Abstract
Acute liver injury results from exposure to toxins, pharmacological agents, or viral infections, contributing to significant morbidity and mortality worldwide. While hepatic inflammation is critical for liver repair, the transcriptional mechanisms required for the recruitment of inflammatory cells to the liver are not understood. Forkhead box M1 (Foxm1) transcription factor is a master regulator of hepatocyte proliferation, but its role in inflammatory cells remains unknown. In this study, we generated transgenic mice in which Foxm1 was deleted from myeloid-derived cells, including macrophages, monocytes, and neutrophils. Carbon tetrachloride liver injury was used to demonstrate that myeloid-specific Foxm1 deletion caused a delay in liver repair. Although Foxm1 deficiency did not influence neutrophil infiltration into injured livers, the total numbers of mature macrophages were dramatically reduced. Surprisingly, Foxm1 deficiency did not influence the proliferation of macrophages or their monocytic precursors but impaired monocyte recruitment during liver repair. Expression of L-selectin and the CCR2 chemokine receptor, both critical for monocyte recruitment to injured tissues, was decreased. Foxm1 induced transcriptional activity of the mouse CCR2 promoter in cotransfection experiments. Adoptive transfer of monocytes to Foxm1-deficient mice restored liver repair and rescued liver function. Foxm1 is critical for liver repair and is required for the recruitment of monocytes to the injured liver.
Acute liver injury and underlying hepatic inflammation result from a variety of insults, including natural and industrial toxins, pharmacological agents, and viral infections, contributing to significant morbidity and mortality worldwide (6). In some patients, acute liver injury progresses to acute (fulminant) hepatic failure (FHF), a severe life-threatening disease characterized by impairment of liver function and hepatic encephalopathy (34). Before the emergence of liver transplantation, the survival rates for patients with FHF were as low as 15% (8). In recent years, 10% of annual liver transplantations are due to FHF (8). Acute liver injury can also trigger chronic inflammatory liver disease characterized by hepatic fibrosis and cirrhosis, leading to liver failure (7). Acute liver injury is characterized by necrosis of hepatocytes surrounding the central vein caused by local expression of a P450 enzyme forming toxic free-radical metabolites (32). Hepatic repair following acute liver injuries involves a proliferation of periportal hepatocytes and transient differentiation of stellate cells into fibrogenic myofibroblasts, which secrete collagen that is essential for reestablishment of normal liver architecture (7). During acute liver injury, recruitment of inflammatory cells such as macrophages, monocytes, and neutrophils to the site of injury and inflammation is an important event in removing necrotic tissue and normal liver repair (1, 7). The transcription factors required for the recruitment of inflammatory cells during liver repair are not understood.
The Forkhead box (Fox) proteins are a large family of transcription factors that share homology in the Winged Helix/Forkhead DNA-binding domain. Foxm1 transcription factor (previously known as HFH-11B, Trident, Win, or MPP2) is expressed in all tissues during embryogenesis, but its expression in adult mice is restricted to the intestinal crypts, thymus, and testes (44). During carbon tetrachloride (CCl4)-mediated liver injury and liver regeneration, Foxm1 expression is induced in a variety of hepatic cell types, including hepatocytes, bile duct epithelial cells, and hepatic stromal and inflammatory cells (40, 44). Foxm1 expression is increased in tumor cells during the progression of liver, lung, and prostate cancers (15, 18, 21). In previous studies we demonstrated that Foxm1−/− mice die in utero between embryonic day 13.5 (E13.5) and E16.5 due to multiple abnormalities in development of the embryonic liver, lung, and heart (22, 26, 35). Abnormal accumulation of polyploid cells, resulting from diminished DNA replication and failure to enter mitosis, was observed in these Foxm1−/− mouse organs (24, 26, 35). Foxm1 is required for differentiation of hepatoblast precursor cells toward the biliary epithelial cell lineage, and Foxm1−/− livers fail to form intrahepatic bile ducts (26). Likewise, Foxm1−/− embryos exhibit defects in differentiation of pulmonary mesenchyme into mature capillary endothelial cells during the canalicular stage of lung development (22).
Recently, several conditional knockout mouse models were generated to address Foxm1 requirements in different cell types. Hepatocyte-specific deletion of Foxm1 in albumin-Cre Foxm1fl/fl mice impaired hepatocyte proliferation during liver regeneration (41). The Foxm1 deletion in hepatocytes prevented formation of hepatocellular carcinoma in adult mice by inducing p27kip1 tumor suppressor (18). Endothelial cell-specific Foxm1 deletion caused increased lung vascular permeability and increased mortality in response to the inflammatory mediator lipopolysaccharide (46). Mice with pancreas-specific deletion of Foxm1 displayed severe abnormalities in postnatal β-cell mass expansion, causing impaired islet function and diabetes (45). Foxm1 inactivation in precursors of cerebellar granule neurons caused a delay in brain development by interfering with Shh-induced neuroproliferation (39). We recently demonstrated that Foxm1 deletion from the respiratory epithelium did not influence epithelial proliferation but inhibited lung maturation, surfactant homeostasis, and the differentiation of type II epithelial cells, causing respiratory failure after birth (16). Although these studies showed that Foxm1 plays distinct roles in differentiation and proliferation in various cell lineages, the specific role of Foxm1 in myeloid-derived inflammatory cells, such as macrophages, neutrophils, and monocytes, remains unknown.
In the present study, we used a CCl4-mediated liver injury model to determine the function of Foxm1 in myeloid inflammatory cells during acute liver injury and liver repair. Transgenic mice with a specific deletion of Foxm1 in these cells were generated (mFoxm1−/−). CCl4 liver injury caused a delay in liver repair in mFoxm1−/− mice. Although Foxm1 did not influence neutrophil infiltration, the total numbers of mature macrophages were dramatically reduced in injured mFoxm1−/− livers. Reduced expression of CCR2, a chemokine receptor critical for monocyte recruitment into injured tissues, was found in mFoxm1−/− livers and isolated mFoxm1−/− monocytes. Foxm1 directly induces CCR2 promoter activity and is required for recruitment of monocytes into injured liver. We demonstrate here a novel function of Foxm1 in inflammatory cells and tissue repair.
MATERIALS AND METHODS
Mouse strains and CCl4 treatment.
We previously described the generation of Foxm1flox/flox (Foxm1fl/fl) mice, which contain LoxP sequences flanking DNA binding and transcriptional activation domains of the Foxm1 gene (26). Foxm1fl/fl female mice were bred with LysM-Cretg/− male mice (purchased from Jackson Laboratories) to generate the LysM-Cretg/− Foxm1fl/fl double transgenic mice (mFoxm1−/−). All mice were maintained in a C57BL/6 genetic background. LysM-Cre deletes Foxm1 in all myeloid cells, including macrophages, monocytes, and granulocytes (4). mFoxm1−/− mice displayed no birth abnormalities and were capable of mating. Foxm1fl/fl littermates lacking the LysM-Cre transgene were used as controls. Further controls included double-heterozygous LysM-Cretg/− Foxm1fl/+ mice. No liver defects were observed in control mice. Carbon tetrachloride (CCl4; Sigma, St. Louis, MO) was dissolved in mineral oil at a 1:10 ratio, and a single intraperitoneal injection of CCl4 (1 μl of CCl4/g of body weight) was administered to mFoxm1−/− mice and their Foxm1fl/fl littermates (17). To determine statistical significance of any observed differences, we used five female mice per time point following CCl4 administration, which included 24, 48, and 72 h. The levels of alkaline phosphatase, albumin, bilirubin, glucose, and the aminotransferases ALT and AST were determined by serological analysis of blood serum. Liver tissue was used to prepare total RNA. For histological studies, livers were fixed, paraffin embedded, and sectioned as described previously (17).
Immunohistochemical staining and flow cytometry.
Paraffin liver sections were stained with hematoxylin and eosin (H&E) or used for immunohistochemical staining as described previously (14, 17). The following antibodies were used for immunohistochemistry: Ki-67 (1:500; clone Tec-3; Dako); activated caspase 3 (1:200; 5A1; Cell Signaling), and Foxm1 (1:2,000; K-19; Santa Cruz). Antibody-antigen complexes were detected by using biotinylated secondary antibody, followed by avidin-horseradish peroxidase complex and DAB substrate (Vector Laboratories, Burlingame, CA). Sections were counterstained with nuclear fast red.
For flow cytometry experiments, hepatic inflammatory cells were prepared from mFoxm1−/− and control Foxm1fl/fl livers as described previously (27). Briefly, livers were perfused with buffered saline, removed, gently minced with scissors, and then placed in RPMI 1640 medium containing Liberase Blendzyme 3 (0.2 U/ml; Roche Diagnostics) and DNase I (0.5 mg/ml; Sigma) for 50 min at 37°C. After digestion, liver tissue was forced through a 40-μm-pore-size cell strainer. Hepatocyte and stromal debris was removed by using 30% (vol/vol) Percoll gradient centrifugation (GE Healthcare). Red blood cells were lysed with ACK lysis buffer (Invitrogen).
Inflammatory cells were stained with fluorescence-labeled antibodies against CD11b, Ly-6C, Ly-6G, F4/80, CD62L, and CD86 as previously described (19). Staining was performed at 4°C after incubation with FcBlock (anti-mouse CD16/32, clone 93; eBiosciences, San Diego, CA) for 30 min. The following antibodies were used: anti-F4/80 (clone BM8; eBiosciences), anti-CD11b (clone M1/70; eBiosciences), anti-Ly-6C (clone HK1.4; BioLegend, San Diego, CA), anti-Ly-6G (clone 1A8; BioLegend), anti-CD62L (clone MEL-14; eBiosciences), and anti-CD86 (clone PO3; BioLegend). Dead cells were excluded by using 7-AAD stain (eBiosciences).
For cell cycle analysis, numbers of cells in the G0/G1, S, and G2/M phases of the cell cycle were identified by using Vybrant DyeCycle violet stain (Invitrogen) according to manufacturer's recommendations. The data were acquired by using an LSRII flow cytometer (BD Biosciences, San Jose, CA). Cell sortings were performed by using a 5-laser FACSAria II flow cytometer (BD Biosciences). Spectral overlap was compensated for by using FACSDiVa software (BD Biosciences) and analyzed using FlowJo software (Treestar, Inc., Ashland, OR).
Isolation and adoptive transfer of monocytes during liver injury.
Monocyte-enriched cell population was isolated from bone marrow of Foxm1fl/fl mice as described previously (36). Erythrocytes and neutrophils were removed by a Ficoll gradient centrifugation. Cells were incubated with biotin-conjugated anti-CD115 (M-CSF receptor) antibodies (eBioscience), followed by anti-biotin MACS beads (Miltenyi Biotech). A positive fraction was obtained through magnetic separation and labeled with carboxyfluorescein succinimidyl ester (CFSE) as described previously (42). The percentage of monocytes in the positive fraction was 85%, as determined by flow cytometric analysis of the monocytic phenotype CD115+/CD11b+/Ly-6G−/Ly-6Chi/F4/80lo. At 24 h after CCl4 injury, 106 labeled cells were injected into tail veins of mFoxm1−/− or control Foxm1fl/fl mice. Recipient mice were harvested at 24 or 48 h after adoptive transfer.
RNA preparation and qRT-PCR.
Total RNA was prepared from either mouse liver or sorted inflammatory cells by using RNA-STAT-60 (Tel-Test “B”, Inc., Friendswood, TX). Quantitative real-time RT-PCR (qRT-PCR) analysis was performed using a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA) as described previously (16). Samples were amplified with TaqMan gene expression master mix (Applied Biosystems) combined with inventoried TaqMan gene expression assays for the gene of interest (Table 1). Reactions were analyzed in triplicates, and the expression levels were normalized to β-actin. Five mice were used in each group.
TABLE 1.
qRT-PCR analysis of total RNA prepared from mFoxm1−/− and Foxm1fl/fl livers
| Mouse TaqMan gene expression assaya | Catalog no. |
|---|---|
| Foxm1 | Mm01184444_g1 |
| β-Actin | Mm00607939_s1 |
| CCR2 | Mm00438270_m1 |
| CCL2 (MCP-1) | Mm99999056_m1 |
| TNF-α | Mm00443258_m1 |
| IL-1β | Mm01336189_m1 |
| c-myc | Mm00487804_m1 |
| Cyclin D1 | Mm00432359_m1 |
| CD62L (L-selectin) | Mm00441291_m1 |
The TaqMan gene expression assays used for qRT-PCR were from Applied Biosystems.
Cotransfection studies.
Mouse CCR2 promoter fragments were amplified by PCR of mouse genomic DNA using following primers: AAG ACA TGT ACC CTT TGT AGC (−269/−249), AAG ATG AAA GTT GAG AGG (−575/−558), TGT GTT AAA GTA CTT GCC GTG (−1428/−1408), and TTC CTT TGA TTC TGT GGT (+5/+22). We transfected human osteosarcoma U2OS cells with either cytomegalovirus promoter (CMV)-Foxm1b or control CMV-empty expression plasmids, as well as with luciferase (LUC) reporters driven by the mouse CCR2 promoter regions. CMV-Renilla was used as an internal control to normalize transfection efficiency. A dual luciferase assay (Promega) was performed 48 h after transfection, as described previously (22, 23, 29).
Statistical analysis.
Student t test was used to determine statistical significance. P values of ≤0.05 were considered significant. Values for all measurements were expressed as the means ± the standard deviations (SD).
RESULTS
Conditional deletion of Foxm1 in myeloid cells.
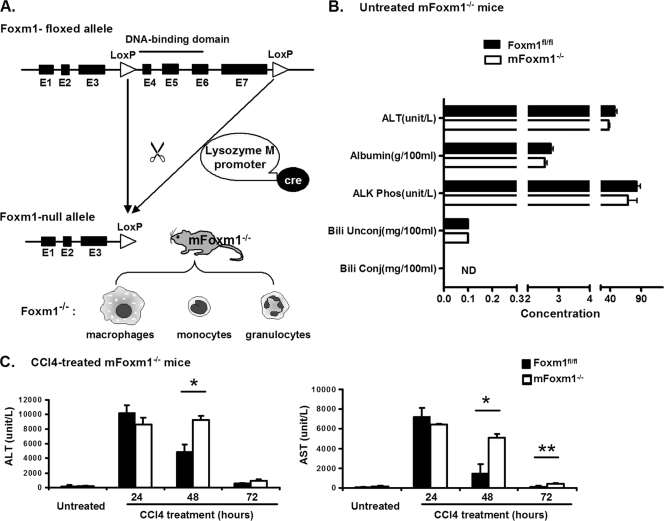
Previous studies demonstrated that Foxm1 is required for hepatocyte proliferation following either partial hepatectomy or carbon tetrachloride (CCl4)-induced liver injury (40, 41). During the liver injury, Foxm1 is expressed in various hepatic cell types, including inflammatory cells (40, 41). To address the role of Foxm1 in myeloid-derived inflammatory cells, such as macrophages, monocytes and neutrophils, a mouse line with a myeloid-specific Foxm1 deletion (mFoxm1−/−) was generated. mFoxm1−/− mice (LysM-Cretg/− Foxm1fl/fl) contain LoxP-flanked Foxm1-floxed gene (Foxm1fl/fl [26]) and the LysM-Cre transgene (Fig. 1A), mediating a conditional deletion of exons 4 to 7 of the Foxm1 gene in myeloid cells (4). These exons encode DNA-binding and C-terminal transcriptional activation domains of the Foxm1 protein (Fig. 1A), both of which are required for Foxm1 transcriptional activity (41). mFoxm1−/− mutant mice were healthy, displaying no obvious abnormalities. The percentages of neutrophils and monocytes were not changed in either the blood or bone marrow of mFoxm1−/− mice (data not shown). Morphological examination of liver sections from mFoxm1−/− mice revealed no differences compared to control Foxm1fl/fl mice (Fig. 2A). To evaluate liver metabolic functions in Foxm1 mutant mice, series of serological tests were carried out. The serum levels of hepatic aminotransferases ALT and AST, alkaline phosphatase, albumin, bilirubin, and glucose were normal (Fig. 1B and data not shown), indicating that Foxm1 deficiency in myeloid cells does not influence liver function.
FIG. 1.
Increased ALT and AST serum levels in CCl4-treated mFoxm1−/− mice. (A) Schematic drawing of Cre-mediated deletion in Foxm1-floxed gene. The Foxm1-floxed allele contains a deletion of exons 4 to 7 of the mouse Foxm1 gene. To delete Foxm1 in myeloid cells (macrophages, monocytes, and granulocytes), Foxm1fl/fl mice were bred with LysM-Cretg/− transgenic mice to generate LysM-Cretg/− Foxm1fl/fl (mFoxm1−/−) mice. (B) Normal liver function in untreated mFoxm1−/− mice. Blood serum was obtained from untreated Foxm1fl/fl and mFoxm1−/− mice and then examined for concentrations of hepatic aminotransferase ALT, alkaline phosphatase (ALK Phos), albumin, and conjugated and unconjugated bilirubin. (C) ALT and AST serum levels in CCl4-treated mFoxm1−/− mice. At 24 h after CCl4 injection, mFoxm1−/− and control Foxm1fl/fl mice displayed similar increases in serum ALT and AST levels. Although hepatic aminotransferases were decreased in control Foxm1fl/fl mice at 48 and 72 h after CCl4 treatment, CCl4-treated mFoxm1−/− mice maintained significantly increased levels of ALT and AST. Means ± the SD were determined with five different mice in each group. Statistically significant differences are indicated (*, P < 0.05; **, P < 0.01).
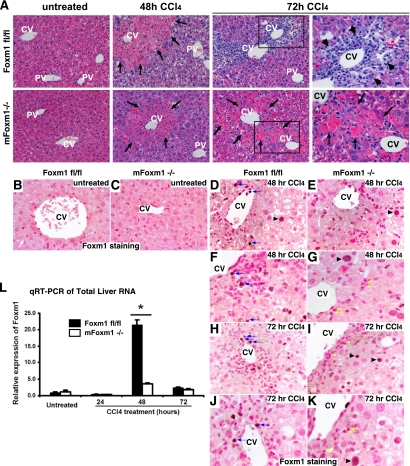
FIG. 2.
Delayed resolution of liver injury in mFoxm1−/− mice. Paraffin sections from mFoxm1−/− mice and their control Foxm1fl/fl littermates were either stained with H&E or used for immunohistochemistry with Foxm1 antibodies. (A) Delayed liver repair in mFoxm1−/− mice. Similar histological structures were observed in untreated Foxm1fl/fl and mFoxm1−/− livers. Liver damage (shown with arrows) was observed around hepatic central veins (CV) but not around portal veins (PV) at 48 h after CCl4 administration. Liver remodeling and leukocyte infiltration were noted in the pericentral regions of Foxm1fl/fl mice at 72 h after liver injury (shown with arrowheads). CCl4-treated mFoxm1−/− livers failed to repair severe necrotic damage around central veins, and the number of inflammatory cells in pericentral regions was decreased. (B to K) Diminished Foxm1 staining in injured mFoxm1−/− livers. Foxm1 staining was not detected in the livers of untreated mice (B and C). In CCl4-treated control Foxm1fl/fl livers, nuclear Foxm1 staining (brown) was detected in a subset of hepatocytes (arrowhead in panel D) and inflammatory cells infiltrating central vein (CV) regions (blue arrows in panels D, F, H, and J). Although CCl4-injured mFoxm1−/− mice displayed normal Foxm1 staining in hepatocytes (arrowheads in panels E, G, I, and K), Foxm1 was not detected in a majority of mFoxm1−/− inflammatory cells (yellow arrows). (L) qRT-PCR shows diminished Foxm1 mRNA in the total liver RNA. Each individual sample was normalized to its corresponding β-actin level. Means ± the SD were determined with five different mFoxm1−/− and Foxm1fl/fl livers. A statistically significant P value of <0.05 is indicated by an asterisk. Magnifications: A to E, H, and I, ×200; A (right panels), F, G, J, and K, ×400.
Delayed liver repair mFoxm1−/− mice.
To determine whether Foxm1 function in myeloid cells is required for liver repair, we used CCl4 liver injury to induce inflammatory response. Adult mFoxm1−/− mice, along with the corresponding control Foxm1fl/fl littermates, were intraperitoneally injected with CCl4 and their livers, peripheral blood, and bone marrow were harvested at different intervals after CCl4 liver injury. At 24 h after CCl4 injection, the serum ALT and AST levels were equally increased in both mFoxm1−/− and control Foxm1fl/fl mice (Fig. 1C), indicating that Foxm1 deficiency does not alter the initial response to liver injury. At 48 h after CCl4 treatment, hepatic aminotransferases were decreased in control Foxm1fl/fl mice, reaching nearly normal serum levels by 72 h (Fig. 1C). In contrast, CCl4-treated mFoxm1−/− mice maintained significantly elevated levels of ALT and AST (Fig. 1C), indicating that the liver repair was delayed in mFoxm1−/− mice. Consistent with serological analysis of liver enzymes, H&E staining showed that hepatic damage was repaired in control Foxm1fl/fl mice at 72 h after liver injury, whereas a severe pericentral necrosis was still observed in mFoxm1−/− livers (Fig. 2A).
Previous studies demonstrated that Foxm1 induces the proliferation of hepatocytes, fibroblasts, endothelial cells, and numerous neoplastic cells (18, 22, 24, 26). Therefore, cellular proliferation in mFoxm1−/− and control livers was assessed by immunostaining with antibody against the cell proliferation marker Ki-67. After the liver injury, Ki-67 staining was detected in hepatocytes and nonhepatocytes (hepatic stromal cells and inflammatory cells) (Fig. 3A). The numbers of Ki-67-positive cells in mFoxm1−/− and control Foxm1fl/fl livers were similar at 48 h after CCl4 treatment (Fig. 3B), indicating that the initial proliferation response was normal in Foxm1 mutants. At 72 h after liver injury, mFoxm1−/− mice displayed significantly reduced numbers of Ki-67-positive cells in both pericentral and parenchymal hepatic regions (Fig. 3A and B), a result consistent with a delay in liver repair. Decreased mRNA levels of c-Myc and cyclin D1 were found in CCl4-treated mFoxm1−/− livers by qRT-PCR (Fig. 3C and D), which is consistent with decreased cellular proliferation. Immunostaining of liver sections with an antibody specific to activated form of caspase 3 demonstrated an increase in pericentral apoptosis in mFoxm1−/− mice at 72 h after liver injury (Fig. 3A). Thus, decreased proliferation and increased hepatic apoptosis may contribute to a delay of liver repair in mFoxm1−/− mice. Interestingly, decreased numbers of infiltrating inflammatory cells were observed in pericentral areas of CCl4-treated mFoxm1−/− livers compared to Foxm1fl/fl controls (Fig. 2A). Altogether, these results demonstrated that deletion of Foxm1 from cells of myeloid lineage delayed liver repair and decreased leukocyte infiltration after CCl4 injury.
FIG. 3.
Diminished cellular proliferation and increased apoptosis in CCl4-injured mFoxm1−/− livers. (A) CCl4-treated mFoxm1−/− livers displayed diminished proliferation in pericentral and parenchymal liver regions. Paraffin sections from the livers of mFoxm1−/− and control Foxm1fl/fl mice were stained with Ki-67 antibody (dark brown nuclei) and counterstained with nuclear fast red (red nuclei). To detect apoptotic cells, paraffin liver sections were stained with antibodies against activated caspase 3 (lower right panels). Increased apoptosis was detected in pericentral regions of mFoxm1−/− livers at 72 h after CCl4 treatment. No apoptosis was observed in livers from control Foxm1fl/fl mice. Magnifications, ×200 and ×400 as indicated. (B) mFoxm1−/− livers exhibited a decreased number of Ki-67-positive cells at 72 h after CCl4 treatment. The Ki-67-stained nuclei were counted in hepatocytes and nonhepatocytes by using 10 random ×400 microscope fields from mFoxm1−/− and control Foxm1fl/fl mice. (C and D) CCl4-injured mFoxm1−/− livers displayed diminished expression of c-myc and cyclin D1. Livers from either untreated or CCl4-treated mFoxm1−/− and control Foxm1fl/fl mice were used to prepare total liver RNA. qRT-PCR analysis showed decreased mRNAs of cyclin D1 and c-myc in mFoxm1−/− livers at 24 h after liver injury. Each individual sample was normalized to its corresponding β-actin level. Means ± the SD were determined using five different mFoxm1−/− and Foxm1fl/fl livers. A statistically significant P value of <0.05 is indicated by an asterisk.
Decreased Foxm1 expression in CCl4-injured mFoxm1−/− livers.
We focused our next experiments to determine the efficiency of Foxm1 deletion in mFoxm1−/− livers. Expression of Foxm1 in the liver was examined by immunohistochemistry using an antibody specific to N-terminal region of the Foxm1 protein. Consistent with previous studies (40, 41, 43), Foxm1 staining was not detected in livers of untreated mice (Fig. 2B and C). At 48 h after CCl4 injury, nuclear staining of Foxm1 became detectable in various hepatic cells of control Foxm1fl/fl livers, including hepatocytes and endothelial cells, as well as in inflammatory cells, which had infiltrated the pericentral liver regions (Fig. 2D and F). Although Foxm1 expression in Foxm1fl/fl hepatocytes had decreased by 72 h after CCl4 injury, Foxm1 protein was still observed in hepatic stromal cells and inflammatory cells (Fig. 2H and J). In mFoxm1−/− livers, Foxm1 staining was detected in hepatocytes (Fig. 2E and G) and endothelial cells (Fig. 2K) but not in a majority of inflammatory cells after the injury (Fig. 2I and K). Quantitative real-time RT-PCR analysis (qRT-PCR) was used to demonstrate that Foxm1 mRNA from CCl4-injured mFoxm1−/− livers was decreased 9-fold compared to Foxm1fl/fl controls (Fig. 2L).
Reduced numbers of infiltrating macrophages in mFoxm1−/− livers after CCl4 injury.
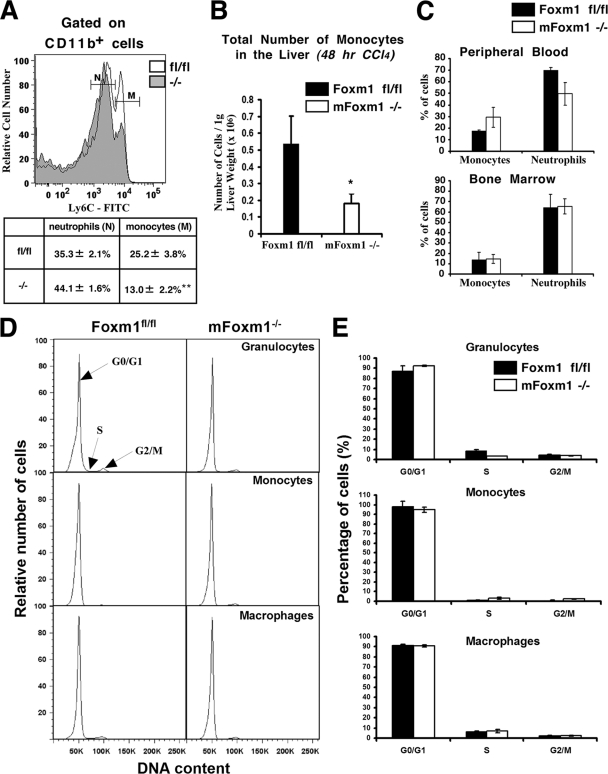
Since histological studies revealed decreased numbers of inflammatory cells in CCl4-treated mFoxm1−/− livers (Fig. 2A), we used flow cytometry to identify specific populations of myeloid cells during liver repair. Inflammatory cells were isolated from either mFoxm1−/− or control Foxm1fl/fl livers by using Percoll gradient centrifugation (2) and then stained the cells with antibodies against CD11b and F4/80. CD11b is an adhesion molecule expressed on the surface of myeloid cells, whereas F4/80 is a specific marker of mature macrophages (28). Untreated mFoxm1−/− and control Foxm1fl/fl livers displayed similar numbers of cells with high CD11b (CD11bhi) expression (Fig. 4A). After CCl4 injury, decreased numbers of CD11bhi myeloid cells were observed in mFoxm1−/− livers (Fig. 4A), whereas the numbers of CD11blo cells remained unchanged (data not shown). To determine which populations of myeloid (CD11bhi) cells were influenced by the Foxm1 deficiency, we examined Ly-6C and F4/80 cell surface markers in hepatic myeloid cells. Consistent with published studies (2, 9, 30, 38), monocyte-derived hepatic macrophages were identified as Ly-6Clo/F4/80hi cells. In control Foxm1fl/fl livers, numbers of hepatic macrophages were dramatically increased at 72 h after liver injury (Fig. 4B). In contrast, mFoxm1−/− livers displayed a significant decrease in numbers of infiltrating macrophages compared to control Foxm1fl/fl livers (Fig. 4B). Altogether, these results indicate that deletion of Foxm1 from cells of myeloid lineage decreased numbers of hepatic macrophages (Kupffer cells) during liver repair.
FIG. 4.
Reduced numbers of macrophages in CCl4-injured mFoxm1−/− livers. Inflammatory cells were isolated from either mFoxm1−/− or control Foxm1fl/fl livers by using Percoll gradient centrifugation. Flow cytometry was used to determine numbers of myeloid cells, macrophages and neutrophils. (A and B) Flow cytometric analysis for surface CD11b, F4/80, and Ly-6C expression. The percentages of CD11bhi myeloid cells are shown for mFoxm1−/− or control Foxm1fl/fl livers at 72 h after the injury (top sections in panel A). mFoxm1−/− livers display a significant reduction in total numbers of CD11bhi cells per 1 g of liver weight at 72 h after liver injury (bottom section in panel A). The cumulative data from three experiments are presented as means ± the SD. Statistically significant differences are indicated (*, P < 0.05). The expression of F4/80 and Ly-6C surface markers was also analyzed in CD11bhi cells (B). The gate shows CD11bhi/Ly-6Clo/F4/80hi hepatic macrophages (top sections in panel B). Reduced numbers of macrophages were found in CCl4-injured mFoxm1−/− livers at 72 h after liver injury (bottom section in panel B). Bars represent total numbers of cells per 1 g of liver weight in mFoxm1−/− and control Foxm1fl/fl livers. (C) Flow cytometry was used to visualize Ly-6G+/Ly-6C+ neutrophils in the population of myeloid cells gated for CD11bhi and F4/80low (top sections in panel C). mFoxm1−/− and control Foxm1fl/fl livers display similar numbers of neutrophils at all time points after the injury (bottom section in panel C).
Foxm1 deficiency does not influence neutrophil recruitment into CCl4-injured liver.
To determine whether Foxm1 deficiency influences neutrophil infiltration during liver injury, inflammatory cells were isolated from either mFoxm1−/− or control Foxm1fl/fl livers and then simultaneously stained with fluorescently labeled antibodies against CD11b, Ly-6C, and Ly-6G, the latter of which is a specific marker of neutrophils (5). Hepatic inflammatory cells expressing all three markers (CD11bhi/Ly-6C+/Ly-6G+) were identified as neutrophils (30). Consistent with published studies (12), total numbers of neutrophils in the liver were increased in the first 24 h after CCl4 injury and then returned to nearly normal levels at 72 h (Fig. 4C). No differences in neutrophil numbers were observed between mFoxm1−/− and control Foxm1fl/fl livers (Fig. 4C). Furthermore, the percentages of activated neutrophils in the liver tissue were determined using antibody against CD62L (L-selectin), a cell adhesion molecule expressed on activated neutrophils (37). Similar percentages of CD62L-positive neutrophils were found in mFoxm1−/− and Foxm1fl/fl livers at all time points after liver injury (data not shown). These results indicate that Foxm1 deficiency in myeloid cells does not influence recruitment or activation of neutrophils during CCl4-mediated liver injury.
Foxm1 deficiency does not influence monocyte proliferation but inhibits monocyte recruitment to the injured liver.
Decreased numbers of macrophages in CCl4-injured mFoxm1−/− livers can be a result of impaired recruitment and/or proliferation of the macrophage precursor cells, monocytes (20). Therefore, we focused on the 48-h time point, which is prior to the accumulation of macrophages in the liver. We used a histogram distribution of Ly-6C expression in hepatic CD11bhi/F4/80lo myeloid cells to distinguish between neutrophil and monocyte cell populations in the liver (Fig. 5A). Consistent with published studies (33), Ly-6C was expressed at higher levels on monocytes compared to neutrophils (Fig. 5A). mFoxm1−/− and control Foxm1fl/fl livers displayed similar percentages of neutrophils among hepatic inflammatory cells (Fig. 5A), confirming our results with Ly-6G antibodies (Fig. 4C). In contrast, significant decreases in the percentages of Ly-6Chi monocytes (Fig. 5A) and the total numbers of Ly-6Chi monocytes in 1 g of liver weight (Fig. 5B) were observed in mFoxm1−/− livers at 48 h after liver injury. The percentages of Ly-6Chi/CD115+ monocytes were not significantly changed in bone marrow of CCl4-treated mFoxm1−/− mice (Fig. 5C). Interestingly, a 40% increase in circulating Ly-6Chi/CD115+ monocytes was observed in the peripheral blood of CCl4-treated mFoxm1−/− mice (Fig. 5C), suggesting that Foxm1 deficiency impairs recruitment of Ly-6Chi/CD115+ monocytes from the blood to injured liver tissue.
FIG. 5.
Reduced numbers of monocytes in mFoxm1−/− livers after injury. Inflammatory cells were isolated from mFoxm1−/− and control Foxm1fl/fl livers at 48 h after CCl4 liver injury. The cells were stained with fluorescence-labeled antibodies against CD11b and Ly-6C and analyzed by flow cytometry. (A to C) Decreased numbers of monocytes in mFoxm1−/− and Foxm1fl/fl livers at 48 h after CCl4 liver injury. A histogram shows a distribution of Ly-6C cell surface expression in CD11bhi hepatic inflammatory cells (A). The percentages of neutrophils (N) and monocytes (M) are shown as means ± the SD (see the inset table in panel A). mFoxm1−/− livers displayed decreased numbers of monocytes per 1 g of liver weight (B). Statistically significant differences are indicated (*, P < 0.05; **, P < 0.01). Although the percentages of neutrophils and Ly-6Chi/CD115+ monocytes were not changed in the bone marrow of CCl4-treated mFoxm1−/− mice, a 40% increase in circulating Ly-6Chi/CD115+ monocytes was observed in the peripheral blood (C). (D and E) mFoxm1−/− and Foxm1fl/fl livers displayed similar numbers of myeloid cells undergoing G0/G1, S, and G2/M of the cell cycle. Hepatic inflammatory cells were stained with Vybrant DyeCycle DNA stain, and cell cycle analysis was performed by flow cytometry. Means ± the SD were calculated from three distinct mice.
Since decreased numbers of macrophages could be influenced by impaired proliferation of monocytes in CCl4-injured mFoxm1−/− livers, cell cycle analysis of hepatic inflammatory cells was performed using Vybrant DyeCycle DNA stain. Similar numbers of monocytes undergoing the G0/G1, S, and G2/M phases of the cell cycle were observed in mFoxm1−/− and control Foxm1fl/fl livers (Fig. 5D and E). Furthermore, Foxm1 deficiency did not influence the G0/G1, S, and G2/M phases in neutrophils and macrophages during liver injury (Fig. 5D and E).
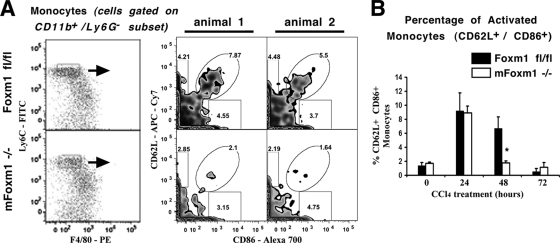
Published studies demonstrated that activated monocytes increased cell surface expression of CD62L (L-selectin) prior to their migration into injured tissues (11, 25). Therefore, we used flow cytometry to examine CD62L expression on monocytes during liver injury (Fig. 6A). Antibodies against costimulatory molecule CD86 (B7-2) were used to further identify activated monocytes. The percentages of CD62L+ CD86+ double-positive monocytes were initially increased in both mFoxm1−/− and control Foxm1fl/fl livers at 24 h after the liver injury (Fig. 6B). A 3-fold reduction in CD62L+/CD86+ monocytes was observed in mFoxm1−/− livers at 48 h after the liver injury (Fig. 6). Thus, Foxm1 is critical for the proper expression of CD62L on monocytes during liver repair. Altogether, impaired recruitment of monocytes may contribute to decreased numbers of macrophages and reduced repair in mFoxm1−/− livers.
FIG. 6.
Reduced percentages of CD62L+ CD86+ monocytes in mFoxm1−/− livers after CCl4 injury. Inflammatory cells were isolated from mFoxm1−/− and control Foxm1fl/fl livers and then stained with fluorescence-labeled antibodies against CD11b, F4/80, Ly-6C, Ly-6G, CD62L, and CD86. Hepatic inflammatory cells were gated on CD11b+/Ly-6G−/Ly-6Chi/F4/80lo cells (left side of panel A) and then analyzed for CD62L and CD86 expression (right side of panel A). The percentages of gated cells are shown for two distinct mice treated with CCl4 for 48 h (right sections of panel A). mFoxm1−/− livers displayed reduced the percentages of CD62L+ CD86+ monocytes at 48 h after CCl4 liver injury (B). The cumulative data from three experiments are presented as means ± the SD. A statistically significant P value of <0.05 is indicated by an asterisk.
Foxm1 induces expression of CCR2 chemokine receptor in monocytes.
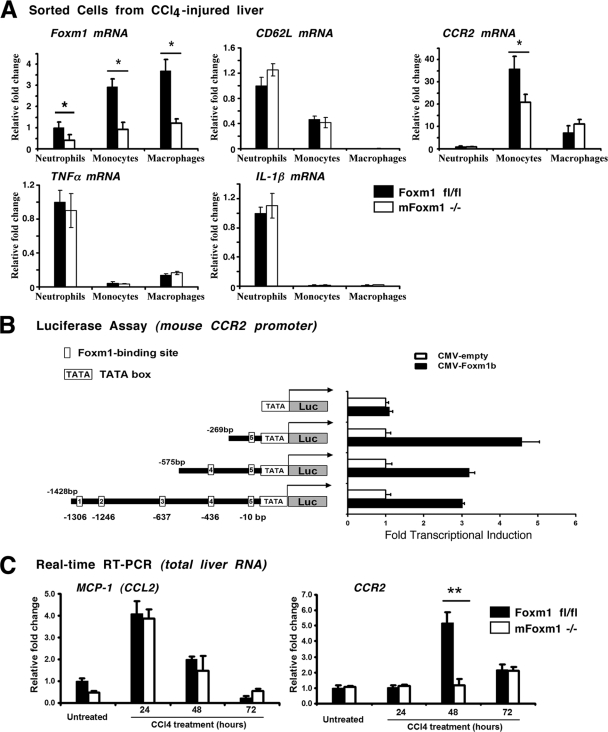
To determine new Foxm1 target genes, flow cytometry-based cell sorting was used to isolate enriched populations of monocytes, neutrophils, and macrophages from the livers of mFoxm1−/− and Foxm1fl/fl mice treated with CCl4 for 48 h. Sorted cells were used to prepare total RNA and examine Foxm1 expression by qRT-PCR. Foxm1 mRNA was significantly reduced in all three populations of mFoxm1−/− myeloid cells (Fig. 7A). CD62L mRNA was not altered in mFoxm1−/− monocytes (Fig. 7A), suggesting that CD62L is not a direct transcriptional target for Foxm1. No changes were observed in the mRNA levels of the proinflammatory cytokines tumor necrosis factor alpha (TNF-α) and interleukin-1β (IL-1β) (Fig. 7A). Interestingly, mFoxm1−/− monocytes displayed a significant reduction in mRNA levels of CCR2 (Fig. 7A), a chemokine receptor critical for monocyte recruitment to the injured liver (9, 31). Furthermore, total liver RNA was used to demonstrate that CCR2 expression was dramatically decreased in mFoxm1−/− livers at 48 h after CCl4 liver injury (Fig. 7C), correlating with decreased Foxm1 mRNA (Fig. 2L). No differences were observed in mRNA levels of chemokine MCP-1, a known ligand for CCR2 receptor (Fig. 7C). These results suggest that Foxm1 deficiency causes reduced expression of CCR2 in monocytes.
FIG. 7.
Foxm1 induces CCR2 expression in monocytes. (A) Decreased expression of CCR2 mRNA in mFoxm1−/− monocytes. Flow cytometry was used to isolate pure cell populations of neutrophils, monocytes, and macrophages from livers of mFoxm1−/− and control Foxm1fl/fl mice treated with CCl4 for 48 h. qRT-PCR shows decreased Foxm1 mRNA levels in all three populations of hepatic myeloid cells. CCR2 mRNA was specifically reduced in mFoxm1−/− monocytes. No differences were observed in TNF-α, IL-1β, and CD62L mRNAs. Each individual sample was normalized to its corresponding β-actin level. Means ± the SD were determined using five mFoxm1−/− and Foxm1fl/fl livers. Statistically significant differences are indicated (*, P < 0.05). (B) Foxm1 directly regulates mouse CCR2 promoter through a −269/+22-bp CCR2 DNA region. Schematically shown are luciferase (LUC) reporter constructs that use either a −1428-bp mouse CCR2 promoter or one of its deletion mutants to drive expression of the LUC reporter. U2OS cells were transiently transfected with CMV-Foxm1b expression vector and CCR2 LUC reporter plasmids. Cells were harvested 36 h after transfection. Dual luciferase assays were used to determine the LUC activity. Transcriptional induction is expressed as fold changes relative to CMV-empty vector (± the SD). (C) Decreased expression of CCR2 mRNA in mFoxm1−/− livers. qRT-PCR analysis shows decreased CCR2 mRNA in total liver RNA from mFoxm1−/− mice at 48 h after liver injury (A, right panel). MCP-1 (CCL2 ligand) mRNA was not changed in mFoxm1−/− livers (A, left panel).
Next, we investigated whether Foxm1 induces transcriptional activity of CCR2 promoter region. Five potential Foxm1 DNA-binding sites were identified in the −1.4-kb promoter region of the mouse CCR2 gene: −10/+3, −436/-425, −637/ -625, −1246/-1234, and −1306/-1294 (Fig. 7B). The −1.4-kb CCR2 promoter region was cloned by using PCR amplification of mouse genomic DNA. Cotransfection experiments were performed in human U2OS cells using CMV-Foxm1b expression vector and luciferase (LUC) reporter driven by the −1.4-kb CCR2 promoter region. Cotransfection of the Foxm1 expression vector increased expression of the −1.4-kb CCR2-LUC reporter plasmid (Fig. 7B), indicating that the Foxm1 protein is a transcriptional activator of CCR2 gene. Although deletion of either the −1428/-575-bp or the −575/-269-bp CCR2 region did not influence LUC activity, deletion of the −269/+22-bp CCR2 promoter region was sufficient to completely abolish the ability of Foxm1 to activate transcription of the CCR2 promoter in cotransfection experiments (Fig. 7B). These results indicate that Foxm1 transcriptionally activates the mouse CCR2 gene via the −269/+22-bp CCR2 promoter region.
Adoptive transfer of Foxm1fl/fl monocytes to mFoxm1−/− mice accelerated liver repair and improved liver function.
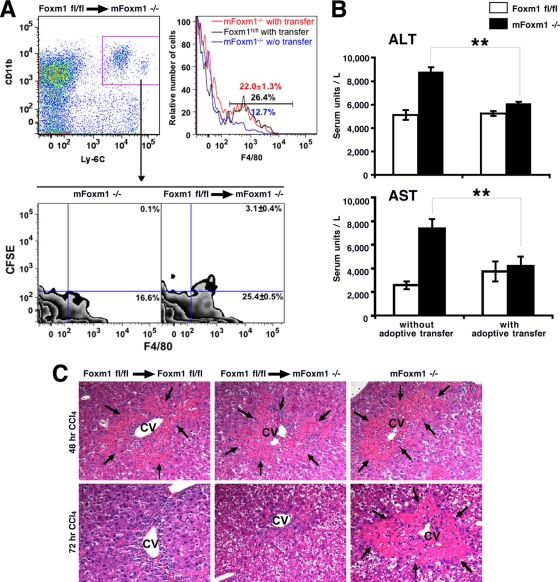
To determine whether diminished monocyte recruitment influences liver repair in mFoxm1−/− mice, adoptive transfer of monocytes to injured mice was performed. A cell population enriched in monocytes was purified from the bone marrow of Foxm1fl/fl mice by using CD115 antibody and then labeled with CFSE fluorescent dye. The cell population contained 85% monocytes and 15% granulocytes (data not shown). CFSE-labeled cells were injected into the tail vein of CCl4-treated mFoxm1−/− or control Foxm1fl/fl mice. Additional controls included mFoxm1−/− mice without adoptive transfer. At 48 h after adoptive transfer, CFSE-labeled cells were observed in mFoxm1−/− livers as demonstrated by flow cytometry (Fig. 8A). The majority of CFSE-labeled cells expressed CD11b and F4/80 surface markers (Fig. 8A), a phenotype of mature macrophages. Thus, transferred Foxm1fl/fl monocytes were effectively recruited to mFoxm1−/− livers and differentiated into mature macrophages. CFSE fluorescence was not found in other cell populations in the liver (data not shown). Histological and functional assessment of liver repair revealed that adoptive transfer of Foxm1fl/fl monocytes to mFoxm1−/− mice accelerated liver repair (Fig. 8C), decreased serum levels of ALT and AST enzymes (Fig. 8B), and increased total numbers of hepatic macrophages to the levels found in control mice (Fig. 8A). Altogether, our results demonstrate that the expression of Foxm1 in cells of monocyte lineage is required for liver repair.
FIG. 8.
Adoptive transfer of Foxm1fl/fl monocytes to injured mFoxm1−/− mice restored liver repair. Monocytes were isolated from the bone marrow of untreated Foxm1fl/fl mice, labeled with CFSE fluorescent dye, and then injected into the tail veins of injured mFoxm1−/− mice. (A) Adoptive transfer of Foxm1fl/fl monocytes increases numbers of macrophages in injured mFoxm1−/− livers. Hepatic inflammatory cells were isolated 48 h after adoptive transfer and stained with fluorescence-labeled antibodies against CD11b, F4/80, and Ly-6C. Stained cells were initially gated on CD11b+ Ly-6C+ (left top panel in panel A) and then analyzed for F4/80 and CFSE (bottom sections in panel A). CFSE-labeled macrophages are observed in mFoxm1−/− livers after the adoptive transfer (bottom right portion of panel A). A histogram shows increased numbers of macrophages in injured mFoxm1−/− livers compared to mFoxm1−/− mice without (w/o) transfer (right top portion of panel A). (B) Adoptive transfer of Foxm1fl/fl monocytes to injured mFoxm1−/− mice decreases the serum concentrations of ALT and AST enzymes to the levels found in control Foxm1fl/fl mice. Statistically significant differences with P values of <0.05 are indicated by asterisks. (C) H&E staining of liver paraffin sections shows that adoptive transfer of Foxm1fl/fl monocytes to injured mFoxm1−/− mice decreases liver necrosis around hepatic central vein (CV; necrosis is indicated by arrows).
DISCUSSION
Macrophages and their precursors, monocytes, play an important role during organ injury and repair. The subset of Ly-6Chi monocytes was increased dramatically in lesions of infection and injury in a variety of inflammatory diseases (2, 47). The Ly-6Chi monocytes migrated to the inflamed/necrotic area and subsequently differentiated into mature macrophages. This process required downregulation of Ly-6C expression and an increase in the expression of the mature macrophage marker F4/80 (2, 38). In the present study, we demonstrated that the loss of Foxm1 in a myeloid lineage reduced numbers of Ly-6Clo/F4/80hi mature macrophages at 72 h after liver injury, coinciding with delayed liver repair. The numbers of monocytic macrophage precursor cells were reduced in mFoxm1−/− livers 48 h after liver injury, a time prior to the accumulation of mature macrophages in the liver. These results suggest that Foxm1 may play an important role in liver repair by inducing migration of Ly-6Chi/F4/80lo monocytes into the injured liver. Consistent with this hypothesis, we found decreased expression of L-selectin in Foxm1-deficient monocytes. L-selectin is a cell adhesion molecule essential for monocyte migration (25). Alternatively, reduced numbers of mature macrophages in injured mFoxm1−/− livers can be a direct consequence of impaired differentiation of monocytes toward the macrophage cell lineage. Interestingly, we found no differences in the total numbers of neutrophils that migrated into an injured liver in mFoxm1−/− mice. Furthermore, Foxm1 did not influence cell surface expression of L-selectin in these cells. These results suggest that Foxm1 is dispensable for neutrophil recruitment to the injured liver.
In the present study, expression of CCR2 was decreased in mFoxm1−/− monocytes. Previous studies demonstrated that CCR2−/− mice exhibited decreased macrophage infiltration following acetaminophen-induced liver injury, indicating that MCP-1/CCR2 signaling is essential for monocyte migration to the injured liver (9). CCR2 gene expression is regulated by several signaling pathways, including calcineurin/NFAT (13), IFN-γ/Stat1 (10), and PPARγ (3). Liver repair was delayed following acetaminophen-mediated liver injury in CCR2−/− mice (9). Therefore, decreased CCR2 expression in mFoxm1−/− monocytes can contribute to impaired monocyte recruitment, causing a delay in liver repair in mFoxm1−/− mice. The finding that Foxm1 directly stimulated transcriptional activity of the CCR2 promoter and that this stimulation required the Foxm1 binding site located between the bp −269 and +22 of the mouse CCR2 promoter provide a potential mechanism by which Foxm1 influences CCR2 levels in monocytes. Our studies suggest that Foxm1 is a critical transcriptional activator of the CCR2 gene during liver injury and repair.
Previous studies focused on the role of Foxm1 protein in hepatocytes, the major cell population in the liver. Liver regeneration and liver injury studies with Alb-Cre Foxm1−/− mice demonstrated that Foxm1 is essential for DNA replication and mitosis in hepatocytes by altering expression of proteins that are required for cell cycle progression (41). Consistent with these studies, a majority of Foxm1−/− embryos died in utero by E16.5 due to a 75% reduction in the number of hepatoblasts because they failed to progress into mitosis causing a polyploid phenotype (26). We demonstrated here that mFoxm1−/− livers displayed a specific Foxm1 deletion in myeloid cells but not in hepatocytes. However, the proliferation of both hepatocytes and nonhepatocytes was decreased in CCl4-injured mFoxm1−/− livers. It is possible that Foxm1 influences the secretion of inflammatory mediators, which in turn indirectly affects proliferative responses and liver repair after CCl4 injury. We also found that adoptive transfer of Foxm1fl/fl monocytes to CCl4-treated mFoxm1−/− mice restored liver repair and rescued liver function after the injury. These results demonstrated a critical role of Foxm1 in monocytes and established a direct link between monocyte migration and liver repair after the injury.
Numbers of cells in the G0/G1, S, and G2/M phases of the cell cycle among all three major populations of hepatic myeloid cells (neutrophils, monocytes, and macrophages) were not influenced by Foxm1 deletion. Considering the important role of Foxm1 in cell cycle regulation of hepatocytes, fibroblasts, endothelial cells, and numerous neoplastic cells (18, 22, 26), these results are surprising and indicate that Foxm1 is not required for proliferation of myeloid cells during either normal hematopoiesis or acute liver injury. Interestingly, recent studies showed that a conditional Foxm1 deletion from respiratory epithelial cells did not alter proliferation of these cells during lung development but caused impaired lung maturation and decreased surfactant production (16). Altogether, these studies indicate that Foxm1 plays distinct roles in different cell populations in vivo. Although the molecular mechanisms of this Foxm1 selectivity are still unknown, various cell-specific transcription factors may function as coactivators with Foxm1, altering a set of Foxm1 transcriptional targets and influencing cellular responses to Foxm1. Alternatively, it is possible that other cellular pathways compensate for Foxm1 deficiency in myeloid inflammatory cells.
In summary, deletion of Foxm1 in myeloid cells caused a delay in liver repair and increased pericentral apoptosis following CCl4-mediated injury. Although the recruitment of neutrophils was not influenced by the Foxm1 deficiency, reduced numbers of infiltrating macrophages were found in mFoxm1−/− livers after injury. Monocyte recruitment during liver repair was dependent upon Foxm1 expression in myeloid cells, a finding that may be related to reduced expression of L-selectin and CCR2 chemokine receptor. Foxm1 is critical for transcriptional activation of the mouse CCR2 promoter and required for the recruitment of monocytes to the injured liver.
Acknowledgments
This study was supported by NIH grant HL 84151 (V.V.K.) and a Research Scholar Grant from the American Cancer Society (V.V.K.).
We thank J. Whitsett, B. Trapnell, I.-C. Wang, C. Bolte, S. Wert, and A. Zorn for critically reviewing the manuscript.
Footnotes
Published ahead of print on 13 September 2010.
REFERENCES
- 1.Antoniades, C. G., P. A. Berry, J. A. Wendon, and D. Vergani. 2008. The importance of immune dysfunction in determining outcome in acute liver failure. J. Hepatol. 49:845-861. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, L., A. Henry, F. Poron, Y. Baba-Amer, N. van Rooijen, A. Plonquet, R. K. Gherardi, and B. Chazaud. 2007. Inflammatory monocytes recruited after skeletal muscle injury switch into anti-inflammatory macrophages to support myogenesis. J. Exp. Med. 204:1057-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, Y., S. R. Green, J. Ho, A. Li, F. Almazan, and O. Quehenberger. 2005. The mouse CCR2 gene is regulated by two promoters that are responsive to plasma cholesterol and peroxisome proliferator-activated receptor gamma ligands. Biochem. Biophys. Res. Commun. 332:188-193. [DOI] [PubMed] [Google Scholar]
- 4.Clausen, B. E., C. Burkhardt, W. Reith, R. Renkawitz, and I. Forster. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8:265-277. [DOI] [PubMed] [Google Scholar]
- 5.Daley, J. M., A. A. Thomay, M. D. Connolly, J. S. Reichner, and J. E. Albina. 2008. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J. Leukoc. Biol. 83:64-70. [DOI] [PubMed] [Google Scholar]
- 6.Fontana, R. J. 2008. Acute liver failure due to drugs. Semin. Liver Dis. 28:175-187. [DOI] [PubMed] [Google Scholar]
- 7.Friedman, S. L. 2000. Molecular regulation of hepatic fibrosis, an integrated cellular response to tissue injury. J. Biol. Chem. 275:2247-2250. [DOI] [PubMed] [Google Scholar]
- 8.Gotthardt, D., C. Riediger, K. H. Weiss, J. Encke, P. Schemmer, J. Schmidt, and P. Sauer. 2007. Fulminant hepatic failure: etiology and indications for liver transplantation. Nephrol. Dial. Transplant. 22(Suppl. 8):viii5-viii8. [DOI] [PubMed] [Google Scholar]
- 9.Holt, M. P., L. Cheng, and C. Ju. 2008. Identification and characterization of infiltrating macrophages in acetaminophen-induced liver injury. J. Leukoc. Biol. 84:1410-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, X., K. H. Park-Min, H. H. Ho, and L. B. Ivashkiv. 2005. IFN-gamma-primed macrophages exhibit increased CCR2-dependent migration and altered IFN-gamma responses mediated by Stat1. J. Immunol. 175:3637-3647. [DOI] [PubMed] [Google Scholar]
- 11.Jacobson, S. H., B. Hylander, P. Thylen, and J. Lundahl. 2001. Monocyte-related determinants of inflammation in patients on peritoneal dialysis. Am. J. Nephrol. 21:40-46. [DOI] [PubMed] [Google Scholar]
- 12.Jaeschke, H., and T. Hasegawa. 2006. Role of neutrophils in acute inflammatory liver injury. Liver Int. 26:912-919. [DOI] [PubMed] [Google Scholar]
- 13.Jung, H., and R. J. Miller. 2008. Activation of the nuclear factor of activated T cells (NFAT) mediates upregulation of CCR2 chemokine receptors in dorsal root ganglion (DRG) neurons: a possible mechanism for activity-dependent transcription in DRG neurons in association with neuropathic pain. Mol. Cell Neurosci. 37:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalin, T. V., L. Meliton, A. Y. Meliton, X. Zhu, J. A. Whitsett, and V. V. Kalinichenko. 2008. Pulmonary mastocytosis and enhanced lung inflammation in mice heterozygous null for the Foxf1 gene. Am. J. Respir. Cell Mol. Biol. 39:390-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalin, T. V., I. C. Wang, T. J. Ackerson, M. L. Major, C. J. Detrisac, V. V. Kalinichenko, A. Lyubimov, and R. H. Costa. 2006. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 66:1712-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalin, T. V., I. C. Wang, L. Meliton, Y. Zhang, S. E. Wert, X. Ren, J. Snyder, S. M. Bell, L. Graf, Jr., J. A. Whitsett, and V. V. Kalinichenko. 2008. Forkhead Box m1 transcription factor is required for perinatal lung function. Proc. Natl. Acad. Sci. U. S. A. 105:19330-19335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalinichenko, V. V., D. Bhattacharyya, Y. Zhou, G. A. Gusarova, W. Kim, B. Shin, and R. H. Costa. 2003. Foxf1+/− mice exhibit defective stellate cell activation and abnormal liver regeneration following CCl4 injury. Hepatology 37:107-117. [DOI] [PubMed] [Google Scholar]
- 18.Kalinichenko, V. V., M. Major, X. Wang, V. Petrovic, J. Kuechle, H. M. Yoder, B. Shin, A. Datta, P. Raychaudhuri, and R. H. Costa. 2004. Forkhead Box m1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 18:830-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalinichenko, V. V., M. B. Mokyr, L. H. Graf, Jr., R. L. Cohen, and D. A. Chambers. 1999. Norepinephrine-mediated inhibition of antitumor cytotoxic T lymphocyte generation involves a β-adrenergic receptor mechanism and decreased TNF-alpha gene expression. J. Immunol. 163:2492-2499. [PubMed] [Google Scholar]
- 20.Karlmark, K. R., R. Weiskirchen, H. W. Zimmermann, N. Gassler, F. Ginhoux, C. Weber, M. Merad, T. Luedde, C. Trautwein, and F. Tacke. 2009. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology 50:261-274. [DOI] [PubMed] [Google Scholar]
- 21.Kim, I. M., T. Ackerson, S. Ramakrishna, M. Tretiakova, I. C. Wang, T. V. Kalin, M. L. Major, G. A. Gusarova, H. M. Yoder, R. H. Costa, and V. V. Kalinichenko. 2006. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 66:2153-2161. [DOI] [PubMed] [Google Scholar]
- 22.Kim, I. M., S. Ramakrishna, G. A. Gusarova, H. M. Yoder, R. H. Costa, and V. V. Kalinichenko. 2005. The Forkhead box M1 transcription factor is essential for embryonic development of pulmonary vasculature. J. Biol. Chem. 280:22278-22286. [DOI] [PubMed] [Google Scholar]
- 23.Kim, I. M., Y. Zhou, S. Ramakrishna, D. E. Hughes, J. Solway, R. H. Costa, and V. V. Kalinichenko. 2005. Functional characterization of evolutionary conserved DNA regions in Forkhead box f1 gene locus. J. Biol. Chem. 280:37908-37916. [DOI] [PubMed] [Google Scholar]
- 24.Korver, W., M. W. Schilham, P. Moerer, M. J. van den Hoff, K. Dam, W. H. Lamers, R. H. Medema, and H. Clevers. 1998. Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor trident. Curr. Biol. 8:1327-1330. [DOI] [PubMed] [Google Scholar]
- 25.Kremer, I. B., K. D. Cooper, M. B. Teunissen, and S. R. Stevens. 1998. Low expression of CD40 and B7 on macrophages infiltrating UV-exposed human skin; role in IL-2Rα-T cell activation. Eur. J. Immunol. 28:2936-2946. [DOI] [PubMed] [Google Scholar]
- 26.Krupczak-Hollis, K., X. Wang, V. V. Kalinichenko, G. A. Gusarova, I.-C. Wang, M. B. Dennewitz, H. M. Yoder, H. Kiyokawa, K. H. Kaestner, and R. H. Costa. 2004. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev. Biol. 276:74-88. [DOI] [PubMed] [Google Scholar]
- 27.Lawton, P., R. Mancassola, M. Naciri, and A. F. Petavy. 2001. Use of Percoll for the infection of cells in vitro with Cryptosporidium parvum oocysts. J. Microbiol. Methods 46:81-84. [DOI] [PubMed] [Google Scholar]
- 28.Leenen, P. J., M. F. de Bruijn, J. S. Voerman, P. A. Campbell, and W. van Ewijk. 1994. Markers of mouse macrophage development detected by monoclonal antibodies. J. Immunol. Methods 174:5-19. [DOI] [PubMed] [Google Scholar]
- 29.Malin, D., I. M. Kim, E. Boetticher, T. V. Kalin, S. Ramakrishna, L. Meliton, V. Ustiyan, X. Zhu, and V. V. Kalinichenko. 2007. Forkhead box F1 is essential for migration of mesenchymal cells and directly induces integrin-β3 expression. Mol. Cell. Biol. 27:2486-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mildner, A., M. Djukic, D. Garbe, A. Wellmer, W. A. Kuziel, M. Mack, R. Nau, and M. Prinz. 2008. Ly-6G+CCR2− myeloid cells rather than Ly-6ChighCCR2+ monocytes are required for the control of bacterial infection in the central nervous system. J. Immunol. 181:2713-2722. [DOI] [PubMed] [Google Scholar]
- 31.Mitchell, C., D. Couton, J. P. Couty, M. Anson, A. M. Crain, V. Bizet, L. Renia, S. Pol, V. Mallet, and H. Gilgenkrantz. 2009. Dual role of CCR2 in the constitution and the resolution of liver fibrosis in mice. Am. J. Pathol. 174:1766-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison, G. R., F. E. Brock, I. E. Karl, and R. E. Shank. 1965. Quantitative analysis of regenerating and degenerating areas within the lobule of the carbon tetrachloride-injured liver. Arch. Biochem. Biophys. 111:448-460. [DOI] [PubMed] [Google Scholar]
- 33.Nahrendorf, M., F. K. Swirski, E. Aikawa, L. Stangenberg, T. Wurdinger, J. L. Figueiredo, P. Libby, R. Weissleder, and M. J. Pittet. 2007. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J. Exp. Med. 204:3037-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramachandran, R., and S. Kakar. 2009. Histological patterns in drug-induced liver disease. J. Clin. Pathol. 62:481-492. [DOI] [PubMed] [Google Scholar]
- 35.Ramakrishna, S., I. M. Kim, V. Petrovic, D. Malin, I. C. Wang, T. V. Kalin, L. Meliton, Y. Y. Zhao, T. Ackerson, Y. Qin, A. B. Malik, R. H. Costa, and V. V. Kalinichenko. 2007. Myocardium defects and ventricular hypoplasia in mice homozygous null for the Forkhead Box M1 transcription factor. Dev. Dyn. 236:1000-1013. [DOI] [PubMed] [Google Scholar]
- 36.Serbina, N. V., T. M. Hohl, M. Cherny, and E. G. Pamer. 2009. Selective expansion of the monocytic lineage directed by bacterial infection. J. Immunol. 183:1900-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon, S. I., and H. L. Goldsmith. 2002. Leukocyte adhesion dynamics in shear flow. Ann. Biomed. Eng. 30:315-332. [DOI] [PubMed] [Google Scholar]
- 38.Templeton, S. P., T. S. Kim, K. O'Malley, and S. Perlman. 2008. Maturation and localization of macrophages and microglia during infection with a neurotropic murine coronavirus. Brain Pathol. 18:40-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueno, H., N. Nakajo, M. Watanabe, M. Isoda, and N. Sagata. 2008. FoxM1-driven cell division is required for neuronal differentiation in early Xenopus embryos. Development 135:2023-2030. [DOI] [PubMed] [Google Scholar]
- 40.Wang, X., N.-J. Hung, and R. H. Costa. 2001. Earlier expression of the transcription factor HFH 11B (FOXM1B) diminishes induction of p21CIP1/WAF1 levels and accelerates mouse hepatocyte entry into S-phase following carbon tetrachloride liver injury. Hepatology 33:1404-1414. [DOI] [PubMed] [Google Scholar]
- 41.Wang, X., H. Kiyokawa, M. B. Dennewitz, and R. H. Costa. 2002. The Forkhead box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc. Natl. Acad. Sci. U. S. A. 99:16881-16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westcott, D. J., J. B. Delproposto, L. M. Geletka, T. Wang, K. Singer, A. R. Saltiel, and C. N. Lumeng. 2009. MGL1 promotes adipose tissue inflammation and insulin resistance by regulating 7/4hi monocytes in obesity. J. Exp. Med. 206:3143-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ye, H., A. Holterman, K. W. Yoo, R. R. Franks, and R. H. Costa. 1999. Premature expression of the winged helix transcription factor HFH-11B in regenerating mouse liver accelerates hepatocyte entry into S-phase. Mol. Cell. Biol. 19:8570-8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ye, H., T. F. Kelly, U. Samadani, L. Lim, S. Rubio, D. G. Overdier, K. A. Roebuck, and R. H. Costa. 1997. Hepatocyte nuclear factor 3/forkhead homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol. Cell. Biol. 17:1626-1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, H., A. M. Ackermann, G. A. Gusarova, D. Lowe, X. Feng, U. G. Kopsombut, R. H. Costa, and M. Gannon. 2006. The FoxM1 transcription factor is required to maintain pancreatic beta-cell mass. Mol. Endocrinol. 20:1853-1866. [DOI] [PubMed] [Google Scholar]
- 46.Zhao, Y. Y., X. P. Gao, Y. D. Zhao, M. K. Mirza, R. S. Frey, V. V. Kalinichenko, I. C. Wang, R. H. Costa, and A. B. Malik. 2006. Endothelial cell-restricted disruption of FoxM1 impairs endothelial repair following LPS-induced vascular injury. J. Clin. Invest. 116:2333-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, B., Y. Bando, S. Xiao, K. Yang, A. C. Anderson, V. K. Kuchroo, and S. J. Khoury. 2007. CD11b+Ly-6C(hi) suppressive monocytes in experimental autoimmune encephalomyelitis. J. Immunol. 179:5228-5237. [DOI] [PubMed] [Google Scholar]