Abstract
Histone modifications are regarded as the carrier of epigenetic memory through cell divisions. How the marks facilitate cell cycle-dependent gene expression is poorly understood. The evolutionarily conserved AAA ATPase ANCCA (AAA nuclear coregulator cancer-associated protein)/ATAD2 was identified as a direct target of oncogene AIB1/ACTR/SRC-3 and a transcriptional coregulator for estrogen and androgen receptors and is strongly implicated in tumorigenesis. We report here that ANCCA directly interacts with E2F1 to E2F3 and that its N terminus interacts with both the N and C termini of E2F1. ANCCA preferentially associates via its bromodomain with H3 acetylated at lysine 14 (H3K14ac) and is required for key cell cycle gene expression and cancer cell proliferation. ANCCA associates with chromosomes at late mitosis, and its occupancy at E2F targets peaks at the G1-to-S transition. Strikingly, ANCCA is required for recruitment of specific E2Fs to their targets and chromatin assembly of the host cell factor 1 (HCF-1)-MLL histone methyltransferase complex. ANCCA depletion results in a marked decrease of the gene activation-linked H3K4me3 mark. Bromodomain mutations disable ANCCA function as an E2F coactivator and its ability to promote cancer cell proliferation, while ANCCA overexpression in tumors correlates with tumor growth. Together, these results suggest that ANCCA acts as a pioneer factor in E2F-dependent gene activation and that a novel mechanism involving ANCCA bromodomain may contribute to cancer cell proliferation.
Histone modifications such as acetylation and methylation at specific lysines of H3 and H4 are important epigenetic mechanisms in the maintenance of active transcription through cell divisions (26, 57). Histone marks such as acetylated H3 Lys-9 and Lys-14 (H3K9K14ac) and trimethylated H3 Lys-4 (H3K4me3) retained at certain levels during mitosis at gene promoters are thought to allow the imminent reactivation of genes upon cells exiting mitosis (25, 47). In early G1, the marks are likely recognized by protein complexes through corresponding histone binding modules. In mammalian cells, such marks are maintained at promoters of constitutively expressed genes, even when transcription is blocked (25, 34, 49, 62). However, for genes activated orderly during cell cycle progression, it is unclear how histone marks play the “bookmarking” role for transcription to occur during different stages of the cell cycle.
Multiple proteins and their associated complexes contain modules to recognize, individually or in combination, specific histone modifications and thus are regarded as epigenetic effectors (46, 51, 57, 64). The acetylation of histone lysines, which is strongly linked to gene activation (50), is recognized by bromodomains embedded in many nuclear proteins. These proteins play critical roles in transcriptional regulation, including histone acetylation (e.g., p300, CBP, GCN5, and PCAF), chromatin remodeling (e.g., RSC, Brg,1 and Brm), and transcription initiation (e.g., TAF1/TAFII250) or elongation (e.g., Brd4) (40, 70). Structural studies show that bromodomains usually contain a conserved fold of four helices and recognize acetyl-lysine through a hydrophobic pocket formed among the interhelical loops (40). Selectivity in binding histones acetylated at different sites is likely conferred by sequence variations in the loop regions, as demonstrated in CBP (binding to H4K20ac) and PCAF (binding to H3K36ac) (73). While Brd2 and Brd4, members of the BET tandem bromodomain protein family, both play important roles in facilitating transcription elongation, Brd4 prefers H3K9K14ac and H4K5K8K12ac, while Brd2 interacts primarily with H4K5K12ac (11, 16, 22, 30, 39, 68, 74).
The E2F-Rb complexes are primarily responsible for the timely expression of genes with functions in DNA synthesis and replication and for the control of cell cycle progression (41, 45, 54, 67) as well as hormone-dependent cancer cell proliferation (2, 6, 52, 66). Loss of Rb and deregulation of E2Fs are common events in tumorigenesis (5, 8). Recent studies indicated that, like gene repression by E2F-Rb, gene activation by E2Fs also requires chromatin regulatory proteins (3). During the G1-to-S transition, the so-called activator E2Fs (E2F1 to E2F3) associate with target genes, and the process is accompanied by increases in H3 and H4 acetylation and H3K4 methylation (54, 56, 60). Histone acetyltransferase (HAT) GCN5 and its related protein PCAF were shown to interact with E2Fs and coactivate E2F-mediated transcription (38, 43). Ectopic E2F expression induces the recruitment of HAT Tip60 and its complex to a subset of E2F target genes at late G1 (56). Likewise, the SET-containing histone lysine methyltransferases (hKMTs) MLL1 and MLL2 were found to directly interact with E2Fs and are required for histone methylation and activation of key cyclin genes (33, 55). Interestingly, host cell factor 1 (HCF-1) protein, a key regulator of mammalian cell cycle progression, was demonstrated to facilitate the recruitment of MLL complexes to chromatin targets through its direct interaction with E2F1 and E2F4, underscoring the critical role of the H3K4 methylation in gene expression control of cell cycle and apoptosis (13, 44, 60, 61, 72). Moreover, recent studies by us and others found that optimal activation of E2F target genes requires other nuclear factors, including the oncogenic, nuclear hormone receptor coactivator ACTR/AIB1/SRC-3, c-myc, and the homeobox protein p110CUX1, suggesting multiple regulator involvement in E2F-dependent gene activation (31, 37, 42, 58). Recently, it was also demonstrated that specific E2F members can associate with either ARID1A or ARID1B, the alternative subunit of the SWI/SNF chromatin-remodeling complexes, to activate or repress E2F target genes (42), underscoring the dynamics of coregulator function in the control of cell cycle gene expression.
AAA+ (ATPases associated with various cellular activities) proteins constitute a large family of evolutionarily conserved enzymes that function primarily to assemble or disassemble their substrate protein complexes (15). AAA+ proteins such as p97/VCP, NSF, and dynein perform diverse cellular functions, including membrane fusion, protein degradation, and microtubule sliding. Others, such as the replication factor C (RFC)-PCNA complex, are involved in chromatin loading of multiprotein complexes during DNA replication (18). They contain one or more AAA+ ATPase domains with the Walker A and B motifs and other structural features distinct from the other ATPases, including the SWI/SNF family of chromatin-remodeling helicases (14). One salient feature is that AAA+ proteins usually function in an oligomeric form. We identified the gene encoding the AAA nuclear coregulator cancer-associated protein (ANCCA, also known as ATAD2) as a target gene of the oncogene ACTR/AIB1 and as a transcriptional coregulator of estrogen receptor (ER) alpha and androgen receptor (17, 75, 76). We found that ANCCA expression is highly induced by estrogen or androgen in the cancer cells and tumor xenografts and that it is required for hormone-induced expression of a subset of receptor target genes (e.g., cyclin D1, c-myc, E2F1, IRS-2, and survivin) and for the proliferation and survival of hormone-responsive cancer cells. Our further study indicated that ANCCA mediates recruitment of other coregulators, such as CBP, and is critical for hormone-induced histone hyperacetylation (75, 76). Intriguingly, orthologs of the ANCCA gene can be found in other eukaryotes (e.g., yta7 in yeast and lex-1 in Caenorhabditis elegans).
Here, we show that ANCCA acts as a crucial coactivator for E2F and is important for the expression of key E2F target genes and hormone-independent cancer cell proliferation. We identified H3 acetylated at lysine 14 (H3K14ac) as a histone mark recognized by the bromodomain of ANCCA and found that chromatin occupancy of ANCCA is a prerequisite for cell cycle-dependent recruitment of E2Fs to their target genes and for the assembly of the HCF-MLL hKMT complex. These results suggest that aberrant reading of epigenetic marks and facilitation of E2F-hKMT complex loading to the chromatin targets can be important mechanisms in promoting cancer cell proliferation.
MATERIALS AND METHODS
Cell culture, cell cycle synchronization, and cell proliferation assay.
SKBR3, MCF7, T98G, HeLa, and HeLa-S3 cells were obtained from ATCC and grown in Dulbecco's modified Eagle's medium (T98G, HeLa, HeLa-S3, and MCF7) or McCoy's medium (SKBR3), supplemented with 10% fetal bovine serum (Omega). HeLa-S3 cells were synchronized in G1/S using double-thymidine block or in G2/M using thymidine-nocodazole block as described previously (35). Cells were collected at the indicated time points and processed for flow cytometry and protein and RNA analysis or fixed with formaldehyde for chromatin immunoprecipitation (ChIP) assay (see below). For cell proliferation assay, cells were plated in 6-well plates in regular growth medium and transfected the next day at a density of about 25% confluence with synthetic small interfering RNA (siRNA) targeting ANCCA or with control siRNA at a 100 nM concentration using Lipofectamine 2000 (Invitrogen) for HeLa and T98G cells or DharmaFECT I (Dharmacon) for SKBR3 cells by following the manufacturer's protocols. The medium was changed every other day, and cell proliferation was measured by cell counting of coded samples.
Transfection with siRNA and expression plasmids and reporter gene assay.
HeLa cells were plated in 24-well plates and cotransfected the next day by using Fugene-HD (Roche) with 200 ng of promoter-luciferase construct (pGL3-cyclin E1 or 3× E2F-tk-luc), 50 ng of pCMX β-galactosidase, 50 ng of pHCMV-ANCCA containing wild-type or mutant bromodomain, and 10 ng of empty pCMV vector or pCMV-E2F1. Luciferase activity was measured 48 h later using the luciferase reporter assay system (Promega) and normalized using the β-galactosidase assay.
For siRNA and plasmid DNA cotransfection, SKBR3 or HeLa cells were plated in 24-well plates in regular growth medium and cotransfected the next day at a density of about 40% confluence using DharmaFECT I, with synthetic siRNA targeting ANCCA or control siRNA at 100 nM, 50 ng of promoter-luciferase constructs, 200 ng of pHCMV vector mediating expression of siRNA-protected ANCCA, and 50 ng of empty pCMV vector or pCMV-E2F1. Luciferase activity was measured 48 h later as described above and normalized to the total protein concentration in the sample. For gene expression analysis, HeLa cells plated in 6-well plates were cotransfected with siRNA at a 100 nM concentration using DharmaFECT I and 0.5 μg of pHCMV vector mediating expression of siRNA-protected ANCCA. Cells were harvested 48 h later for Western analysis.
ChIP assay.
ChIP assay was performed as previously described (7, 36) with the following modifications. Approximately 5 × 106 cells were fixed with 1% formaldehyde for 8 min at room temperature (RT), washed, and harvested in phosphate-buffered saline (PBS). Cells were then lysed and sonicated for 20 min using Bioruptor (Diagenode Inc.) in buffer containing 50 mM HEPES (pH 7.9), 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor cocktail (Roche). After centrifugation, the crude chromatin solutions were incubated overnight at 4°C with the following antibodies (for antibody sources, see below): anti-ANCCA rabbit serum (75) at 10 μg/ml and at 2 μg/ml with anti-E2F1 (1:1 mixture of C-20 and KH-95), anti-E2F2 (C-20), E2F3 (1:1 mixture of C-18, PG-14), and anti-E2F4 (1:1 mixture of C-20 and WUF-2), anti-acetyl H3K9K14 at 5 μg/ml, anti-acetyl H3K9 at 5 μg/ml, anti-acetyl H3K14 at 5 μg/ml, anti-acetyl H4K5K8K12K16 rabbit serum at 10 μl/ml, anti-trimethyl H3K4 at 5 μg/ml, anti-HCF-1 rabbit serum (60) at 5 μl/ml, anti-MLLC at 5 μg/ml, or antihemagglutinin (anti-HA) tag at 2 μg/ml. The solutions were then incubated with preblocked protein A-Sepharose, 0.1 mg/ml salmon sperm DNA, and 0.1 mg/ml bovine serum albumin (BSA) for 1 h at 4°C. Immunoprecipitates were washed sequentially first with lysis buffer as described above, then with lysis buffer containing 500 mM NaCl, LiCl buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% Na-deoxycholate, 1 mM PMSF, and protease inhibitor cocktail [Roche]), and finally two times with TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). After the final wash, precipitated chromatin was eluted by incubation in buffer containing 0.1 M NaHCO3 and 1% SDS overnight at 65°C. Genomic DNA was purified using the QIAquick PCR purification kit (Qiagen) and analyzed by PCR with specific primers. For quantitative PCR analysis, ChIP DNA was analyzed by real-time PCR with the SYBR green kit on ICycler (Bio-Rad). PCR threshold cycle (CT) values from purified normal IgG or preimmune serum were used to subtract the values obtained from ChIP with test antibodies, while input DNA CT values were used to normalize the values obtained from ChIP samples.
Nuclear extracts, coimmunoprecipitation, and glutathione S-transferase (GST) pulldown assay.
Asynchronously growing MCF-7 cells were processed for nuclear extracts as described previously (76). The nuclear extracts were diluted 1:3 with buffer containing 20 mM HEPES (pH 7.9), 0.2 mM EDTA, 1 mM NaF, and 10 mM Na-pyrophosphate. After clearance, diluted nuclear extracts were incubated with anti-E2F antibodies at 1 μg/ml for 2 h at 4°C, followed by incubation with protein A-Sepharose beads (Amersham) for 1 h at 4°C. After being washed extensively, the immunoprecipitates were analyzed by Western blotting with anti-ANCCA antibody.
For the GST pulldown assay, 100 ng of Flag-tagged ANCCA protein purified from baculovirus-infected Sf9 cells or 50 ng of Flag-tagged E2F1 purified from transiently transfected Ad293 cells was incubated with 5 μg of bead-bound GST fusion proteins at 4°C for 1 h in a binding buffer containing 20 mM HEPES (pH 7.9), 150 mM NaCl, 1 mM EDTA, 4 mM MgCl2, 1 mM dithiothreitol (DTT), 0.05% NP-40, 10% glycerol, 1 mM PMSF, protease inhibitor cocktail (Sigma), and 2 mg/ml of BSA. Beads were then washed four times with the binding buffer containing 300 mM KCl. Flag-ANCCA or Flag-E2F1 proteins retained on the beads were separated by SDS-PAGE and detected by immunoblotting with anti-Flag antibody. GST-ANCCA and GST-E2F fusion proteins were expressed and purified from Escherichia coli cells transformed by the corresponding pGEX-KG constructs.
Recombinant adenoviruses and recombinant proteins.
Plasmid vectors and adenovirus expressing the full-length human ANCCA were described previously (76). Viruses were purified by centrifugation in CsCl step gradients. Viral titers were determined by the Adeno-X rapid titer kit (Clontech). Production of full-length, recombinant ANCCA protein was also as described previously (76), except that Sf9 cells were infected with baculoviruses generated using the Bac-to-Bac baculovirus system (Invitrogen) with pFast-Bac-Flag-ANCCA vector. Recombinant E2F1 protein was produced in Ad293 cells by transfection with the pCMV E2F1-Flag plasmid. ANCCA and E2F1 proteins were purified using anti-Flag M2 affinity gel (Sigma-Aldrich) as previously described (7). Recombinant GST proteins were expressed in E. coli and purified using glutathione-conjugated agarose (Sigma) as previously described (7).
Histone peptide binding assay.
Nuclear extracts from HeLa S3 cells infected with adenovirus vectors for expression of HA-tagged ANCCA proteins were prepared as described above. GST-ANCCA bromodomain (amino acids [aa] 985 to 1090) proteins were expressed and purified from E. coli transformed by pGEX-KG-ANCCA-bromo. Nuclear extracts (500 μg) or purified GST-ANCCA bromodomain proteins (10 to 50 ng) were incubated with 1 μg of biotinylated histone peptides (Upstate/Millipore) and 15 μl of NeutrAvidin-conjugated agarose beads (Pierce/Thermo Scientific) in 300 μl of binding buffer (20 mM HEPES [pH 7.9], 200 to 400 mM KCl, 1 mM EDTA, 4 mM MgCl2, 1 mM DTT, 0.05% NP-40, 10% glycerol, 1 mM PMSF, protease inhibitor cocktail [Sigma], and 1 mg/ml of BSA) for 4 h at 4°C. After extensive washes in binding buffer, ANCCA proteins retained on the beads were resolved by SDS-PAGE and analyzed by Western blotting with anti-GST or anti-HA antibodies.
Immunofluorescence (IF) microscopy and immunohistochemistry (IHC).
HeLa cells were treated with nocodazole at 100 ng/ml for 14 h, washed with regular medium, and plated on glass chamber slides (Nunc). One hour later, cells were fixed by 4% formaldehyde in PBS for 10 min at 4°C. After fixation, slides were rinsed with PBS. Cells were permeabilized for 10 min at RT with 0.1% Triton X-100 in PBS and blocked with 5% fetal bovine serum (FBS) in PBS (blocking solution) for 30 min at RT. After 2 h of incubation with primary antibodies diluted 1:100 in the blocking solution and being rinsed three times in PBS, slides were incubated for 1 h with the appropriate secondary antibodies diluted 1:1,000 in blocking solution. Following rinses (four times in PBS), slides were mounted with Vectashield mounting medium (Vector Laboratories) and sealed with clear nail polish. Images were acquired using a Zeiss LSM510 scanning confocal microscope or Olympus BX61 at the same exposure settings.
For IHC, sections (5-μm thickness) of formalin-fixed, paraffin-embedded breast carcinomas were deparaffinized and subjected to antigen retrieving in 0.01 M sodium citrate buffer (pH 6.0) in a microwave oven for 5 min at 1,000 W followed by 30 min at 100 W. Nonspecific immunoglobulin binding was blocked with 10% FBS in PBS for 30 min. They were incubated with primary antibodies (anti-ANCCA at 1:300 and anti-Ki67 at 1:1,000) diluted in blocking solution for 30 min at RT. Then sections were washed and incubated with the appropriate biotin-conjugated secondary antibodies for 30 min, followed by incubation with avidin DH-biotinylated horseradish peroxidase complex for 30 min (Vectastain ABC Elite kit, Vector Laboratories). At the end, the sections were developed with the diaminobenzidine (DAB) substrate kit (Vector Laboratories) and counterstained with Gill's hematoxylin (Vector Laboratories), followed by dehydration and mounting with Permount medium (Sigma-Aldrich). Tissue sections were obtained from the Department of Pathology, UC Davis School of Medicine, in accordance with the Institutional Review Board on Biomedical Specimen Usage.
Antibodies.
The antibodies used in this study were obtained from the following sources: ANCCA, described previously (75); HCF-1 (60), a kind gift from Winship Herr, University of Lausanne, Switzerland; and E2F1 (C-20, KH-95), E2F2 (C-20), E2F3 (C-18, PG-14), E2F4 (C-20, WUF2), cyclin E1 (E-4), Cdc6 (D-1), Cdk2 (M2), Cdc2 (B-6), MCM7 (G-7), and HA probe (F-7) were all from Santa Cruz Biotechnology. Beta-actin (AC-74) and α-tubulin (DM1A) were from Sigma. Acetyl H3K9K14 (06-599), acetyl H3K9 (06-942), acetyl H3K14 (06-911), acetyl H4K5K8K12K16 (06-866), phospho-H3S10 (06-570), histone H3-CT pan (07-690), histone H4 pan, clone 62-141-13 (05-858), MLLC/HRX clone 9-12 (05-765), acetyl-lysine, and clone 4G12 (05-515) were from Upstate/Millipore. Acetyl H3K9 (ab61231), acetyl H3K14 (ab46984), trimethyl H3K4 (ab8580), GST, and clone 3G10/1B3 (ab92) were from Abcam. Cyclin A2 (611268) was from Beckton Dickinson. Ki-67 (SP-6) was from NeoMarkers. Alexa 488-conjugated anti-rabbit and Texas Red-conjugated anti-mouse antibodies were from Invitrogen/Molecular Probes.
Plasmids, primers, and siRNA.
pGL3-cyclin E1 promoter constructs, 3× E2F-tk-Luc, pCMX β-galactosidase, pCMV E2F1, and pCMV E2F1-Flag were described previously (37). The plasmid vector expressing the full-length human ANCCA was described previously (76). Point mutations in ANCCA bromodomain or the siRNA target sequence were introduced using the quick site-directed mutagenesis kit (Stratagene). Mutated bromodomain had four amino acid substitutions: V1013 to A, Y1021 to A, Y1063 to A, and L1073 to A. ANCCA siRNA-protected mutants were generated by introduction of two silent mutations in the siRNA target sequence. Expression plasmids for GST fusion proteins were constructed through the insertion of PCR fragments from human cDNAs of E2F1, E2F2, E2F3, E2F4, and ANCCA into the pGEX-KG vector. The following constructs were used in the study: pGEX-KG-E2F1 aa 1 to 437 (full length), pGEX-KG-E2F1 aa 1 to 284, pGEX-KG-E2F1 aa 284 to 437, pGEX-KG-E2F1 aa 284 to 368, pGEX-KG-E2F1 aa 368 to 437, pGEX-KG-E2F1 aa 121 to 284, pGEX-KG-E2F1 aa 1 to 120, pGEX-KG-E2F2, pGEX-KG-E2F3, pGEX-KG-E2F4, pGEX-KG-ANCCA aa 1 to 250, pGEX-KG-ANCCA aa 293 to 415, pGEX-KG-ANCCA aa 412 to 668, pGEX-KG-ANCCA aa 848 to 1115, pGEX-KG-ANCCA aa 1110 to 1390, pGEX-KG-ANCCA aa 985 to 1090 (Bromo-WT), and pGEX-KG-ANCCA aa 985 to 1090 (Bromo-mut).
Primer sequences of reverse transcription (RT)-PCR and ChIP used in the study were described previously (35, 37, 75, 76). The siRNA targeting ANCCA (5′-GCTACTGTTTACTATCAGGCT-3′) and siGENOME SMARTpools targeting E2F1 and E2F4 were purchased from Dharmacon.
RESULTS
ANCCA is required for the proliferation of cancer cells and the expression of E2F target genes.
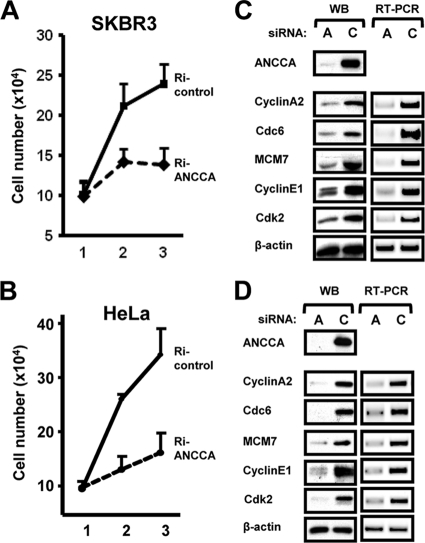
Our analysis of clinical samples revealed that ANCCA is highly overexpressed in different hormone-nonresponsive cancers, including ER-negative breast cancer (21a). To examine whether ANCCA plays a role in cell proliferation of these types of cancer, we knocked down ANCCA in SKBR3 (ER-negative breast cancer), HeLa (cervical carcinoma), and T98G (glioblastoma) cells and observed strong inhibition of proliferation of all three cancer cell lines (Fig. 1 A and B and data not shown). Strikingly, gene expression analysis demonstrated that high levels of ANCCA are required for the expression, at transcript and protein levels, of key cell cycle/proliferation genes such as cyclin A2, cyclin E1, cdk2, cdc6, and MCM7 (Fig. 1C and D). Further analysis demonstrated that ANCCA is important for the expression of genes that are involved in cell cycle progression and DNA synthesis and replication, which include cyclin B1, cdc2, PCNA, RFC2, MCM2, geminin, and RRM2 (data not shown).
FIG. 1.
ANCCA is required for cancer cell proliferation and E2F target gene expression. (A, B) Asynchronously growing SKBR3 and HeLa cells were transfected with siRNAs targeting ANCCA or a control sequence and harvested different days after transfection for cell numeration. Data are representative of results from triplicate experiments. (C, D) Cells transfected as above were harvested for Western blotting (WB) or RT-PCR 48 h after transfection.
ANCCA directly interacts with E2F1 to E2F4, and its interaction with E2F1 and E2F3 is mediated by both N- and C-terminal domains of the E2Fs and its N terminus.
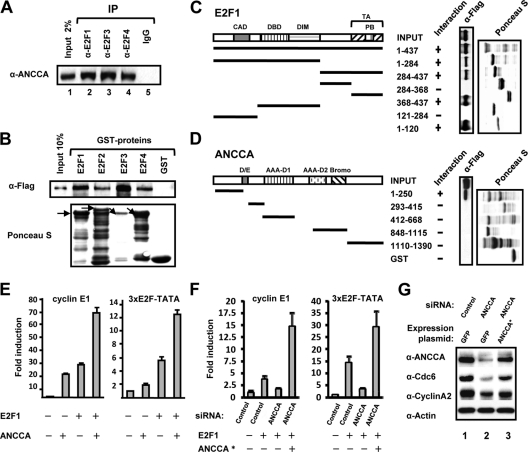
Key cell cycle and DNA replication genes, such as the ones described above, are regulated primarily by the pRb-E2F complexes. Given our previous finding that ANCCA is a novel nuclear coregulator and the remarkable inhibitory effect of ANCCA knockdown on the cell cycle gene expression, we next examined whether ANCCA is involved in E2F function through direct interaction. As shown in Fig. 2 A, endogenous ANCCA in breast cancer cells could be coprecipitated by antibodies recognizing E2F1, E2F3, and E2F4 but not the control IgG, indicating that a subpool (more than 2% of the input) of ANCCA protein is in complex with the E2Fs. GST pulldown experiments with purified, Flag-tagged ANCCA demonstrated that 10 to 30% of ANCCA protein was pulled down by E2F1, E2F2, E2F3, and E2F4 (Fig. 2B), suggesting a rather strong and direct interaction between ANCCA and the E2Fs examined. Together, these results suggest that ANCCA strongly and directly interacts with several members of the E2F family.
FIG. 2.
ANCCA interacts with E2Fs and mediates E2F-dependent gene expression. (A) Nuclear extracts prepared from asynchronously growing breast cancer cells were immunoprecipitated with the indicated antibodies. The presence of ANCCA in the precipitates was analyzed by Western blotting with anti-ANCCA antibody. (B, C) Purified, recombinant Flag-tagged ANCCA protein was used in the pulldown experiment with bead-bound GST-E2F proteins (B) or GST-E2F1 fragments (C), which were displayed by Ponceau S staining. ANCCA retained on the beads was detected by Western blotting with anti-Flag antibody. Amino acid number and domains contained in the fragments of human E2F1 are indicated. CAD, cyclin A binding domain; DBD, DNA binding domain; DIM, dimerization domain; TA, transactivation domain; and PB, pocket protein binding domain. Arrows in panel B indicate the full lengths of different E2F-GST fusion proteins. (D) Recombinant Flag-tagged E2F1 protein was used in pulldown experiments with bead-bound GST-ANCCA fragments. E2F1 protein retained on glutathione beads was detected by immunoblotting with anti-Flag antibody. Loading of GST proteins was visualized by Ponceau S staining. D/E, aspartate- and glutamate-rich region; AAA D1 or D2, AAA+ domains; Bromo, bromodomain. (E) HeLa cells were transfected with a luciferase reporter construct containing the human cyclin E1 promoter or synthetic promoter bearing 3 copies of E2F binding sites. Cells were also cotransfected with E2F1- and/or ANCCA-expressing plasmids as indicated. Luciferase activity data are the means of results from three independent experiments. (F) Cells were cotransfected with the reporters, ANCCA or control siRNA, E2F1, and/or siRNA-protected ANCCA-expressing (ANCCA*) plasmids as indicated. Fold induction was obtained by comparison of normalized luciferase activities from different cells with the one transfected only with control siRNA set as 1. Data are representative of results from three independent experiments. (G) HeLa cells were cotransfected with ANCCA or control siRNA and GFP or siRNA-protected ANCCA expression plasmid (ANCCA*). Two days after transfection, cells were harvested for Western analysis with the indicated antibodies.
To identify the interaction interfaces between ANCCA and E2F, we performed additional pulldown experiments with subdomains or fragments of E2F1 and ANCCA. As shown in Fig. 2C, for E2F1, the N-terminal 120-amino-acid residue fragment containing the cyclin A binding domain (CAD) and the C-terminal 70-amino-acid region containing the transactivation (TA) domain (aa 368 to 437) specifically mediate its interaction with ANCCA (Fig. 2C), while its DNA binding domain (DBD) and the so-called dimerization and marked box (DIM) showed no significant interaction in the assay. Interestingly, like E2F1, the N terminus (aa 1 to 155) of E2F3 displayed strong binding to ANCCA (data not shown). Using GST fusions of ANCCA fragments and Flag-tagged E2F1 protein, we identified the N terminus (aa 1 to 250) of ANCCA as the primary region that mediates its interaction with E2F1 (Fig. 2D).
ANCCA is required for transcriptional activation by the activator E2Fs.
Next, we performed reporter gene assays to examine whether ANCCA can facilitate E2F-mediated transactivation. As shown in Fig. 2E, left, ectopic expression of ANCCA strongly stimulated cyclin E1 promoter reporter activity, and coexpression of E2F1 and ANCCA induced the reporter activity synergistically, suggesting that ANCCA indeed acts as a coactivator on the E2F target gene promoter. To confirm that the coactivator activity was exerted through E2F, we repeated the experiment with a reporter driven by synthetic E2F binding sites and obtained similar synergistic activation (Fig. 2E, right). To demonstrate that endogenous ANCCA possesses the coactivator activity, we performed a knockdown and rescue experiment and found that E2F1-mediated transactivation was almost completely blocked if the endogenous ANCCA protein was depleted through siRNA (Fig. 2F). Remarkably, when cells were simultaneously exposed to ANCCA-siRNA and a vector carrying an siRNA-protected ANCCA cDNA (marked with an asterisk), the reporter activities were even higher than those obtained from cells without ANCCA knockdown, indicating that the ectopic ANCCA was more than sufficient to restore the full transactivation activity of E2F1. Importantly, similar ANCCA rescue restored the expression of endogenous E2F target genes cyclin A2 and cdc6 (Fig. 2G, compare lanes 2 and 3). Further analysis using a mutant form of ANCCA that lacks its N-terminal E2F interaction domain (del 1, aa 1 to 250) indicated that the physical interaction with E2Fs is required for ANCCA to act as an E2F coactivator, as the mutant ANCCA was unable to stimulate the transactivation of E2F1 and E2F3 (data not shown). Together with our finding that ANCCA directly interacts with E2Fs, these results strongly suggest that ANCCA is a new bona fide coactivator for E2F1 and E2F3 of the E2F family.
ANCCA is recruited to the E2F-responsive chromatin region at late mitosis, and its occupancy peaks at late G1.
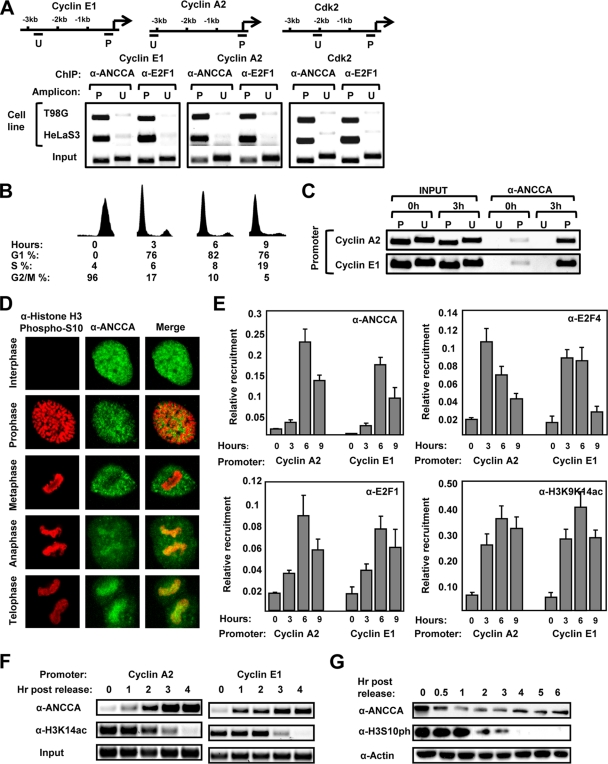
To determine the functional mechanism of ANCCA as a coactivator of E2F, we first performed ChIP assay to examine whether the coregulator occupies E2F target genes. We found that in asynchronously growing HeLa and T98G cells, a robust occupancy of ANCCA, like that of E2F1, can be specifically detected at the promoter (P) but not at a region 2 to 3 kb upstream (U) of E2F target genes cyclin E1, cyclin A2, and cdk2 (Fig. 3 A). To examine whether ANCCA occupies E2F-responsive promoters in a cell cycle-regulated manner, we synchronized HeLa cells by treating them with nocodazole and harvested them at different hours after the release for analysis of the cell cycle profile and ChIP. As shown in Fig. 3B (0-h time point), nocodazole treatment led to the arrest of cells in mitosis. Nine hours after its removal, a significant proportion (19%) of cells passed G1 and entered S phase. Consistent with previous studies (25), we observed that in mitotic cells a significant level of H3K9K14 double acetylation was preserved from interphase on E2F-responsive cyclin A2 and cyclin E1 promoters but not on a transcriptionally inactive prostate-specific antigen (PSA) gene promoter (data not shown). Interestingly, a readily detectable level of ANCCA occupancy at E2F target gene promoters but not at the upstream regions was observed with mitotic cells (Fig. 3C, 0-h point), suggesting that, in mitosis, at least a fraction of ANCCA protein might associate with chromatin. Importantly, ANCCA occupancy at the promoters increased markedly as cells entered G1 (Fig. 3C, 3-h point), implying that ANCCA recruitment to E2F target genes is cell cycle regulated.
FIG. 3.
ANCCA is recruited to E2F-responsive sites at late mitosis, and its occupancy peaks at late G1. (A) Asynchronously growing HeLa S3 and T98G cells were harvested for ChIP assay with anti-ANCCA or anti-E2F1 antibody. ChIP DNA was analyzed by semiquantitative PCR for ANCCA occupancy at the promoter (P) or an upstream region (U) as indicated in the diagrams. (B, C) HeLa cells released from mitotic arrest by nocodazole treatment were harvested at the indicated hours after the release for flow cytometry analysis (B) or ChIP with ANCCA antibody (C). (D) HeLa cells released from mitotic block were processed for immunofluorescence staining followed by confocal microscopic analysis of the localization of H3S10ph (red) and ANCCA (green) at different stages of the cell cycle. (E) HeLa cells released from mitotic arrest were harvested at the indicated hours for ChIP with the indicated antibodies. The anti-H3K9K14ac antibody used here recognizes primarily K9 and K14 double acetylated H3. ChIP DNA was analyzed by real-time PCR for enrichment of H3K9K14ac, E2F, and ANCCA occupancy at the indicated gene promoters with PCR data from triplicate experiments and calculated as percentages of the input. (F, G) HeLa cells released from mitotic arrest were harvested at the indicated hours after the release for ChIP (F) or Western analysis (G) with the indicated antibodies. The anti-H3K14ac antibody used here recognizes only K14-acetylated H3.
Since most transcription factors, including E2Fs, are excluded from condensed chromosomes during mitosis (10, 12), we wondered if ANCCA indeed associates with mitotic chromosomes. We thus performed immunofluorescence staining analysis of ANCCA (with ANCCA stained in green and chromosomes in red). As shown in Fig. 3D, in prophase and metaphase, ANCCA was largely excluded from the condensed chromosomes. However, in anaphase and telophase, a significant pool of ANCCA protein became associated with the chromosomes. To determine whether any fraction of the ANCCA protein was recruited to E2F-responsive genes, we performed ChIP assay with cells harvested shortly after release from a prometaphase blockade and detected a significant occupancy of ANCCA at cyclin A2 and cdc2 promoters at late mitosis (Fig. 3F, 1-h and 2-h points), when levels of mitotic phosphorylated H3-serine 10 remained high (Fig. 3G). Intriguingly, at the 1-h and 2-h points, chromatin regions occupied by ANCCA also displayed relatively high levels of acetylated H3 at lysine 14 (H3K14ac) (Fig. 3F; also see Discussion for the dynamics of ANCCA occupancy versus H3K14ac). Further ChIP analysis demonstrated that ANCCA occupancy at the E2F-responsive regions peaked at late G1 and that a robust occupancy by E2F1, E2F3, and ANCCA remained in early S phase (Fig. 3E, 6 h and 9 h, respectively, and data not shown). Together, these results suggest that ANCCA associates with chromatin at its target gene promoter region at the end of mitosis and that its recruitment increases as the cell cycle progresses from G1 to S.
ANCCA via its bromodomain associates preferentially with acetylated H3K14.
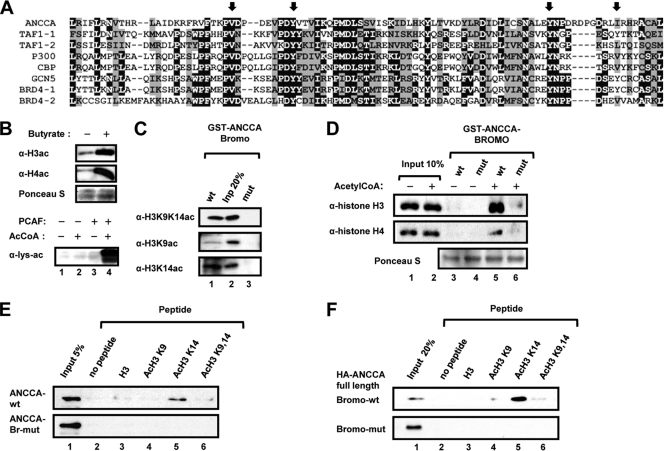
Since E2Fs are excluded from condensed chromosomes during mitosis (10, 12), we postulated that one way ANCCA is recruited to chromatin at late mitosis could be through its bromodomain (see Fig. 4 A for amino acid sequences) association with histone marks such as H3K9K14ac or H3K14ac at the mitotic stage. To examine whether ANCCA associates with chromatin via recognition of acetylated histones by its bromodomain, we performed pulldown assays with purified histones from HeLa cells (Fig. 4B, top) or with recombinant histones acetylated in vitro by PCAF (bottom), which acetylates free histones in vitro with a strong preference to H3K14 (48). As shown in Fig. 4C, wild-type ANCCA-bromodomain (lane 1), but not the mutant form (lane 3), can effectively pull down H3K14ac and H3K9K14ac from HeLa cells (about 20% of input; compare lane 1 and lane 2 in Fig. 4C). Importantly, the ANCCA bromodomain protein showed no significant binding to purified, nonacetylated H3 or H4 that can be readily detected in the input by antihistone antibodies (Fig. 4D, compare lanes 3 and 4 with lanes 1 and 2). Interestingly, approximately 20% of the acetylated H3 was bound by ANCCA bromodomain, while less than half of the input acetylated H4 was detected in the pulldown (compare lane 5 with lane 2), suggesting that ANCCA bromodomain may prefer to associate with acetylated H3 rather than H4. We thus focused our analysis on H3 with acetylation at different lysines. Importantly, we found that among the H3 N-terminal tail peptides examined, H3K14ac displayed the strongest binding to the full-length ANCCA protein expressed in HeLa cells (Fig. 4F, lane 5) or to purified ANCCA bromodomain expressed in E. coli (Fig. 4E, lane 5). Taken together with the data demonstrating that ANCCA associates with late mitotic chromosomes and occupies E2F target promoters, these results support a hypothesis that ANCCA bromodomain may act as a “histone code” reader with a preferential recognition of H3K14ac on the E2F chromatin targets at late mitosis.
FIG. 4.
Bromodomain of ANCCA binds preferentially to H3K14ac. (A) Amino acid sequence alignment of bromodomains from ANCCA and other human proteins. Arrows indicate amino acid sequences changed in the mutant form of the ANCCA bromodomain.(B) Histones either acid extracted from butyrate-treated HeLa S3 cells (top) or purified from E. coli and in vitro acetylated by PCAF (bottom) were subjected to Western analysis for acetylation with the indicated antibodies. (C) GST-ANCCA bromodomain proteins, wild type or mutant, were incubated with histones from HeLa cells. (D) GST-ANCCA bromodomain proteins were incubated with histones in vitro acetylated by PCAF. Histones retained on the glutathione beads were detected by Western analysis with the indicated antibodies. (E, F) GST-ANCCA bromodomain proteins (E) or nuclear extract from HeLa cells expressing HA-tagged full-length ANCCA (F) was incubated with biotinylated histone tail peptides with the indicated acetylation. After being washed extensively, ANCCA proteins retained on the beads were analyzed by Western analysis with anti-GST (E) or anti-HA (F) antibodies, respectively.
Association with acetylated histones is crucial for ANCCA function as an E2F coactivator and for its role in cancer cell proliferation.
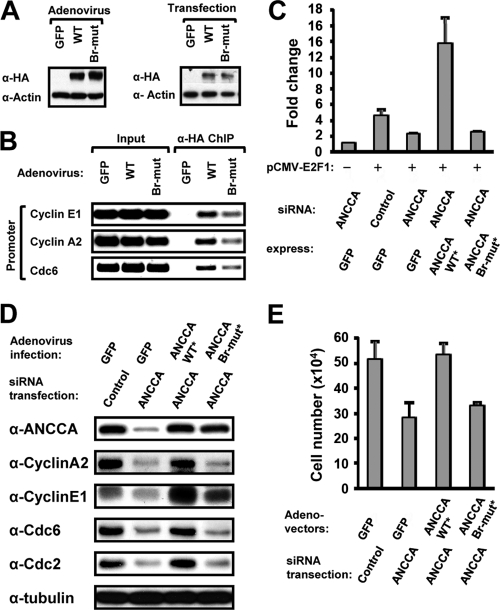
We expressed a wild-type and a bromodomain mutant form of ANCCA by adenovirus vector (Fig. 5 B) or via transfection (Fig. 5C) in cell cycle-synchronized cells and analyzed ANCCA recruitment upon cells exiting mitosis. ChIP using anti-HA antibody that detects ectopically expressed ANCCA (Fig. 5A) demonstrated a dramatic decrease in chromatin association of the mutant ANCCA at the E2F target genes examined compared to that of wild-type ANCCA (Fig. 5B). It is important to note that, like most AAA+ proteins, ANCCA protein has a high tendency to oligomerize in vivo (unpublished observation). Thus, the low-level chromatin occupancy observed with the bromodomain mutant ANCCA may be derived from its oligomerization with endogenous ANCCA protein. Next, reporter gene assays with endogenous ANCCA depleted by siRNA and ectopic ANCCA expressed from an siRNA-protected ANCCA cDNA clearly indicated that the bromodomain-mutated ANCCA was completely inactive in coactivating E2F1 and E2F3 at the cyclin E1 promoter (Fig. 5C and data not shown). Importantly, ANCCA with mutated bromodomain was largely inactive in rescuing the inhibitory effects of ANCCA knockdown on E2F-regulated endogenous protein expression that includes cyclin A2, cyclin E1, cdc2, and cdc6 (Fig. 5D). More importantly, although ectopic expression of the wild-type ANCCA protein displayed an ability to fully restore the reduced proliferation of breast cancer cells due to ANCCA siRNA treatment, expression of the bromodomain mutant ANCCA failed to show any significant rescuing effect on the cell proliferation (Fig. 5E), indicating a crucial role of ANCCA bromodomain in mediating expression of the cell cycle genes and cell proliferation.
FIG. 5.
ANCCA bromodomain is essential for ANCCA chromatin association and for mediating E2F target gene expression and cancer cell proliferation. (A, B) HeLa-S3 cells ectopically expressing HA-tagged ANCCA with wild-type or mutant bromodomain (Br-mut) or GFP via adeno-vector were released from mitotic block for 4 h and then harvested for Western analysis (A) or ChIP (B) with anti-HA antibody. (C) HeLa cells were cotransfected with ANCCA or control siRNA and plasmid constructs for expression of GFP, siRNA-protected ANCCA (marked with an asterisk) with wild-type or mutant bromodomain, E2F1, and the 1.2-kb cyclin E1 promoter reporter. Luciferase activities were measured as described in the legend to Fig. 2. (D, E) SKBR3 cells were transfected with ANCCA or control siRNA and infected 24 h later with adeno-vectors for the indicated expression. Cells were harvested 48 h after infection for Western analysis with the indicated antibodies (D) or cell numerations (E). Data are representative of results from three independent experiments.
ANCCA mediates chromatin loading of activator E2Fs and histone methyltransferase complexes and is required for trimethylation of H3K4.
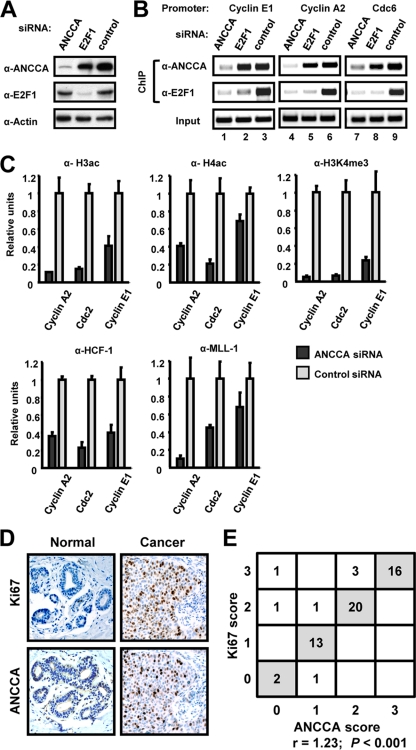
We previously demonstrated that ANCCA is required for estrogen-induced chromatin recruitment of HAT protein CBP but not ER (76). Therefore, we examined the effect of ANCCA depletion on the recruitment of the E2F-coregulator complex at E2F-responsive sites. As shown in Fig. 6 A, E2F1 knockdown dramatically decreased E2F1 levels with no significant effect on ANCCA at the experimental condition. Likewise, silencing ANCCA did not significantly affect E2F1 expression under the same condition. As expected, E2F1 knockdown strongly diminished its occupancy at the E2F target genes cyclin E1, cyclin A2, and cdc6 (Fig. 6B, anti-E2F1 ChIP, compare lanes 3, 6, and 9 with lanes 2, 5, and 8). Interestingly, no significant effect on ANCCA occupancy was observed (anti-ANCCA ChIP, compare the same lanes), suggesting that E2F1 alone did not play a major role in ANCCA recruitment to E2F targets. Unexpectedly, knocking down ANCCA dramatically inhibited the recruitment of E2F1 at the key E2F target genes, with a blocking effect equal to that of E2F1 silencing (Fig. 6B, anti-E2F1 ChIP, compare lanes 3, 6, and 9 with lanes 1, 4, and 7). Similar blocking effects by ANCCA depletion were also observed on target gene occupancy by E2F2 and E2F3 (data not shown). Intriguingly, we observed no significant negative impact on E2F4 chromatin occupancy in the cells examined (data not shown), indicating that the essential role of ANCCA in mediating E2F loading to the chromatin is E2F family member specific.
FIG. 6.
Overexpressed ANCCA is required for chromatin assembly of E2Fs and the HCF-1-MLL hKMT complex and correlates with cancer cell proliferation. (A to C) SKBR3 cells were transfected with siRNA targeting ANCCA, E2F1, or the control sequence and harvested at 24 h after transfection for Western blotting (A) or ChIP (B and C) with the indicated antibodies. (D, E) Sections of breast tissues from 58 patients were subjected to IHC with ANCCA or Ki67 antibodies. The correlation between the IHC scores of ANCCA and Ki67 was analyzed by a Student t test with the coefficient of correlation and P values indicated.
Previous studies suggest that gene activation by E2Fs involves histone modifications at the promoters; one major event is trimethylation of H3 at lysine 4 (H3K4me3) by the HCF-MLL histone methyltransferase complex (3, 60). As demonstrated in Fig. 6C, ANCCA depletion resulted in a marked decrease in H3ac, H4ac, and H3K4me3. Since the most striking impact of ANCCA depletion was on H3K4me3, we determined whether chromatin occupancy of the HCF-MLL complex was affected. Indeed, ANCCA knockdown significantly attenuated promoter occupancy of the intermediary factor HCF-1 and its associated hKMT protein MLL1 (Fig. 6C, bottom). Western analysis indicated that ANCCA depletion did not affect the global level of histone modifications or HCF-1 and MLL1 protein expression (data not shown). Together, these results strongly suggest that binding to chromatin targets by the activator E2Fs (E2F1, E2F2, and E2F3) is a facilitated process and requires ANCCA. Given that ANCCA depletion impacted histone acetylation and methylation, these results also indicate that ANCCA not only is essential for chromatin occupancy by the E2Fs but also plays an important role in the chromatin assembly of histone-modifying enzyme complexes such as HCF-MLL.
Overexpression of ANCCA correlates with a high proliferation index for tumors of human breast cancer.
The essential role of ANCCA in mediating E2F-dependent gene activation and cell proliferation prompted us to examine whether ANCCA expression in human tumors correlates with cancer cell proliferation. We thus performed IHC analysis of ANCCA and the cell proliferation marker Ki67 in a cohort of randomly selected human breast carcinoma tissue. While very little anti-ANCCA antibody staining was observed in the normal tissue, strong staining in the nuclei of tumor cells was observed in the majority of breast cancer specimens examined (Fig. 6D and E). When adjacent tumor sections were stained with either ANCCA or Ki67 antibody and analyzed statistically, positive ANCCA staining correlated strongly with Ki67 staining (Fig. 6E; r = 1.23, P < 0.001), suggesting that overexpressed ANCCA may drive cell proliferation in the tumor.
DISCUSSION
Deregulated E2Fs are thought to play pivotal roles in cancer cell proliferation by their direct association with target gene promoters. We revealed here that the knockdown of coregulator ANCCA strongly impedes E2F1 and E2F3 occupancy at their chromatin targets, suggesting that chromatin association by specific E2Fs is a facilitated process. Our results also suggest that ANCCA can act as a “pioneer” factor for the E2Fs by anchoring itself at specific chromatin locations during mitosis and G1 and then loading activator E2Fs and the MLL-hKMT complex. These findings support a new notion that DNA binding transcription factors such as E2Fs utilize the coregulators to read histone marks for their proper recruitment to chromatin targets.
Histone marks H3K4me3 and H3K9K14ac are associated with transcriptionally active genes and therefore have been regarded as common chromatin cures for imminent activation of gene programs that are constitutively expressed. Interestingly, we identified H3K14ac as a mark that is present abundantly at promoters of E2F-responsive genes in late mitotic cells that are poised to be activated as the cells enter G1, while the level of H3K9K14ac is low in mitotic cells and reaches the highest level at late G1. These data suggest that distinctive histone marks exist for coordinated regulation of cell cycle-specific gene programs. How acetyl-H3K14ac levels change during the cell cycle on the E2F target chromatin is unclear at this point. Given that neither HATs nor histone deacetylases (HDACs) are shown to display specific histone acetylation in a cell cycle-regulated fashion, it is more likely that concerted activities from HATs, HDACs, and specific chromatin-binding proteins that can modulate histone modification levels are in play (21). For instance, HMGN1 has been shown to enhance global H3K14 acetylation by PCAF and the expression of immediate early genes (32). Myc has been shown to recruit TRRAP-GCN5 or Tip60 HAT complexes (28, 38), which may mediate histone acetylation at its target genes during S or G2. Many bromodomain-containing proteins may also be involved, as they associate dynamically with acetylated chromatin in interphase and mitosis and may modulate histone acetylation levels at specific loci (19, 22, 23, 27, 63). It is well demonstrated that the double bromodomain protein Brd4 associates with chromosomes throughout the different stages of mitosis and appears to mark specific genes for expression as cells enter the next cell cycle (12, 71). Interestingly, Brd4 does not appear to play a key role in the control of ANCCA-E2F target genes such as cyclin A, cyclin E, cyclin B, and cdc2. Likewise, a recent genome-wide, comparative analysis of MLL occupancy in interphase versus mitosis indicates that although MLL is widely distributed in interphase on a large number of genes, including E2F targets, in mitosis MLL associates with only a specific subset of genes, such as the ones of the Hox family in the condensed mitotic chromatin for their high levels of imminent induction as cells exit the M phase (4). However, the cell cycle-regulated, E2F target genes were not noted for MLL occupancy during mitosis. These findings suggest that different chromatin regulators, including bromodomain-containing proteins, may mark distinct subsets of genes in mitosis for induction in interphase.
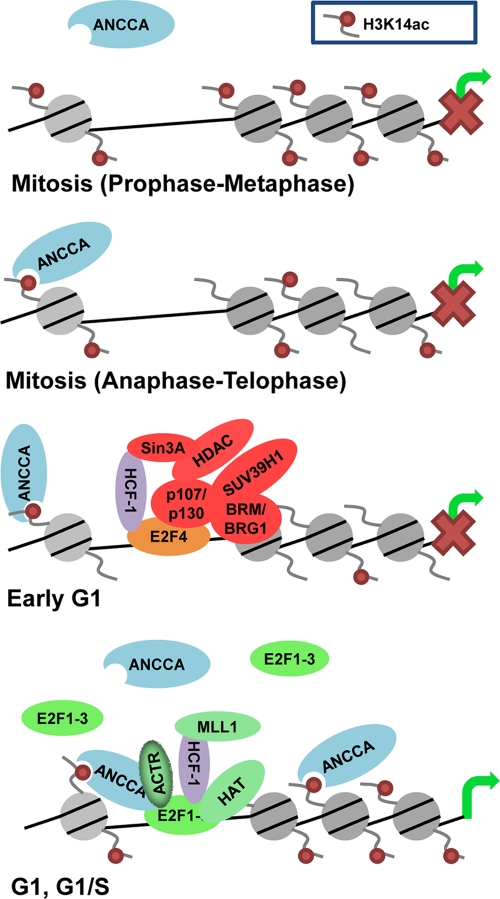
We propose the following model for ANCCA-mediated, E2F-dependent activation of genes in proliferating cells (Fig. 7). At the end of mitosis, E2F coregulator proteins such as ANCCA associate with specific chromatin loci through their histone binding modules. By binding H3K14ac and possibly H3K9K14ac via its bromodomain, ANCCA anchors itself at a subset of its targets, including some of the E2F target genes. As cells enter G1, activator E2Fs, through interaction with ANCCA, are recruited to their targets. During G1-to-S progression, recruited E2Fs may facilitate additional ANCCA recruitment, generating a feed-forward loop for chromatin loading of the ANCCA-E2F complex and assembly of histone-modifying complexes such as HCF-1-MLL. This model is consistent with our results that mutations in ANCCA bromodomain that inactivate its recognition of acetylated histones severely compromised the activity of ANCCA to associate with chromatin at E2F promoters and its ability to serve as an E2F coactivator. The model also takes into consideration that high levels of the H3K14ac mark overlap with ANCCA occupancy at the E2F target promoters, primarily at the late mitosis and beginning of G1 (Fig. 3G). As cells move from early G1 to late G1, it is possible that the E2F-ANCCA-positive loop plays a more important role in mediating ANCCA recruitment, while the recognition of H3K14ac by ANCCA bromodomain becomes less crucial for ANCCA recruitment. Alternatively, ANCCA recruitment at the later points may be facilitated by the H3K9K14 double-acetylated mark at the E2F target chromatin, which ANCCA bromodomain recognizes and increases dramatically as the cell cycle progresses. The other possibility is that as more ANCCA occupies the E2F target chromatin, less H3K14ac epitope at the chromatin becomes accessible for the anti-H3K14ac antibody to recognize, resulting in lower ChIP signal at the later time points. In any event, it is likely that other mechanisms are at play in conjunction with H3K14ac in ANCCA occupancy of the E2F targets as cells exit mitosis and in enforcement of the E2F-ANCCA feed-forward loop for the complex assembly. Given our previous study indicating that the coregulator ACTR acts as an E2F coactivator and that ACTR interacts with ANCCA (1, 37, 76), it is tempting to speculate that proteins such as ACTR may be important in the positive feed-forward loop.
FIG. 7.
A proposed model of ANCCA functional mechanism in mediating transcriptional activation of E2F target genes (see Discussion for details). It needs to be noted that the depicted association of ANCCA to the chromatin at early G1 does not imply a functional interaction between ANCCA and the HCF-1 complex.
The notion of ANCCA as an E2F chromatin loader is also consistent with ANCCA being an AAA ATPase protein. One major biochemical activity of the AAA family proteins is their ability to assemble or disassemble protein complexes. Our previous study with estrogen induction of gene expression revealed that ANCCA is crucial for the recruitment of coactivator CBP to ER target genes (76), supporting the role of ANCCA in protein complex assembly. As described in the introduction, several E2F coregulators have been identified. Interestingly, while HCF-1 interacts with the N terminus of E2Fs, ANCCA interacts with both the N terminus and the C-terminal transactivation/pocket binding domain of E2Fs. Since several cofactors, including ANCCA, ACTR, ARID1 in SWI/SNF complexes, HCF-1, MLL, p300/CBP, Tip60, and TRRAP, may interact with E2Fs at overlapping sites, it is possible that an ordered assembly of these coregulators may take place at E2F-responsive promoters as the cell cycle progresses. A similar process has been observed in hormone induction of gene expression by nuclear receptor-coregulator complexes (20, 24, 29, 65, 69). Intriguingly, ANCCA appears to play a more fundamental role in mediating the E2F transcription program than in the hormonal programs, as it acts at the initial steps of transcriptional activation, namely, the assembly of DNA binding E2Fs at their target promoters. Since ANCCA promoter occupancy increases from G1 to S, it is tempting to speculate that ANCCA may not only serve as an anchoring factor for the initial assembly but may also play a role in the later reorganization of E2F-coregulator complexes. Examining the functional association of ANCCA with the other E2F coregulators will likely provide relevant new insights. Finally, our finding that ANCCA is a key coregulator of E2F is also supported by genetic evidence obtained for lex-1, the ortholog of the ANCCA gene in C. elegans. Although its exact function is still poorly understood, lex-1 was identified as one of the class B synthetic Multivulva genes, which include the C. elegans homologs of Rb, E2F, and DP transcription factors, therefore indicating a genetic interaction between lex-1 and the Rb-E2F pathway (53, 59).
In this study, we provide evidence for a novel mechanism of E2F activation of gene expression in cancer cell proliferation. Our data reveal a crucial role of the bromodomain in a newly identified chromatin coregulator ANCCA in E2F-dependent gene expression and identify H3K14ac as a distinct histone mark recognized in mitosis and early G1 by ANCCA bromodomain for expression of genes crucial for cell cycle progression. Our results also demonstrate that while ANCCA anchors itself at the E2F chromatin targets likely via association with H3K14ac, the coregulator is in turn required for chromatin occupancy of E2Fs and the HCF-1-MLL hKMT complex. Thus, these findings highlight a unique function of ANCCA in directly coupling histone code reading with histone code writing. Given the prevalent defects of the Rb-E2F pathway and the aberration of ANCCA expression detected in multiple cancers (9, 75), the unveiled mechanism may represent a novel molecular underpinning of chromatin coregulator involvement in tumorigenesis.
Acknowledgments
We thank W. Herr, T. Christie, J. Nevins, K. Helin, and W. Xu for reagents, Alexander Borowsky for histopathology evaluations, E. Kalashnikova for confocal microscopy, and P. Farnham, H. Kung, and X. Chen for suggestions.
This work was supported by grants from the NIH (R01-CA113860, R01-CA134766, and R01-DK060019) to H.-W.C. E.V.K. is a recipient of a DoD BCRP predoctoral fellowship award (W81XWH-0810689).
Footnotes
Published ahead of print on 20 September 2010.
REFERENCES
- 1.Acevedo, M. L., and W. L. Kraus. 2003. Mediator and p300/CBP-steroid receptor coactivator complexes have distinct roles, but function synergistically, during estrogen receptor alpha-dependent transcription with chromatin templates. Mol. Cell. Biol. 23:335-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhat-Nakshatri, P., G. Wang, H. Appaiah, N. Luktuke, J. S. Carroll, T. R. Geistlinger, M. Brown, S. Badve, Y. Liu, and H. Nakshatri. 2008. AKT alters genome-wide estrogen receptor alpha binding and impacts estrogen signaling in breast cancer. Mol. Cell. Biol. 28:7487-7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blais, A., and B. D. Dynlacht. 2007. E2F-associated chromatin modifiers and cell cycle control. Curr. Opin. Cell Biol. 19:658-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blobel, G. A., S. Kadauke, E. Wang, A. W. Lau, J. Zuber, M. M. Chou, and C. R. Vakoc. 2009. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol. Cell 36:970-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosco, E. E., Y. Wang, H. Xu, J. T. Zilfou, K. E. Knudsen, B. J. Aronow, S. W. Lowe, and E. S. Knudsen. 2007. The retinoblastoma tumor suppressor modifies the therapeutic response of breast cancer. J. Clin. Invest. 117:218-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldon, C. E., C. M. Sergio, J. Schutte, M. N. Boersma, R. L. Sutherland, J. S. Carroll, and E. A. Musgrove. 2009. Estrogen regulation of cyclin E2 requires cyclin D1 but not c-Myc. Mol. Cell. Biol. 29:4623-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H. Z., S. Y. Tsai, and G. Leone. 2009. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer. 9:785-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciro, M., E. Prosperini, M. Quarto, U. Grazini, J. Walfridsson, F. McBlane, P. Nucifero, G. Pacchiana, M. Capra, J. Christensen, and K. Helin. 2009. ATAD2 is a novel cofactor for MYC, overexpressed and amplified in aggressive tumors. Cancer Res. 69:8491-8498. [DOI] [PubMed] [Google Scholar]
- 10.Delcuve, G. P., S. He, and J. R. Davie. 2008. Mitotic partitioning of transcription factors. J. Cell. Biochem. 105:1-8. [DOI] [PubMed] [Google Scholar]
- 11.Dey, A., F. Chitsaz, A. Abbasi, T. Misteli, and K. Ozato. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. U. S. A. 100:8758-8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dey, A., A. Nishiyama, T. Karpova, J. McNally, and K. Ozato. 2009. Brd4 marks select genes on mitotic chromatin and directs postmitotic transcription. Mol. Biol. Cell 20:4899-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dou, Y., T. A. Milne, A. J. Tackett, E. R. Smith, A. Fukuda, J. Wysocka, C. D. Allis, B. T. Chait, J. L. Hess, and R. G. Roeder. 2005. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell 121:873-885. [DOI] [PubMed] [Google Scholar]
- 14.Erzberger, J. P., and J. M. Berger. 2006. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 35:93-114. [DOI] [PubMed] [Google Scholar]
- 15.Hanson, P. I., and S. W. Whiteheart. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519-529. [DOI] [PubMed] [Google Scholar]
- 16.Hargreaves, D. C., T. Horng, and R. Medzhitov. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138:129-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsia, E. Y., E. V. Kalashnikova, A. S. Revenko, J. X. Zou, A. D. Borowsky, and H. W. Chen. 2010. Deregulated E2F and the AAA+ coregulator ANCCA drive proto-oncogene ACTR/AIB1 overexpression in breast cancer. Mol. Cancer Res. 8:183-193. [DOI] [PubMed] [Google Scholar]
- 18.Indiani, C., and M. O'Donnell. 2006. The replication clamp-loading machine at work in the three domains of life. Nat. Rev. Mol. Cell Biol. 7:751-761. [DOI] [PubMed] [Google Scholar]
- 19.Jang, M. K., K. Mochizuki, M. Zhou, H. S. Jeong, J. N. Brady, and K. Ozato. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 19:523-534. [DOI] [PubMed] [Google Scholar]
- 20.Jeong, K. W., Y. H. Lee, and M. R. Stallcup. 2009. Recruitment of the SWI/SNF chromatin remodeling complex to steroid hormone-regulated promoters by nuclear receptor coactivator flightless-I. J. Biol. Chem. 284:29298-29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnsson, A., M. Durand-Dubief, Y. Xue-Franzen, M. Ronnerblad, K. Ekwall, and A. Wright. 2009. HAT-HDAC interplay modulates global histone H3K14 acetylation in gene-coding regions during stress. EMBO Rep. 10:1009-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Kalashnikova, E. V., A. S. Revenko, A. T. Gemo, N. P. Andrews, C. G. Tepper, J. X. Zou, R. D. Cardiff, A. D. Borowsky, and H. W. Chen. 23 september 2010. ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer Res. [Epub ahead of print.] doi: 10.1158/0008-5472. CAN-10-1199. [DOI] [PMC free article] [PubMed]
- 22.Kanno, T., Y. Kanno, R. M. Siegel, M. K. Jang, M. J. Lenardo, and K. Ozato. 2004. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell 13:33-43. [DOI] [PubMed] [Google Scholar]
- 23.Kasten, M., H. Szerlong, H. Erdjument-Bromage, P. Tempst, M. Werner, and B. R. Cairns. 2004. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 23:1348-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kininis, M., G. D. Isaacs, L. J. Core, N. Hah, and W. L. Kraus. 2009. Postrecruitment regulation of RNA polymerase II directs rapid signaling responses at the promoters of estrogen target genes. Mol. Cell. Biol. 29:1123-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kouskouti, A., and I. Talianidis. 2005. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24:347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128:693-705. [DOI] [PubMed] [Google Scholar]
- 27.Ladurner, A. G., C. Inouye, R. Jain, and R. Tjian. 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 11:365-376. [DOI] [PubMed] [Google Scholar]
- 28.Lang, S. E., S. B. McMahon, M. D. Cole, and P. Hearing. 2001. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem. 276:32627-32634. [DOI] [PubMed] [Google Scholar]
- 29.Lee, Y. H., S. A. Coonrod, W. L. Kraus, M. A. Jelinek, and M. R. Stallcup. 2005. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl. Acad. Sci. U. S. A. 102:3611-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeRoy, G., B. Rickards, and S. J. Flint. 2008. The double bromodomain proteins Brd2 and Brd3 couple histone acetylation to transcription. Mol. Cell 30:51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung, J. Y., G. L. Ehmann, P. H. Giangrande, and J. R. Nevins. 2008. A role for Myc in facilitating transcription activation by E2F1. Oncogene 27:4172-4179. [DOI] [PubMed] [Google Scholar]
- 32.Lim, J. H., K. L. West, Y. Rubinstein, M. Bergel, Y. V. Postnikov, and M. Bustin. 2005. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J. 24:3038-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, H., E. H. Cheng, and J. J. Hsieh. 2007. Bimodal degradation of MLL by SCFSkp2 and APCCdc20 assures cell cycle execution: a critical regulatory circuit lost in leukemogenic MLL fusions. Genes Dev. 21:2385-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loomis, R. J., Y. Naoe, J. B. Parker, V. Savic, M. R. Bozovsky, T. Macfarlan, J. L. Manley, and D. Chakravarti. 2009. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol. Cell 33:450-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Louie, M. C., A. S. Revenko, J. X. Zou, J. Yao, and H. W. Chen. 2006. Direct control of cell cycle gene expression by proto-oncogene product ACTR, and its autoregulation underlies its transforming activity. Mol. Cell. Biol. 26:3810-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Louie, M. C., H. Q. Yang, A. H. Ma, W. Xu, J. X. Zou, H. J. Kung, and H. W. Chen. 2003. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc. Natl. Acad. Sci. U. S. A. 100:2226-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Louie, M. C., J. X. Zou, A. Rabinovich, and H. W. Chen. 2004. ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol. Cell. Biol. 24:5157-5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 39.Mochizuki, K., A. Nishiyama, M. K. Jang, A. Dey, A. Ghosh, T. Tamura, H. Natsume, H. Yao, and K. Ozato. 2008. The bromodomain protein Brd4 stimulates G1 gene transcription and promotes progression to S phase. J. Biol. Chem. 283:9040-9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mujtaba, S., L. Zeng, and M. M. Zhou. 2007. Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26:5521-5527. [DOI] [PubMed] [Google Scholar]
- 41.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagl, N. G., Jr., X. Wang, A. Patsialou, M. Van Scoy, and E. Moran. 2007. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 26:752-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nagy, Z., and L. Tora. 2007. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene 26:5341-5357. [DOI] [PubMed] [Google Scholar]
- 44.Narayanan, A., W. T. Ruyechan, and T. M. Kristie. 2007. The coactivator host cell factor-1 mediates Set1 and MLL1 H3K4 trimethylation at herpesvirus immediate early promoters for initiation of infection. Proc. Natl. Acad. Sci. U. S. A. 104:10835-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rabinovich, A., V. X. Jin, R. Rabinovich, X. Xu, and P. J. Farnham. 2008. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res. 18:1763-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruthenburg, A. J., H. Li, D. J. Patel, and C. D. Allis. 2007. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8:983-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarge, K. D., and O. K. Park-Sarge. 2009. Mitotic bookmarking of formerly active genes: keeping epigenetic memories from fading. Cell Cycle 8:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schiltz, R. L., C. A. Mizzen, A. Vassilev, R. G. Cook, C. D. Allis, and Y. Nakatani. 1999. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 274:1189-1192. [DOI] [PubMed] [Google Scholar]
- 49.Schneider, R., A. J. Bannister, F. A. Myers, A. W. Thorne, C. Crane-Robinson, and T. Kouzarides. 2004. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 6:73-77. [DOI] [PubMed] [Google Scholar]
- 50.Shahbazian, M. D., and M. Grunstein. 2007. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76:75-100. [DOI] [PubMed] [Google Scholar]
- 51.Sims, R. J., III, S. Millhouse, C. F. Chen, B. A. Lewis, H. Erdjument-Bromage, P. Tempst, J. L. Manley, and D. Reinberg. 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol. Cell 28:665-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stender, J. D., J. Frasor, B. Komm, K. C. Chang, W. L. Kraus, and B. S. Katzenellenbogen. 2007. Estrogen-regulated gene networks in human breast cancer cells: involvement of E2F1 in the regulation of cell proliferation. Mol. Endocrinol. 21:2112-2123. [DOI] [PubMed] [Google Scholar]
- 53.Tackett, A. J., D. J. Dilworth, M. J. Davey, M. O'Donnell, J. D. Aitchison, M. P. Rout, and B. T. Chait. 2005. Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 169:35-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 55.Takeda, S., D. Y. Chen, T. D. Westergard, J. K. Fisher, J. A. Rubens, S. Sasagawa, J. T. Kan, S. J. Korsmeyer, E. H. Cheng, and J. J. Hsieh. 2006. Proteolysis of MLL family proteins is essential for taspase1-orchestrated cell cycle progression. Genes Dev. 20:2397-2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taubert, S., C. Gorrini, S. R. Frank, T. Parisi, M. Fuchs, H. M. Chan, D. M. Livingston, and B. Amati. 2004. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol. Cell. Biol. 24:4546-4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taverna, S. D., H. Li, A. J. Ruthenburg, C. D. Allis, and D. J. Patel. 2007. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14:1025-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Truscott, M., R. Harada, C. Vadnais, F. Robert, and A. Nepveu. 2008. p110 CUX1 cooperates with E2F transcription factors in the transcriptional activation of cell cycle-regulated genes. Mol. Cell. Biol. 28:3127-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tseng, R. J., K. R. Armstrong, X. Wang, and H. M. Chamberlin. 2007. The bromodomain protein LEX-1 acts with TAM-1 to modulate gene expression in C. elegans. Mol. Genet. Genomics 278:507-518. [DOI] [PubMed] [Google Scholar]
- 60.Tyagi, S., A. L. Chabes, J. Wysocka, and W. Herr. 2007. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol. Cell 27:107-119. [DOI] [PubMed] [Google Scholar]
- 61.Tyagi, S., and W. Herr. 2009. E2F1 mediates DNA damage and apoptosis through HCF-1 and the MLL family of histone methyltransferases. EMBO J. 28:3185-3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Valls, E., S. Sanchez-Molina, and M. A. Martinez-Balbas. 2005. Role of histone modifications in marking and activating genes through mitosis. J. Biol. Chem. 280:42592-42600. [DOI] [PubMed] [Google Scholar]
- 63.VanDemark, A. P., M. M. Kasten, E. Ferris, A. Heroux, C. P. Hill, and B. R. Cairns. 2007. Autoregulation of the rsc4 tandem bromodomain by gcn5 acetylation. Mol. Cell 27:817-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vermeulen, M., K. W. Mulder, S. Denissov, W. W. Pijnappel, F. M. van Schaik, R. A. Varier, M. P. Baltissen, H. G. Stunnenberg, M. Mann, and H. T. Timmers. 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131:58-69. [DOI] [PubMed] [Google Scholar]
- 65.Vicent, G. P., R. Zaurin, A. S. Nacht, A. Li, J. Font-Mateu, F. Le Dily, M. Vermeulen, M. Mann, and M. Beato. 2009. Two chromatin remodeling activities cooperate during activation of hormone responsive promoters. PLoS Genet. 5:e1000567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wade, H. E., S. Kobayashi, M. L. Eaton, M. S. Jansen, E. K. Lobenhofer, M. Lupien, T. R. Geistlinger, W. Zhu, J. R. Nevins, M. Brown, D. C. Otteson, and D. P. McDonnell. 2010. Multimodal regulation of E2F1 gene expression by progestins. Mol. Cell. Biol. 30:1866-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wells, J., K. E. Boyd, C. J. Fry, S. M. Bartley, and P. J. Farnham. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20:5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu, S. Y., A. Y. Lee, S. Y. Hou, J. K. Kemper, H. Erdjument-Bromage, P. Tempst, and C. M. Chiang. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 20:2383-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu, W., H. Cho, S. Kadam, E. M. Banayo, S. Anderson, J. R. Yates III, B. M. Emerson, and R. M. Evans. 2004. A methylation-mediator complex in hormone signaling. Genes Dev. 18:144-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang, X. J., and E. Seto. 2007. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26:5310-5318. [DOI] [PubMed] [Google Scholar]
- 71.Yang, Z., N. He, and Q. Zhou. 2008. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell. Biol. 28:967-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yokoyama, A., Z. Wang, J. Wysocka, M. Sanyal, D. J. Aufiero, I. Kitabayashi, W. Herr, and M. L. Cleary. 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol. Cell. Biol. 24:5639-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng, L., Q. Zhang, G. Gerona-Navarro, N. Moshkina, and M. M. Zhou. 2008. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure 16:643-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zippo, A., R. Serafini, M. Rocchigiani, S. Pennacchini, A. Krepelova, and S. Oliviero. 2009. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138:1122-1136. [DOI] [PubMed] [Google Scholar]
- 75.Zou, J. X., L. Guo, A. S. Revenko, C. G. Tepper, A. T. Gemo, H. J. Kung, and H. W. Chen. 2009. Androgen-induced coactivator ANCCA mediates specific androgen receptor signaling in prostate cancer. Cancer Res. 69:3339-3346. [DOI] [PubMed] [Google Scholar]
- 76.Zou, J. X., A. S. Revenko, L. B. Li, A. T. Gemo, and H. W. Chen. 2007. ANCCA, an estrogen-regulated AAA+ ATPase coactivator for ERalpha, is required for coregulator occupancy and chromatin modification. Proc. Natl. Acad. Sci. U. S. A. 104:18067-18072. [DOI] [PMC free article] [PubMed] [Google Scholar]