Abstract
Members of the Paramyxovirinae subfamily rely on the concerted action of two envelope glycoprotein complexes, attachment protein H and the fusion (F) protein oligomer, to achieve membrane fusion for viral entry. Despite advances in X-ray information, the organization of the physiological attachment (H) oligomer in functional fusion complexes and the molecular mechanism linking H receptor binding with F triggering remain unknown. Here, we have applied an integrated approach based on biochemical and functional assays to the problem. Blue native PAGE analysis indicates that native H complexes extract predominantly in the form of loosely assembled tetramers from purified measles virus (MeV) particles and cells transiently expressing the viral envelope glycoproteins. To gain functional insight, we have established a bimolecular complementation (BiC) assay for MeV H, on the basis of the hypothesis that physical interaction of H with F complexes, F triggering, and receptor binding constitute distinct events. Having experimentally confirmed three distinct H complementation groups, implementation of H BiC (H-BiC) reveals that a high-affinity receptor-to-paramyxovirus H monomer stoichiometry below parity is sufficient for fusion initiation, that F binding and fusion initiation are separable in H oligomers, and that a higher relative amount of F binding-competent than F fusion initiation- or receptor binding-competent H monomers per oligomer is required for optimal fusion. By capitalizing on these findings, H-BiC activity profiles confirm the organization of H into tetramers or higher-order multimers in functional fusion complexes. Results are interpreted in light of a model in which receptor binding may affect the oligomeric organization of the attachment protein complex.
Enveloped viruses gain access to target cells through fusion of viral and cellular membranes. This involves viral fusogenic envelope glycoprotein complexes that mediate membrane merger in a series of conformational rearrangements. For members of the Paramyxovirinae subfamily, fusion is accomplished by the concerted action of two glycoprotein complexes, the fusion (F) protein and the attachment protein (protein H, HN, or G, depending on the Paramyxovirinae genus) (21). Receptor binding by the attachment protein is thought to trigger refolding of the metastable F complex into the thermodynamically stable postfusion conformation and thus initiate the fusion event (20).
Most paramyxoviruses require coexpression of homotypic envelope glycoproteins for efficient F triggering and membrane fusion (17, 45). This implicates specific protein-protein interactions between the F and the attachment protein in functional fusion complexes. All Paramyxovirinae attachment proteins form homo-oligomers. Their ectodomains are organized in a globular head domain that shows the six-blade propeller fold typical of sialidase structures (3, 5, 8, 15, 40, 49, 52) and a stalk domain connecting the head region to the transmembrane domain and short lumenal tail. Although no crystal information on the stalk domain is available, circular dichroism analyses of parainfluenza virus type 5 (PIV 5) HN (51) and structure predictions of measles virus (MeV) H and PIV 5 HN (22, 51) support an α-helical coiled-coil configuration of the stalk. Regions in the stalk have, furthermore, been implicated for several paramyxovirus HN proteins to mediate specificity for their homotypic F proteins (9, 10, 25, 45, 47). We have demonstrated that this extends to MeV H (22, 30), supporting the view that F-interacting residues may reside in the stalk region of the attachment protein (18, 30).
Despite these advances, the effect of receptor binding on the attachment protein oligomer and the molecular mechanism linking receptor binding with F-protein refolding are poorly understood. MeV H head domains have been crystallized both free and complexed with soluble receptor in monomeric and dimeric forms (5, 15). Data available for attachment proteins of related Paramyxovirus family members, such as henipavirus G, and several HN proteins suggest that the tetramer (dimer of dimers) constitutes the physiological oligomer (2, 51, 52). By extension, this may equally apply to all attachment proteins of viruses of the Paramyxovirinae subfamily. Elucidating the oligomeric status of the attachment protein engaged in functional fusion complexes in situ will likely be paramount for the mechanistic understanding of paramyxovirus F triggering, given that no major conformational differences were observed between crystal structures of PIV 5 HN, human parainfluenza virus type 3 HN, henipavirus G, and MeV H solved alone or in complex with their receptor (3, 5, 8, 15, 40, 49, 52). It was, rather, hypothesized that receptor binding may affect the quaternary status of the attachment protein homo-oligomer, which could ultimately trigger F refolding (52). If a general theme applies to paramyxovirus entry, this brings into focus the question of whether a dimeric organization represents the physiological oligomer of native MeV H.
Beyond the physical organization of the H oligomer, we have only begun to uncover some of the basic dynamics that govern the complex protein machineries required for membrane fusion and virus infection. For instance, it is unknown whether physical interaction of the attachment protein complex with F and induction of F triggering are separable events within an H oligomer, whether interaction of an H oligomer with multiple F complexes is required for optimal fusion activity, or even whether membrane fusion initiation requires engagement of every protein H monomer by the receptor.
In the study described here, we have employed an integrated biochemical and functional approach to better understand some of the basic structural and mechanistic features of the MeV fusion complex. Blue native PAGE (BN-PAGE) analysis was used to test the organization of the native attachment protein oligomer extracted from purified viral particles and transiently transfected cells under different stringency conditions.
Bimolecular protein complementation (BiC) allows the mechanistic assessment of multisubunit protein complexes. Application to the retrovirus envelope, for instance, has elucidated the interactions between HIV-1 gp120 and gp41 and the stoichiometry of the HIV-1 envelope trimer during entry (39, 50). By applying this concept to the paramyxovirus glycoprotein system, we have newly developed an H BiC (H-BiC) assay for MeV protein H that complements BN-PAGE data with functional information and probes the stoichiometric requirements for the organization of biologically active fusion complexes and the receptor-mediated initiation of fusion. Results are interpreted in light of current hypotheses regarding the possible effects of receptor binding on paramyxovirus attachment proteins.
MATERIALS AND METHODS
Cell lines, transfections, and virus stocks.
All cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37°C and 5% CO2. Vero-SLAM cells (29), African green monkey kidney epithelial (Vero) cells stably expressing human signaling lymphocytic activation molecule (SLAM), were incubated in the presence of G-418 (100 μg/ml) every fifth passage. Lipofectamine 2000 (Invitrogen) was used for all cell transfections. MeV stocks were amplified in Vero-SLAM cells at a multiplicity of infection (MOI) of 0.001 PFU/cell and incubated at 37°C. Infected cells were scraped in Opti-MEM medium (Invitrogen), virus was released by two freeze-thaw cycles, and titers were determined by 50% tissue culture infective dose (TCID50) titration according to the Spearman-Karber method (43), as described previously (34). Modified vaccinia virus Ankara expressing T7 polymerase (44) was amplified in DF-1 (chicken embryo fibroblast) cells (ATCC CRL-12203) at an MOI of 1.0 PFU/cell, and cell-associated particles were harvested at 40 h postinfection.
Virus purification.
Vero-SLAM cells were infected with the MeV Alaska isolate (MVi/Alaska.USA/16.00 [37]) at an MOI of 0.001 PFU/cell and incubated until the cytopathic effect reached approximately 80 to 100%. Cell-associated viral particles were then released through Dounce homogenization of cells in TNE buffer (10 mM Tris, pH 7.8, 100 mM sodium chloride, 1 mM EDTA) at 4°C, followed by low-speed pelleting (5,000 × g, 20 min, 4°C) of intact cell nuclei and cell debris. Particles were concentrated in two consecutive ultracentrifugation steps at the interphase of a 10% and 30% iodixanol gradient in TNE buffer (100,000 × g, 90 min, 4°C), followed by loading on a continuous 10% to 30% iodixanol gradient and ultracentrifugation (100,000 × g, 14 h, 4°C). Gradient fractions were subjected to microdialysis (50-kDa-molecular mass cutoff) against TNE buffer, and the fractions with the highest concentrations of infectious particles (yields were typically ≥1 × 107 TCID50 units/ml) were used for further experimentation.
BN-PAGE analysis.
All MeV glycoprotein expression vectors used in this study are based on pCG-F and pCG-H, encoding glycoproteins derived from the MeV Edmonston (MeV-Edm) strain under the control of the constitutive cytomegalovirus promoter (4). For BN-PAGE analysis of transiently expressed glycoproteins, receptor-negative CHO cells were transfected with MeV H and/or F expression constructs. At 36 h posttransfection, the medium was removed, the cells were washed in phosphate-buffered saline (PBS), and BN-PAGE sample buffer (Invitrogen) containing digitonin (0.5%), n-dodecyl-β-d-maltoside (DDM; 0.25%), or Triton X-100 (TX-100; 0.25%) was added to the wells. Cells were extracted for 30 min at 4°C while they were shaken. Purified viral particles were mixed with BN-PAGE sample buffer directly. Cell debris was removed by spinning at 20,000 × g for 30 min at 4°C. Coomassie G-250 was added to the samples, followed by separation on 4 to 12% NativePAGE gels (Invitrogen). Samples were transferred onto polyvinylidene difluoride (PVDF) membranes, and proteins were detected with specific antisera directed against the cytosolic tails of MeV H or F or with α-Flag monoclonal antibodies (M2; Sigma), as indicated.
2D native SDS-PAGE.
For two-dimensional (2D) native SDS-PAGE, single-lane first-dimension (BN-PAGE) strips were excised from native gels, followed by incubation in reducing NuPAGE lithium dodecyl sulfate (LDS) sample buffer (Invitrogen) supplemented with 50 mM dithiothreitol (30 min, 20°C), then alkylating NuPAGE LDS sample buffer supplemented with 50 mM N,N-dimethylacrylamide (30 min, 20°C), and lastly, quenching NuPAGE LDS sample buffer supplemented with 5 mM dithiothreitol and 20% ethanol (15 min, 20°C). Equilibrated gel strips were applied to Novex Tris-glycine gels (Invitrogen) with 2D wells and subjected to SDS-PAGE and immunoblotting to PVDF membranes. H antigenic material was visualized using a specific antiserum directed against the H cytosolic tail.
Site-directed mutagenesis and epitope tagging.
Site-directed mutagenesis was performed following the QuikChange protocol (Stratagene) and using pCG-H as the template. Some mutant constructs were, in addition, epitope tagged, resulting in a set of constructs that contained an amino-terminal double HA tag and an additional set harboring a triple Flag epitope tag (53) on the carboxy terminus. The changes were confirmed by DNA sequencing.
Flow cytometry for analysis of H expression and receptor binding capacity.
To determine surface expression levels of MeV H and the ability of MeV H variants to bind the SLAM receptor, SLAM- and CD46-negative CHO cells were transfected in a six-well plate format with MeV H constructs (5 μg) and an enhanced green fluorescent protein (eGFP) expression plasmid (1 μg) as a marker for transfection, as described previously (30). At 36 h posttransfection, cells were lifted and washed twice in PBS supplemented with 3% fetal calf serum. Cell populations were split into two equal aliquots and incubated with either soluble SLAM-murine IgG (sSLAM-mIgG) or a monoclonal antibody mixture directed against epitopes in the MeV H ectodomain (Millipore) for 1 h on ice. Following absorption of cells with anti-mouse Fc-specific allophycocyanin conjugate (Jackson Immunoresearch) for 30 min on ice, samples were analyzed using a FACSCanto II cytometer and FlowJo software. Fluorescence intensity was determined for the GFP-positive cell population.
Quantitative cell-to-cell fusion assays.
To quantify fusion activity, an effector cell population (3 × 105 cells/well) was cotransfected with 2 μg each of H and F expression plasmids. To inhibit fusion until the cell overlay, the effector cells are incubated in the presence of 100 μM fusion-inhibitory peptide (Bachem). Single transfections of plasmids encoding MeV F served as controls. Target cells (3 × 105 cells/well) were transfected with 2 μg of the reporter plasmid encoding firefly luciferase under the control of the T7 promoter. Two hours posttransfection, modified vaccinia virus Ankara expressing T7 polymerase at an MOI of 1.0 PFU/cell was added to the effector cells. Following incubation for 16 h at 37°C, target cells were detached and overlaid on washed effector cells at a 1:1 ratio, and the mixture was incubated at 37°C. At 5 h postoverlay, cells were lysed using Bright Glo lysis buffer (Promega), and the luciferase activity was determined in a luminescence counter (PerkinElmer), after addition of Britelite substrate (PerkinElmer). The instrument's arbitrary values were analyzed by subtracting the relative background provided by the values for the controls, and these values were normalized against the reference constructs indicated in the figure legends. On average, background values were <1% of the values obtained for reference constructs. For qualitative assessment, transfected Vero-SLAM cells were photographed at 15 h posttransfection at a magnification of ×200.
Envelope glycoprotein heterodimer coimmunoprecipitation.
Vero-SLAM cells were transfected with 6 μg of MeV H-encoding total DNA, 3 μg of an HA-tagged H variant (H-HA), and 3 μg of a Flag-tagged H (H-Flag) counterpart, as indicated. A 36 h posttransfection, the cells were washed and lysed in RIPA buffer (1% sodium deoxycholate, 1% NP-40, 150 mM NaCl, 50 mM Tris-Cl, pH 7.2, 10 mM EDTA, 50 mM NaF, 0.05% SDS, protease inhibitors [Roche], 1 mM phenylmethylsulfonyl fluoride). Lysates were cleared (20,000 × g, 30 min, 4°C) and incubated with a monoclonal antibody directed against the HA epitope tag (Covance 16b12) at 4°C. After precipitation with immobilized protein G at 4°C, samples were washed two times in buffer A (100 mM Tris, pH 7.6, 500 mM lithium chloride, 0.1% Triton X-100) and then buffer B (20 mM HEPES, pH 7.2, 2 mM EGTA, 10 mM magnesium chloride, 0.1% Triton X-100), eluted in urea buffer, and subjected to SDS-PAGE and immunoblotting. Total precipitated HA-tagged H material was visualized with α-HA antibody (16b12) and normalized for HA signal intensities, and coprecipitated Flag-tagged H material was detected on parallel immunoblots with α-Flag antibody (M2). Independent of normalization, the total Flag-tagged H available in the individual samples prior to immunoprecipitation was assessed through direct Western blot analysis of the cleared lysates with α-Flag antibody (M2). Immunostained PVDF membranes were developed using a ChemiDoc XRS digital imaging system (Bio-Rad) and a horseradish peroxidase conjugate directed against mouse IgG light chains. For densitometry, signals were quantified using the QuantityOne software package (Bio-Rad). To compare H heterodimer assortment ratios, the relative amounts of coimmunoprecipitated Flag-tagged H material, adjusted for the total HA-tagged H material that was directly precipitated, were determined proportionally to the total Flag-tagged H material available in the respective sample: [H-Flag variant (IP)/H-Flag reference (IP)]/[H-Flag variant (total lysate)/H-Flag reference (total lysate)]/[H-HA variant (IP)/H-HA reference (IP)], where IP is immunoprecipitation.
RESULTS
To biochemically probe the organization of native MeV H oligomers, we established a BN-PAGE assay (48) for nondenaturing analysis of the envelope glycoprotein complexes. By affording the gentle separation of intact multisubunit membrane protein complexes, BN-PAGE typically yields a higher resolution and better preservation than traditional approaches, such as gel filtration or isopycnic gradient ultracentrifugation (35, 36, 48).
BN-PAGE analysis of native MeV glycoprotein oligomers.
For extraction of intact envelope glycoprotein complexes, gradient ultracentrifugation-purified MeV Alaska particles were gently lysed with a mild detergent, digitonin, DDM, or, for comparison, TX-100. Both digitonin and DDM reportedly remove multisubunit membrane-associated proteins from the lipid bilayer without denaturation or dissociation of noncovalent protein-protein interactions (36, 42, 48). However, digitonin better preserves native, fragile complexes, whereas the harsher DDM more frequently induces separation into subcomplexes (35, 36).
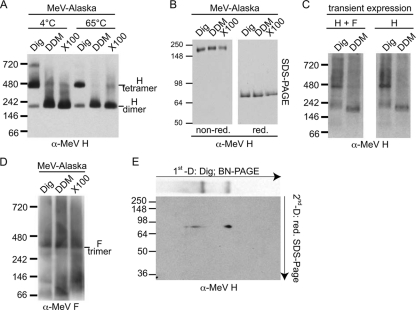
When extracted material was separated on native gels, digitonin extraction indeed predominantly preserved higher-order H oligomers with a relative molecular mass (approximately 460 kDa, according to the native protein standard) approximately twice that (approximately 230 kDa) observed after DDM or TX-100 extraction (Fig. 1A). Only after digitonin extraction did we in addition detect a discrete H subpopulation of approximately three to four times higher molecular mass (migration at approximately the 720-kDa marker position). The temperature sensitivity of this complex, which was completely destroyed by a 10-min incubation at 65°C prior to extraction (Fig. 1A), indicates that digitonin preserves preexisting fragile protein complexes rather than induces nonspecific higher-order aggregates. SDS-PAGE analysis of the extracted material under reducing and nonreducing conditions confirmed the migration of H as a covalently linked homodimer (33) under denaturing conditions independent of the detergent used for solubilization (Fig. 1B).
FIG. 1.
BN-PAGE analysis of MeV glycoproteins reveals an organization of H oligomers in loosely assembled higher-order complexes. (A) Purified MeV particles were extracted with digitonin (0.5%; Dig), DDM (0.25%), or Triton X-100 (0.25%; X100), subjected to BN-PAGE, and probed for MeV H antigenic material using a specific antiserum directed against the H cytosolic tail (α-MeV H). Numbers indicate the migration pattern of a native, globular protein standard. (B) Material extracted for the experiment whose results are presented in panel A was denatured with urea and subjected to conventional SDS-PAGE under nonreducing (non-red.) and reducing (red.) conditions. (C) MeV receptor-negative CHO cells transiently coexpressing MeV H and F or expressing H only were subjected to solubilization with digitonin or DDM, as described for panel A, and BN-PAGE analysis of MeV H. (D) BN-PAGE analysis of glycoprotein extracts from viral particles, as described for panel A, with antiserum directed against the MeV F cytosolic tail (α-MeV F). (E) 2D native SDS-PAGE analysis of digitonin extracts. After the first, native dimension, a single lane was excised, subjected to reducing and alkylating treatment, and loaded on a conventional, reducing SDS gel.
Digitonin versus DDM extraction of receptor-negative CHO cells transiently transfected with MeV glycoprotein expression plasmids revealed essentially the same dual mobility pattern (discrete bands migrating at 460 kDa and 230 kDa, respectively) observed when purified MeV particles were analyzed (Fig. 1C). This was independent of whether MeV H and F were coexpressed or cells were transfected with an H expression plasmid alone, indicating that the higher-molecular-weight material does not represent H and F hetero-oligomers. Parallel analysis of F proteins extracted from purified MeV particles, furthermore, returned, independent of the detergent applied, discrete, predominantly higher-order complexes of equal mobility, which most likely correspond to noncovalently associated F trimers (Fig. 1D). In addition, some F monomeric and dimeric material that was essentially absent from the digitonin preparations appeared after solubilization with DDM or TX-100. Superimposition of the BN-PAGE H and F mobility patterns did not reveal discrete comigrating glycoprotein complexes, again confirming that MeV H and F hetero-oligomers are not preserved by digitonin or DDM extraction and BN-PAGE.
Deconvolution of digitonin-extracted H complexes through two-dimensional native SDS-PAGE confirmed the equal, H monomer-like electrophoretic mobility of the material originating from the different BN-PAGE H complexes under the reducing and denaturing conditions of the second dimension (Fig. 1E). Overall, these data reveal that digitonin predominantly solubilizes fragile, noncovalently associated H complexes.
MeV H predominantly extracts in loosely assembled tetramers.
The smallest MeV H oligomer that can emerge in a native gel should be a covalently linked homodimer with a predicted molecular mass of approximately 150 kDa, while the smallest H oligomer seen experimentally migrated at approximately 230 kDa. Since BN-PAGE separates proteins in their native state, the structure, a lack of an ideal globular fold, and the pI values of individual proteins can substantially influence their mobility profiles (36, 42). A certain discrepancy between the theoretically calculated size of native H and F complexes and mobility relative to those of the native standard was therefore not unexpected.
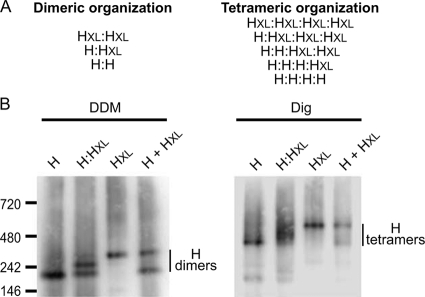
To experimentally probe whether the material migrating at 230 kDa in the native gels represents H homodimers, we took advantage of an H Edm variant (H-Edm) of substantially larger molecular weight due to a C-terminally attached single-chain antibody moiety (14). This variant (termed here HXL) is reportedly capable of heterodimerizing with standard H-Edm (14). Consequently, DDM solubilization of cells coexpressing both standard H and HXL should return three discrete bands, if the DDM-extracted H material represents homodimers (schematically illustrated in Fig. 2A). When samples were analyzed by BN-PAGE, three bands were indeed found, confirming the dimeric nature of the 230-kDa material (Fig. 2B, left panel). To test whether the central band represents bona fide heterodimers or simply postextraction aggregates, DDM extracts of cell populations individually expression H-Edm or HXL were mixed prior to BN-PAGE. This procedure returned H complexes corresponding to H-Edm or HXL homodimers (Fig. 2B, H + HXL), demonstrating the specificity of the assay.
FIG. 2.
DDM predominantly extracts MeV H dimers, digitonin preserves H tetramers. (A) Schematic representation of possible homo- and hetero-oligomer combinations of standard MeV H (H) and an H-single-chain antibody fusion protein (HXL) for a dimeric and tetrameric organization of the H complex. Three discrete oligomers are predicted in the case of H dimers, while the tetramer returns five possible combinations. (B) BN-PAGE analysis of CHO cells transiently transfected with plasmid DNA encoding MeV H (H) or MeV HXL (HXL) or cotransfected with both plasmids at an equimolar ratio (H:HXL). Both H constructs harbor an N-terminal Flag epitope tag. For the control, extracts of cell populations expressing each of the H variants individually were mixed postextraction (H + HXL). Cells were solubilized with DDM (left panel) or digitonin (right panel), extracts were subjected to BN-PAGE, and immunoblots were stained with α-Flag antibodies.
Conversely, digitonin solubilization of cells coexpressing H-Edm and HXL is anticipated to resolve into five different complex species (Fig. 2A), if the 460-kDa H material predominantly extracted by digitonin corresponds to H tetramers. While the size difference between the different species was too small to support detection of distinct bands in this case, BN-PAGE revealed migration of multiple H species between the mobility boundaries established by standard H and HXL, respectively (Fig. 2B, right panel). Individual expression of H-Edm and HXL in separate cell populations followed by postextraction mixing again confirmed the bona fide nature of these H complexes.
Taken together, biochemical analysis through BN-PAGE thus demonstrates that MeV H predominantly migrates in the form of loosely assembled heterotetramers. Higher-order complexes are preexisting and do not represent postsolubilization aggregates.
Identification of independent functional domains in MeV H.
Toward establishing an assay system to explore the mechanistic relevance of these findings, we examined whether the paramyxovirus attachment protein can be dissected into discrete functional domains that, when mutated, can be restored in trans and thus support H-BiC studies. To conceptually test the feasibility of the approach, we identified suitable H mutant candidates with distinct phenotypic profiles: an H-I98A variant reportedly interacts efficiently with F but is incapable of efficient F fusion initiation (7), while we have shown previously that mutation of H stalk residue 111 impairs physical interaction of H with F (30).
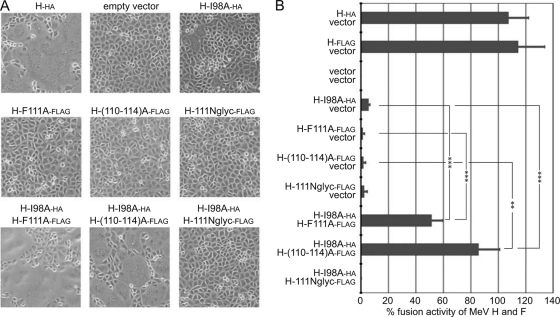
H-I98A was thus coexpressed with standard MeV F and either H harboring a 5-residue alanine substitution of stalk residues 110 to 114 [H-(110-114)A], H carrying an engineered N-glycosylation site at position 111 (H-111Nglyc), or an H F111A (H-F111A) variant; and the effect on F fusion initiation was determined in qualitative and quantitative cell-to-cell fusion assays. Consistent with previous reports, expression of each of these constructs alone with standard MeV F did not result in detectable cell-to-cell fusion [H-(110-114)A, H-111Nglyc, or H-F111A] or caused only minimal fusion (H-I98A). However, combination of H-I98A with H-F111A or H-(110-114)A significantly restored fusion activity, confirming effective bimolecular complementation (Fig. 3A and B). Different epitope tags (Flag or HA) were added to individual H constructs to facilitate their biochemical differentiation in subsequent experiments. The tags showed only small effects on H functionality in quantitative fusion assays (Table 1). The surface expression levels and receptor-binding competence of these and all subsequently discussed H variants were quantified in parallel and in nearly all cases showed less than 30% variation from those of standard H (summarized with the results of quantitative fusion assays in Table 1).
FIG. 3.
F fusion initiation and F binding mutants in the MeV H stalk complement each other in trans. (A) Microphotographs of Vero-SLAM cells cotransfected with 2 μg each of plasmid DNA encoding MeV F and an H variant, as specified. For combinations of different H mutants, 1 μg plasmid DNA of each H construct was transfected. All other samples received, in addition, 1 μg noncoding vector DNA (empty vector). Cell-to-cell fusion was assessed 15 h posttransfection at a magnification of ×200. Representative fields of view are shown. (B) Quantitative firefly luciferase-based H-BiC fusion assay of the constructs and H mutant combinations shown in panel A. Cells were transfected as described for panel A, and values are expressed relative to those observed for MeV F and unmodified protein H and represent averages of at least four independent experiments ± standard errors of the means. Symbols indicate statistically significant divergence (**, P < 0.01; ***, P < 0.001).
TABLE 1.
Characterization of MeV H variants analyzed in this studya
| MeV H variant | Surface expression (% of that for MeV H) | SLAM receptor binding (SLAM/MeV H ratio) | Fusion activity(% of MeV H and F levels) with the following target: |
|
|---|---|---|---|---|
| Vero | Vero-SLAM cells | |||
| H | 100 ± 0 | 1.0 ± 0.04 | 100 ± 0 | 100 ± 0 |
| H-Flag | 82.1 ± 7.8 | 1.1 ± 0.03 | 87.2 ± 4.1 | 114.4 ± 19.7 |
| H-HA | 95.7 ± 16.2 | 1.0 ± 0.2 | 105.6 ± 3.4 | 107.4 ± 14.6 |
| H-I98Ab | 121.0 ± 7.2 | 1.0 ± 0.02 | 0.2 ± 0.1 | 0.3 ± 0.1 |
| H-I98A-HA | 113.1 ± 9.1 | 0.9 ± 0.03 | 4.9 ± 0.9 | 6.5 ± 1.1 |
| H-F111Ac | 110.7 ± 8.7 | 101.3 ± 5.6 | 0.4 ± 0.3 | 0.01 ± 0.01 |
| H-F111A-Flag | 70.2 ± 2.6 | 1.06 ± 0.04 | 0.2 ± 0.1 | 1.3 ± 1.3 |
| H-F111A-HA | 85.3 ± 3.2 | 1.0 ± 0.1 | 0.2 ± 0.1 | 1.1 ± 0.4 |
| H-(110-114)A-Flag | 75.9 ± 6.4 | 1.1 ± 0.1 | ND | 1.73 ± 1.73 |
| H-111Nglyc-Flag | 80.8 ± 11.8 | 1.1 ± 0.1 | ND | 2.05 ± 2.05 |
| H-ΔCD46d | 148.7 ± 0.4 | 0.9 ± 0.03 | 15.7 ± 1.0 | 103.8 ± 11.0 |
| H-ΔCD46-Flag | 109.1 ± 3.3 | 1.0 ± 0.03 | 10.6 ± 0.8 | 113.0 ± 20.6 |
| H-I98A/F111A-HA | 113.1 ± 10.5 | 0.8 ± 0.1 | 0.1 ± 0.1 | 0.03 ± 0.02 |
| H-ΔCD46/I98A-Flag | 104.1 ± 2.4 | 1.0 ± 0.03 | 0.02 ± 0.02 | 0.5 ± 0.1 |
| H-ΔCD46/F111A-Flag | 147.3 ± 4.1 | 1.1 ± 0.1 | 0.1 ± 0.1 | 0.1 ± 0.03 |
All constructs are based on MeV H-Edm, and values are expressed relative to the value for H-Edm. To quantify fusion activity, cells were cotransfected with plasmid DNA encoding MeV F-Edm and individual H variants. Values represent averages of at least four independent experiments ± standard errors of the means. In addition to functional mutations, epitope (Flag or HA) tags were added to individual constructs as specified to differentiate between discrete H variants in the coexpression experiments summarized in Fig. 4B and 6C. ND, not determined.
Construct first described in reference 7.
Construct first described in reference 30.
Construct first described in reference 6.
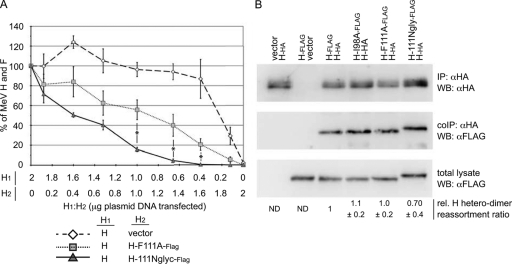
Only the H-111Nglyc variant failed to be complemented by H-I98A, suggesting a dominant negative effect of the engineered N-glycan at position 111 on the formation of functional fusion complexes. This was further accentuated when we generated fusion profiles of H-111Nglyc and H-F111A by diluting them against standard H-Edm (Fig. 4A). Increasing amounts of the former yielded a steep reduction of fusion activity, whereas the latter returned an intermediate profile compared to that for a control series in which levels of standard protein H were gradually decreased by replacing the H-Edm-encoding plasmid incrementally with vector DNA. Consistent with a previous analysis of PIV 5 HN (12), fusion activity and the amount of attachment protein did not follow a linear correlation (Fig. 4A), but activity remained above 80% that of the reference until an H-Edm/vector ratio of approximately 1:4 was reached (5-fold reduction in the amount of the H-Edm-encoding plasmid compared to that for the reference).
FIG. 4.
H-111Nglyc suppresses H-Edm-mediated F triggering in mixed H oligomers. (A) For fusion activity profiles, cells were cotransfected with plasmid DNA encoding MeV F (2 μg) and H-Edm and an H variant in different relative ratios ranging from 16:1 to 1:16 (the total amount of H-encoding plasmid DNA was 2 μg in all cases). Fusion activity was assessed with the quantitative firefly luciferase-based fusion assay and plotted relative to that observed for standard H alone (2 μg of MeV H-encoding DNA). For comparison, the standard H-encoding plasmid was diluted against vector DNA to assess the reduction in fusion as a consequence of the incrementally smaller amounts of standard H expressed. Data represent averages of at least four independent experiments ± standard errors of the means. Symbols indicate statistically significant divergence of the H/H-F111A-Flag and H/H-111Nglyc-Flag data sets (*, P < 0.05). (B) Assessment of relative dimerization of H variants with standard H. Cells cotransfected with standard H-HA and Flag-tagged H variants, as indicated (3 μg plasmid DNA each), were subjected to immunoprecipitation (IP) using α-HA antibodies. Precipitated HA-tagged material was visualized through α-HA Western blotting (WB). In duplicate blots, coimmunoprecipitated (coIP) Flag-tagged H variants were detected through α-Flag immunostaining. Prior to immunoprecipitation, aliquots of each sample lysate (total lysate) were subjected to direct α-Flag Western blotting to determine the relative amounts of totally available Flag-tagged material. Numbers are derived from normalization of the samples for the amount of HA-tagged material precipitated and then assessment of the relative amounts (rel.) of coimmunoprecipitated Flag-tagged H variants, adjusted for total Flag-tagged H available in the respective sample. They represent averages of four independent experiments ± standard errors of the means. ND, not determined.
Since the interpretation of these bioactivity data assumes random reassortment of H monomers into homo- and heterodimers, we compared the ability of H-111Nglyc and H-F111A and also H-I98A to oligomerize with standard H through coprecipitation analysis. The relative coprecipitation efficiencies of the H-F111A and H-I98A constructs were virtually indistinguishable from each other and the coprecipitation efficiency of standard H (Fig. 4B), demonstrating that these point mutations in the H stalk domain do not interfere with free assortment of H monomers into covalently linked dimers. Only the H-1111Nglyc variant showed an approximately 30% reduction in dimerization efficiency, which likely suggests that the supernumerous N-glycan affects somewhat H dimer formation.
Taken together, these results confirm the general feasibility of BiC studies with paramyxovirus attachment proteins. They reveal a recessive-negative phenotype of H variants H-I98A, H-F111A, and H-(110-114)A, which stands in contrast to the phenotype of H-111Nglyc, and demonstrate that these neighboring regions in the H stalk belong to functionally distinct domains.
Receptor to H stoichiometry requirements for paramyxovirus fusion initiation.
To appreciate the full potential of the H-BiC assay, we next explored whether receptor binding-deficient variants constitute a third, independent complementation group. A 5-residue alanine substitution of residues 473 to 477 reportedly interferes with H interaction with cellular CD46 (6, 31), resulting in impaired CD46-dependent F triggering while leaving SLAM-mediated fusion promotion intact. Consistent with these reports, coexpression of the CD46 binding-deficient H variant (termed here H-ΔCD46) with standard MeV F resulted in little cell-to-cell fusion of CD46-positive Vero cells (Fig. 5A and Table 1). Efficient fusion was observed upon expression in Vero-SLAM cells stably expressing human SLAM, confirming that overall F triggering competence of H-ΔCD46 was intact (Fig. 5A and Table 1).
FIG. 5.
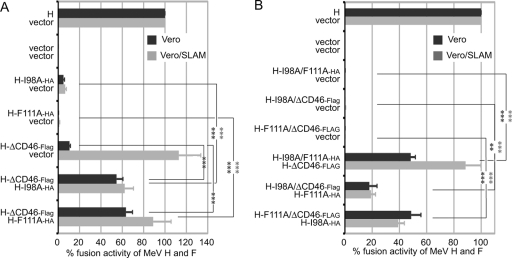
The CD46 receptor binding site in the H head domain and the F fusion initiation and F binding domains in the H stalk region constitute three functionally distinct complementation groups. Quantitative H-BiC assays were carried out as before (see the legend to Fig. 3) for dual (A) or triple (B) combination pairs. Vero and Vero-SLAM cells were analyzed in parallel to ensure the basic functionality of the H-ΔCD46 variant. Values are expressed relative to those observed for MeV F and unmodified protein H and represent averages of at least four independent experiments ± standard errors of the means. Symbols indicate statistically significant divergence for Vero (dark asterisks) or Vero-SLAM (light asterisks) cell-based data sets (**, P < 0.01; ***, P < 0.001).
Coexpression of H-ΔCD46 with either H-I98A or H-F111A in Vero cells significantly restored F fusion initiation in quantitative fusion assays, demonstrating efficient bimolecular complementation (Fig. 5A). To probe the robustness of the phenotype and determine the extent of H-BiC possible, we next engineered H proteins that contained two of the three mutation classes in all possible combinations. All double mutants remained, as anticipated, individually defective in fusion promotion, indicating that the mutations are not compensatory (Fig. 5B and Table 1). When these variants were subjected to the H-BiC assay, however, successful 3-way complementation was observed in all combination settings (Fig. 5B).
These data demonstrate that we have identified three mechanistically distinct, recessive H-BiC groups that are located in different physical domains (head versus stalk) of the protein. Successful complementation of the receptor binding-deficient H-ΔCD46 variant by either H-I98A or H-111A reveals that not every monomer of the paramyxovirus attachment protein complex needs to tightly engage receptor, and thus, a receptor-to-protein H stoichiometry below parity is sufficient to initiate membrane fusion.
The tetramer or a higher-order multimer constitutes the functional paramyxovirus H oligomer.
Productive H-BiC can conceptually occur on an H dimer or tetramer/higher-order multimer level. The BiC assay enables us to differentiate between these alternatives and thus determine the organization of the physiological paramyxovirus H oligomer present in functional fusion complexes, if the distinct complementation groups differ in their individual stoichiometric requirements for productive BiC. To test this, we varied the relative ratios of the expression plasmids encoding the H complementation variants again over a range of from 16:1 to 1:16 for each combination and generated fusion complementation activity profiles. Provided that assortment of the complementation variants into H heterodimers is unbiased, this should return identical activity profiles for each combination, approximately following a second-degree optimum curve, if the dimers constitute the physiological MeV H oligomer. Profiles differing from a symmetric optimum curve can, in contrast, emerge only if functional H exists in a higher-order oligomer organization in situ (illustrated schematically for an H dimer-versus-tetramer scenario in Fig. 6A).
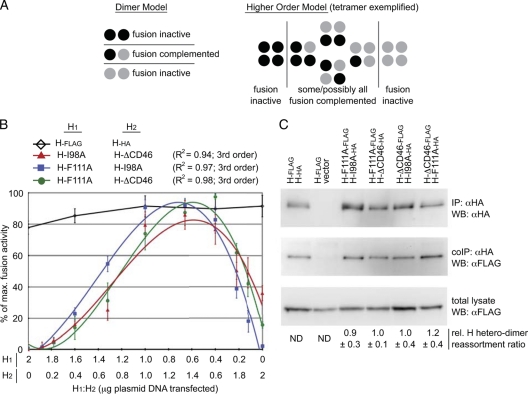
FIG. 6.
The tetramer or a higher-order multimer constitutes the physiological H oligomer in functional fusion complexes. (A) Representation of H fusion complementation on the basis of a dimeric or tetrameric organization of the H oligomer. The schematic is adapted from reference 38. (B) Complementation activity profiles show third-order optimum curves for different H-BiC combinations. Cells were cotransfected with plasmid DNA encoding MeV F (2 μg) and two untagged H variants in different relative ratios ranging from 16:1 to 1:16 (the total amount of H-encoding plasmid DNA was 2 μg in all cases). Fusion activity was assessed with the quantitative firefly luciferase-based fusion assay and plotted as a percentage of the maximal fusion observed for each combination. H-BiC activity of the H-F111A/H-I98A, H-F111A/H-ΔCD46, and H-I98A/H-ΔCD46 pairs showed BiC peaks at approximately 1:2 ratios. Data points represent averages of four independent experiments ± standard errors of the means. Regression curves of all pairs involving the F111A mutation tightly follow a third-degree order (H-F111A/H-I98A, H-F111A/H-ΔCD46); R2 values are given in the key. A pair of standard MeV H proteins harboring Flag (H-Flag) or HA (H-HA) epitope tags, respectively, was analyzed in parallel. (C) Assessment of relative H variant reassortment. Cells cotransfected with the different epitope-tagged constructs as indicated (3 μg plasmid DNA each) were subjected to IP using α-HA antibodies and Western blotting (WB) using specific α-HA or α-Flag antibodies, as detailed in the legend to Fig. 4B. For densitometric quantification, the H-I98A-HA/H-ΔCD46-Flag pair was used as a reference. Numbers are derived from normalizing samples for HA-tagged material precipitated and then assessing the relative amounts (rel.) of coimmunoprecipitated (coIP) Flag-tagged H variants adjusted for total Flag-tagged H available in the respective sample. They represent averages of four independent experiments ± standard errors of the means. ND, not determined.
When experimentally obtained data sets were subjected to regression analysis, we found that the activity curves of each combination involving the F binding-incompetent H-F111A variant in particular show digressions toward a third-order profile (R2 = 0.97 to 0.98, depending on the individual combination; Fig. 6B), which was characterized by fusion activity peaks in the presence of a higher relative amount of H-I98A or H-ΔCD46 compared to that of the H-F111A variant. For comparison, second-degree regression fitting for these data returned R2 values of 0.76 and 0.85. In the case of the H-I98A/H-ΔCD46 pair, the phenotype appeared to be slightly less robust, since we observed a somewhat lower R2 value of 0.94 for a third-order regression fitting. The fusion activity of this pair nevertheless peaked in the presence of a higher relative amount of the H-ΔCD46 variant compared to the amount of H-I98A.
Plasma membrane steady-state levels of all complementation constructs were overall very similar and, if anything, were slightly lower rather than higher for the H-F111A variant (Table 1), and the relative coprecipitation efficiencies of H-F111A and H-I98A were indistinguishable from each other and the coprecipitation efficiency of standard H, as shown in Fig. 4B. To experimentally assess relative assortment of the three H complementation variants, we next compared the extent of H heterodimer formation through selective H coimmunoprecipitation, using H variants harboring distinct Flag and HA epitope tags, as specified. When HA-tagged H variants were immunoprecipitated from cells coexpressing complementation pairs, a virtually identical portion of the available Flag-tagged H-BiC counterpart pool was coprecipitated for each BiC combination tested (Fig. 6C). Taken together, this indicates unbiased heterodimer formation between the H complementation group mutants. The epitope tags themselves did not affect H oligomerization, as evidenced by an essentially linear fusion profile of an H-Flag/H-HA pair tested in parallel (Fig. 6B). Exemplified by swapping the epitope tag distribution in the H-ΔCD46/H-F111A pair, the coprecipitation efficiency was, furthermore, independent of the relative position of the tags (Fig. 6C).
Third-order fusion profiles of different H-BiC pairs indicate that the minimal paramyxovirus H functional unit in situ consists of a tetramer or higher-order multimer. For the F binding-defective H variant, the most effective H-BiC is achieved when a higher relative amount of functional F binding sites (H-F111) than H-I98A or receptor binding-competent H is present in the complemented H oligomers. For the H-I98A/H-ΔCD46 pair, optimal complementation occurs in the presence of a higher relative amount of standard H-I98. Since assortment of the individual H variants was found to be random, these observations are not compatible with a dimeric organization of the minimal functional H unit.
DISCUSSION
To advance our structural and mechanistic insight into paramyxovirus fusion complexes, we have in this study applied novel assays to the MeV envelope glycoprotein system in an integrated biochemical and functional design. Both approaches have confirmed that the tetramer or a higher-order multimer constitutes the physiological H oligomer present in functional fusion protein complexes. Multiple F binding-competent monomers per H oligomer are required for optimal fusion, but a receptor-to-H stoichiometry below parity is sufficient for F fusion initiation.
In particular, the BN-PAGE study reveals that digitonin, but not the slightly more stringent DDM or TX-100, predominantly extracts tetrameric H complexes. This is consistent with the conclusion of multiple studies that digitonin best preserves native membrane complexes (35, 36). Even higher-molecular-mass complexes (migration at approximately the 720-kDa marker position) may represent a subpopulation of H hexamers or a dimer of tetramers. Complete dissociation of the tetramers into covalently linked homodimers by DDM suggests a small intermolecular dimer-dimer interface. A recent biochemical analysis of soluble variants of the related PIV 5 HN protein has supported a tetrameric four-helix bundle organization of the stalk. On the basis of our BN-PAGE results, such a superhelix arrangement is unlikely for MeV H in situ, given that the large internal surface of the resulting coiled coil should withstand DDM solubilization. Assuming that the interface is restricted to the H head domains alone, however, molecular modeling on the basis of coordinates published for MeV H head dimers and PIV 5 tetramers predicts a buried intermolecular surface of approximately 575 Å2 for the MeV H dimer-dimer pair, which appears to be consistent with a fragile, easily disruptable arrangement.
Relative to the mostly globular native protein standard, MeV H and also F complexes appeared to be overproportionally retarded in the native gels. Especially in the case of H oligomers, we consider the overall acidic pI of the protein and the predicted α-helical structure of the H stalk domain (22, 30) to account for this discrepancy, in accordance with previous observations for irregular migration of nonglobular proteins in BN-PAGE (36, 42). Comparative analysis of H variants with considerable size differences has further confirmed the dimeric nature of the DDM-solubilized material. We did not, however, detect any discrete H and F hetero-oligomers in BN-PAGE independent of the detergent used for glycoprotein extraction. This was unexpected, given that both protein complexes efficiently coimmunoprecipitate (22, 32, 34), and likely reflects separation of the hetero-oligomers in the electric field. Hetero-oligomers could also be buried in very large multimers, which would likely result in a high-molecular-weight smear rather than a defined migration pattern in BN-PAGE. Attempts to elucidate the organization of the functional paramyxovirus glycoprotein hetero-oligomer are complicated thus far by the density of the glycoproteins on the virion (23, 24). Independent novel assays were therefore required to examine the applicability of our biochemical data to functional fusion complexes.
Protein bimolecular complementation studies have advanced the understanding of protein complexes of both cellular (16, 19, 26, 28) and viral (1, 11, 13, 39) origin. We have transferred this principle to the paramyxovirus glycoprotein system to parallel the BN-PAGE analysis with independent functional insight. By identifying two mechanistically distinct microdomains within a short physical distance of each other in the H stalk, H-BiC clearly demonstrates that the physical interaction of the paramyxovirus envelope glycoproteins and actual F fusion initiation constitute discrete, experimentally separable events. Discovery of the receptor binding site in the H head as a third complementation group may be less surprising per se but sets the stage for a comparative analysis of the relative ratios of functional triggering and F binding domains in the stalk that are required for fusion initiation. These studies have yielded three major conclusions.
First, a high-affinity receptor-to-paramyxovirus attachment protein monomer stoichiometry below parity is sufficient for fusion to occur. Successful complementation of H-I98A/ΔCD46 and H-F111A/ΔCD46 double mutants by H-F111A and H-I98A, respectively, furthermore underscores that the signal induced by receptor binding is transferable to neighboring H monomers within the H complex that are F fusion initiation competent or that are capable of physical contact with F. Cocrystals of CD46 and MeV H show docking of the receptor moieties to the side of the MeV H β-propeller domain (40). Provided that H oligomers exist in a tetrameric or higher-order multimeric organization, our findings may reflect that steric constraints preclude direct receptor binding by every H monomer even if all binding sites are functional. Clearly, this reduces the receptor density threshold for MeV infection. We have not extended our analysis in this study to the SLAM receptor, since reports concerning the SLAM binding site are mixed (27, 40, 46). Presuming that CD46 and SLAM binding sites are partially overlapping, as suggested by Casasnovas and colleagues (40), however, we predict that a SLAM-to-H stoichiometry below 1:1 will likewise be sufficient for SLAM-mediated MeV infection.
Second, tightly fitting third-order fusion profiles obtained in particular for the H-F111A/H-ΔCD46 and H-F111A/H-I98A BiC pairs demonstrate a tetrameric or higher-order multimeric organization of the physiological paramyxovirus H oligomer in functional fusion complexes. These third-order profiles cannot be explained by differences in intracellular transport competence or a bias in reassortment of the three H complementation variants. Cell surface steady-state levels were very similar overall and, if anything, slightly lower for H-F111A than for the H-I98A variant, which should dampen rather than accentuate the shift in the fusion profiles observed. Selective coimmunoprecipitation of epitope-tagged versions of the H variants furthermore revealed unbiased assembly into heterodimers for the different complementation pairs. Since F was kept consistent in all quantitative fusion assays (MeV F Edmonston), we furthermore do not anticipate that the results are influenced by distinct kinetics of F refolding.
Lastly, the fusion profiles obtained for the different H-BiC pairs suggest a hierarchy of the different complementation groups, since a higher relative amount of F binding-competent (F111) than F fusion initiation-competent (I98) and receptor binding-competent H variants is required in functional fusion complexes for optimal fusion. Thus, loss of receptor binding sites can best be complemented for, followed by a functional I98 domain and, lastly, disruption of physical interaction with F. This implies that the physiological paramyxovirus H tetramer or higher-order multimer may likely need to simultaneously interact with multiple F-protein trimers for optimal fusion, which presumably results in concerted triggering of all associated F moieties after receptor binding. Most likely due to steric constraints of the bulky N-glycan, the H-111Nglyc variant not only fails in H-BiC but also asserts a dominant negative effect on F triggering when it is coexpressed with standard H.
In toto, we conclude that current crystals of soluble monomeric or dimeric MeV H head domains have substantially advanced our structural insight into paramyxovirus H (5, 15, 41). Native gel analysis and biomolecular complementation reveal an organization of native, membrane-embedded full-length H in loosely arranged tetramers or higher-order multimers in functional fusion complexes, making conceivable a conserved basic mechanism of paramyxovirus fusion initiation in which high-affinity receptor binding by a subset of H monomers may induce modifications to the higher-order oligomer status of the attachment protein, which in turn constitutes the mechanistic link to F triggering.
Acknowledgments
We thank A. Prussia for calculating the dimensions of hypothetical MeV H dimer-dimer interfaces and A. L. Hammond for critical reading of the manuscript.
M.A.B. was funded by Molecular Mechanism of Microbial Pathogenesis training grant T32 AI007470 from the NIH. This work was supported, in part, by U.S. Public Health Service grants AI071002 and AI083402 (to R.K.P.) from the NIH/NIAID and a seed grant from the Childrens Healthcare of Atlanta Vaccines & Immunology Center (to R.K.P.).
Footnotes
Published ahead of print on 22 September 2010.
REFERENCES
- 1.Atanasiu, D., J. C. Whitbeck, M. P. de Leon, H. Lou, B. P. Hannah, G. H. Cohen, and R. J. Eisenberg. 2010. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J. Virol. 84:3825-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bossart, K. N., G. Crameri, A. S. Dimitrov, B. A. Mungall, Y. R. Feng, J. R. Patch, A. Choudhary, L. F. Wang, B. T. Eaton, and C. C. Broder. 2005. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J. Virol. 79:6690-6702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowden, T. A., A. R. Aricescu, R. J. Gilbert, J. M. Grimes, E. Y. Jones, and D. I. Stuart. 2008. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 15:567-572. [DOI] [PubMed] [Google Scholar]
- 4.Cathomen, T., H. Y. Naim, and R. Cattaneo. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colf, L. A., Z. S. Juo, and K. C. Garcia. 2007. Structure of the measles virus hemagglutinin. Nat. Struct. Mol. Biol. 14:1227-1228. [DOI] [PubMed] [Google Scholar]
- 6.Corey, E. A., and R. M. Iorio. 2009. Measles virus attachment proteins with impaired ability to bind CD46 interact more efficiently with the homologous fusion protein. Virology 383:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corey, E. A., and R. M. Iorio. 2007. Mutations in the stalk of the measles virus hemagglutinin protein decrease fusion but do not interfere with virus-specific interaction with the homologous fusion protein. J. Virol. 81:9900-9910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068-1074. [DOI] [PubMed] [Google Scholar]
- 9.Deng, R., A. M. Mirza, P. J. Mahon, and R. M. Iorio. 1997. Functional chimeric HN glycoproteins derived from Newcastle disease virus and human parainfluenza virus-3. Arch. Virol. Suppl. 13:115-130. [DOI] [PubMed] [Google Scholar]
- 10.Deng, R., Z. Wang, A. M. Mirza, and R. M. Iorio. 1995. Localization of a domain on the paramyxovirus attachment protein required for the promotion of cellular fusion by its homologous fusion protein spike. Virology 209:457-469. [DOI] [PubMed] [Google Scholar]
- 11.Donzella, G. A., O. Leon, and M. J. Roth. 1998. Implication of a central cysteine residue and the HHCC domain of Moloney murine leukemia virus integrase protein in functional multimerization. J. Virol. 72:1691-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dutch, R. E., S. B. Joshi, and R. A. Lamb. 1998. Membrane fusion promoted by increasing surface densities of the paramyxovirus F and HN proteins: comparison of fusion reactions mediated by simian virus 5 F, human parainfluenza virus type 3 F, and influenza virus HA. J. Virol. 72:7745-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, W., H. Granzow, B. G. Klupp, A. Karger, K. Michael, C. Maresch, R. Klopfleisch, and T. C. Mettenleiter. 2007. Relevance of the interaction between alphaherpesvirus UL3.5 and UL48 proteins for virion maturation and neuroinvasion. J. Virol. 81:9307-9318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hammond, A. L., R. K. Plemper, J. Zhang, U. Schneider, S. J. Russell, and R. Cattaneo. 2001. Single-chain antibody displayed on a recombinant measles virus confers entry through the tumor-associated carcinoembryonic antigen. J. Virol. 75:2087-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashiguchi, T., M. Kajikawa, N. Maita, M. Takeda, K. Kuroki, K. Sasaki, D. Kohda, Y. Yanagi, and K. Maenaka. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. U. S. A. 104:19535-19540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu, C. D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789-798. [DOI] [PubMed] [Google Scholar]
- 17.Hu, X. L., R. Ray, and R. W. Compans. 1992. Functional interactions between the fusion protein and hemagglutinin-neuraminidase of human parainfluenza viruses. J. Virol. 66:1528-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iorio, R. M., and P. J. Mahon. 2008. Paramyxoviruses: different receptors—different mechanisms of fusion. Trends Microbiol. 16:135-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnsson, N., and A. Varshavsky. 1994. Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. U. S. A. 91:10340-10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb, R. A., and T. S. Jardetzky. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17:427-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamb, R. A., and G. D. Parks. 2007. Paramyxoviridae: the viruses and their replication, p. 1449-1496. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1, 5th ed. Wolters Kluwer/Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 22.Lee, J. K., A. Prussia, T. Paal, L. K. White, J. P. Snyder, and R. K. Plemper. 2008. Functional interaction between paramyxovirus fusion and attachment proteins. J. Biol. Chem. 283:16561-16572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loney, C., G. Mottet-Osman, L. Roux, and D. Bhella. 2009. Paramyxovirus ultrastructure and genome packaging: cryo-electron tomography of Sendai virus. J. Virol. 83:8191-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ludwig, K., B. Schade, C. Bottcher, T. Korte, N. Ohlwein, B. Baljinnyam, M. Veit, and A. Herrmann. 2008. Electron cryomicroscopy reveals different F1+F2 protein states in intact parainfluenza virions. J. Virol. 82:3775-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melanson, V. R., and R. M. Iorio. 2006. Addition of N-glycans in the stalk of the Newcastle disease virus HN protein blocks its interaction with the F protein and prevents fusion. J. Virol. 80:623-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monnot, C., C. Bihoreau, S. Conchon, K. M. Curnow, P. Corvol, and E. Clauser. 1996. Polar residues in the transmembrane domains of the type 1 angiotensin II receptor are required for binding and coupling. Reconstitution of the binding site by co-expression of two deficient mutants. J. Biol. Chem. 271:1507-1513. [DOI] [PubMed] [Google Scholar]
- 27.Navaratnarajah, C. K., S. Vongpunsawad, N. Oezguen, T. Stehle, W. Braun, T. Hashiguchi, K. Maenaka, Y. Yanagi, and R. Cattaneo. 2008. Dynamic interaction of the measles virus hemagglutinin with its receptor signaling lymphocytic activation molecule (SLAM, CD150). J. Biol. Chem. 283:11763-11771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obara, M., M. S. Kang, and K. M. Yamada. 1988. Site-directed mutagenesis of the cell-binding domain of human fibronectin: separable, synergistic sites mediate adhesive function. Cell 53:649-657. [DOI] [PubMed] [Google Scholar]
- 29.Ono, N., H. Tatsuo, Y. Hidaka, T. Aoki, H. Minagawa, and Y. Yanagi. 2001. Measles viruses on throat swabs from measles patients use signaling lymphocytic activation molecule (CDw150) but not CD46 as a cellular receptor. J. Virol. 75:4399-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paal, T., M. A. Brindley, C. St. Clair, A. Prussia, D. Gaus, S. A. Krumm, J. P. Snyder, and R. K. Plemper. 2009. Probing the spatial organization of measles virus fusion complexes. J. Virol. 83:10480-10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patterson, J. B., F. Scheiflinger, M. Manchester, T. Yilma, and M. B. Oldstone. 1999. Structural and functional studies of the measles virus hemagglutinin: identification of a novel site required for CD46 interaction. Virology 256:142-151. [DOI] [PubMed] [Google Scholar]
- 32.Plemper, R. K., and R. W. Compans. 2003. Mutations in the putative HR-C region of the measles virus F2 glycoprotein modulate syncytium formation. J. Virol. 77:4181-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plemper, R. K., A. L. Hammond, and R. Cattaneo. 2000. Characterization of a region of the measles virus hemagglutinin sufficient for its dimerization. J. Virol. 74:6485-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Plemper, R. K., A. L. Hammond, D. Gerlier, A. K. Fielding, and R. Cattaneo. 2002. Strength of envelope protein interaction modulates cytopathicity of measles virus. J. Virol. 76:5051-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reisinger, V., and L. A. Eichacker. 2007. How to analyze protein complexes by 2D blue native SDS-PAGE. Proteomics 7(Suppl. 1):6-16. [DOI] [PubMed] [Google Scholar]
- 36.Reisinger, V., and L. A. Eichacker. 2008. Solubilization of membrane protein complexes for blue native PAGE. J. Proteomics 71:277-283. [DOI] [PubMed] [Google Scholar]
- 37.Rota, P. A., S. L. Liffick, J. S. Rota, R. S. Katz, S. Redd, M. Papania, and W. J. Bellini. 2002. Molecular epidemiology of measles viruses in the United States, 1997-2001. Emerg. Infect. Dis. 8:902-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakaguchi, T., Q. Tu, L. H. Pinto, and R. A. Lamb. 1997. The active oligomeric state of the minimalistic influenza virus M2 ion channel is a tetramer. Proc. Natl. Acad. Sci. U. S. A. 94:5000-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salzwedel, K., and E. A. Berger. 2000. Cooperative subunit interactions within the oligomeric envelope glycoprotein of HIV-1: functional complementation of specific defects in gp120 and gp41. Proc. Natl. Acad. Sci. U. S. A. 97:12794-12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santiago, C., M. L. Celma, T. Stehle, and J. M. Casasnovas. 2010. Structure of the measles virus hemagglutinin bound to the CD46 receptor. Nat. Struct. Mol. Biol. 17:124-129. [DOI] [PubMed] [Google Scholar]
- 41.Santiago, C., A. Gutierrez-Rodriguez, P. A. Tucker, T. Stehle, and J. M. Casasnovas. 2010. Crystallization and preliminary crystallographic analysis of the measles virus hemagglutinin in complex with the CD46 receptor. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66:91-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schagger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 43.Spearman, C. 1908. The method of right and wrong cases (constant stimuli) without Gauss's formula. Br. J. Psychol. 2:227-242. [Google Scholar]
- 44.Sutter, G., M. Ohlmann, and V. Erfle. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9-12. [DOI] [PubMed] [Google Scholar]
- 45.Tanabayashi, K., and R. W. Compans. 1996. Functional interaction of paramyxovirus glycoproteins: identification of a domain in Sendai virus HN which promotes cell fusion. J. Virol. 70:6112-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vongpunsawad, S., N. Oezgun, W. Braun, and R. Cattaneo. 2004. Selectively receptor-blind measles viruses: identification of residues necessary for SLAM- or CD46-induced fusion and their localization on a new hemagglutinin structural model. J. Virol. 78:302-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, Z., A. M. Mirza, J. Li, P. J. Mahon, and R. M. Iorio. 2004. An oligosaccharide at the C-terminus of the F-specific domain in the stalk of the human parainfluenza virus 3 hemagglutinin-neuraminidase modulates fusion. Virus Res. 99:177-185. [DOI] [PubMed] [Google Scholar]
- 48.Wittig, I., H. P. Braun, and H. Schagger. 2006. Blue native PAGE. Nat. Protoc. 1:418-428. [DOI] [PubMed] [Google Scholar]
- 49.Xu, K., K. R. Rajashankar, Y. P. Chan, J. P. Himanen, C. C. Broder, and D. B. Nikolov. 2008. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Acad. Sci. U. S. A. 105:9953-9958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, X., S. Kurteva, X. Ren, S. Lee, and J. Sodroski. 2006. Subunit stoichiometry of human immunodeficiency virus type 1 envelope glycoprotein trimers during virus entry into host cells. J. Virol. 80:4388-4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yuan, P., G. P. Leser, B. Demeler, R. A. Lamb, and T. S. Jardetzky. 2008. Domain architecture and oligomerization properties of the paramyxovirus PIV 5 hemagglutinin-neuraminidase (HN) protein. Virology 378:282-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan, P., T. B. Thompson, B. A. Wurzburg, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2005. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure 13:803-815. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, L., R. Hernan, and B. Brizzard. 2001. Multiple tandem epitope tagging for enhanced detection of protein expressed in mammalian cells. Mol. Biotechnol. 19:313-321. [DOI] [PubMed] [Google Scholar]