. Immunol. 19:281-286. [DOI] [PubMed] [Google Scholar]
- 77.Stockinger, B., M. Veldhoen, and B. Martin. 2007. Th17 T cells: linking innate and adaptive immunity. Semin. Immunol. 19:353-361. [DOI] [PubMed] [Google Scholar]
- 78.Subramanian, A., P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, and J. P. Mesirov. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102:15545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Theocharidis, A., S. van Dongen, A. J. Enright, and T. C. Freeman. 2009. Network visualization and analysis of gene expression data using BioLayout Express(3D). Nat. Protoc. 4:1535-1550. [DOI] [PubMed] [Google Scholar]
- 80.Thomas, S. M., L. F. Garrity, C. R. Brandt, C. S. Schobert, G. S. Feng, M. W. Taylor, J. M. Carlin, and G. I. Byrne. 1993. IFN-gamma-mediated antimicrobial response. Indoleamine 2,3-dioxygenase-deficient mutant host cells no longer inhibit intracellular Chlamydia spp. or Toxoplasma growth. J. Immunol. 150:5529-5534. [PubMed] [Google Scholar]
- 81.Thylefors, B., C. R. Dawson, B. R. Jones, S. K. West, and H. R. Taylor. 1987. A simple system for the assessment of trachoma and its complications. Bull. World Health Organ. 65:477-483. [PMC free article] [PubMed] [Google Scholar]
- 82.Trifari, S., C. D. Kaplan, E. H. Tran, N. K. Crellin, and H. Spits. 2009. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat. Immunol. 10:864-871. [DOI] [PubMed] [Google Scholar]
- 83.Tseng, C. T., and R. G. Rank. 1998. Role of NK cells in early host response to chlamydial genital infection. Infect. Immun. 66:5867-5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Uhrberg, M., N. M. Valiante, B. P. Shum, H. G. Shilling, K. Lienert-Weidenbach, B. Corliss, D. Tyan, L. L. Lanier, and P. Parham. 1997. Human diversity in killer cell inhibitory receptor genes. Immunity 7:753-763. [DOI] [PubMed] [Google Scholar]
- 85.van Dongen, S. 2000. Graph clustering by flow simulation. University of Utrecht, Utrecht, Netherlands.
- 86.Virok, D., A. Loboda, L. Kari, M. Nebozhyn, C. Chang, C. Nichols, V. Endresz, E. Gonczol, K. Berencsi, M. K. Showe, and L. C. Showe. 2003. Infection of U937 monocytic cells with Chlamydia pneumoniae induces extensive changes in host cell gene expression. J. Infect. Dis. 188:1310-1321. [DOI] [PubMed] [Google Scholar]
- 87.Wang, S., Y. Fan, R. C. Brunham, and X. Yang. 1999. IFN-gamma knockout mice show Th2-associated delayed-type hypersensitivity and the inflammatory cells fail to localize and control chlamydial infection. Eur. J. Immunol. 29:3782-3792. [DOI] [PubMed] [Google Scholar]
- 88.Wilson, M. J., M. Torkar, A. Haude, S. Milne, T. Jones, D. Sheer, S. Beck, and J. Trowsdale. 2000. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. U. S. A. 97:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xia, M., R. E. Bumgarner, M. F. Lampe, and W. E. Stamm. 2003. Chlamydia trachomatis infection alters host cell transcription in diverse cellular pathways. J. Infect. Dis. 187:424-434. [DOI] [PubMed] [Google Scholar]
- 90.Yang, X., J. Gartner, L. Zhu, S. Wang, and R. C. Brunham. 1999. IL-10 gene knockout mice show enhanced Th1-like protective immunity and absent granuloma formation following Chlamydia trachomatis lung infection. J. Immunol. 162:1010-1017. [PubMed] [Google Scholar]
- 91.Yang, X., K. T. Hayglass, and R. C. Brunham. 1998. Different roles are played by alpha beta and gamma delta T cells in acquired immunity to Chlamydia trachomatis pulmonary infection. Immunology 94:469-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yoneda, C., C. R. Dawson, T. Daghfous, I. Hoshiwara, P. Jones, M. Messadi, and J. Schachter. 1975. Cytology as a guide to the presence of chlamydial inclusions in Giemsa-stained conjunctival smears in severe endemic trachoma. Br. J. Ophthalmol. 59:116-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zaharik, M. L., T. Nayar, R. White, C. Ma, B. A. Vallance, N. Straka, X. Jiang, J. Rey-Ladino, C. Shen, and R. C. Brunham. 2007. Genetic profiling of dendritic cells exposed to live- or ultraviolet-irradiated Chlamydia muridarum reveals marked differences in CXC chemokine profiles. Immunology 120:160-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.