Abstract
The mechanisms by which signals are transmitted across the plasma membrane to regulate signaling are largely unknown for receptors with single-pass transmembrane domains such as the epidermal growth factor receptor (EGFR). A crystal structure of the extracellular domain of EGFR dimerized by epidermal growth factor (EGF) reveals the extended, rod-like domain IV and a small, hydrophobic domain IV interface compatible with flexibility. The crystal structure and disulfide cross-linking suggest that the 7-residue linker between the extracellular and transmembrane domains is flexible. Disulfide cross-linking of the transmembrane domain shows that EGF stimulates only moderate association in the first two α-helical turns, in contrast to association throughout the membrane over five α-helical turns in glycophorin A and integrin. Furthermore, systematic mutagenesis to leucine and phenylalanine suggests that no specific transmembrane interfaces are required for EGFR kinase activation. These results suggest that linkage between ligand-induced dimerization and tyrosine kinase activation is much looser than was previously envisioned.
Fundamental to cellular physiology is the ability to transmit extracellular signals across the cell membrane to trigger intracellular responses. Although the extracellular and intracellular portions of cell surface receptors are responsible for detecting ligands and initiating signal cascades, respectively, transmembrane (TM) domains are thought to play critical roles by specifically associating and propagating signals across the phospholipid bilayer. However, the mechanisms by which single-pass TM domains associate and conduct signals are poorly understood.
The epidermal growth factor receptor (EGFR) is the prototypical type I TM receptor tyrosine kinase. EGFR and related members of the ErbB family—ErbB2, ErbB3, and ErbB4—contain a glycosylated extracellular ligand binding domain; a single-pass TM domain; and intracellular juxtamembrane, tyrosine kinase, and autophosphorylation domains. The extracellular domain of EGFR binds polypeptide growth factor ligands, such as epidermal growth factor (EGF), to stimulate an array of intracellular signaling cascades that regulate normal and oncogenic cellular growth and proliferation (3, 17, 36). In one model of growth factor-dependent EGFR activation, ligand binding promotes receptor dimerization and activation of intracellular protein tyrosine kinase activity (35); other models suggest that receptors are predimerized on the cell surface and ligand binding alters the equilibrium between inactive and active dimeric (or higher-order oligomeric) configurations (9, 29).
Structural mechanisms of growth factor-mediated receptor dimerization and allosteric kinase domain activation have been proposed from recent crystal structures of isolated extracellular ligand binding domains (7) and intracellular tyrosine kinase domains (37). The orientation between the four extracellular domains is dramatically altered upon ligand binding, which frees interfaces that are masked in tethered, unliganded monomers to mediate dimer formation (7). Furthermore, an unusual asymmetric interface between two kinase domain monomers is linked to rearrangement of the kinase site to the active conformation (37). However, neither the position of the last extracellular domain, domain IV, nor association between the TM domains is well-defined experimentally in liganded receptors. The approximate location of domain IV has been suggested by models based on the orientation between domains III and IV in unliganded monomers (7, 12) and two-dimensional negative-stain electron microscopy (EM) averages (27); however, the position of domain IV in the liganded dimer has not been determined in previous crystal structures (13, 30). Thus, it is not known how the extracellular domain positions the TM domains for transmembrane signaling.
Several lines of evidence suggest that the TM domain contributes directly to receptor dimerization and signaling. The neu oncogene encodes a Val → Glu substitution in the TM domain of ErbB2 that results in constitutive activation (34). Recombinant EGFR fragments consisting of the extracellular and TM domains have a 105-fold higher affinity for dimerization than the isolated soluble extracellular domains (31). The TM domains of all four ErbB family members self-associate when expressed in bacterial inner membranes (26). A dimeric structure for isolated ErbB2 TM peptides in bicelles has been defined by nuclear magnetic resonance (NMR) imaging (4). However, ErbB2 does not bind ligand and does not physiologically homodimerize (17). Moreover, different ErbB family member TM domains utilize potentially distinct GxxxG sequence motifs to dimerize, as shown with fusion proteins in bacterial membranes (26). However, it is not clear how the TM domains contribute to dimerization and signaling in intact receptors on the cell surface.
Here, we characterize the structural basis for EGFR transmembrane signaling. An improved crystal structure of the EGF-bound EGFR extracellular domain resolves domain IV in electron density maps and identifies a small domain IV dimerization interface, the mutation of which does not abolish signaling. The crystal structure and disulfide cross-linking demonstrate a flexible, dimeric linker between the extracellular and transmembrane domains. EGF-induced dimerization of the TM domains involves an interface far less extensive than that found in two receptors that dimerize in the absence of activation. Furthermore, mutagenesis shows that no unique interface is required for transmembrane signaling. Thus, we propose that signal transmission through the EGFR is communicated much more loosely than was previously thought.
MATERIALS AND METHODS
Refinement of EGFR extracellular domain crystal structure.
Calculation of a simulated annealing-composite omit map with the native 3.3-Å diffraction data set and coordinates of residues 2 to 512 (30) visualized the electron density for domain IV. Domain IV (residues 480 to 614) from the unliganded, tethered conformation (Protein Data Bank [PDB] accession number 1YY9) (24) was placed in density using Coot tools (11). The model was subjected to rigid-body and group B factor refinement, energy minimization, and simulated annealing using the CNS program (6). Subsequently, multiple rounds of model building with Coot tools, refinement with TLS, and individual isotropic ADP refinement with the PHENIX program (1) were carried out. To guide rebuilding, domains from high-resolution EGFR structures were superimposed, and alterations were made when these suggested better alternatives for fitting the density. Furthermore, the MOLPROBITY server was used to find clashes and rotamer and backbone outliers that could be repaired and side chains that could be flipped. During refinement, Noncrystallographic symmetry (NCS) and some secondary-structure main-chain hydrogen-bonding restraints were applied; these restraints were released in the final round of refinement. The final model contains 1,314 amino acids (2 EGFR extracellular domains plus 2 EGF molecules), 12 N-acetylglucosamines, 5 polyethylene glycols, and 24 water molecules.
Reagents.
Recombinant human EGF was from PeproTech, Inc. (Rocky Hill, NJ). Antibodies to protein C epitope and to phosphotyrosine (clone 4G10) were from Roche Applied Science and Millipore, respectively. The anti-EGFR monoclonal antibody (MAb) 528 (Ab528) hybridoma cell line was provided by ATCC, and antibody was purified by protein G affinity chromatography from culture supernatant. Anti-EGFR antibodies R-1 and 1005 were from Santa Cruz.
Construction of EGFR mutants.
Full-length EGFR cDNAs were in the ExpressTag-1 (ET-1) mammalian expression vector, which contains C-terminal protein C epitope, His6, and streptavidin binding peptide (SBP) tags (27). Single cysteine substitutions were introduced into the EGFR C3M and C6M mutants, in which 3 and 6 endogenous cysteine residues in the cytoplasmic region were replaced by serine or alanine, respectively (27). Leucine and phenylalanine substitutions in TM were in the EGFR wild-type background. Site-directed mutagenesis using a QuikChange II XL site-directed mutagenesis kit (Stratagene) was confirmed by DNA sequencing.
Generation of Ba/F3 stable transfectants.
Ba/F3 cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 10% WEHI-3 cell conditioned medium as a source of IL-3. EGFR cDNA was transfected into Ba/F3 cells by electroporation (33). Transfected cells were selected in culture medium containing 1 mg/ml G418. Populations of EGFR-expressing cells were further selected by fluorescence-activated cell sorting (FACS) by cell surface staining with anti-EGFR antibody 528 and phycoerythrin-conjugated secondary antibody. Sorted cells were expanded and maintained in culture medium supplemented with 1 mg/ml G418.
Transient transfection of HEK 293T cells.
HEK 293T cells (16) were maintained in Dulbecco modified Eagle medium supplemented with 10% FBS and 4 mM l-glutamine. Cells were seeded in 12-well plates at 3 × 105 cells/well 24 h prior to transfection. Cultures were transfected with 0.5 μg of DNA/well using a 10-fold greater weight of linear polyethyleneimine (PEI) or 2 μl of Lipofectamine 2000 reagent (Invitrogen). The amount of DNA was pretitrated to give optimal EGF-stimulated activation. After incubation at 37°C for 4 h, culture medium was changed to ExCell 293 serum-free medium (SAFC Biosciences), and the cells were cultured for an additional 20 h.
EGF stimulation, lysate preparation, and Western blot analysis.
Ba/F3 transfectants were collected, washed, and resuspended at 2 × 106 cells/ml in RPMI 1640 medium without serum or interleukin-3 (IL-3) and plated in 12- or 6-well plates. After 4 h at 37°C (5% CO2), cells were treated with or without EGF at a final concentration of 100 nM for 5 min at 37°C; HEK 293T transient transfectants were treated with or without EGF 24 h after transfection. Cells were then washed 2 times with cold phosphate-buffered saline (PBS) and solubilized in Triton X-100 lysis buffer (25 mM HEPES, pH 7.4, 150 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM sodium orthovanadate, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1× complete protease inhibitor cocktail [Roche]) containing 5 mM β-mercaptoethanol for reducing SDS-polyacrylamide gel electrophoresis (PAGE) or 10 mM N-ethylmaleimide (NEM) for nonreducing SDS-PAGE.
Following SDS-PAGE, the proteins were blotted to a PVDF membrane using a Trans-Blot SD semidry transfer cell (Bio-Rad), and the membrane was incubated with appropriate primary antibodies and horseradish peroxidase (HRP)-conjugated secondary antibody. Blots were developed with chemiluminescence substrate (ECL detection reagents; GE Healthcare) and exposed to film (HyBlot CL; Denville Scientific, Inc.) or were analyzed on a chemiluminescence imager (LAS-3000; Fujifilm). The chemiluminescence intensity of the protein bands was quantitated using Fujifilm Image Gauge software.
Cell surface disulfide cross-linking.
Cross-linking procedures for EGFR cysteine mutants followed closely those for cross-linking of integrins (38). Ba/F3 transfectants were starved of serum and IL-3 for 4 h and treated without or with EGF as described above. Cells were collected, washed with cold PBS, and resuspended in 45 μl of HS buffer (25 mM HEPES, pH 7.4, 150 mM NaCl) per 4 × 106 cells. For mutants with cysteine substitutions in the extracellular domain, Ba/F3 transfectants were lysed in Triton X-100 lysis buffer without cell permeabilization and oxidant treatment. Cross-linking was compared in the absence and presence of redox buffer (5 mM cysteamine, 1 mM cystamine, 25 mM Tris, pH 8.2, 150 mM NaCl) for 2 h at 4°C. No significant difference was found for each cysteine substitution (data not shown). For disulfide cross-linking of EGFR TM cysteine mutants, cells were pretreated with 2-bromopalmitate for 16 h to inhibit palmitylation of cysteines, permeabilized, and then treated with oxidant as described previously for GpA and integrin αIIbβ3 TM domains (38) with the following modifications. Cells were permeabilized by 2 cycles of freezing on dry ice for 10 min and thawing at room temperature. Permeabilization was monitored as uptake of trypan blue by >99% of cells. Permeabilized cells were treated with 5 μl of 10× CuSO4-o-phenanthroline (final concentrations, 500 μM Cu2+ and 2 mM o-phenanthroline) and incubated for 10 min at room temperature. Cells were solubilized by the addition of 50 μl of 2× Triton X-100 lysis buffer containing NEM for 30 min on ice. Lysates were subjected to nonreducing SDS-PAGE and Western blotting with anti-protein C monoclonal antibody to determine the extent of disulfide cross-linking. The disulfide cross-linking achieved with Cu2+-o-phenanthroline at room temperature for 10 min, with Cu2+-o-phenanthroline at 0°C for 30 min, and without Cu2+-o-phenanthroline was compared. Similar cross-linking efficiencies were achieved under the three conditions for cysteine substitution mutants I622C and A623C, which are near the extracellular face. For cysteine substitutions from T624 to A629, Cu2+-o-phenanthroline treatment at room temperature resulted in slightly higher cross-linking than that achieved at 0°C (data not shown).
Initially, a panel of reagents that included EGFR-specific antibodies 528, R-1, and 1005 or protein C antibody and streptavidin to detect C-terminal tags were tested for their ability to immunoprecipitate EGFR from radiolabeled (38) EGF-treated EGFR T624C lysates. The R-1 and 1005 antibodies and streptavidin appeared to preferentially detect dimeric EGFR, whereas Ab528 and anti-protein C MAb reacted with monomeric and dimeric EGFR equally well (data not shown). Because of its higher sensitivity, protein C antibody was chosen over Ab528 for determining the extent of disulfide cross-linking. The cross-linking efficiency determined by Western blotting with anti-protein C antibody was similar to that determined by immunoprecipitation of radiolabeled material using anti-protein C antibody or Ab528.
The disulfide cross-linking efficiency for each cysteine substitution was determined as the ratio of dimeric EGFR to total EGFR (monomer plus dimer) detected by Western blotting of detergent cell extracts. Since intracellular EGFR is inaccessible to EGF treatment, it was subtracted from the amount of monomeric EGFR. The amount of intracellular receptor was estimated by cell surface staining of transfectants with Ab528, extensive washing to remove unbound antibody, and preparation of detergent-solubilized cell lysates. Antibody-bound EGFR (representing the cell surface population) was depleted by three rounds of incubation with immobilized protein G; the EGFR retained in the detergent lysate was considered to represent the intracellular receptor pool. An average of 21% of the EGFR in 3 representative Ba/F3 stable cell lines was found to be intracellular and was used to adjust the disulfide cross-linking efficiency.
Flow cytometry.
Cells were incubated with EGFR-specific Ab528 at 10 μg/ml in PBS containing 1% FBS for 30 min on ice, followed by incubation with phycoerythrin-conjugated secondary antibody. After the cells were washed, they were resuspended in PBS and analyzed by flow cytometry on a FACScan flow cytometer (Becton Dickinson). EGFR cell surface expression levels were expressed as the mean fluorescence intensity of cells.
Rosetta-based modeling of EGFR TM domains.
EGFR TM structures were calculated with Membrane Rosetta and disulfide cross-linking-based Cα-Cα distance constraints, as described previously (38). All TM residues with cross-linking efficiencies of greater than 18%, i.e., I622, A623, T624, G625, and V627, were used for calculation of Cα-Cα distance constraints. Docking and two steps of chain growing in a membrane slab environment were used. First, two ideal helices with residues I622 to L633 were docked and the 10,000 models with low all-atom energies were selected (2). Then, the 1,000 models with the lowest symmetry violations were selected, and of these, the 100 models with the lowest disulfide constraint violations were selected as starting structures for chain growing. Symmetry was not selected for in subsequent steps. In the first chain-growing step, coordinates from two C-terminal residues were kept, and the remaining N-terminal residues were rebuilt using low-resolution Monte Carlo-based peptide fragment insertion and then refined at the all-atom level; disulfide distance constraints were implemented in both the low-resolution and all-atom potentials. Of 10,000 models, 100 models were independently selected for low restraint violation and low energy scores (in this case, energy did not include the restraint violation penalty). These were used as seeds for the final step, in which only the two N-terminal residues were kept and the remaining C-terminal residues were rebuilt by chain growing. Models with the 10% lowest energy were clustered on the basis of the Cα root mean square deviation (RMSD) with a 1.0-Å cutoff for all 46 TM residues (5). The center models of the top 5 most-populated clusters were selected as representative of model 1.
Because the amount of EGF-dependent cross-linking never reached 100%, even for the Y602C in the domain IV interface, and few cross-linking restraints were available, we used a second method to obtain model 2. Cross-linking efficiency was normalized by setting the efficiency of Thr-624, the most strongly cross-linked residue, to 100%, increasing the cross-linking efficiencies of other residues proportionally, and using these values for calculation of distance restraints. The modeling procedure was as described above, except the weight of distance restraint violations was increased 5-fold. The final 5,000 models were clustered on the basis of the Cα RMSD with a 2.0-Å cutoff. The center models of the top 5 most-populated clusters were selected to be representative of model 2.
RESULTS
EGF-bound dimeric EGFR extracellular domain crystal structure containing domain IV.
Using the 3.3-Å native diffraction data set reported previously (30), we observed a clear density for domain IV in a simulated annealing-composite omit map calculated with the published coordinates of domains I to III and the beginning of domain IV (residues 2 to 512). The structure of domain IV (residues 480 to 614) from the unliganded, tethered EGFR extracellular domain (24) was placed in this density. After many cycles of refinement and rebuilding of the entire ectodomain, an improved model was obtained (Table 1). The current structure contains residues 1 to 614; residues 615 to 619 are present in the EGFR protein construct but are disordered in the crystal. The TM domain is predicted to contain 23 residues beginning at Ile-622; thus, only a 7-residue linker, residues 615 to 621, is missing from the ectodomain crystal structure.
TABLE 1.
X-ray data collection and refinement statistics
| Parameter | Data statistics | Refinement statistics |
|
|---|---|---|---|
| PDB accession no. 3NJP | PDB accession no. 1IVO | ||
| Wavelength (Å) | 1.0000 | ||
| Rsym (%)b | 7.8 (35.8)a | ||
| Resolution (Å) | 50-3.3 | 50.0-3.3 | 10.0-3.3 |
| Completeness (%) | 98.2 (92.6)a | ||
| Unique reflections | 46,667 | ||
| I/σ(I)g | 21.5 (2.9)a | ||
| Total no. of reflections | 320,883 | ||
| Rfactor/Rfree (%)c | 26.3/29.8 | 25.5/32.6 | |
| RMSD from ideality | |||
| Bond length (Å) | 0.002 (0.003)d | 0.006 | |
| Bond angles (degrees) | 0.54 (1.67)d | 1.40 | |
| Dihedrals (degrees) | 13.7 (26.3)d | 25.10 | |
| Ramachandran plot (%)e (favored/allowed/outliers) | 89.4/10.1/0.5 | 75.0/18.8/6.2 | |
| No. of B factors (protein/other/water/I/II/III/IV)f | 128/126/77/107/118/108/199 | 82/126/57/76/88/79/113 | |
| No. of atoms (protein/other/water) | 10,235/308/24 | 8,673/140/79 | |
| EGFR residue no. range | 1-614 | 2-512 | |
Values in parentheses refer to data in the highest-resolution shell.
Rsym = ΣhΣi|Ih,i − <Ih>|/ΣhΣi|Ih,i|, where <Ih> is the mean intensity of a set of equivalent reflections, h is the unique reflection index, and i is the symmetry equivalent index.
Rfactor = Σ|Fobs − Fcalc|/ΣFobs, where Fobs and Fcalc are observed and calculated structure factor amplitudes, respectively. Rfree was calculated for R factor with a random 5% subset from all reflections.
The current structure was refined against PHENIX restraints, which differ from those in CNS. Deviations from CNS restraints are shown in parentheses, for comparison to the previous structure.
Ramachandran plot was calculated with the MOLPROBITY server (http://molprobity.biochem.duke.edu).
I to IV, domains I to IV. Isotropic B factors from the current model are calculated from the anisotropic B factors that include TLS. The protein with PDB accession no. 1IVO was refined without TLS.
I/σ(I), ratio of the mean intensity of all reflections to the mean standard uncertainty of intensity of all reflections.
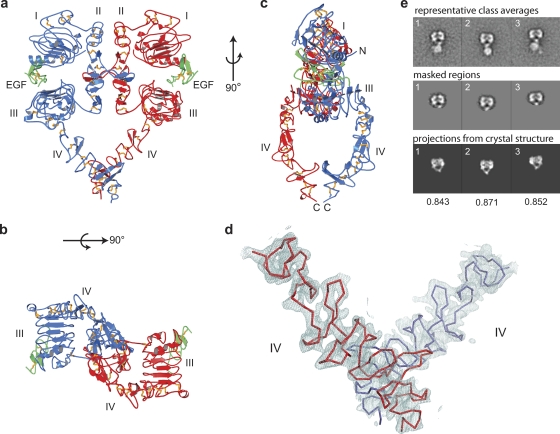
In the asymmetric unit of the crystal, two 1:1 EGF-EGFR complexes interact in a receptor-mediated dimer (Fig. 1a), as reported previously (13, 30). The overall structure is heart shaped. EGF binds between domains I and III of each monomer, and the major dimer interface is formed by domain II. Domain IV of each receptor monomer extends from the base of domain III and curves inward toward the center of the molecule (Fig. 1c), with the C termini of each domain IV converging to form the vertex at the bottom of the heart. Although the density is not as strong as it is for domains I to III, the density for domain IV clearly establishes its overall structure and orientation (Fig. 1d). Furthermore, the arrangement of domain IV in crystals correlates well with that in detergent-soluble intact receptor dimers in negative-stain EM (27) (Fig. 1e).
FIG. 1.
Structure of EGF-bound EGFR extracellular domain. (a to c) Ribbon diagrams. Different EGFR monomers are blue and red, and EGF is green. Gold sticks are disulfide bonds. (d) The 2Fobs − Fcalc (where Fobs and Fcalc are observed and calculated structure factor amplitudes, respectively) electron density map contoured at 1σ (blue mesh) is overlaid on the Cα trace of domain IV monomers in a dimer (red and blue lines). (e) Comparison to negative-stain EM class averages of intact receptors. Previously published (27) class averages of EGF-bound EGFR in dodecylmaltoside (upper panel) were masked (middle panel) and showed excellent cross-correlation with projections from the current crystal structure (bottom panel with correlation coefficients below).
The first two disulfide-linked modules of domain IV, modules 1 and 2, tuck tightly against domain III. This portion of the domain III/IV interface, including Trp-492 in module 1 of domain IV, which plugs into a hydrophobic pocket in domain III, is conserved in the I-II interface and in interfaces of related receptors (13, 14, 30). Modules 3, 4, 5, 6, and 7 of domain IV form a slender rod extending away from the much more compact domain I to III-EGF complex (Fig. 1a and 2b).
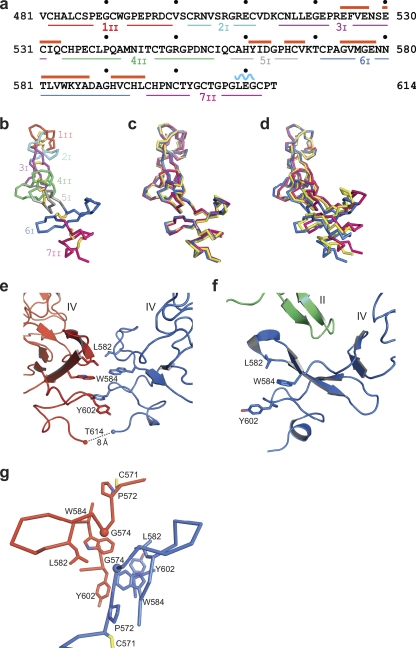
FIG. 2.
Domain IV structure. (a) Sequence of domain IV. Cysteine-rich modules 1 to 7 have subscripts denoting one (I) or two (II) disulfide bonds. The β-strand and α-helix have red and blue overlines, respectively. (b) Backbone Cα trace of domain IV with cysteine-rich modules in different colors and disulfide bonds as gold sticks. (c and d) Superpositions of domain IV from the present dimeric structure (blue) and monomeric tethered EGFR structures (red, PDB accession no. 1YY9 [24]; yellow, PDB accession no. 1NQL [12]). Superposition analyses were performed using all domain IV residues (c) or cysteine-rich module I (residues 48 to 501) (d). (e and f) Cartoon representations of the intermolecular domain IV-domain IV interface in the current dimeric structure (e) and the intramolecular domain II-domain IV interface in the monomeric structure (PDB accession no. 1YY9) (f). (g) Residues mutated in glioblastoma in relation to the domain IV interface. Gly-574 (G574V mutation) is shown as a Cα sphere, and Pro-572 (P572L mutation) is shown in stick form. Other side chains in the interface and Cys-571 are shown in stick forms.
There are no significant structural rearrangements in domain IV in the dimeric receptor compared to the structure in the monomeric receptor (Fig. 2c). However, there is gradual bending along the length of domain IV, so that when superposition is conducted on module 1 of domain IV, the domain IV C termini of the unliganded monomeric receptor (24) (Fig. 2d, red) and dimeric receptor (Fig. 2d, blue) are displaced by 10 Å, and a monomeric, tethered receptor cocrystallized with ligand at acidic pH (12) (Fig. 2d, yellow) is intermediate in position. Bending along the length of domain IV is necessary for equilibration between the monomeric, unliganded EGFR and dimeric, liganded EGFR. The same orientation as in the monomer would result in steric overlap, i.e., clashes at the domain IV-domain IV interface in the dimer. On the other hand, the same orientation as in the dimer would result in disruption of domain II-domain IV tethering interactions in the monomer.
The domain IV-domain IV interface (Fig. 2e) is modest in size, with a total solvent-accessible surface area of 434 Å2. Nonetheless, the hydrophobic and aromatic character of the interface with residues Leu-582, Trp-548, and Tyr-602 supports a structural role. These residues are distinct from those that participate in the interface of domain IV with domain II in the intramolecular tether in the monomeric receptor (Fig. 2f) (12). The domain IV dimer interface places the two C-terminal Thr-614 residues visible in density in each monomer only 8 Å apart. A 7-residue linker, N615 to S621, intervenes between the ectodomain and transmembrane domain.
Disulfide cross-linking of EGFR on cell surface.
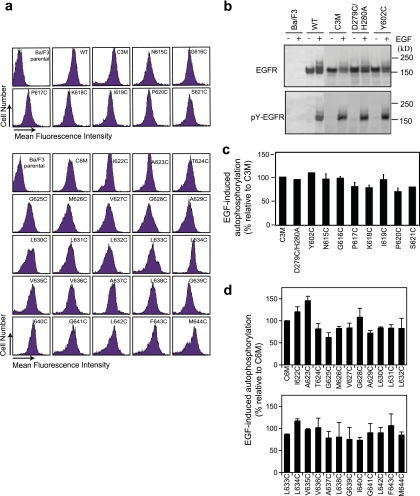
Disulfide cross-linking studies utilized full-length EGFR in which either 3 or 6 cysteines in the cytoplasmic domain were mutated to serine or alanine (C3M and C6M mutants) to eliminate background cross-linking. Transfectants were established in Ba/F3 cells, a murine bone marrow-derived IL-3-dependent cell line that does not express endogenous ErbB receptors and that, when it is transfected with EGFR, shows EGF-induced activation of the EGFR kinase and downstream pathways (33). Transfectants expressed comparable levels of EGFR on the cell surface (Fig. 3a). A 125I-labeled EGF binding assay showed that the transfectants expressed (2.2 to 4.2) × 104 ligand binding sites per cell (data not shown). All cysteine substitution mutants exhibited levels of EGF-induced autophosphorylation comparable to the level exhibited by the wild type (Fig. 3b, c, and d).
FIG. 3.
Expression and activation of EGFR cysteine mutants in Ba/F3 stable transfectants. Cysteine substitutions were on the EGFR C3M or EGFR C6M background shown in the same subpanel. (a) Cell surface expression detected with EGFR Ab528, phycoerythrin-conjugated secondary antibody, and flow cytometry. (b) Autophosphorylation of EGFR wild type (WT) and representative mutants. Lysates from transfectants were treated without or with EGF and subjected to reducing SDS-5% PAGE and Western blotting with protein C antibody (upper) or phosphotyrosine antibody 4G10 (lower). Results for the rest of the mutants are not shown. (c and d) EGF-induced autophosphorylation relative to that for background C3M and C6M EGFR constructs, respectively. Phosphorylated EGFR (pY-EGFR) and total EGFR were quantitated, and the ratio of phosphorylated EGFR/total EGFR of each mutant was normalized to that of the C3M or C6M EGFR.
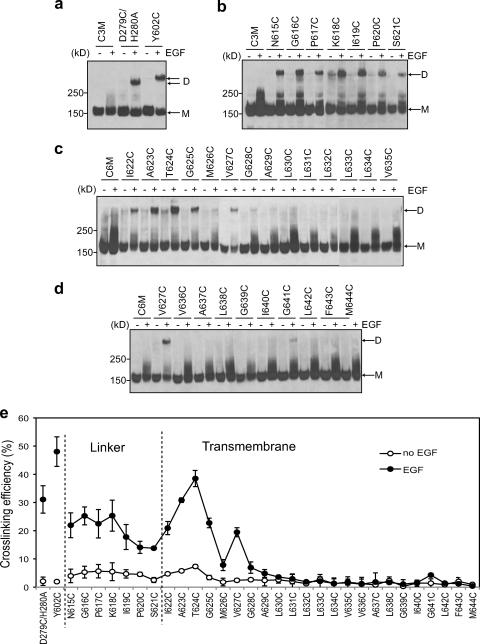
Cross-linking procedures for EGFR cysteine mutants closely followed those for integrin and GpA (38). Results for extracellular residues were similar with and without Cu(II)-o-phenanthroline or in redox buffer (data not shown); TM cysteine mutants required both freeze-thaw and Cu(II)-o-phenanthroline to achieve cross-linking that was independent of burial depth in the membrane, as previously described for integrin and GpA (38) (data not shown). Ba/F3 transfectants were treated with or without EGF for 5 min at 37°C and subjected to cross-linking, and detergent cell extracts were subjected to nonreducing SDS-PAGE and Western blotting (Fig. 4a to d). As positive controls, we introduced cysteines into the domain II dimer interface (D279C with H280A to prevent steric hindrance of cysteine cross-linking) and into the domain IV dimer interface defined here (Y602C). There was essentially no cross-linking in the absence of EGF and substantial cross-linking in the presence of EGF (Fig. 4a and e). Mobility in SDS-polyacrylamide gels showed some dependence on the distance of the cross-link from the end of the polypeptide chain, as expected from the model of proteins in SDS as extended polypeptide chain rods surrounded by SDS. Notably, cysteine substitutions in domains II and IV, which lead to 30 and 50% cross-linking upon addition of EGF (Fig. 4e) under the same conditions used for autophosphorylation assays, respectively, are fully active (Fig. 3b and c). This demonstrates that when the domain IV interface is locked in an orientation similar to that seen in crystals (Fig. 2f), the receptor has full autophosphorylation activity.
FIG. 4.
Disulfide cross-linking of EGFR cysteine mutants in Ba/F3 transfectants treated with or without EGF. Lysates were subjected to nonreducing SDS-5% PAGE and Western blotting with protein C antibody. (a) Positive controls with substitutions in the domain II (D279C/H280A) and domain IV (Y602C) interfaces; (b) linker residues; (c and d) TM domain residues; (e) cross-linking efficiency quantitated as the percentage of EGFR dimer with respect to total cell surface EGFR. Error bars represent the standard deviations of 3 independent cross-linking experiments.
Each of the 7 linker residues between domain IV and the TM domain was scanned with cysteine. EGF markedly stimulated cross-linking at each position (Fig. 4b and e). These results demonstrate that EGF-stimulated receptor dimerization brings into close proximity linker residues 615 to 621 in each monomer, consistent with the close opposition of residue 614 in the crystal structure. However, no cross-linking peaks were seen in the linker, consistent with the lack of a well-defined structure in the receptor dimer.
The entire EGFR transmembrane domain was subjected to cysteine scanning. Substantial EGF-stimulated cross-linking was observed in the first α-helical turn, with peaks being observed at Ala-623 and Thr-624 (Fig. 4c and e). Another smaller peak was observed at Val-627. However, after the end of the second helical turn, at Gly-628, essentially no EGF-stimulated cross-linking was seen, except for a very small, reproducible peak at Gly-641 (Fig. 4d and e).
The transmembrane domain cross-linking results were very surprising to us, because in both integrin αIIbβ3 and GpA, cross-linking peaks with α-helical periodicity were seen throughout the transmembrane domains and the peaks in the middle and C-terminal regions were as high as those in the N-terminal region (38). As an additional control, we prepared Ba/F3 transfectants stably expressing an integrin αIIbβ3 mutant. Cross-linking under the same conditions used for EGFR, with freeze-thaw to permeabilize cells and Cu(II)-o-phenanthroline, resulted in cross-linking of αIIb(V984C)β3(L712C) at a position deeply buried in the inner membrane leaflet (data not shown). The cross-linking efficiency in Ba/F3 cells was 27%, whereas 50% cross-linking was observed in HEK 293T cells (38). We therefore conclude that in EGF-bound EGFR dimers on the cell surface, the TM domains associate only through their N-terminal portions, in strong contrast to integrin αIIbβ3 and GpA.
Disulfide-Rosetta models of EGFR TM domain interfaces.
To visualize a potential structural mechanism for TM domain association, the disulfide cross-linking results were used together with physiochemical and knowledge-based potentials. Membrane Rosetta builds structures in a membrane environment that is modeled as three slabs (an inner hydrophobic slab and two outer slabs of intermediate hydrophobicity). The potentials in these slabs differ from those in the aqueous environments on either side of the slabs, and the orientation and embedding of the transmembrane regions in these slabs are varied to achieve the lowest energy (2). The procedure and relationship between percent cross-linking and Cα-Cα atom distance restraints used for TM model 1 were the same as those previously reported for an integrin and validated by comparison between GpA membrane cross-linking results and NMR structures in detergent and lipid environments (38). The final 1,000 models with the 10% lowest energy were structurally clustered. The structures corresponding to the center of the largest cluster (Fig. 5a) show a right-handed association of two α-helices, with the crossing angle of their axes being −56° ± 6° over the five representative models. The helices associate near the extracellular side of the TM domain. The Cα atoms of the N-terminal Ile-622 residues are in close proximity (4.7 ± 0.5 Å), whereas the helices splay apart as they cross the plasma membrane, with the Cα atoms of the C-terminal Met-644 residues being separated by 27 ± 7 Å.
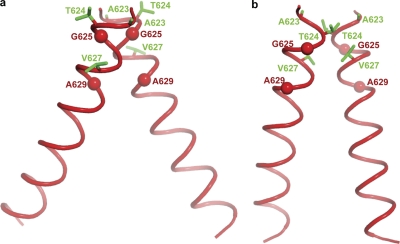
FIG. 5.
Disulfide-Rosetta models of EGF-dependent dimerization of the EGFR TM domain. (a) Model 1; (b) model 2. Each model represents the center of the largest cluster of models. Residues in cross-linking peaks are shown as green sticks. The Cα atoms of G625 and A629 in the GxxxG-like motif are shown as red spheres.
In TM model 1 (Fig. 5a), association of EGFR TM helices is mediated by a GxxxG-like motif, similar to GpA TM dimerization (21-23, 25). The small side chains on the helical face formed by G625 and A629 allow the close approach of the helical backbone (the Cα atoms of G625 and A629 are separated by 4.5 ± 0.1 Å and 6.3 ± 0.2 Å, respectively).
In our previous integrin and GpA TM domain structures, a much larger number of disulfide restraints were available, and these were available throughout the length of the membrane. The residues with peak cross-linking values were in the interface between the two TM helices. This was not the case in EGFR TM model 1; the residues in the cross-linking peaks, Thr-624 and Val-627, faced outward away from the interface (Fig. 5a); nonetheless, the cross-linking restraints dictated association in the outer portion of the TM domain. Thus, the physicochemical and knowledge-based restraints in Rosetta had sufficiently more weight than the cross-linking-derived constraints to dictate the orientation at the TM domain interface. We therefore tested the effect of strengthening the distance restraints by normalizing the cross-linking using 100% for the peak at Thr-624 and increasing the weight of the cross-linking restraints by 5-fold (see Materials and Methods). TM model 2 also shows an association in the first two turns of the α-helix and a right-handed crossing angle of −20° ± 5° (Fig. 5b); however, the residues which gave cross-linking peaks are now located at the TM interface. Furthermore, the C-terminal residues of the TM domain are not separated as much as in model 1.
Association through specific domain IV interfaces and a specific TM interface is not required for EGFR transmembrane signaling.
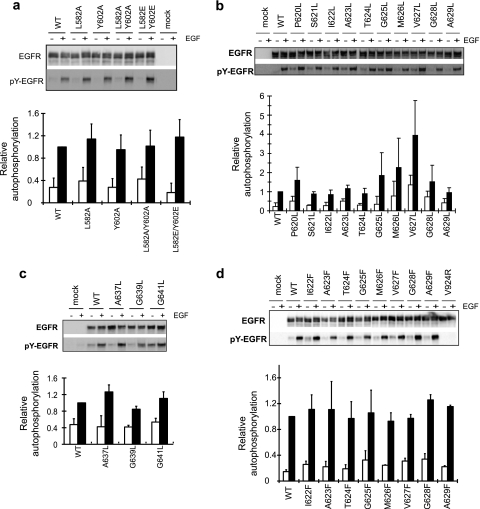
The importance of the domain IV interface in EGFR kinase activation was tested by mutating interface residues Leu-582, Trp-584, and Tyr-602. The W584A mutation inhibited cell surface expression and processing during biosynthesis, whereas other mutants were well expressed (data not shown). Individual and double mutations of residues 582 and 602, including L582E/Y602E, had no effect on EGF-stimulated autophosphorylation (Fig. 6a).
FIG. 6.
Mutagenesis of domain IV and TM dimerization interfaces and EGFR kinase activation. (a) Domain IV interface mutations; (b) leucine substitutions in the last two linker residues and in the TM domain; (c) leucine substitutions in the C-terminal GxxxG-like motif; (d) phenylalanine substitutions in the TM domain. Transiently transfected HEK 293T cells were treated with or without EGF, and detergent extracts were subjected to reducing SDS-7.5% PAGE and Western blotting with protein C antibody for detection of total EGFR and phosphotyrosine antibody for detection of phosphorylated EGFR (pY-EGFR). Bars represent the averages and standard deviations of the ratio of phosphorylated EGFR/EGFR relative to that for wild-type (WT) EGFR with EGF from 3 independent transfection experiments. White bars, without EGF; black bars, with EGF.
We used leucine scanning to test if the GxxxG-like interface in the EGFR transmembrane domain is required for signaling. Substitution of small interfacial residues with leucine has been shown to completely block dimerization through the GxxxG motif in the GpA TM domain (21, 22). Thus, residues P620 to A629 of EGFR, which include the GxxxA motif at residues 625 to 629, were individually mutated to leucine. However, leucine substitutions do not impair EGF-stimulated autophosphorylation (Fig. 6b) and in some cases result in increased autophosphorylation. These results suggest that formation of a specific GpA-like interface is not required for EGFR transmembrane signaling.
We also examined the role of a second, C-terminal GxxxG-like motif (Fig. 7). Small residues A637, G639, and G641 were individually mutated to Leu. None of the mutations significantly affected EGFR autophosphorylation (Fig. 6c), suggesting that the C-terminal GxxxG-like motif is also not required for EGFR transmembrane signaling.
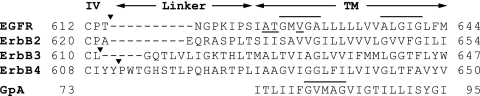
FIG. 7.
Linker and TM domain sequences of human ErbB receptors and the TM domain sequence of GpA. GxxxG-like motifs are overlined, and residues in cross-linking peaks are underlined. Arrowheads follow the last domain IV residue visible in density (EGFR, present structure and tethered, PDB accession no. 1YY9; tethered ErbB3, PDB accession no. 1M6B) or the last residue with interactions with domain IV (ErbB4, PDB accession no. 2AHX, in which the linker engages in lattice contacts with symmetry-related molecules).
A second type of interface was predicted by model 2 (Fig. 5b). In this interface, residues of moderate size, including Thr-624 and Val-627, are present; and Leu, which is essentially one methyl group larger, might not be sufficiently large to disrupt the interface. Therefore, we replaced the first 8 TM residues, residues 622 to 629, with Phe, which is larger than any of the Ile, Ala, Gly, Val, or Met residues that were replaced. All of the substitutions showed EGF-dependent autophosphorylation that was comparable to that for the wild type (Fig. 6d). The requirement of the asymmetric kinase domain interface for activation was demonstrated by the V924R mutation (Fig. 6d). Therefore, we conclude that no specific TM interface is required for EGFR signaling.
DISCUSSION
The mechanisms by which signals are transmitted across the plasma membrane have not been resolved at the structural level for EGFR or, more generally, for any receptor with a single-pass TM domain. We have filled in part of the missing structural information for EGFR, but also find that the signaling pathway between the ligand binding site and the kinase domains in the cytoplasm is surprisingly flexible. Thus, the regions in between come into close apposition but are not precisely aligned by ligand binding and instead loosely link extracellular dimerization to close association of the kinase domains.
In our improved EGFR extracellular domain crystal structure, ligand-induced dimerization is mediated by the previously described intermolecular interface involving the dimerization arm of domain II (13, 30). Furthermore, we identified an additional dimerization interface in domain IV that brings the C termini of the extracellular domains into close proximity. The domain IV-IV interface is composed mainly of hydrophobic and van der Waals interactions between side chains of moderately conserved residues and buries a total solvent-accessible surface area of 430 Å2, considerably less than the 2,800 Å2 buried by the domain II-II interface. The domain IV interface is therefore likely to make a comparably smaller energetic contribution to dimerization, consistent with only a 3-fold decrease in dimerization affinity upon its removal (10) and a lack of an effect on kinase activation of mutating Leu-582 and Tyr-602 in the interface. The primary function of domain IV may be to provide orientation between domain III and the TM domain to bring the two TM domains of a dimer into close proximity.
Interestingly, two residues mutated in glioblastoma (20), Gly-574 and Pro-572, map in and near the domain IV interface, respectively (Fig. 2g). G574V and P572L are thus far the only substitutions within EGFR domains III and IV to be implicated in glioblastoma (20). Furthermore, the G574V substitution has been tested in astrocyte and Ba/F3 transfectants and found to activate autophosphorylation (20). The Gly-574 residues in two monomers are only 6 Å apart and are adjacent to Leu-582 in the hydrophobic domain IV dimerization interface. Their mutation to Val would extend the hydrophobic interface. Pro-572 connects the disulfide-bonded Cys-571 to a loop bearing residues Gly-574, Leu-582, and Trp-584 in the interface (Fig. 2g). Thus, mutation of Pro-572 to Leu might also enhance the interface. Although the precise mechanism for the oncogenic effect of these mutations is unclear, our structure suggests that the mutations could promote stronger interactions at the domain IV interface.
In contrast to a possible role for domain IV in perturbing interactions of the unliganded receptor in glioblastoma, we found no evidence of a requirement for a specific domain IV interface in EGF-stimulated kinase activation. Single or double mutation of Leu-582 and Tyr-602 to Ala and Glu did not inhibit EGF-stimulated autophosphorylation. Although Leu-582 and Tyr-602 are on the periphery and Trp-584 is at the center of the small domain IV interface (Fig. 2e), we expected that the L582E/Y602E mutation would weaken the interface by introducing charge repulsion and/or would require a shift in the interface to accommodate the mutations. Consistent with flexibility within domain IV between the liganded and unliganded EGFR, the mutational results suggest that the small domain IV interface can adjust in response to mutations, without perturbing signal transmission.
EGFR residues 615 to 621 form a 7-residue linker between the extracellular residues visible in density in the crystal structure and the TM domain. Our crystallography and cross-linking results are consistent with close proximity of the linker residues in the dimer, with the absence of a stable structure for the linker, and with a flexible connection between the ectodomain and transmembrane domain in the dimerized receptor. An additional linker of 9 residues, including 8 Ala residues and 1 cysteine residue varying in position from residues 1 to 9, were previously inserted between the EGFR linker and TM domain (29). Spontaneous cross-linking independent of EGF was obtained. Although the results were interpreted with respect to the hypothesis that the introduced linker formed an α-helix continuous with the TM α-helix, helix formation was not tested; furthermore, cross-linking peaks on opposite sides of a helical wheel did not support a unique mode of ligand-independent association.
We examined EGF-dependent disulfide cross-linking of cysteine substitutions across the entire EGFR TM domain. The results show that the TM domains associate, in the presence of EGF, in the exofacial leaflet of the lipid bilayer, with the closest approach being in the first α-helical turn, and then separate before they approach the cytoplasmic face of the membrane. Two cross-linking peaks at Ala-623/Thr-624 and Val-627 were separated by 3.5 residues and thus demonstrated a clear signature for two associating α-helices. However, the lack of any other cross-linking peaks deeper in the membrane was a complete surprise to us. This is quite different from the results for the GpA and integrin TM domains, which associate throughout their traversal of the membrane (19, 25, 38) and show five strong cross-linking peaks, with the most C-terminal peak being similar in efficiency to the most N-terminal peak (38).
Because the energetics of association are to a first approximation proportional to the amount of buried surface area, these differences immediately suggest that the association between the EGFR TM domains is markedly weaker than that between GpA or integrin TM domains. The GpA TM domains are constitutively associated. Integrin TM domains are basally associated and, upon activation, dissociate. In contrast, EGFR TM domains are constitutively unassociated, as shown here by the lack of cross-linking in the absence of EGF, and associate only upon stimulation with EGF. The much more modest association between the EGFR TM domains may thus be appropriate for a receptor where the equilibrium in the absence of activation favors the dissociated state. This may be a mechanism for decreasing the possibility of inappropriate activation. We propose that small interfaces may be characteristic of receptors with single-pass TM domains that associate only upon activation.
Using our cross-linking restraints, we calculated two types of TM domain association models. Model 2 is most consistent with our data, because the helices associate through the same residues that appear in the EGF-stimulated cross-linking peaks. This mode of association may be driven by receptor dimerization. On the other hand, model 1, which involves association through GxxxG-like motifs, was favored by the Rosetta energy function with lower cross-linking restraint weights. The TM domains of ErbB family members contain N-terminal and C-terminal GxxxG-like motifs (Fig. 7); however, the G residues are not well conserved, and the motifs are less buried in the membrane than they are in GpA (Fig. 7). In fusion proteins in bacterial membranes, different ErbB GxxxG-like motifs can drive dimerization (15, 26). In the NMR structure of the isolated ErbB2 TM domain in bicelles, the ErbB2 TM helices associate through the N-terminal GxxxG-like motif (4).
Quite recently, an NMR structure appeared for EGFR and ErbB2 TM peptides associating in bicelles (28). The region of EGFR TM association, from Thr-624 to Ala-629, is very similar to that identified here. The crossing angle of −46 ± 5° is similar to that of our model 1. The EGFR residues most buried in the interface, Thr-624, Gly-625, Gly-628, and Ala-629, are shifted by 1 reside from our first peak at Ala-623 and Thr-624 and 1.5 residues from our second peak at Val-627. The interface with ErbB2 is very similar to that in our model 1, except that it is translated deeper in the membrane along the same face of the EGFR TM domain.
We used mutations to test the hypothesis that a specific TM interface is required for EGFR signaling. None of the cysteine substitutions across the entire EGFR TM domain impaired receptor transmembrane signaling. This contrasts with integrins, where a Gly-to-Cys mutation broke the TM domain association, as shown by activation (38), and with GpA, where Cys mutations can impair dimerization (22). Furthermore, leucine-scanning mutagenesis of the two residues preceding the TM domain and TM domain residues 622 to 629, 637, 639, and 641, which include all putative G-like residues in GxxxG-like motifs, failed to impair EGFR signaling. Moreover, phenylalanine scanning through residues 622 to 629 showed that no unique interface, including the interface showing EGF-dependent cross-linking, was required for EGFR-stimulated autophosphorylation. These results are in agreement with those of previous studies that showed that activation by EGF failed to be prevented by shortening or lengthening the N-terminal TM boundary; mutation of Val-627, Gly-628, or Leu-642; or combined mutation of Leu-633, Ala-637, and Gly-641 to proline (8, 18). Interestingly, the second cross-linking peak at Val-627 corresponds to the Val residue in ErbB2 that, when it is mutated to Glu, is transforming in the neu oncogene; however, the same substitution in EGFR has no effect (8, 18). We conclude that, to our surprise, no specific interface is required for stimulating autophosphorylation by EGFR. We have used Ba/F3 transfectants and HEK 293T cells transfected with limiting amounts of DNA to obtain EGF-dependent autophosphorylation. One caveat is that the use of other cell types or measurement of signals downstream of autophosphorylation might enable detection of the effects of mutations in the domain IV interface or the TM domain.
We expected the work described here to yield a detailed understanding of how signals are transmitted from the EGFR ectodomain and across the plasma membrane to the kinase domain. In a sense it has, in that we find that the connection between the ligand binding and kinase domains in EGFR is surprisingly loose. Our data show that signal transmission is not via a rigid connection between these domains, as it would be if signaling were mediated by a classic allosteric mechanism involving closely coordinated changes in the relative positions of extracellular and cytoplasmic domains. Although cross-linking identifies a preferred orientation between the TM domains in EGFR dimers, this orientation is not required for autophosphorylation. Comparison to the NMR structure of the EGFR/ErbB2 TM heterodimer (24) raises the possibility that the EGFR TM dimer and heterodimer interfaces may differ, consistent with the loose linkage between EGFR ectodomain dimerization and kinase domain autophosphorylation found here. Similarly, although we find an interface at domain IV in EGFR dimers by crystallography and confirm it by cross-linking, this interface also appears to be loose. The interface is small, and the lower quality of the electron density in domain IV is consistent with motion of this domain. The long, thin, rod-like shape of domain IV and the ability to mutate residues in the domain IV interface without affecting signaling are also consistent with flexibility. Furthermore, there is a 7-residue linker between the last residue visible in density in domain IV and the first EGFR TM residue. Cross-linking revealed no evidence for structure in this region. Moreover, the length of this linker region and its sequence is highly variable among ErbB family members (Fig. 7). These features suggest that the linker is too flexible to transmit a specific orientation between the ectodomain monomers to the kinase domain monomers.
Our results demonstrate that autophosphorylation requires only a loose tether between the ectodomain and the kinase domain in EGFR. Because attention on single-pass TM domains has focused on motifs that mediate association and specific interfaces have long been thought to be required for signal transmission across the membrane, our results are heretical. However, we believe that these results are of wider biological significance and are more important than if a specific interface had been found. In EGFR, many types of events, including phosphorylation on Ser and Thr in the long juxtamembrane region between the TM and kinase domains, are thought to modulate kinase activity after receptor ligation (see reference 32 and references therein). The lack of a tight connection between the extracellular and kinase domains provides more scope for regulation of receptor activity. Recently, using negative-stain EM and image classification, we have obtained projection averages of EGF-bound EGFR dimers that show simultaneously the conformation of the extracellular and kinase domains (L.-Z. Mi, C. Lu, Z. Li, N. Nishida, T. Walz, and T. A. Springer, unpublished data). These projections show that the EGFR ectodomain dimer can associate with more than one arrangement of kinase domains within a dimer. These results are compatible with the loose linkage between ectodomain dimerization and autophosphorylation found here.
Acknowledgments
M.J.G. was supported by a postdoctoral fellowship from the American Cancer Society (PF-07-055-01-GMC). This work was supported by NIH grant HL-48675.
Footnotes
Published ahead of print on 13 September 2010.
REFERENCES
- 1.Adams, P. D., R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter, and T. C. Terwilliger. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948-1954. [DOI] [PubMed] [Google Scholar]
- 2.Barth, P., J. Schonbrun, and D. Baker. 2007. Toward high-resolution prediction and design of transmembrane helical protein structures. Proc. Natl. Acad. Sci. U. S. A. 104:15682-15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blume-Jensen, P., and T. Hunter. 2001. Oncogenic kinase signalling. Nature 411:355-365. [DOI] [PubMed] [Google Scholar]
- 4.Bocharov, E. V., K. S. Mineev, P. E. Volynsky, Y. S. Ermolyuk, E. N. Tkach, A. G. Sobol, V. V. Chupin, M. P. Kirpichnikov, R. G. Efremov, and A. S. Arseniev. 2008. Spatial structure of the dimeric transmembrane domain of the growth factor receptor ErbB2 presumably corresponding to the receptor active state. J. Biol. Chem. 283:6950-6956. [DOI] [PubMed] [Google Scholar]
- 5.Bonneau, R., C. E. Strauss, C. A. Rohl, D. Chivian, P. Bradley, L. Malmstrom, T. Robertson, and D. Baker. 2002. De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol. 322:65-78. [DOI] [PubMed] [Google Scholar]
- 6.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 7.Burgess, A. W., H. S. Cho, C. Eigenbrot, K. M. Ferguson, T. P. Garrett, D. J. Leahy, M. A. Lemmon, M. X. Sliwkowski, C. W. Ward, and S. Yokoyama. 2003. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol. Cell 12:541-552. [DOI] [PubMed] [Google Scholar]
- 8.Carpenter, C. D., H. A. Ingraham, C. Cochet, G. M. Walton, C. S. Lazar, J. M. Sowadski, M. G. Rosenfeld, and G. N. Gill. 1991. Structural analysis of the transmembrane domain of the epidermal growth factor receptor. J. Biol. Chem. 266:5750-5755. [PubMed] [Google Scholar]
- 9.Chung, I., R. Akita, R. Vandlen, D. Toomre, J. Schlessinger, and I. Mellman. 2010. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature 464:783-787. [DOI] [PubMed] [Google Scholar]
- 10.Dawson, J. P., M. B. Berger, C. C. Lin, J. Schlessinger, M. A. Lemmon, and K. M. Ferguson. 2005. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol. Cell. Biol. 25:7734-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 12.Ferguson, K. M., M. B. Berger, J. M. Mendrola, H. S. Cho, D. J. Leahy, and M. A. Lemmon. 2003. EGF activates its receptor by removing interactions that autoinhibit ectodomain dimerization. Mol. Cell 11:507-517. [DOI] [PubMed] [Google Scholar]
- 13.Garrett, T. P., N. M. McKern, M. Lou, T. C. Elleman, T. E. Adams, G. O. Lovrecz, H. J. Zhu, F. Walker, M. J. Frenkel, P. A. Hoyne, R. N. Jorissen, E. C. Nice, A. W. Burgess, and C. W. Ward. 2002. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor α. Cell 110:763-773. [DOI] [PubMed] [Google Scholar]
- 14.Garrett, T. P. J., N. M. McKern, M. Lou, M. J. Frenkel, J. D. Bentley, G. O. Lovrecz, T. C. Elleman, L. J. Cosgrove, and C. W. Ward. 1998. Crystal structure of the first three domains of the type-1 insulin-like growth factor receptor. Nature 394:395-399. [DOI] [PubMed] [Google Scholar]
- 15.Gerber, D., N. Sal-Man, and Y. Shai. 2004. Two motifs within a transmembrane domain, one for homodimerization and the other for heterodimerization. J. Biol. Chem. 279:21177-21182. [DOI] [PubMed] [Google Scholar]
- 16.Heinzel, S. S., P. J. Krysan, M. P. Calos, and R. B. DuBridge. 1988. Use of simian virus 40 replication to amplify Epstein-Barr virus shuttle vectors in human cells. J. Virol. 62:3738-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hynes, N. E., and H. A. Lane. 2005. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat. Rev. Cancer 5:341-354. [DOI] [PubMed] [Google Scholar]
- 18.Kashles, O., D. Szapary, F. Bellot, A. Ullrich, J. Schlessinger, and A. Schmidt. 1988. Ligand-induced stimulation of epidermal growth factor receptor mutants with altered transmembrane regions. Proc. Natl. Acad. Sci. U. S. A. 85:9567-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lau, T. L., C. Kim, M. H. Ginsberg, and T. S. Ulmer. 2009. The structure of the integrin αIIbβ3 transmembrane complex explains integrin transmembrane signalling. EMBO J. 9:1351-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, J. C., I. Vivanco, R. Beroukhim, J. H. Huang, W. L. Feng, R. M. Debiasi, K. Yoshimoto, J. C. King, P. Nghiemphu, Y. Yuza, Q. Xu, H. Greulich, R. K. Thomas, J. G. Paez, T. C. Peck, D. J. Linhart, K. A. Glatt, G. Getz, R. Onofrio, L. Ziaugra, R. L. Levine, S. Gabriel, T. Kawaguchi, K. O'Neill, H. Khan, L. M. Liau, S. F. Nelson, P. N. Rao, P. Mischel, R. O. Pieper, T. Cloughesy, D. J. Leahy, W. R. Sellers, C. L. Sawyers, M. Meyerson, and I. K. Mellinghoff. 2006. Epidermal growth factor receptor activation in glioblastoma through novel missense mutations in the extracellular domain. PLoS Med. 3:e485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemmon, M. A., J. M. Flanagan, J. F. Hunt, B. D. Adair, B. J. Bormann, C. E. Dempsey, and D. M. Engelman. 1992. Glycophorin A dimerization is driven by specific interactions between transmembrane α-helices. J. Biol. Chem. 267:7683-7689. [PubMed] [Google Scholar]
- 22.Lemmon, M. A., J. M. Flanagan, H. R. Treutlein, J. Zhang, and D. M. Engelman. 1992. Sequence specificity in the dimerization of transmembrane α-helices. Biochemistry 31:12719-12725. [DOI] [PubMed] [Google Scholar]
- 23.Lemmon, M. A., H. R. Treutlein, P. D. Adams, A. T. Brunger, and D. M. Engelman. 1994. A dimerization motif for transmembrane α-helices. Nat. Struct. Biol. 1:157-163. [DOI] [PubMed] [Google Scholar]
- 24.Li, S., K. R. Schmitz, P. D. Jeffrey, J. J. Wiltzius, P. Kussie, and K. M. Ferguson. 2005. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell 7:301-311. [DOI] [PubMed] [Google Scholar]
- 25.MacKenzie, K. R., J. H. Prestegard, and D. M. Engelman. 1997. A transmembrane helix dimer: structure and implications. Science 276:131-133. [DOI] [PubMed] [Google Scholar]
- 26.Mendrola, J. M., M. B. Berger, M. C. King, and M. A. Lemmon. 2002. The single transmembrane domains of ErbB receptors self-associate in cell membranes. J. Biol. Chem. 277:4704-4712. [DOI] [PubMed] [Google Scholar]
- 27.Mi, L. Z., M. J. Grey, N. Nishida, T. Walz, C. Lu, and T. A. Springer. 2008. Functional and structural stability of the epidermal growth factor receptor in detergent micelles and phospholipid nanodiscs. Biochemistry 47:10314-10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mineev, K. S., E. V. Bocharov, Y. E. Pustovalova, O. V. Bocharova, V. V. Chupin, and A. S. Arseniev. 2010. Spatial structure of the transmembrane domain heterodimer of ErbB1 and ErbB2 receptor tyrosine kinases. J. Mol. Biol. 400:231-243. [DOI] [PubMed] [Google Scholar]
- 29.Moriki, T., H. Maruyama, and I. N. Maruyama. 2001. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J. Mol. Biol. 311:1011-1026. [DOI] [PubMed] [Google Scholar]
- 30.Ogiso, H., R. Ishitani, O. Nureki, S. Fukai, M. Yamanaka, J. H. Kim, K. Saito, A. Sakamoto, M. Inoue, M. Shirouzu, and S. Yokoyama. 2002. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 110:775-787. [DOI] [PubMed] [Google Scholar]
- 31.Tanner, K. G., and J. Kyte. 1999. Dimerization of the extracellular domain of the receptor for epidermal growth factor containing the membrane-spanning segment in response to treatment with epidermal growth factor. J. Biol. Chem. 274:35985-35990. [DOI] [PubMed] [Google Scholar]
- 32.Thiel, K. W., and G. Carpenter. 2007. Epidermal growth factor receptor juxtamembrane region regulates allosteric tyrosine kinase activation. Proc. Natl. Acad. Sci. U. S. A. 104:19238-19243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker, F., M. L. Hibbs, H. H. Zhang, L. J. Gonez, and A. W. Burgess. 1998. Biochemical characterization of mutant EGF receptors expressed in the hemopoietic cell line BaF/3. Growth Factors 16:53-67. [DOI] [PubMed] [Google Scholar]
- 34.Weiner, D. B., J. Liu, J. A. Cohen, W. V. Williams, and M. I. Greene. 1989. A point mutation in the neu oncogene mimics ligand induction of receptor aggregation. Nature 339:230-231. [DOI] [PubMed] [Google Scholar]
- 35.Yarden, Y., and J. Schlessinger. 1987. Self-phosphorylation of epidermal growth factor receptor: evidence for a model of intermolecular allosteric activation. Biochemistry 26:1434-1442. [DOI] [PubMed] [Google Scholar]
- 36.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2:127-137. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, X., J. Gureasko, K. Shen, P. A. Cole, and J. Kuriyan. 2006. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell 125:1137-1149. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, J., B. H. Luo, P. Barth, J. Schonbrun, D. Baker, and T. A. Springer. 2009. The structure of a receptor with two associating transmembrane domains on the cell surface: integrin αIIbβ3. Mol. Cell 34:234-249. [DOI] [PMC free article] [PubMed] [Google Scholar]