Abstract
Cks1, Cdk1 (Cdc28), and the proteasome are required for efficient transcriptional induction of GAL1 and other genes in Saccharomyces cerevisiae. We show here that one function of these proteins is to reduce nucleosome density on chromatin in a gene induction-specific manner. The transcriptional requirement for Cks1 can be bypassed if nucleosome density is reduced by an alternative pathway, indicating that this is the primary function of Cks1 in the context of gene induction. We further show that Cks1, Cdk1, and the 19S subunit of the proteasome are recruited to chromatin by binding directly to the histone H4 amino-terminal tail. However, this activity of the proteasome does not require the protease activity associated with the 20S subunit. These data suggest a model where binding of a complex consisting of Cks1, Cdk1, and the 19S proteasome to histone H4 leads to removal of nucleosomes via a nonproteolytic activity of the proteasome.
Proteasomes are large multiprotein assemblies that are thought to function primarily in the turnover of cellular proteins (29, 39). In most cases, proteins destined for proteasomal degradation are tagged with covalent multiubiquitin chains (11). These chains are then recognized by regulatory caps at either end of the barrel-shaped catalytic core of the proteasome. Once bound to the proteasomal cap, substrates are stripped of their multiubiquitin chains and then threaded into the internal chamber of the proteasome barrel where three distinct protease activities cleave polypeptide chains into small peptides. The regulatory cap of the proteasome contains a ring of ATPases that abut the catalytic core and function to unwind and translocate folded proteins and to open an aperture into the catalytic core (37). Recently, proteasomal function has been linked to transcription in a number of different contexts. In Saccharomyces cerevisiae, genetic studies have yielded two classes of mutations in the ATPase subunits of the proteasome regulatory cap. The first class was identified as suppressors of a defective transcription factor, Gal4, and defined two genes, SUG1 and SUG2, which are now known to be identical to proteasomal ATPase genes RPT6 and RPT1, respectively (5, 9, 32, 33). Subsequently, it has been shown that proteasomal function is required for effective transactivation by Gal4 (5, 9). The second class was identified in a screen for suppressors of cdc68-1 (spt16), a temperature-sensitive (TS) mutation in a component of the FACT transcriptional elongation complex (named FACT for facilitates transcription elongation) (40). Interestingly, mutations capable of suppressing cdc68 mutations mapped to the same loci as gal4 suppressor mutations SUG1 and SUG2 did, but the individual classes were distinct in that each mutation was capable of suppressing either gal4 or cdc68 mutations, but not both (5, 6). This suggests that the proteasome may have multiple roles in transcription, one associated with transactivation and another involved in elongation. Recently, it was reported that the proteasome regulatory particle is required to efficiently recruit the SAGA (Spt-Ada-Gcn5-acetyltransferase) coactivator to transcriptional activators (19). This activity most likely accounts for at least one role of the proteasome in transactivation. However, the role of the proteasome in transcriptional elongation remains to be determined. Another unresolved issue is whether protease activity of the proteasome is required for either of these functions, as papers coming to divergent conclusions have been published (21, 26). At least for some transcription factors, proteasome-mediated proteolysis has been proposed as an essential part of the transcription initiation cycle (12).
Genetic interactions have also been observed between proteasome mutations and yeast cdk1 (cdc28) mutations, as well as between proteasome mutations and mutations in the gene encoding the small Cdk-binding protein, Cks1 (15). In addition, Cdk1 and Cks1 copurify with and interact physically with proteasomes (15). We have reported that, like the proteasome, Cdk1 and Cks1 are necessary for efficient induction of the GAL1 gene in response to activating signals (41). Furthermore, both Cdk1 and Cks1 are required for recruitment of the proteasome to the GAL1 open reading frame (ORF), suggesting that Cdk1/Cks1 and the proteasome function in a common pathway required for efficient induction of galactose-responsive and, most likely, other inducible genes (25, 41). We report here that at least one function of Cdk1/Cks1 and the 19S subunit of the proteasome is to evict nucleosomes from chromatin in the context of gene induction. We propose that the failure of Cdk1/Cks1 to recruit 19S proteasomes to chromatin at the GAL1 ORF results in excessive nucleosomal density and therefore impairment of transcription. We further report that the function of the proteasome in this context does not require protease function, and consistent with this, only the 19S regulatory subunit is actively recruited upon gene induction.
MATERIALS AND METHODS
Yeast strains.
All S. cerevisiae strains and genotypes are shown in Table 1 and were constructed by standard yeast genetic methods.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotypea | Reference |
|---|---|---|
| BF264-15DU | aura2 ade1 his2 leu2 trp1 | 31 |
| Δcks1 mutant | BF264-15DU cks1Δ::KANR | 41 |
| cdc28-1N mutant | BF264-15DU cdc28-1N | 41 |
| W303a | aade2 ura3 his3 trp1 leu2 | |
| sug1-20 mutant | W303a sug1-20 | 5 |
| UKY403 (HHFΔ GAL::HHF) | aade2 his3 leu2 lys2 trp1 ura3 arg4 hhf1::HIS3 hhf2::LEU2 [pTRP1 GAL1::HHF2] | 16 |
| HHFΔ GAL::HHF Δcks1 mutant | UKY403 cks1Δ::KANR | This study |
| JHY86 (HHTΔ HHFΔ [pTRP1 HHT2 HHF2]) | ahis3 leu2 trp1 ura3 hht1 hhf1Δ hht2 hhf2Δ [pTRP1 HHT2 HHF2] | 2 |
| HHTΔ HHFΔ [pTRP1 HHT2(Δ4-30) HHF2] mutant | Same as JHY86 but with [pTRP1 HHT2(Δ4-30) HHF2] | This study |
| GFY428 (HHTΔ HHFΔ [pTRP1 HHT2 HHF2Δ4-28]) | Same as JHY86 but with [pTRP1 HHT2 HHF2(Δ4-28)] | This study |
| Cks1-Flag strain | BF264-15DU CKS1-Flag | 41 |
| Rpt1-Flag strain | BF264-15DU RPT1-Flag | 41 |
| Pre1-HA strain | BF264-15DU PRE1-HA | 25 |
| Cln2-Flag strain | BF264-15DU CLN2-Flag | 13 |
Genotypes in brackets indicate a replicating plasmid with designated markers.
Yeast growth.
For GAL1 induction experiments, yeast cells were grown in yeast extract-peptone (YEP) medium with 2% raffinose, and galactose (Gal) was added to a final concentration of 2% for 60 min. For phosphate depletion experiments, yeast cells were grown overnight in YEP medium with Gal at room temperature. Yeast cells grown to an optical density at 600 nm (OD600) of 50 were collected by centrifugation, washed with water, and suspended in 100 ml of synthetic medium containing 2% galactose and 100 μM inorganic phosphate. The cells were grown at room temperature for 1.5 h and then shifted to the inactivating temperature (38°C for sug1-20 mutant cells, 30°C for cdc28-1N mutant cells, and 30°C for cks1Δ mutant cells) for an additional 1.5 h. The culture was split into four. The cells were collected, washed, and suspended in one of four conditions at the inactivating temperature with dextrose or galactose with or without 7.34 mM Pi. The cells were cultured under these conditions for an additional 3 h. The cells were then collected and frozen at −80°C.
Chromatin immunoprecipitation (ChIP).
Cells were cross-linked by the addition of formaldehyde to a final concentration of 1% for 30 min at room temperature (RT). Glycine was then added to a final concentration of 125 mM for 5 min at RT. The cells were washed twice with ice-cold Tris-buffered saline (TBS), and the pellets were snap-frozen in liquid nitrogen. Thawed cells were resuspended in 500 μl of lysis buffer (50 mM HEPES [pH 7.5], 140 mM NaCl, 1% Triton X-100, 0.1% deoxycholate [DOC], 1 mM EDTA, and protease inhibitors) and lysed with glass beads in a Fastprep for four 30-s intervals at 4.5 output. Lysed cells were centrifuged for 15 min at 14,000 rpm at 4°C. Pellets were resuspended in 500-μl portions of lysis buffer and sonicated for 10 times for 30 s each time with a Cup-Horn Sonicator 3000, with 2-min intervals between pulses. After centrifugation, lysates were incubated overnight at 4°C with histone H3 or histone H4 antibodies (Abcam) or FLAG antibodies (Sigma) followed by 2 h of incubation with protein A- or G-coupled Dynabeads (Invitrogen). The beads were washed twice with lysis buffer, twice with lysis buffer containing 0.5 M NaCl, twice with wash buffer (10 mM Tris [pH 8.0], 250 mM LiCl, 0.75% NP-40, 0.75% DOC, 1 mM EDTA, and protease inhibitors), and once with TE buffer (10 mM Tris [pH 7.5], 1 mM EDTA). Bound proteins were eluted with 100 μl elution buffer (50 mM Tris [pH 8.0], 10 mM EDTA, 1% SDS) for 30 min at 65°C. DNA-protein cross-links were reversed by overnight incubation at 65°C, and DNA was purified using QIAquick PCR purification kit (Qiagen) per the manufacturer's instructions. Real-time PCRs were performed using 1/25 of the immunoprecipitated fraction or 1/500 of the input fraction as the template. Primer locations for GAL1 have been described previously (41), and all primer sequences are given in Table 2. The control primers amplify a nontranscribed region on chromosome VI. The amount of immunoprecipitated DNA, as related to the amount of input DNA, was calculated for the different genomic regions and then normalized to the control region. All reactions were carried out at least three times, and all experiments were carried out at least twice. Statistical data are provided in Table 3.
TABLE 2.
PCR oligonucleotide primer sequences
| Primer | Templatea | Functionb | Sequence |
|---|---|---|---|
| SC05 | GAL1 | ChIP qPCR 1f | CTCCGACGGAAGACTCTCCTC |
| SC06 | GAL1 | ChIP qPCR 1r | AGGCACATCTGCGTTTCAGG |
| SC07 | GAL1 | ChIP qPCR 2f | AGTGCCCGATAATTAAGAAAT |
| SC08 | GAL1 | ChIP qPCR 2r | TTGACTCTACCAGGCGATCTAGC |
| SC09 | GAL1 | ChIP qPCR 3f | AACTTGCACCGGAAAGGTTTG |
| SC010 | GAL1 | ChIP qPCR 3r | TGCCAGTTGGTACATCACCCT |
| SC011 | GAL1 | ChIP qPCR 4f | GCCCAAATGGCAACATAGAAA |
| SC012 | GAL1 | ChIP qPCR 4r | AGAACTCATTGGCAAGGGCTT |
| SC013 | CHRVI | ChIP qPCRf | CTCGTTAGGATCACGTTCGAATCC |
| SC014 | CHRVI | ChIP qPCRr | GCGTAACAAAGCCATAATGCCTCC |
| SC017 | ACT1 | RT-qPCRf | ACGTTCCAGCCTTCTACGTTTCCA |
| SC018 | ACT1 | RT-qPCRr | TCGAAGTCCAAGGCGACGTAACAT |
| SC019 | GAL1 | RT-qPCRf | ACACCCTGGAACGGCGATATTGAA |
| SC020 | GAL1 | RT-qPCRr | TGAGACTCGTTCATCAAGCGACCA |
| SC043 | PHO5 | RT-qPCRf | AAGGTGCTCGTGTCTACACCGAAA |
| SC044 | PHO5 | RT-qPCRr | AAGCTGCTGACGTTACAGACCTTT |
| SC049 | PHO5 | ChIP qPCR UASf | GAATAGGCAATCTCTAAATGAATCGA |
| SC050 | PHO5 | ChIP qPCR UASr | GAAAACAGGGACCAGAATCATAAATT |
| SC055 | PHO5 | ChIP qPCR 2f | AGCAAAGTGAGATTCAAGACC |
| SC056 | PHO5 | ChIP qPCR 2r | GCGGTGGTCAAAAAGTTTAGG |
| SC057 | PHO5 | ChIP qPCR 1f | AGATTGGTACCCAAAAAGATATCT |
| SC058 | PHO5 | ChIP qPCR 1r | TTGATAGTCTTAGCCAGACTGACA |
CHRVI, chromosome VI.
ChIP, chromatin immunoprecipitation; qPCR, quantitative PCR, 1f, reaction 1 forward; 1r, reaction 1 reverse; qPCRf, forward quantitative PCR; QPCRr, reverse quantitative PCR; RT-qPCRf, reverse transcription-forward quantitative PCR; UASf and UAS, forward and reverse upstream activation sequences, respectively.
TABLE 3.
P values (Student's t test) determined for histone chromatin immunoprecipitationsa
| Comparisonb |
P valuec |
|||||||
|---|---|---|---|---|---|---|---|---|
| H3 ChIP |
H4 ChIP |
|||||||
| Gal1 | Gal2 | Gal3 | Gal4 | Gal1 | Gal2 | Gal3 | Gal4 | |
| WT 30′/cks1 mutant 30′ | 0.0965 | 0.0562 | 0.0154 | 0.0864 | 0.6238 | 0.0813 | 0.0412 | 0.1126 |
| WT 60′/cks1 mutant 60′ | 0.0061 | 0.5194 | 0.2904 | 0.032 | 0.0052 | 0.4777 | 0.0919 | 0.0132 |
| WT 60′/cdc28-1N mutant 60′ | 0.0072 | 0.0224 | 0.0085 | 0.0163 | 0.0121 | 0.0047 | 0.0106 | 0.0038 |
| WT 60′/sug1-20 mutant 60′ | 0.0031 | 0.0322 | 0.0013 | 0.0037 | 0.0154 | 0.124 | 0.0369 | 0.0576 |
| WT 60′/ΔH3 tail mutant 60′ | 0.1471 | 0.1378 | 0.1735 | 0.4142 | ||||
| WT 60′/ΔH4 tail mutant 60′ | 0.0796 | 0.2113 | 0.0407 | 0.0847 | ||||
| ΔH3 tail mutant 60′/ΔH4 tail mutant 60′ | 0.0231 | 0.0925 | 0.0213 | 0.0512 | ||||
P values (Student's t test) determined for histone H3 and H4 chromatin immunoprecipitations shown in Fig. 1 and 5.
The values for two strains were compared, and the P values are given. The two strains were usually the wild type (WT) and a mutant strain (e.g., cks1 mutant, cdc28-1N mutant, and sug1-20 mutant) but strains in which the histone 3 or histone 4 tail had been deleted (ΔΗ3 tail or ΔH4 tail mutant, respectively) were also compared. In the first row of the table, WT 30′/cks1 mutant 30′ indicates that the wild type (WT) and the cks1 mutant were compared 30 min (30′) after the addition of galactose. Strains were also compared 60 min after the addition of galactose.
Gal1 to Gal4 refer to regions spanning the GAL1 promoter and open reading frame analyzed by chromatin immunoprecipitation.
RNA purification and RT-PCR.
Cells were resuspended in TRIzol (Invitrogen), and RNA was purified per the manufacturer's instructions. Fifty nanograms of RNA was used as template for real-time reverse transcription-PCRs (RT-PCRs) using the iScript one-step RT-PCR kit with SYBR green (Bio-Rad). Primer sequences are shown in Table 2. All reactions were carried out at least three times, and all experiments were carried out at least twice.
Superhelicity assay.
Cells were transformed with pCMB3-55 (a 9-kb plasmid containing the 2μm origin, TRP1, GAL1 ORF and flanking sequences, and bacterial replication and selection sequences) and cultured in dextrose-based liquid medium without tryptophan overnight. Cells grown in liquid medium to an OD600 of 400 were collected by centrifugation, washed with water, and cultured in 400 ml of raffinose-based medium without tryptophan for 3 h. Cultures were split between two flasks containing 25 ml of 20% galactose or 20% raffinose and grown for an additional 2 h at 30°C prior to harvesting. Cell pellets were lysed with zymolyase, and plasmids were isolated using 10 columns (per 200-ml culture) of Zymoprep Yeast Plasmid Miniprep II (Zymo Research). Eluted plasmids were concentrated and exchanged into TE buffer using an Amicon Ultra 0.5-ml 10,000 concentrator (Millipore). Plasmid DNA was separated on a 15-cm 0.7% agarose-1.0 μg/ml chloroquine-TBE gel at 1.25 V/cm for 72 h (16). DNA was transferred to Immobilon N+ membrane using 0.25 M HCl for denaturing and 0.4 M NaOH for transfer. Radioactive probe was generated by nick translation of pBlueScript II SK+ (Applied Biosystems) in the presence of 40 μCi [α-32P]dCTP (Perkin Elmer) and purified on a 2-ml Sephadex G-50 column equilibrated with TNE buffer (10 mM Tris [pH 7.4], 1 mM EDTA, 200 mM NaCl). The probe was boiled and snap cooled on ice just prior to use. The blot was prehybridized and probed with APH/H buffer (5× SSC [1× SSC is 150 mM NaCl plus 15 mM trisodium citrate [pH 7.0], 5× Denhardt's solution, 1% SDS, 100 μg/ml denatured salmon sperm DNA) containing 10% polyethylene glycol 8000 (PEG 8000), washed twice with 50 ml of 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% SDS, and then washed twice with 0.2× SSC plus 0.1% SDS. Radioactive bands were detected using a Pharos FX Plus molecular imager.
Protein purification.
Escherichia coli BL21 cells expressing FLAG-tagged Cdc28 (FLAG-Cdc28) from pRSFDuet1 were induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 30°C. The cells were harvested, resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM EDTA, 10% glycerol, 0.2% Triton X-100, and protease inhibitors) and sonicated 4 times for 30 s each time. Cleared lysates were incubated with agarose-conjugated FLAG antibody (Sigma) overnight. Bound proteins were washed and eluted with FLAG peptide in binding buffer (20 mM Tris-HCl [pH 7.5], 10% glycerol, 200 mM NaCl). E. coli BL21 cells expressing histidine-tagged Cks1 (His-Cks1) from pRSFDuet1 were induced with 1 mM IPTG for 3 h at 30°C. The cells were harvested, resuspended in NETN buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% NP-40, and protease inhibitors) and sonicated 4 times for 30 s. Cleared lysates were incubated with nickel-nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen) overnight. Bound proteins were washed in binding buffer containing 100 mM NaCl and eluted by incubation with biotinylated thrombin (Novagen) overnight at 4°C.
Binding assay.
Streptavidin-agarose beads (Novagen) were incubated with biotinylated histone H3 or histone H4 amino-terminal peptides (Upstate) in TBS for 1 h at room temperature. The beads were washed with binding buffer and incubated with FLAG-Cdc28 for 45 min at RT. The beads were washed with binding buffer and eluted with sample buffer. Samples were run on a 10% Bis-Tris gel (Invitrogen) and transferred to polyvinylidene difluoride (PVDF). The membrane was probed with anti-FLAG antibody (Sigma) or streptavidin conjugated to horseradish peroxidase (streptavidin-HRP) (Pierce). Magnetic streptavidin beads (Invitrogen) were incubated with biotinylated histone H3 or histone H4 amino-terminal peptides (Upstate) in TBS for 1 h at RT. The beads were washed with 100 mM binding buffer, and incubated with Cks1 for 1 h at RT. The beads were washed with 100 mM binding buffer and eluted with sample buffer. Samples were run on a 10% Bis-Tris gel (Invitrogen) and transferred to PVDF. The membrane was probed with anti-Cks1 antibody or streptavidin-HRP (Pierce).
Proteasome inhibition.
Proteasome inhibition was performed by the method of Liu et al. (22). For GAL1 induction experiments, yeast cells were grown in synthetic medium (0.17% yeast nitrogenous base without ammonium sulfate, 0.1% proline, appropriate amino acids) with 2% raffinose as the carbon source. When the cells reached an OD600 of 1, 0.003% SDS was added, and the cells were grown for an additional 3 h at 30°C. The cells were then incubated with 75 μM MG132 or the equivalent volume of dimethyl sulfoxide (DMSO) for 30 min prior to the addition of 2% galactose. Samples were taken for RNA purification, and cultures were prepared for chromatin immunoprecipitation as described bellow.
Yeast cells expressing Cln2 tagged with FLAG (FLAG-Cln2) under the control of the GAL1 promoter were grown as described above except the initial cultures were grown in 2% galactose as a carbon source. After incubation with MG132 or DMSO, 2% dextrose was added to shut down expression of FLAG-Cln2.
Samples were collected at the indicated time points, and yeast cells were resuspended in sample buffer and incubated at 95°C for 5 min. After centrifugation, soluble proteins were run on a 10% Bis-Tris gel (Invitrogen) and transferred to PVDF. The membrane was probed with anti-FLAG antibody and reprobed with anti-PSTAIRE peptide antibody, which detects Cdk1 in yeast, as a loading control.
RESULTS
Cks1, Cdk1, and the proteasome regulate nucleosome density.
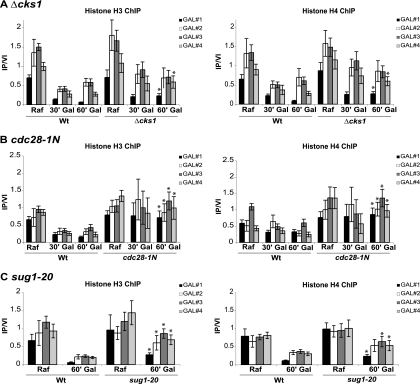
In the course of investigating the role of Cks1 in transcription of the GAL1 gene, histone chromatin immunoprecipitations (ChIPs) were carried out and compared for the wild-type strain and a cks1 mutant strain using four primer sets that span GAL1. In the wild-type strain, the densities of two histones analyzed (histone H3 and histone H4) were significantly reduced upon induction of the GAL1 gene by the addition of galactose, indicative of nucleosome eviction as has been observed previously (Fig. 1 A) (4, 34). In the cks1 mutant strain, this histone density was modestly elevated under the uninduced condition (although not statistically significant) and remained somewhat elevated upon induction (Fig. 1A). The comparison between the wild type and cks1 mutant was particularly apparent and reached significance in the promoter and 3′ ORF regions 60 min after induction, although a trend was observed over the entire gene in multiple experiments. Therefore, the cks1 mutant appears to exhibit an abnormally high density of histones associated with the GAL1 gene specifically under inducing conditions. Since Cks1 and Cdk1 loading onto chromatin are mutually dependent (41), we determined whether regulation of histone density also depended on Cdk1. A Cdk1 mutant shown to be defective in Cks1 binding, cdc28-1N, was compared to the wild type. The GAL1 promoter and ORF exhibited an abnormally high density of histone H3 and histone H4 in the cdc28-1N mutant both under induced and noninduced conditions (Fig. 1B). The histone density phenotype was consistently more severe for the cdc28-1N mutant, with significant differences relative to the wild type observed over the entire GAL1 ORF. Therefore, Cks1 and Cdk1 are both required for proper maintenance of histone density across the GAL1 promoter and ORF under noninducing and, more dramatically, for nucleosome eviction under inducing conditions.
FIG. 1.
cks1, cdk1, and proteasome mutations confer an abnormally high histone density at the GAL1 promoter and open reading frame (ORF). Chromatin immunoprecipitation (ChIP) was performed using histone H3 and histone H4 antibodies, respectively. In each case, GAL1 was induced for the indicated time (30 min [30′] or 60 min [60′]) by the addition of galactose (Gal) to cultures growing on raffinose (Raf). Chromatin immunoprecipitations were analyzed using four primer sets where GAL#1 is within the GAL1 promoter region and GAL#2, GAL#3, and GAL#4 span the GAL1 ORF. Immunoprecipitated signals were normalized using a primer set corresponding an untranscribed region of chromosome VI (IP/VI). (A) The wild type (Wt) and cks1 deletion mutant (Δcks1) were compared in an experiment carried out at 30°C. (B) The wild type and a cdk1 mutant specifically defective in Cks1 binding (cdc28-1N) were compared in an experiment carried out at 25°C. (C) sug1-20 is a temperature-sensitive mutation of the RPT6 gene encoding a proteasomal ATPase subunit. The experiment was carried out after shifting cultures from 25°C to 30°C for 5 h. Values are means ± standard deviations (SDs) (error bars). Asterisks indicate statistically significant differences (Student's t test) when comparing the values for the wild type and mutant 60 min after the addition of galactose. P values are given in Table 3. PCR primer sequences are provided in Table 2.
Since the transcriptional function of Cks1/Cdk1 is linked to the proteasome (41), we carried out similar histone ChIPs for a temperature-sensitive proteasomal ATPase mutant (sug1-20 mutant). Even at a permissive temperature for growth, the histone density at the GAL1 gene was slightly elevated compared to that of the wild type under noninducing conditions, but again, a statistically significant defect was observed under inducing conditions (Fig. 1C). These data are consistent with Cks1, Cdk1, and proteasome recruitment and function being required for maintenance of normal histone density at the GAL1 ORF, particularly under inducing conditions.
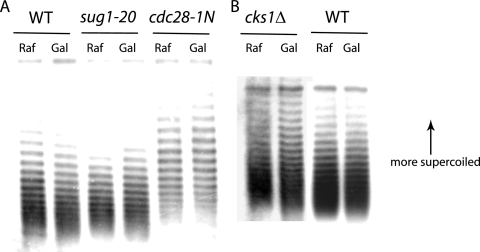
In contrast, no change in histone density was observed in the ORF of a non-galactose-responsive gene, ACT1, which encodes actin, and there was no apparent effect of mutating either CDK1, CKS1, or SUG1 (data not shown). To confirm that increased detection of histones by ChIP, as reported in Fig. 1, is a result of increased nucleosome density, the number of nucleosomes was determined directly based on the superhelical density of a circular plasmid containing the GAL1 gene (16). Whereas the GAL1-containing plasmid in wild-type cells exhibited a small but reproducible shift (approximately 1 supercoil) to lower superhelical density upon gene induction, no such shift was observed in the cks1, cdk1 (cdc28-1N), and sug1-20 mutant cells (Fig. 2). Since the plasmid used is 9 kb and the GAL1 gene is approximately 1.5 kb, only a small shift is anticipated in response to nucleosome eviction at the GAL1 ORF. Furthermore, the cks1 and cdc28-1N mutant cells exhibited elevated average superhelical densities than the wild-type cells did under all conditions, suggesting non-GAL1-associated changes mediated by Cks1 and Cdk1. Since lower superhelical density is associated with a reduction in nucleosome density, these data are consistent with the results based on histone ChIP, indicating that cks1, sug1, and cdk1 mutations do indeed confer a defect in transcription-associated nucleosome eviction and, possibly, basal nucleosome density.
FIG. 2.
Mutation of cks1, cdk1, or sug1 prevents induction-dependent reduction of nucleosome density at the GAL1 gene. (A) Autoradiograph showing distributions of supercoiled GAL1-containing plasmids extracted from the wild-type (WT) cells and sug1-20 and cdc28-1N mutant cells before and after induction. (B) Autoradiograph showing distributions of supercoiled GAL1-containing plasmids extracted from wild-type and cks1Δ mutant cells before and after induction. DNA was separated by agarose gel electrophoresis in the presence of chloroquine. Under the conditions used, plasmids with greater numbers of supercoils migrate more slowly, with each band corresponding to a species containing a specific number of supercoils. The shift of the distribution downward (most noticeable at the top of the distribution) for plasmids derived from wild-type cells is consistent with reduced supercoiling due to nucleosome eviction. The 9-kb plasmid used contains the GAL1 ORF and flanking regions, the TRP1 ARS1 marker, the 2μm origin, and bacterial replication and selection sequences.
Although these results establish a link between the function of Cks1, Cdk1, and proteasomal ATPases and modulation of histone density upon transcriptional induction, they do not establish direct causality, as mutations that block transcription also indirectly prevent a reduction in histone density at the affected loci (4, 17, 27, 34, 35, 42).
Ectopic reduction in nucleosome density bypasses the requirement of Cks1 and proteasomes in transcriptional induction.
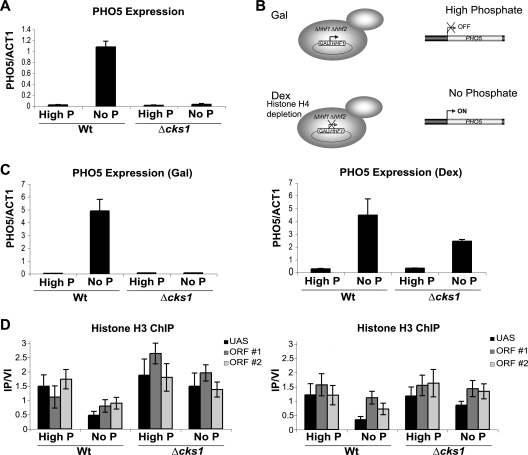
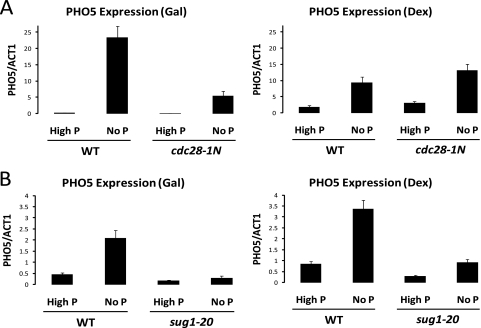
As stated above, an inability to reduce the nucleosome density associated with transcriptional induction could be the result of a direct effect on regulation of nucleosome density per se or could be the result of an indirect effect mediated through some other defect in the transcriptional machinery. If excessive nucleosome density and an inability to evict nucleosomes are responsible for the defect in GAL1 transcription observed in cks1, cdk1, and proteasome mutants, then depletion of nucleosomes by other means should obviate the requirement for the products of these genes in the context of GAL1 transcription. To deplete nucleosomes, we utilized a strain deleted for both endogenous histone H4 genes (HHF1 and HHF2) and kept alive by means of a plasmid containing a copy of the HHF2 coding region under the control of the GAL1 promoter (16). Therefore, when grown on galactose-containing medium, this strain synthesizes histone H4 and can produce nucleosomes but when shifted to dextrose-containing medium, synthesis of histone H4 is terminated and nucleosome assembly ceases, leading to a decrease in nucleosome density (Fig. 3 B). Because depletion of nucleosomes in this system is dependent on repression of the GAL1 promoter, we could not use conditional expression of histone H4 to investigate effects on GAL1 transcription. However, we have observed that induction of the PHO5 gene in response to phosphate depletion is dependent on Cks1 (Fig. 3A). PHO5 encodes a secreted acid phosphatase used for scavenging phosphate from the extracellular environment when intracellular phosphate is depleted (1). We therefore compared PHO5 induction in the wild-type strain versus a cks1 mutant strain under conditions of nucleosome depletion by culturing cells in medium containing dextrose. In medium containing galactose, which supports histone H4 expression, depletion of phosphate led to induction of PHO5 only in wild-type cells and not in cks1 mutant cells (Fig. 3C). Induction of PHO5 was accompanied by nucleosome depletion at both the PHO5 promoter region and ORF (Fig. 3D). However, induction of PHO5 by shifting the cells to phosphateless medium after ectopic nucleosome depletion (induction after shifting the cells to dextrose-containing medium) no longer depends on Cks1 (Fig. 3C). Importantly, induction under these conditions still depends on depletion of phosphate, indicating that it is not simply artifactual derepression due to histone depletion. Histone chromatin immunoprecipitation of the PHO5 promoter region and ORF confirmed that termination of histone H4 biosynthesis led to depletion of nucleosomes even in the absence of Cks1 (Fig. 3D). Although the ectopic nucleosome depletion was designed to be modest so as not to undermine regulatory functions, it was reproducible and approached statistical significance, i.e., P = 0.08 by Student's t test, comparing the promoter region of PHO5 in the cks1 mutant strain on medium containing dextrose versus galactose. Therefore, depletion of nucleosomes at the PHO5 promoter and ORF allows induction regardless of the status of CKS1. Similar bypass results were obtained comparing cdc28-1N and sug1-20 mutants to wild-type cells (Fig. 4). Thus, one direct role of Cks1, Cdk1, and the proteasome in the context of transcription is to enable a nucleosome density environment that is permissive for induction.
FIG. 3.
Depleting nucleosomes by alternative means can bypass the transcriptional requirement for Cks1. (A) Induction of PHO5 in response to phosphate (P) depletion. Wild-type and cks1Δ cells were shifted from complete synthetic medium supplemented with 2% dextrose to the same medium without phosphate (No P) for 7 h. PHO5 mRNA levels were determined by real-time PCR and normalized to ACT1 (actin) mRNA levels. (B) Schematic of the histone depletion scheme. Dex, dextrose. (C) PHO5 induction under conditions of histone depletion. Wild-type and cks1Δ strains were grown with galactose (histone H4 synthesis on) or dextrose (histone synthesis off) in either high-phosphate or no-phosphate medium for 7 h. PHO5 mRNA levels relative to ACT1 mRNA levels were determined by real-time PCR. (D) Histone H3 chromatin immunoprecipitations corresponding to the experiment described above for panel C. Primer sets correspond to the PHO5 promoter region (upstream activation sequence [UAS]) and the PHO5 ORF (ORF #1 and ORF #2). Signals were normalized using a primer set corresponding to an untranscribed region of chromosome VI. Values are means ± SDs (error bars). PCR primer sequences are provided in Table 2.
FIG. 4.
Depleting nucleosomes by alternative means can bypass the transcriptional requirement for Cdk1 and 19S proteasome function. PHO5 induction under conditions of histone depletion is shown. (A) Wild-type and cdc28-1N strains were grown with galactose (histone H4 synthesis on) or dextrose (histone synthesis off) in either high-phosphate or no-phosphate medium for 7 h. (B) Wild-type and sug1-20 strains were grown with galactose (histone H4 synthesis on) or dextrose (histone synthesis off) in either high-phosphate or no-phosphate medium for 7 h. PHO5 mRNA levels were determined by real-time PCR and normalized to ACT1 (actin) mRNA levels. Values are means plus SDs (error bars). PCR primer sequences are provided in Table 2.
Deletion of the histone H4 tail increases nucleosome density at the GAL1 gene.
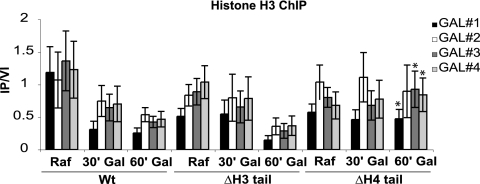
Both histones H3 and H4 possess amino-terminal tails that are dynamically modified and are thought to be involved in recruiting chromatin-modifying enzymes and complexes (14). Yeast cells with either tail deleted are viable, but deletion of both tails is lethal (20), suggesting that redundant or parallel pathways mediated by each tail individually are required for viability. Interestingly, deletion of the histone H4 tail confers a defect in GAL1 transcription, whereas deletion of the histone H3 tail does not (3). Instead, deletion of the H3 tail promotes derepression of GAL1 transcription (24), consistent with each tail responding to different regulatory pathways that can maintain viability. Furthermore, it has previously been shown that deletion of the histone H4 tail increases nucleosome occupancy at the GAL1 promoter (7). We sought to determine whether a similar effect occurred over the entire GAL1 open reading frame, as with cks1, cdk1, and proteasome mutants. Histone H3 chromatin immunoprecipitations were carried out for wild-type, histone H3 tail deletion, and histone H4 tail deletion strains under uninduced (raffinose-containing medium) and induced conditions (galactose present) and analyzed with primers spanning the GAL1 promoter and open reading frame (Fig. 5). As with cks1, cdc28, and proteasome mutations, the histone H4 tail deletion strain had a statistically significant elevated density of histone under induced conditions, indicative of increased nucleosome density across the GAL1 open reading frame, whereas the histone H3 tail deletion strain did not. Thus, the histone H4 tail deletion confers a similar chromatin phenotype to cks1, cdk1, and proteasome mutants at the GAL1 locus and may directly impair the ability to evict nucleosomes upon transcriptional induction.
FIG. 5.
Deletion of the histone H4 amino-terminal tail confers the same chromatin phenotype as deletion of Cks1 does. Histone H3 chromatin immunoprecipitations across the GAL1 promoter and ORF upon induction comparing the wild-type (Wt) strain and strains in which the histone 3 or histone 4 tail was deleted (ΔΗ3 tail or ΔH4 tail, respectively). Primer sets and normalization are as described in the legend to Fig. 1. Values are means ± SDs (error bars). Asterisks indicate statistically significant differences (Student's t test) when comparing ΔΗ3 tail and ΔH4 tail strains 60 min after the addition of galactose. P values are provided in Table 3. PCR primer sequences are provided in Table 2.
The histone H4 tail is required for transcription-associated recruitment of Cks1 and the proteasome to the GAL1 gene.
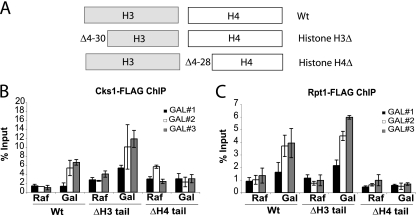
On the basis of the phenotypic similarities between the cks1/cdk1/proteasome mutants and histone H4 tail deletion mutants, we speculated that the histone H4 tail might be important for recruitment of Cks1, Cdk1, and proteasomes to chromatin. Therefore, we carried out chromatin immunoprecipitation experiments to determine the histone H3 and H4 tail requirements for Cks1 and proteasome recruitment, respectively (Fig. 6). The histone tail mutant genotypes are shown schematically in Fig. 6A. Whereas histone H3 tail deletion mutants were as competent as wild-type cells to recruit Cks1 and the proteasome under GAL1 induction conditions, histone H4 tail deletion mutants were completely defective in recruiting Cks1 and the proteasome (Fig. 6B). Therefore, Cks1 and the proteasome (and most likely Cdk1) are recruited to chromatin in a manner dependent on the amino-terminal tail of histone H4.
FIG. 6.
Recruitment of Cks1 and the proteasome depend on the histone H4 amino-terminal tail. (A) Schematic of the histone genotypes used. (B and C) Chromatin immunoprecipitation of endogenously tagged Cks1 (B) and Rpt1 (C) upon GAL1 induction. Primer sets correspond to GAL#1, GAL#2, and GAL#3 in Fig. 1. Signal is presented as a percentage of the signal obtained from input chromatin. Values are means ± SDs (error bars). PCR primer sequences are provided in Table 2.
Cdk1 and Cks1 bind specifically to the histone H4 tail.
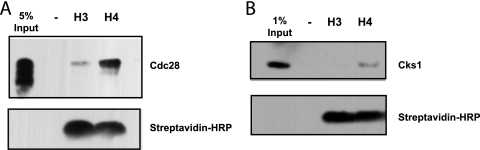
In order to determine whether recruitment of Cks1 and the proteasome are mediated through direct interaction with the histone H4 tail, synthetic peptides corresponding to the histone H3 and H4 tails, respectively, were linked to streptavidin beads via a C-terminal biotin moiety. After incubation with purified Cks1, Cdk1, or Cks1-Cdk1 heterodimers, the fraction retained on the beads after washing was determined by SDS-PAGE followed by Western blotting. As can be seen in Fig. 7 A, Cdk1 was retained on beads containing the H4 tail peptide, but much less efficiently on beads containing the H3 tail peptide. This result was reproducible using yeast Cdk1 purified either from yeast or E. coli. Purified Cks1 also bound preferentially to the histone H4 tail (Fig. 7B). Surprisingly, though, Cks1-Cdk1 heterodimers did not interact directly and specifically with the histone H4 amino-terminal tail under the conditions employed. These data suggest that Cdk1 and Cks1 might assemble onto the histone H4 tail independently prior to interacting with each other to form a high-affinity complex. Alternatively, additional factors or histone tail modifications are necessary to efficiently load Cdk1-Cks1 dimers. This might also explain the relatively low efficiency of binding of Cks1 and Cdk1. It has previously been shown that Cdk1 and Cks1 interact physically with the proteasome (15). Thus, it is possible that proteasomes are required to reconstitute efficient binding of the complete complex.
FIG. 7.
Cdk1 and Cks1 bind directly to the histone H4 amino-terminal tail. (A) FLAG-tagged yeast Cdk1 (Cdc28) expressed and purified from E. coli was incubated with peptides corresponding to the biotinylated histone H3 and histone H4 amino-terminal tails bound to streptavidin beads or with unbound streptavidin beads. Eluates subjected to SDS-PAGE and blotting were assayed for FLAG-Cdk1 using anti-FLAG antibody and for biotinylated peptide using streptavidin-HRP. Lane −, no input and no histone H3 or H4 added (negative control). (B) Cks1 purified from E. coli was incubated and analyzed as described above for panel A using anti-Cks1 antibodies.
The protease activity of the proteasome is not required for transcription-associated nucleosome reduction.
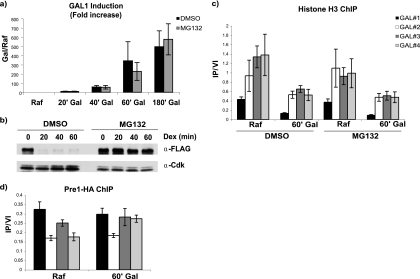
Although it has been shown that transcriptional elongation of the GAL1 gene does not depend on proteolytic function of the proteasome (5), this has not yet been established for nucleosome eviction per se. The role of the proteasome in transcription-associated nucleosome eviction could be a function of direct proteolysis of histones by protease activities of the 20S subunit of the proteasome. Alternatively, mechanical energy generated by the ATPases of the 19S regulatory subunit might be responsible for removing nucleosomes. Indeed, there are conflicting data in the literature concerning whether the protease activity of the proteasome is required for GAL1 transcription at all (21, 26). We therefore determined whether inhibition of the protease activity of the proteasome would have an effect on the transcription-associated depletion of nucleosomes at the GAL1 locus. The cells were treated with MG132 in DMSO or an equivalent volume of DMSO, and the induction of GAL1 was determined after a shift from raffinose-containing medium to galactose-containing medium (Fig. 8 a). Under these conditions, treatment with MG132 had little, if any, effect on induction of GAL1, as in both instances, a 500- to 600-fold induction was observed after 3 h. To ensure that the procedures used were effective at inhibiting the proteasome, a congenic strain containing a CLN2 gene under the control of the GAL1 promoter (13) was shifted from galactose-containing medium to dextrose-containing medium to terminate Cln2 synthesis in the presence of MG132 (Fig. 8b). Without MG132, Cln2, an unstable protein due to proteasomal turnover (38), decreased to basal levels in significantly less than 20 min. In the presence of MG132, however, turnover of Cln2 was barely detectable, consistent with efficient inhibition of proteasomal protease activity. To determine whether inhibition with proteasomal proteases affects transcription-associated nucleosome depletion, histone H3 ChIP was carried out across the GAL1 promoter and ORF in the presence and absence of MG132 (Fig. 8c). Inhibition of proteasomal protease activity using MG132 had no effect on reduction of nucleosome density either at the GAL1 promoter or ORF regions. This then raises the question of whether the 20S subunit of the proteasome is even recruited to the GAL1 promoter and ORF during induction. We have previously shown that Rpt1, a component of the 19S subunit, is recruited in a transcription-dependent manner across the GAL1 promoter and ORF (41). Chromatin immunoprecipitations targeting the 20S subunit Pre1 were therefore carried out before and after induction of GAL1 (Fig. 8d). Although immunoprecipitation of hemagglutinin-tagged Pre1 (HA-Pre1) was efficient based on Western blotting, GAL1 locus DNA precipitation was barely detectable above background (data not shown). However, there was a small reproducible induction-dependent increase in Pre1 binding to the 3′ end of the gene, as has been reported previously (8). Therefore, it is unlikely that functions of the proteasome associated with nucleosome eviction during transcription depend on the protease activities of the proteasome.
FIG. 8.
Transcriptional induction and nucleosome depletion at the GAL1 locus do not require the protease activities of the 20S proteasome. (a) Real-time PCR analysis of GAL1 induction over a 180-min time course in the presence and absence of the proteasome inhibitor MG132. (b) Analysis of turnover of the unstable protein Cln2 using the same conditions and genetic background used in panel a. FLAG-tagged Cln2 (FLAG-Cln2) under the control of the GAL1 promoter was analyzed by Western blotting after promoter shutoff by the addition of dextrose at time zero. FLAG-Cln2 was detected using anti-FLAG antibody (α-FLAG), and Cdk1, used as a loading control, was detected using anti-PSTAIRE peptide antibody. Note that smaller amounts of the MG132-treated samples were loaded so as not to produce an excessive signal. (c) Histone H3 chromatin immunoprecipitations across the GAL1 promoter and ORF in the presence and absence of MG132. The cells were grown in raffinose (Raf) and then induced by the addition of galactose(Gal) for 60 min. (d) Pre1-HA chromatin immunoprecipitations across the GAL1 promoter and ORF before and after induction. Anti-HA antibody was used to precipitate HA-tagged Pre1, a subunit of the 20S proteasome particle. Values are means ± SDs (error bars).
DISCUSSION
The function of Cdk1/Cks1 and the proteasome is to reduce nucleosome density.
In eukaryotic organisms, genomic DNA is packaged by winding it around histone octamers to produce chromatin. Each histone-DNA unit is referred to as a nucleosome (28). The existence of DNA as chromatin poses a number of topological problems for enzymes that must modify, transcribe, replicate, or repair DNA. In general, it is thought that nucleosomes restrict access to DNA and therefore must be moved or temporarily disassembled in order for the many facets of DNA metabolism to be carried out (30). It is therefore not surprising that a number of complexes capable of context-specific remodeling of chromatin have been identified.
We have shown here that the 19S subunit of the proteasome in conjunction with Cks1 and Cdk1 functions in a chromatin remodeling capacity. Specifically, proteasomes are required to maintain an appropriate density of nucleosomes on transcriptionally induced chromatin. It has been shown in several studies that nucleosomes are evicted and their density reduced upon gene induction in yeast, presumably to facilitate movement of transcription complexes (18). Taken together, these findings suggest that the role of Cdk1-Cks1 is to recruit 19S proteasomes to chromatin in order to lower nucleosome density, thereby facilitating transcriptional initiation and elongation. Surprisingly, this function has been shown not to require the kinase activity of Cdk1 (41). Since many mutations that impair transcription initiation result in failure to reduce nucleosome density (4, 17, 27, 34, 35, 42), the fact that cks1, cdk1, and proteasome mutants are defective in this capacity does not mean per se that the encoded proteins have a direct role in chromatin remodeling. However, the observation that reducing nucleosome density in a Cks1-independent manner allows efficient signal-dependent transcriptional induction of the PHO5 gene in strains with CKS1 deleted or carrying CDK1 (cdc28-1N) or RPT6 (sug1-20) mutations strongly supports this conclusion. One unresolved question is the manner in which proteasomal reduction of nucleosome density occurs. Nucleosome particles might simply be disassembled via the protein unfolding activities of the 19S subunit ATPases of the proteasome regulatory cap, similar to the unfolding of ubiquitylated proteins prior to degradation. Alternatively, the mechanical energy potentially generated by ATP hydrolysis might be utilized in a novel fashion in this context. The assumption that protease activity of the proteasome is not involved in transcription-associated regulation of nucleosome density is based both on the lack of an effect of the proteasome inhibitor MG132 (26) (Fig. 8) and a failure to actively recruit 20S subunits to the GAL1 promoter and most of the ORF in response to induction, although 20S subunits are found associated with the GAL1 gene (36). Previous in vitro work and experiments with MG132 suggest that protease activity of the proteasome is not required for efficient transcriptional elongation (6, 26). It is therefore likely that the 19S proteasomal function required for transcriptional elongation and that identified in that work is the eviction of nucleosomes. At this time, it remains to be determined whether the entire 19S subunit is recruited or whether this transcriptional function is carried out by a subcomplex known as APIS (named APIS for AAA ATPase proteins independent of 20S) (9).
Cdk1-Cks1 and proteasome affect nucleosome density both before and after induction.
A surprising result of our study is that even though the phenotype of cdk1, cks1, and proteasomal ATPase mutations is exhibited as a defect in induction of specific genes and these proteins are recruited to ORFs in an induction-specific manner, the same mutations confer a defect in nucleosome density at the same loci, even prior to induction. In cks1, cdk1, and proteasome ATPase mutants, nucleosome density is somewhat higher than normal at the GAL1 ORF under noninducing conditions. However, this difference may still be due to transcriptional differences, as these mutants exhibit a reduced level of basal transcription under noninducing conditions (41). The histone H4 tail deletion mutant did not exhibit this phenotype, possibly because survival without the histone H4 tail requires compensatory adjustments in chromatin metabolism. Under inducing conditions, nucleosome density is partially reduced, at least for the cks1 and proteasomal mutants. However, the cdc28-1N mutant of CDK1 consistently showed little or no reduction in histone density upon the addition of galactose and higher nucleosome density under noninduced conditions. Consistent with this, we found that Cdk1 bound to unmodified histone H4 tails with greater efficiency than Cks1 in vitro. Cks1 may serve as a facilitating cofactor. Alternatively, one or more parallel pathways regulating nucleosome density in the context of gene induction may require Cdk1, only one of which may also require Cks1. For the sug1-20 mutation of RPT6 encoding one of the proteasomal ATPases, there was also a partial decrease in nucleosome density upon GAL1 induction. However, since this is a temperature-sensitive mutation, the partial reduction in nucleosome density might be a result of incomplete inactivation of the gene product at 30°C, which is a semipermissive temperature. Interestingly, the chloroquine gel experiments showed a significant plasmid-wide increase in nucleosome density for the cks1 and cdc28-1N mutations (not shown by the sug1-20 mutation). Since GAL1 makes up only a small part of the plasmid, these results suggest a more global function in regulation of nucleosome density for Cks1 and Cdc28, possibly not shared by Rpt6, although further analysis will be required to verify this. In any case, the results reported here suggest that gene-specific recruitment of Cdk1-Cks1-proteasome complexes upon transcriptional induction is required for achieving a nucleosome density sufficiently low for efficient transcription.
Cdk1-Cks1-proteasome function and the histone H3 and H4 amino-terminal tails.
Phenotypic and physical interaction studies suggest that the Cdk1-Cks1-proteasome complex is recruited to chromatin via the amino-terminal tail of histone H4. Since the Cdk1-Cks1-proteasome complex and histone H4 tails presumably function in the same pathway, their individual mutant phenotypes should not be additive and therefore not synthetically lethal. This was found to be true for the sug1-20 proteasomal ATPase mutation, which was synthetically lethal with a histone H3 tail deletion mutant, but not with a histone H4 tail deletion mutant (data not shown). However, surprisingly, we found that mutation of cks1 is synthetically lethal in the context of both the histone H3 and H4 tail deletion mutants (data not shown). This is consistent with the earlier observation that a cdk1 (cdc28) mutation is also synthetically lethal with both histone H3 and H4 tail deletions (23). These results suggest that whereas the Cdk1-Cks1-proteasome pathway acts directly through the histone H4 tail, Cdk1 and Cks1 may also function, albeit indirectly, in the histone H3 tail pathway. Alternatively, these synthetic lethalities may result from mechanisms unrelated to chromatin regulatory functions of Cdk1-Cks1. Indeed, previous genetic analysis has suggested that Cdk1 kinase activity may be compromised in the context of the histone H3 and H4 tail deletions, in that mutations that decrease Cdk1 kinase activity are also synthetically lethal in combination with the histone H3 and H4 tail deletions (23). One explanation, consistent with these data, is that histone tail deletions trigger a checkpoint that delays the G2/M transition via inhibition of Cdk1 kinase activity. Since histone tail deletions have been shown to impair assembly of nucleosomes (20), this putative checkpoint may be sensitive to the efficiency of nucleosome assembly during S phase. Compromising either Cdk1 activity or Cks1 function may render cells incapable of recovering from checkpoint-mediated arrest. This would suggest that Cks1 is required for maintaining Cdk1-kinase activity or reversing Cdk1 inhibition in the context of this putative chromatin assembly checkpoint. Consistent with this interpretation, CKS1 was originally isolated as a high-copy suppressor of a TS cdk1 (cdc28-4) mutation (10), suggesting that Cks1 can enhance Cdk1 kinase activity. Future studies should determine whether a Cdk1-Cks1-based nucleosome checkpoint exists and how it functions.
Acknowledgments
We thank Michael Grunstein (UCLA) and David Allis (Rockefeller University) for strains and plasmids and Rob de Bruin (The Scripps Research Institute) for help with optimizing chromatin immunoprecipitation protocols.
S.C. and V.Y. were supported by fellowships from the Lance Armstrong Foundation and the Wellcome Trust, respectively. This work was supported by NIH grant R37 GM38328 to S.I.R.
Footnotes
Published ahead of print on 20 September 2010.
REFERENCES
- 1.Almer, A., H. Rudolph, A. Hinnen, and W. Horz. 1986. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5:2689-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheung, W. L., F. B. Turner, T. Krishnamoorthy, B. Wolner, S. H. Ahn, M. Foley, J. A. Dorsey, C. L. Peterson, S. L. Berger, and C. D. Allis. 2005. Phosphorylation of histone H4 serine 1 during DNA damage requires casein kinase II in S. cerevisiae. Curr. Biol. 15:656-660. [DOI] [PubMed] [Google Scholar]
- 3.Durrin, L. K., R. K. Mann, P. S. Kayne, and M. Grunstein. 1991. Yeast histone H4 N-terminal sequence is required for promoter activation in vivo. Cell 65:1023-1031. [DOI] [PubMed] [Google Scholar]
- 4.Ercan, S., M. J. Carrozza, and J. L. Workman. 2004. Global nucleosome distribution and the regulation of transcription in yeast. Genome Biol. 5:243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferdous, A., F. Gonzalez, L. Sun, T. Kodadek, and S. A. Johnston. 2001. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell 7:981-991. [DOI] [PubMed] [Google Scholar]
- 6.Ferdous, A., T. Kodadek, and S. A. Johnston. 2002. A nonproteolytic function of the 19S regulatory subunit of the 26S proteasome is required for efficient activated transcription by human RNA polymerase II. Biochemistry 41:12798-12805. [DOI] [PubMed] [Google Scholar]
- 7.Fisher-Adams, G., and M. Grunstein. 1995. Yeast histone H4 and H3 N-termini have different effects on the chromatin structure of the GAL1 promoter. EMBO J. 14:1468-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillette, T. G., F. Gonzalez, A. Delahodde, S. A. Johnston, and T. Kodadek. 2004. Physical and functional association of RNA polymerase II and the proteasome. Proc. Natl. Acad. Sci. U. S. A. 101:5904-5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonzalez, F., A. Delahodde, T. Kodadek, and S. A. Johnston. 2002. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296:548-550. [DOI] [PubMed] [Google Scholar]
- 10.Hadwiger, J. A., C. Wittenberg, M. D. Mendenhall, and S. I. Reed. 1989. The Saccharomyces cerevisiae CKS1 gene, a homolog of the Schizosaccharomyces pombe suc1+ gene, encodes a subunit of the Cdc28 protein kinase complex. Mol. Cell. Biol. 9:2034-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershko, A. 1983. Ubiquitin: roles in protein modification and breakdown. Cell 34:11-12. [DOI] [PubMed] [Google Scholar]
- 12.Hilt, W. 2004. Targets of programmed destruction: a primer to regulatory proteolysis in yeast. Cell. Mol. Life Sci. 61:1615-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsiung, Y. G., H. C. Chang, J. L. Pellequer, R. La Valle, S. Lanker, and C. Wittenberg. 2001. F-box protein Grr1 interacts with phosphorylated targets via the cationic surface of its leucine-rich repeat. Mol. Cell. Biol. 21:2506-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 15.Kaiser, P., V. Moncollin, D. J. Clarke, M. H. Watson, B. Bertolaet, S. I. Reed, and E. Bailly. 1999. Cyclin-dependent kinase and Cks/Suc1 interact with the proteasome in yeast to control proteolysis of M-phase targets. Genes Dev. 13:1191-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, U. J., M. Han, P. Kayne, and M. Grunstein. 1988. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 7:2211-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, Y., N. McLaughlin, K. Lindstrom, T. Tsukiyama, and D. J. Clark. 2006. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol. Cell. Biol. 26:8607-8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 19.Lee, D., E. Ezhkova, B. Li, S. G. Pattenden, W. P. Tansey, and J. L. Workman. 2005. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell 123:423-436. [DOI] [PubMed] [Google Scholar]
- 20.Ling, X., T. A. Harkness, M. C. Schultz, G. Fisher-Adams, and M. Grunstein. 1996. Yeast histone H3 and H4 amino termini are important for nucleosome assembly in vivo and in vitro: redundant and position-independent functions in assembly but not in gene regulation. Genes Dev. 10:686-699. [DOI] [PubMed] [Google Scholar]
- 21.Lipford, J. R., G. T. Smith, Y. Chi, and R. J. Deshaies. 2005. A putative stimulatory role for activator turnover in gene expression. Nature 438:113-116. [DOI] [PubMed] [Google Scholar]
- 22.Liu, C., J. Apodaca, L. E. Davis, and H. Rao. 2007. Proteasome inhibition in wild-type yeast Saccharomyces cerevisiae cells. Biotechniques 42:158-162. [DOI] [PubMed] [Google Scholar]
- 23.Ma, X. J., Q. Lu, and M. Grunstein. 1996. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10:1327-1340. [DOI] [PubMed] [Google Scholar]
- 24.Mann, R. K., and M. Grunstein. 1992. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 11:3297-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris, M. C., P. Kaiser, S. Rudyak, C. Baskerville, M. H. Watson, and S. I. Reed. 2003. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 423:1009-1013. [DOI] [PubMed] [Google Scholar]
- 26.Nalley, K., S. A. Johnston, and T. Kodadek. 2006. Proteolytic turnover of the Gal4 transcription factor is not required for function in vivo. Nature 442:1054-1057. [DOI] [PubMed] [Google Scholar]
- 27.Nourani, A., R. T. Utley, S. Allard, and J. Cote. 2004. Recruitment of the NuA4 complex poises the PHO5 promoter for chromatin remodeling and activation. EMBO J. 23:2597-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pederson, D. S., F. Thoma, and R. T. Simpson. 1986. Core particle, fiber, and transcriptionally active chromatin structure. Annu. Rev. Cell Biol. 2:117-147. [DOI] [PubMed] [Google Scholar]
- 29.Pickart, C. M., and R. E. Cohen. 2004. Proteasomes and their kin: proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5:177-187. [DOI] [PubMed] [Google Scholar]
- 30.Pollard, K. J., and C. L. Peterson. 1998. Chromatin remodeling: a marriage between two families? Bioessays 20:771-780. [DOI] [PubMed] [Google Scholar]
- 31.Richardson, H. E., C. Wittenberg, F. Cross, and S. I. Reed. 1989. An essential G1 function for cyclin-like proteins in yeast. Cell 59:1127-1133. [DOI] [PubMed] [Google Scholar]
- 32.Rubin, D. M., O. Coux, I. Wefes, C. Hengartner, R. A. Young, A. L. Goldberg, and D. Finley. 1996. Identification of the gal4 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature 379:655-657. [DOI] [PubMed] [Google Scholar]
- 33.Russell, S. J., U. G. Sathyanarayana, and S. A. Johnston. 1996. Isolation and characterization of SUG2. A novel ATPase family component of the yeast 26 S proteasome. J. Biol. Chem. 271:32810-32817. [DOI] [PubMed] [Google Scholar]
- 34.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwabish, M. A., and K. Struhl. 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 27:6987-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sikder, D., S. A. Johnston, and T. Kodadek. 2006. Widespread, but non-identical, association of proteasomal 19 and 20 S proteins with yeast chromatin. J. Biol. Chem. 281:27346-27355. [DOI] [PubMed] [Google Scholar]
- 37.Smith, D. M., N. Benaroudj, and A. Goldberg. 2006. Proteasomes and their associated ATPases: a destructive combination. J. Struct. Biol. 156:72-83. [DOI] [PubMed] [Google Scholar]
- 38.Willems, A. R., S. Lanker, E. E. Patton, K. L. Craig, T. F. Nason, N. Mathias, R. Kobayashi, C. Wittenberg, and M. Tyers. 1996. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell 86:453-463. [DOI] [PubMed] [Google Scholar]
- 39.Wolf, D. H., and W. Hilt. 2004. The proteasome: a proteolytic nanomachine of cell regulation and waste disposal. Biochim. Biophys. Acta 1695:19-31. [DOI] [PubMed] [Google Scholar]
- 40.Xu, Q., R. A. Singer, and G. C. Johnston. 1995. Sug1 modulates yeast transcription activation by Cdc68. Mol. Cell. Biol. 15:6025-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu, V. P., C. Baskerville, B. Grunenfelder, and S. I. Reed. 2005. A kinase-independent function of Cks1 and Cdk1 in regulation of transcription. Mol. Cell 17:145-151. [DOI] [PubMed] [Google Scholar]
- 42.Zhao, J., J. Herrera-Diaz, and D. S. Gross. 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 25:8985-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]