Abstract
Understanding the lack of disease progression in nonpathogenic simian immunodeficiency virus (SIV) infections is essential for deciphering the immunopathogenesis of human AIDS. Yet, in vivo studies have been hampered by a paucity of infectious molecular clones (IMCs) of SIV suitable to dissect the viral and host factors responsible for the nonpathogenic phenotype. Here, we describe the identification, cloning, and biological analysis of the first transmitted/founder (T/F) virus representing a nonpathogenic SIV infection. Blood was collected at peak viremia from an acutely infected sabaeus monkey (Chlorocebus sabaeus) inoculated intravenously with an African green monkey SIV (SIVagm) strain (Sab92018) that had never been propagated in vitro. To generate IMCs, we first used conventional (bulk) PCR to amplify full-length viral genomes from peripheral blood mononuclear cell (PBMC) DNA. Although this yielded two intact SIVagmSab genomes, biological characterization revealed that both were replication defective. We then performed single-genome amplification (SGA) to generate partially overlapping 5′ (n = 10) and 3′ (n = 13) half genomes from plasma viral RNA. Analysis of these amplicons revealed clusters of nearly identical viral sequences representing the progeny of T/F viruses. Synthesis of the consensus sequence of one of these generated an IMC (Sab92018ivTF) that produced infectious CCR5-tropic virions and replicated to high titers in Molt-4 clone 8 cells and African green monkey PBMCs. Sab92018ivTF also initiated productive infection in sabaeus monkeys and faithfully recapitulated the replication kinetics and nonpathogenic phenotype of the parental Sab92018 strain. These results thus extend the T/F virus concept to nonpathogenic SIV infections and provide an important new tool to define viral determinants of disease nonprogression.
Simian immunodeficiency viruses (SIVs) have been identified in over 40 African nonhuman primate (NHP) species (reviewed in references 40 and 64), although to date only SIVs infecting sooty mangabeys (SIVsmm), African green monkeys (SIVagm) and mandrills (SIVmnd), have been studied in detail (reviewed in references 39, 40, 61, and 64). Natural SIV infections share many similarities with human immunodeficiency virus (HIV) infections, including high levels of viral replication and marked depletion of mucosal CD4+ T cells during acute infection (reviewed in references 39, 40, 61, and 64). However, fundamental differences have also been identified. Most importantly, African green monkey (AGM) and sooty mangabey (SM) hosts of SIV show only an early transient increase in immune activation and do not develop AIDS (6-8, 10, 16, 20, 21, 28, 37, 41, 44, 50, 58, 59).
Comparisons of nonpathogenic and pathogenic primate lentiviral infections have become an important area of AIDS research (reviewed in references 27, 39, 40, and 61). It is now clear that host factors can play an important role in the clinical outcome of infection. This is perhaps best illustrated by the fact that SIV from SMs is usually nonpathogenic in its natural mangabey host, but highly virulent in experimentally infected macaques (reviewed in references 27, 39, 40, and 61). Features that distinguish SMs and AGMs from macaques that may allow them to avoid chronic immune activation and disease progression include effective regulatory T-cell responses that establish an anti-inflammatory milieu, reduced levels of CCR5 on the surface of CD4+ T cells, and the maintenance of an intact mucosal barrier that prevents microbial translocation (reviewed in references 27, 39, 40, and 61). However, viral properties that differentiate SIVagm and SIVsmm from HIV-1 and its chimpanzee (CPZ) precursor, SIVcpz, such as the lack of a vpu gene and the expression of Nef proteins that block the activation of virally infected T cells by T-cell receptor (TCR)-CD3 down-modulation (3, 55, 56), may also contribute to an anti-inflammatory environment (reviewed in reference 27). The recent finding that SIVcpz is pathogenic in naturally infected chimpanzees in the wild (26) is consistent with this possibility.
One approach to explore viral determinants of in vivo pathogenicity is to generate infectious proviral clones and mutants that differ in virus-specific properties, such as coreceptor tropism or accessory gene functions. Site-specific mutants of SIV have been used extensively in the pathogenic SIV of macaques (SIVmac)/macaque model (reviewed in reference 2). However, this has not been done for nonpathogenic SIV infections, such as SIVagm infection of AGMs and SIVsmm infection of SMs, which represent the best-studied model systems (reviewed in references 39, 40, 61, and 64). Although some infectious SIVsmm clones exist (18, 63), SMs are classified as endangered and can thus not be used for invasive studies. In contrast, AGMs are available in large numbers in captivity and there is no restriction concerning in vivo experimentation. To date a number of infectious molecular clones (IMCs) from vervet (TYO-1, 155, 3, and 9063), grivet (GRI-1), tantalus (TAN-1), and sabaeus (SAB-1) monkeys have been described (1, 4, 13, 19, 22, 23, 60). However, all of these were obtained after extensive passage in human cell lines, and many exhibit features selected by in vitro propagation, such as a truncated transmembrane envelope glycoprotein (gp41) or nonsense mutations in accessory genes (4, 13, 22, 60). Since it is unknown to what extent in vitro culture alters in vivo viral properties, existing SIVagm clones may not be suitable for in vivo pathogenesis experiments.
To generate a physiologically relevant SIVagm proviral clone, we thus selected a virus stock (Sab92018) that had never been propagated in vitro. This strain was originally identified in a wild-caught sabaeus monkey from Senegal and thus represents a viral lineage that circulated in the wild (10). Plasma from this chronically infected animal was used to infect two captive sabaeus monkeys to generate high-titer viral stocks without in vitro propagation. In each case, this was done by intravenous inoculation of plasma, followed by the harvest of plasma from the experimentally infected animal at peak viral replication (10, 41). The plasma stocks of Sab92018 have already been studied extensively in vitro and in vivo, and the biological properties of this strain are thus well known (10, 14, 15, 17, 41-45). To clone Sab92018, we first generated two full-length SIVagmSab proviruses (A5 and E2), using conventional PCR and cloning techniques. Since this approach yielded replication defective clones, we next used single-genome amplification (SGA) and direct sequencing of plasma viral RNA to infer the sequence of a transmitted/founder (T/F) virus that had established a productive infection. We have used this approach in the past to generate replication-competent clones of HIV-1 and SIVmac (32, 53; M. J. Lopker, B. Hahn, and G. Shaw, unpublished data). Chemical synthesis of a Sab92018 T/F genome generated a biologically active molecular clone that replicated to high titers both in vitro and in vivo. Our results thus show that the same principles of T/F virus inference and analysis previously established for HIV-1 and SIVmac also apply to nonpathogenic SIV infections and that this strategy can thus be used to generate molecular clones that recapitulate the in vivo replication properties of the parental virus strains.
MATERIALS AND METHODS
Passage history of Sab92018.
The original Sab92018 strain was identified in a wild-caught adult sabaeus monkey from Senegal (10). Plasma from this index animal was used to infect a captive sabaeus monkey (92018) originating from Senegal by intravenous inoculation to generate a high-titer virus stock without in vitro propagation (10). This plasma stock was used for various in vitro and in vivo experiments (10, 28) and subsequently replenished by infecting a second sabaeus monkey originating from St. Kitts. This Caribbean AGM, termed EI43, was also infected by intravenous inoculation with uncultured plasma with 300 50% tissue culture infectious doses (TCID50s) (41). Sab92018 thus represents a naturally occurring SIVagm strain that was never adapted in tissue culture. Sab92018 was passaged only twice, and then only in its natural host. Moreover, in vivo passage was performed using sufficiently large inocula to maintain the quasispecies complexity of the original viral strain. Importantly, there was no evidence of increased viral loads or altered pathogenicity following the two passages (10, 28, 41). Animal EI43 was euthanized 9 days postinoculation and maximum volumes of blood were collected. Plasma and peripheral blood mononuclear cells (PBMCs) were separated and frozen in aliquots at −80°C. The new plasma virus stock was named SIVagmSab92018 (EI43) and quantified by real-time PCR, and the titer was determined on SupT1 as described previously (10, 41).
Generation of near-full-length SIVagmSab92018 genomes by conventional PCR.
High-molecular-weight DNA was isolated by phenol-chloroform extraction from the PBMCs of animal EI43 9 days postinfection (p.i.). Near-full-length genomes were amplified by nested PCR using the expand long-template PCR system (Roche) according to the manufacturer's instructions. Long terminal repeat (LTR) regions were amplified separately in a single-round PCR The resulting 768-bp amplicon was cloned into pCR-Script SK+ and used to generate a complete provirus.
Single-genome amplifications of 5′ and 3′ genomic halves.
Viral RNA was extracted from plasma of animal EI43, collected 9 days p.i. using the QIAamp viral RNA minikit (Qiagen). cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) and strain-specific primers a829r (5′-CCAAAGAGGTCTGACTATCCAAAGCTTTTC-3′) for the 5′ half, and a796r (5′-CTCCTCCCTGGAAAGTCCCGCT-3′) for the 3′ half, respectively. Single genome amplification was performed as described previously (5, 32, 52). Briefly, cDNA was serially diluted and distributed in replicates of eight PCRs in 96-well plates. Dilutions in which positive wells constituted less than 30% of PCRs were identified and used for additional amplifications. At these dilutions, most wells contained amplicons from single cDNA molecules. PCR amplifications were carried out using 1× Platinum Taq high-fidelity PCR buffer, 2 mM MgSO4, 0.2 mM each deoxynucleoside triphosphate, 0.2 μM each primer, and 0.02 U/μl of Platinum high-fidelity polymerase in 20-μl reactions for the first round and 50-μl reactions for the second round. The nested primers for generating 5′-half-genome amplicons (covering U5, gag, and pol) included 798f (5′-CAAGTGTGTGCCCATTTATTCCTCAG-3′) and a829r (5′-CCAAAGAGGTCTGACTATCCAAAGCTTTTC-3′) in the first round and a799f (5′-GTAAAACCCTGGTTTACTAAGGATCCCTG-3′) and a828r (5′-TTCCTGTATCACCACTGCTCCTTCTCCTTTCCA-3′) in the second round, respectively. The nested primers for generating 3′-half-genome amplicons (covering vif, vpr, rev, tat, env, nef, and U3) included a787f (5′-TGYTGGTGGGGAAAIATAGAGCACAC-3′) and a796r (5′-CTCCTCCCTGGAAAGTCCCGCT-3′) for the first round, and a788f (5′-CACAATTTTAAAAGAAARGGRGGRATTGGGG-3′) and a797r (5′-GGATGTGGTTTTGTGGTTAGGCAGA-3′) for the second round, respectively. The PCR conditions were as follows: 94°C for 1 min and then 35 cycles of 94°C for 15 s, 55°C for 30 s, and 68°C for 5 min and 30 s, followed by a final extension of 15 min at 68°C. One microliter of the first-round reaction was transferred as template to the second round. Amplicons were gel purified and sequenced directly using an ABI 3730 DNA analyzer.
Molecular cloning of an Sab92018 transmitted/founder virus.
To clone a full-length SIVagmSab T/F virus, we first inferred its sequence from 5′- and 3′-half-genome SGA amplicons and 426 bp from the LTR sequence obtained following conventional PCR. This sequence was then synthesized (Blue Heron Biotechnology, Bothell, WA) as two fragments, 4,707 bp and 5,322 bp in length, which were joined at a unique BglI site (Fig. 1). NotI and MluI cloning sites attached during synthesis at the 5′ and 3′ ends of the half-genome fragments, respectively, facilitated cloning into the pCR-XL-TOPO vector. The clone, designated pSab92018ivTF, was grown in XL2-MRF cells at 30°C (Stratagene).
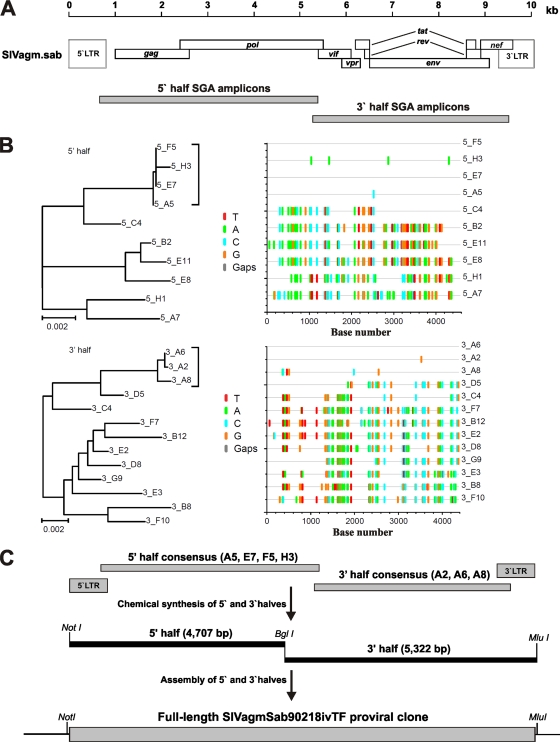
FIG. 1.
Molecular cloning of a SIVagmSab transmitted/founder virus. (A) Single-genome amplification was used to generate 5′ (n = 10)- and 3′ (n = 13)-half-genome sequences (drawn to scale) from the plasma of a sabaeus monkey acutely infected with a high-titer stock of isolate SIVagmSab92018. (B) Phylogenetic trees and Highlighter plots of 5′ (top row)- and 3′ (bottom row)-half-genome sequences. Tick marks indicate differences compared to the top sequence, which also represents the inferred transmitted/founder sequence (red, T; green, A; blue, C; and orange, G). Trees were inferred by the neighbor-joining method and are midpoint rooted. The scale bar represents 0.002 substitution per site. Discrete low-diversity lineages representing the progeny of a transmitted founder virus are indicated by brackets (C) The consensus sequence of low-diversity lineages was used to synthesize the Sab92018ivTF genome as proviral halves. Flanking NotI and MluI restriction sites and an internal BglI site allowed cloning into the pCR-XL TOPO vector.
Phylogenetic analyses.
The Sab92018 T/F virus was compared to representative SIVagm strains and other primate lentiviruses in the viral Env region. Deduced Env amino acid sequences were used from the following SIV strains (GenBank accession numbers are in parentheses) (29): SIVagmSab (SAB1, U04005), SIVagmSab (P1, U59187), SIVagmGri (GRI1, M66437), SIVagmVer (155, M29975; and 9063, L40990), SIVagmTan (Tan1, U58991), SIVsmm (SL92B, AF334679), SIVrcm (GAB1, AF382829), SIVcpz (TAN1, AF447763), SIVmus-1 (CM1085, AY340700), SIVtal (00CM266, AY655744), SIVsyk (KE51, AY523867), SIVdeb (CM40, AY523865), SIVden (CD1, AJ580407), SIVmnd-2 (CM16, AF367411), and SIVcol (CGU1, AF301156). Sequences were aligned using ClustalW (30), and the resulting alignments were visually adjusted. Sites that could not be aligned unambiguously or sites with a gap in any sequence were excluded. Phylogenetic trees, along with support using 1,000 bootstrap replicates (12), were inferred by the neighbor-joining method (51), as implemented in ClustalW.
Cell culture and virus stocks.
TZM-bl cells, which express CD4, CXCR4, and CCR5 and contain Tat-responsive reporter genes for β-galactosidase and the firefly luciferase under the control of an HIV-1 long terminal repeat (LTR) sequence (47, 65), were obtained from the NIH ARRRP, as contributed by John Kappes and Xiaoyun Wu. These cells and 293T cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. For the generation of virus stocks, 293T cells were transfected with proviral constructs by the calcium phosphate method as described previously (35). The medium was changed after overnight incubation, and virus was harvested 24 h later. Residual cells in the supernatants were pelleted, and the supernatants were stored at −80°C. The content of viral p27 capsid antigen was quantified by SIV core p27 antigen capture assay kit (Zeptometrix Corp., Buffalo, NY). Quantification was done according to the manufacturer's instructions.
Western blotting.
To assess viral gene expression, 293T cells were transfected with 5 μg of DNA of the SIVagmSab constructs A5, E2, Sab92018ivTF, and SAB-1. Two days posttransfection, cells were lysed in 500 μl radioimmunoprecipitation assay (RIPA) buffer. To analyze viral particles produced from the molecular clones, 500 μl of the culture supernatants was centrifuged at 14,000 rpm for 30 min, and the resulting pellets were resuspended in 25 μl RIPA buffer. Proteins were separated on a 4 to 12% NuPAGE Novex Bis-Tris precast gel (Invitrogen) and transferred to polyvinylidene difluoride (PVDF) microporous membranes (Millipore, Bedford, MA) by electroblotting. Viral proteins were detected using serum from an AGM infected with SIVagmsab92018 and probed with horseradish peroxidase-conjugated goat anti-human immunoglobulin G (γ-chain-specific) secondary antibody (Sigma). Antibody complexes were detected by DAB (3, 3′-diaminodbenzidine; Vector Labs).
Viral infectivity and coreceptor usage.
To determine the infectivity of virions produced by the various SIVagmSab clones, TZM-bl cells were seeded in 96-well plates at a density of 5,000 cells/well and infected after overnight incubation with virus stocks containing 10 ng of p27 capsid antigen produced by transiently transfected 293T cells. Two days p.i., viral infectivity was detected using a galactosidase screen kit from Tropix as recommended by the manufacturer. β-Galactosidase activities were quantified as relative light units (RLU) per second with an Orion Microplate luminometer. TZM-bl cells were also used to determine the coreceptor preference of the SIVagmSab92018ivTF clone. Cells were seeded at 5,000 cells/well in 96-well plates overnight and then treated with Maraviroc (10 nM) and/or AMD3100 (10 μM) for 1 h. Infections were performed using virus stocks containing normalized quantities of p24 or p27 antigen (10 ng). After 48 h of incubation at 37°C, supernatant was removed, and cells were processed for detection of luciferase activity as described above. To further analyze the coreceptor usage of the SIVagmSab92018ivTF clone, GHOST cells were used as previously described (34). The six GHOST cell lines used express CD4 alone or together with the viral coreceptors CCR5, CXCR4, BOB/GPR15, and Bonzo/STRL33 and contain the gene encoding the green fluorescent protein (GFP) under the control of the HIV LTR (34). A total of 5 × 104 cells were exposed to virus stocks containing 10 ng of p24 or p27 antigen produced by transient transfection of 293T cells. Three days after infection, the percentage of virally infected GFP-positive cells was analyzed by fluorescence-activated cell sorting (FACS). Uninfected cells were used as a negative control.
Viral replication in African green monkey PBMCs.
PBMCs from SIV-uninfected AGMs (n = 4), were isolated by density gradient centrifugation over lymphocyte separation medium (MP Biomedical, Irvine, CA). Briefly, blood was layered over density gradient medium in a ratio of 2:1 and centrifuged at 18°C at 400 × g for 25 min. The monolayer containing PBMCs was resuspended with RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin (1 mg/ml; Invitrogen, CA) for subsequent studies. Freshly isolated PBMCs were stimulated with 10 μg phytohemagglutinin (PHA) per ml of medium for 2 days followed by overnight incubation in interleukin-2 (IL-2) medium. Activated PBMCs (5 × 106) were infected with virus stocks containing 4 ng of p27 capsid antigen of the SIVagmSab92018ivTF clone produced by transiently transfected 293T cells or the parental SIVagmSab92018 (EI43) plasma stock at 37°C for 4 h; cells were then washed extensively to remove any cell-free virus. Cells were maintained in IL-2 medium for 4 weeks. Virus production in culture supernatants was monitored weekly by SIV P27 antigen capture assay. An aliquot of PBMCs was also depleted of CD8+ cells by a positive selection procedure (CD8 microbead kit; Miltenyi Biotech, Auburn, CA). This was done to improve the levels of viral replication since the CD4/CD8 T-cell ratio is relatively low (about 1/4) in uninfected AGMs. The CD8+-depleted cells were then stimulated with 10 μg/ml PHA for 2 days, followed by overnight incubation in IL-2 medium. A total of 5 × 105 CD8+-depleted cells were infected as described above for the PBMCs. For 4 weeks, one-half of the supernatant was collected every third day and replaced with fresh IL-2-containing medium. Virus production in culture supernatants was monitored by SIV p27 antigen capture assay.
Infection of AGMs.
One Caribbean AGM housed at the Tulane National Primate Research Center (TNPRC), an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) International facility, was infected with a virus stock of Sab92018ivTF derived from the supernatant of transiently transfect 293T cells containing 4 ng of p27 capsid antigen. Additionally, viral replication was compared to that recorded in five AGMs infected with 300 TCID50 of the SIVagmSab92018 (EI43) plasma stock. The animals were fed and housed according to regulations set forth by the Guide for the Care and Use of Laboratory Animals (36) and the Animal Welfare Act. This study was approved by the Tulane University Institutional Animal Care and Use Committee (IACUC). Blood was collected from the animals at two preinfection times (days −15 and −7) at the time of virus inoculation, biweekly for the first 2 weeks, weekly for the next 4 weeks, and monthly thereafter, up to 100 days p.i. Another three Caribbean AGMs imported from Barbados were infected intravenously with 500 ng of p27 capsid antigen of the SIVagmSab92018ivTF viral clone at the German Primate Centre. This animal experiment was licensed by the ethical board enacted through the German Animal Welfare Act under no. 33.11.42502-04-094/08 issued by the Lower Saxony State Office for Consumer Protection and Food Safety. Blood was collected from all animals twice to three times before infection and at regular intervals after infection. Plasma viral loads were determined as previously described (41).
FACS analysis for CD4+ T cells.
CD4+ T-cell proportions were determined by staining whole-blood leukocytes with a pretitrated antibody cocktail comprising anti-CD11a-allophycocyanin (APC) (clone HI 111), CD3-Alexa700 (clone SP34-2) and CD4-Horizon V450 (clone L200), all obtained from Becton Dickinson (BD). Following staining with the antibody mixture, red blood cells were lysed with BD FACS lysing solution and labeled lymphocytes were analyzed for their expression of cell surface markers by flow cytometry on an LSR II flow cytometer (Becton Dickinson, Heidelberg, Germany). Lymphocyte populations were gated based on forward and side scatter characteristics and then exclusion of doublets and expression of CD11a, followed by that of CD3. Data were generated with BD FACS Diva 6.1.3 Software before analysis with FlowJo 8.8 Software (Treestar).
Nucleotide sequence accession number.
The sequence of the molecular SIVagmSab92018ivTF clone generated in this study has been submitted to GenBank under accession no. HQ378594.
RESULTS
Generation of two replication-defective SIVagmSab proviral clones by conventional PCR.
Our first attempt to generate infectious molecular clones of Sab92018 involved conventional bulk amplification and cloning of near-full-length genome fragments from PBMC DNA. Two such clones, termed E2 and A5, were selected for proviral construction because sequence analysis revealed that both carried uninterrupted gag, pol, vif, vpr, tat, rev, env, and nef genes, as well as intact cis-regulatory sequence elements, such as the TATA box, the primer-binding site, the ribosomal frameshift region at the gag-pol junction, and the polypurine tract. Both clones were thus reconstructed to contain full-length LTRs. However, biological analysis showed that the two clones produced virions that were only poorly infectious and failed to replicate in CD4+ target cells in vitro (data not shown). Thus, conventional PCR and cloning approaches failed to generate biologically active clones of SIVagmSab92018, despite yielding fragments with a seemingly intact genomic organization.
SGA analysis identifies a transmitted/founder SIVagmSab genome.
Since plasma was collected from animal EI43 during the acute phase of infection, we reasoned that SGA of plasma viral RNA would allow us to infer the sequence of viruses that had established the infection 9 days earlier (24, 25). We thus used SGA to generate 5′ (n = 10) and 3′ (n = 13) genome halves and sequenced them directly (Fig. 1A). As shown in Fig. 1B, the resulting amplicons exhibited a substantial amount of sequence diversity, consistent with the fact that monkey EI43 was infected with virus from a chronically infected animal. However, phylogenetic analysis also revealed one low-diversity lineage among both the 5′ and 3′ amplicons, which comprised sequences that differed by only few nucleotides (indicated by brackets in Fig. 1B). Such discrete low-diversity lineages represent the progeny of T/F viruses, whose sequences can be inferred by determining their consensus sequences (24, 25, 31, 53, 62). Since both 5′ and 3′ consensus sequences were identical in the 298-bp region of sequence overlap, we reasoned that they represented the same T/F virus. Missing LTR sequences were derived by conventional PCR (using a high-fidelity polymerase) and direct sequencing, and the entire T/F virus genome was synthesized as two fragments, which were joined at a unique BglI site (Fig. 1C).
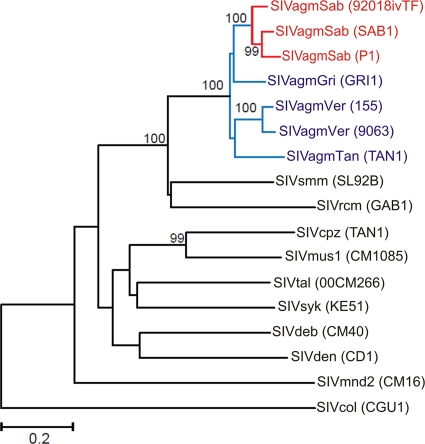
The full-length Sab92018 T/F virus was 10,004 bp in length and carried intact gag, pol, vif, vpr, tat, rev, env, and nef genes. Like the previously reported SAB-1 molecular clone and the genomes of SIVsmm and HIV-2 (22), the Sab92018 T/F virus had a single NF-κb binding site in its LTR, three NF-AT interaction sites and duplicated TAR sequences. Known cis-regulatory sequence elements were also preserved. Unlike other molecular clones of SIV obtained after extensive in vitro propagation, the Sab92018ivTF env gene encoded a full-length gp41 with a cytoplasmic tail of 157 amino acids (aa). The predicted length of the Sab92018ivTF Vpr protein is 138 aa, which is similar to that of SAB-1 (140 aa) but longer than those of SIVagmVer and SIVagmGri (120 aa) (22). As expected, in phylogenetic trees of Env amino acid sequences, Sab92018ivTF clustered closely with SAB-1 and P1, forming a species-specific clade within the SIVagm radiation (Fig. 2).
FIG. 2.
Phylogenetic relationship of the SIVagmSab transmitted/founder virus to other primate lentiviruses. The tree was inferred from Env amino acid sequences. Numbers at nodes are percent bootstrap support (only values of 70% or greater are shown). The scale represents 0.2 substitution per site.
The SIVagmSab transmitted/founder virus is replication competent.
To determine whether the Sab92018ivTF virus produces infectious virus, we infected TZM-bl indicator cells with 293T cell-derived virus stocks containing 10 ng of p27 capsid antigen and determined the β-galactosidase activities 2 days later. Previously described SAB-1 and TAN-1 clones (22, 60) were used as positive controls. In contrast to the SIVagmSab A5 and E2 proviruses derived by conventional PCR, the Sab92018ivTF clone yielded highly infectious viral particles (Fig. 3A) that replicated efficiently in Molt-4 clone 8 cells (Fig. 3B). To determine whether this was due to altered protein expression or particle assembly, we performed Western blot analyses. The results showed that A5, E2, and Sab92018ivTF proviral constructs expressed all major viral proteins at levels similar to those of the SAB-1-positive control (Fig. 3C). Moreover, transfection-derived supernatants contained viral particles that could be pelleted (Fig. 3C). Thus, the replication block in the A5 and E2 proviruses must be due to defects following protein expression and virion assembly.
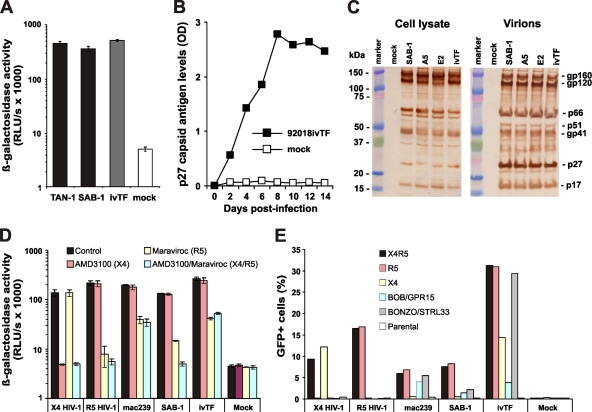
FIG. 3.
Functional characterization of the transmitted/founder Sab92018ivTF clone. (A) TZM-bl indicator cells were infected with the indicated SIVagm molecular clones. Infections were performed with virus stocks containing 10 ng of p27 antigen. Panels A and D show average values of triplicate infections ± standard deviations (SD). ivTF, SIVagmSab92018ivTF. (B) Replication of SIVagmSab92018ivTF in Molt-4 clone 8 cells. OD, optical density. (C) Western blot analysis of cellular extracts of 293T cells transfected with the indicated molecular clones of SIVagmSab (left) and virions pelleted from the culture supernatant (right). (D) TZM-bl cells were left untreated (control) or were pretreated with specific inhibitors of CCR5 (Maraviroc), CXCR4 (AMD3100), or a combination thereof prior to infection with the indicated molecular clones of HIV-1 or SIV. (E) Coreceptor usage by SIVagmSab92018ivTF and the indicated control viruses tested in GHOST cells expressing CD4 alone (parental) or together with CCR5, CXCR4, or the orphan receptors BOB/GPR15 and BONZO/STRL33. All infectivity data were confirmed in one or two independent experiments.
Next, we analyzed the coreceptor tropism of SIVagm 90218ivTF using two specific small-molecule coreceptor antagonists: i.e., Maraviroc, which specifically blocks CCR5(R5)-tropic HIV-1 infection; and AMD3100, a specific inhibitor of CXCR4(X4)-mediated HIV-1 entry (reviewed in reference 49). The X4-tropic wild-type HIV-1 NL4-3 strain, an R5-tropic derivative thereof containing the gp120 V3 loop of HIV-1 TH014 (46), SIVmac239, and SIVagm SAB-1 were analyzed in the same experiment for control. As expected, AMD3100 blocked wild-type NL4-3 infection, whereas Maraviroc specifically inhibited the R5-tropic HIV-1 derivative (Fig. 3D). Infection of SIVmac239, SIVagmSAB-1 and SIVagm90218ivTF was inhibited by Maraviroc but not affected by AMD3100 (Fig. 3D), demonstrating that all three molecular clones of SIV utilize R5 but not X4 for entry into TZM-bl cells. However, infection by SIVagm92018ivTF and SIVmac239 was not blocked entirely, suggesting that these viruses may utilize alternative coreceptors to gain entry into TZM-bl cells. To further investigate the coreceptor tropism of SIVagm92018ivTF, we infected GHOST cells, which stably express CD4 alone or together with different coreceptors and contain the GFP reporter gene under the control of the viral LTR promoter (34), with virus stocks containing normalized amounts of p24 or p27 antigen. In agreement with published data (9, 11), SIVmac239 infected cells expressing CCR5 or the orphan receptors BOB/GPR15 and BONZO/STRL33 (Fig. 3E). Unexpectedly, Sab92018ivTF utilized BONZO/STRL33 about as efficiently as R5 for entry into GHOST target cells (Fig. 3E). Sab92018ivTF also infected GHOST cells expressing X4 and BOB/GPR15, albeit with lower efficacy. Thus, like most primary SIV and HIV-1 strains, Sab92018ivTF utilizes R5 for infection but is also able to enter cells via alternative coreceptors, particularly BONZO/STRL33.
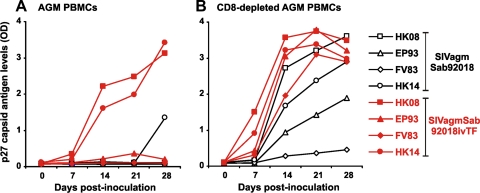
We next compared the replicative capacities of the Sab92018ivTF clone and its parental SIVagmSab92018 strain in AGM PBMCs. In the presence of CD8+ T cells, Sab92018ivTF replicated efficiently only in the PBMCs from two of four animals. Replication in PBMCs from the remaining two AGMs was markedly reduced, but titers were higher than those of the parental SIVagmSab92018 isolate, which did not replicate to detectable levels (Fig. 4A). In contrast, both cloned and uncloned SIVagm strains were capable of establishing a productive infection in AGM PBMCs in the absence of CD8+ T cells (Fig. 4B). Again, the Sab92018ivTF clone consistently replicated with higher efficiency than the parental SIVagmSab92018 isolate in cells derived from all four animals examined. These results demonstrate that the Sab92018ivTF clone shows high replication fitness in primary cells derived from its natural AGM host.
FIG. 4.
Replication of SIVagmSab molecular clones in AGM PBMCs. PBMCs (left) or PBMCs depleted of CD8+ T cells (right) derived from four AGMs were infected with the parental SIVagmSab92018 strain or the 92018ivTF molecular clone. Virus production was monitored by p27 antigen ELISA.
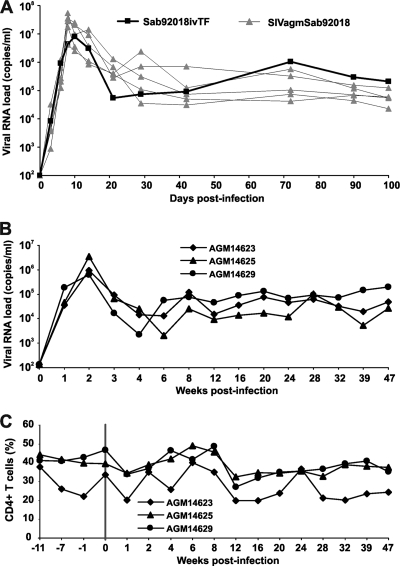
To evaluate whether the T/F clone was also able to establish a productive infection in vivo, we initially infected one Caribbean AGM (Chlorocebus sabaeus) intravenously with a virus stock produced by transfection of 293T cells. For comparison, five other animals received the parental SIVagmSab92018 strain. We found that the Sab92018ivTF clone replicated about as efficiently in vivo as the parental SIVagmSab92018 strain (Fig. 5A). Peak levels of viral RNA were observed at 10 days p.i. for the molecular Sab92018ivTF clone (7.91 × 106 copies/ml) and at 8 days p.i. for the uncloned Sab92018 virus [(2.81 ± 1.91) × 107 copies/ml]. Most importantly, Sab92018ivTF established a set-point viral load by day 42 p.i., ranging from 105 to 106 copies per ml and which was maintained up to day 100 p.i. (Fig. 5A). Thus, the levels of Sab92018ivTF replication during the chronic phase of infection were as high as or even higher than those detected in the wild-type SIVagmSab92018-infected animals. To examine further the replication fitness of Sab92018ivTF in vivo, three additional AGMs were infected at the German Primate Research Center. The results showed that the SIVagm Sab92018ivTF molecular clone replicated persistently at high levels for almost 1 year of follow-up (Fig. 5B). Furthermore, the three infected AGMs maintained stable CD4+ T-cell counts (Fig. 5C) and did not show signs of disease progression (data not shown).
FIG. 5.
Replication of the SIVagmSab92018ivTF clone in African green monkeys. (A) Viral RNA loads in one AGM infected with SIVagmSab92018ivTF and five animals that received the uncloned parental SIVagmSab92018 strain at the TNPRC. (B and C) Levels of plasma viremia (B) and CD4+ T-cell counts (C) in three sabaeus monkeys infected with SIVagmSab92018ivTF at the German Primate Center. Individual animal numbers are indicated by five-digit numbers. The vertical line marks the time point of infection. The limit of viral RNA detection is approximately 100 copies/ml of plasma. Values were determined as described in Materials and Methods.
DISCUSSION
In the present study, we generated and functionally characterized three full-length molecular clones of a strain of SIVagmSab that has only been propagated in its natural host. We found that two clones (A5 and E2) derived by conventional PCR produced poorly infectious virions that were replication defective. In contrast, a proviral clone (Sab92018ivTF) representing a T/F virus was highly infectious and replicated efficiently both in vitro and in vivo. Virus generated from the T/F IMC exhibited biological properties that were very similar to those of the parental SIVagmSab92018 strain, including CCR5 tropism and the ability to establish a productive and persistent infection in AGMs in vivo. Thus, we have generated the first IMC that exhibits the properties of nonpathogenic SIV infection but is not compromised by interim propagation in tissue culture.
Conventional PCR can generate in vitro artifacts, especially when used to amplify genetically diverse sequence mixtures such as viral quasispecies (52). Single-genome amplification precludes Taq polymerase-induced recombination (template switching) and nucleotide misincorporation, thereby ensuring an accurate representation of viral variants as they exist in vivo. Given that animal EI43 was infected with SIVagm from a chronically infected monkey, it was not surprising that the two proviruses derived by conventional PCR were replication defective. Notably, most biologically active SIVagm clones available thus far (3, 155, 9063, TYO, GRI-1, SAB-1, and TAN-1) were generated by lambda phage cloning of proviral DNA (1, 4, 13, 22, 23, 60), which is not prone to these same artifacts. To avoid bulk PCR shortcomings, we opted to clone a T/F provirus inferred from SGA amplicons, a strategy that has yielded replication-competent molecular clones in the past (32, 53). T/F viruses are responsible for initiating productive clinical infections. It is thus not surprising that the T/F IMC of SIVagmSab replicated to high titers both in vitro and in vivo.
Sabaeus monkey EI43 was infected by intravenous inoculation with a plasma stock originally derived from a chronically infected sabaeus monkey. Although in this study, we focused on cloning only one T/F IMC, it is clear that additional transmitted founder viruses would have been identified had we generated additional SGA amplicons. Recent studies in SIVmac-infected rhesus macaques have shown that intravenous infection is more than 2,000-fold more efficient than intrarectal infection and is frequently associated with multivariant transmissions (25). Similarly, humans infected with HIV-1 by intravenous routes are more likely to acquire multiple variants than humans exposed by sexual routes (5). Thus, the T/F virus that we identified in the plasma of animal EI43 is likely one of many. Since we cloned a T/F virus that was not subjected to a mucosal bottleneck, we also examined AGMs that were infected intrarectally with the same plasma stock used to derive SAB92018ivTF. Characterizing the transmitted founder viruses in four such animals, we found one that differed from SAB92018ivTF by only a single synonymous substitution (I. Pandrea, C. Apetrei, and B. Hahn, unpublished observations). These results suggest that SAB92018ivTF will also be capable of infecting AGMs by mucosal transmission routes.
In vivo analyses suggest that Sab92018ivTF faithfully reproduced the properties of its parental strain, which has been extensively characterized (10, 14, 15, 17, 41-45). All four Sabaeus monkeys infected with the Sab92018ivTF clone maintained high viral loads without developing immunodeficiency during follow-up. Thus, the newly generated Sab92018ivTF strain will be useful to characterize viral determinants of nonprogression in the natural NHP hosts. For example, it has been suggested that the lack of a vpu gene and the ability of Nef to block T-cell activation by down-modulation of T-cell receptor (TCR)-CD3 may contribute to the low levels of immune activation (27). To examine these hypotheses, we have already created derivatives of the Sab92018ivTF clone expressing the SIVgsn Vpu, which antagonizes AGM tetherin (54, 66) and the HIV-1 Nef protein that is unable to down-modulate CD3 (55). Preliminary results show that this chimeric SIVagmSab clone is capable of establishing a persistent infection in sabaeus monkeys (U. Sauermann, C. Stahl-Hennig and F. Kirchhoff, unpublished data). Future studies will thus be able to address whether this “HIV-1-like” SIVagmSab derivative causes higher levels of immune activation and is more pathogenic than the original Sab92018ivTF clone. The Sab92018ivTF clone will also be useful to evaluate other determinants of virus transmission and pathogenicity, such as the role of viral properties in the frequency of vertical transmissions (38, 43) or the importance of coreceptor tropism for the maintenance of stable CD4+ T-cell counts (33).
Like the parental SIVagmSab92018 strain, as previously reported (44), the Sab92018ivTF molecular clone mainly uses CCR5 for viral entry but is capable of infecting GHOST cells engineered to express high levels of CXCR4. Unexpectedly, Sab92018ivTF entered GHOST cells expressing BONZO/STRL33 as efficiently as those expressing CCR5. It has been reported that STRL33 is used by a variety of SIVs and by some HIV-1 strains and is expressed on specific subsets of CD4+-naive T cells, B lymphocytes, and natural killer cells (48, 57). It has been suggested that STRL33 may be a relevant coreceptor for HIV-1 in vivo (57), but experimental evidence is missing. Studies using derivatives of Sab92018ivTF that maintain the capability to use CCR5 as an entry cofactor but are unable to utilize BONZO/STRL33 will provide novel insights into the role of this orphan receptor for viral replication in vivo.
In summary, we have generated the first replication-competent molecular clone of SIVagm that has not been adapted to growth in cell culture. Our results suggest that Sab92018ivTF recapitulates the biological properties of the naturally occurring parental SIVagmSab90218 strain and will thus be an important resource to identify those viral properties that contribute to the lack of disease progression in the natural AGM hosts.
Acknowledgments
We thank Daniela Krnavek for expert technical assistance and Dre van der Merwe and Kristina Wohllaib for critical reading of the manuscript.
This work was supported by grants from the NIH (R01 AI064066 and R37 AI 050529) and the Deutsche Forschungsgemeinschaft. Nicholas Parrish is supported by the UAB Medical Scientist Training Program (T32 GM008361).
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.Allan, J. S., M. Short, M. E. Taylor, S. Su, V. M. Hirsch, P. R. Johnson, G. M. Shaw, and B. H. Hahn. 1991. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J. Virol. 65:2816-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambrose, Z., V. N. KewalRamani, P. D. Bieniasz, and T. Hatziioannou. 2007. HIV/AIDS: in search of an animal model. Trends Biotechnol. 25:333-337. [DOI] [PubMed] [Google Scholar]
- 3.Arhel, N., M. Lehmann, K. Clauss, G. U. Nienhaus, V. Piguet, and F. Kirchhoff. 2009. The inability to disrupt the immunological synapse between infected human T cells and APCs distinguishes HIV-1 from most other primate lentiviruses. J. Clin. Invest. 119:2965-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baier, M., A. Werner, K. Cichutek, C. Garber, C. Müller, G. Kraus, F. J. Ferdinand, S. Hartung, T. S. Papas, and R. Kurth. 1989. Molecularly cloned simian immunodeficiency virus SIVagm3 is highly divergent from other SIVagm isolates and is biologically active in vitro and in vivo. J. Virol. 63:5119-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bar, K. J., H. Li, A. Chamberland, C. Tremblay, J. P. Routy, T. Grayson, C. Sun, S. Wang, G. H. Learn, C. J. Morgan, J. E. Schumacher, B. F. Haynes, B. F. Keele, B. H. Hahn, and G. M. Shaw. 2010. Wide variation in the multiplicity of HIV-1 infection among injection drug users. J. Virol. 84:6241-6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beer, B., J. Denner, C. R. Brown, S. Norley, J. zur Megede, C. Coulibaly, R. Plesker, S. Holzammer, M. Baier, V. M. Hirsch, and R. Kurth. 1998. Simian immunodeficiency virus of African green monkeys is apathogenic in the newborn natural host. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 18:210-220. [DOI] [PubMed] [Google Scholar]
- 7.Bosinger, S. E., Q. Li, S. N. Gordon, N. R. Klatt, L. Duan, L. Xu, N. Francella, A. Sidahmed, A. J. Smith, E. M. Cramer, M. Zeng, D. Masopust, J. V. Carlis, L. Ran, T. H. Vanderford, M. Paiardini, R. B. Isett, D. A. Baldwin, J. G. Else, S. I. Staprans, G. Silvestri, A. T. Haase, and D. J. Kelvin. 2009. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J. Clin. Invest. 119:3556-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broussard, S. R., S. I. Staprans, R. White, E. M. Whitehead, M. B. Feinberg, and J. S. Allan. 2001. Simian immunodeficiency virus replicates to high levels in naturally infected African green monkeys without inducing immunologic or neurologic disease. J. Virol. 75:2262-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng, H. K., D. Unutmaz, V. N. KewalRamani, and D. R. Littman. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296-300. [DOI] [PubMed] [Google Scholar]
- 10.Diop, O. M., A. Gueye, M. Dias-Tavares, C. Kornfeld, A. Faye, P. Ave, M. Huerre, S. Corbet, F. Barre-Sinoussi, and M. C. Muller-Trutwin. 2000. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in African green monkeys. J. Virol. 74:7538-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farzan, M., H. Choe, K. Martin, L. Marcon, W. Hofmann, G. Karlsson, Y. Sun, P. Barrett, N. Marchand, N. Sullivan, N. Gerard, C. Gerard, and J. Sodroski. 1997. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J. Exp. Med. 186:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 13.Fukasawa, M., T. Miura, A. Hasegawa, S. Morikawa, H. Tsujimoto, K. Miki, T. Kitamura, and M. Hayami. 1988. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature 333:457-461. [DOI] [PubMed] [Google Scholar]
- 14.Gaufin, T., R. M. Ribeiro, R. Gautam, J. Dufour, D. Mandell, C. Apetrei, and I. Pandrea. 2010. Experimental depletion of CD8+ cells in acutely SIVagm-infected African green monkeys results in increased viral replication. Retrovirology 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaufin, T., M. Pattison, R. Gautam, C. Stoulig, J. Dufour, J. MacFarland, D. Mandell, C. Tatum, M. H. Marx, R. M. Ribeiro, D. Montefiori, C. Apetrei, and I. Pandrea. 2009. Effect of B-cell depletion on viral replication and clinical outcome of simian immunodeficiency virus infection in a natural host. J. Virol. 83:10347-10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein, S., I. Ourmanov, C. R. Brown, B. E. Beer, W. R. Elkins, R. Plishka, A. Buckler-White, and V. M. Hirsch. 2000. Wide range of viral load in healthy African green monkeys naturally infected with simian immunodeficiency virus. J. Virol. 74:11744-11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gueye, A., O. M. Diop, M. J. Ploquin, C. Kornfeld, A. Faye, M. C. Cumont, B. Hurtrel, F. Barre-Sinoussi, and M. C. Muller-Trutwin. 2004. Viral load in tissues during the early and chronic phase of non-pathogenic SIVagm infection. J. Med. Primatol. 33:83-97. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch, V. M., D. Adger-Johnson, B. Campbell, S. Goldstein, C. Brown, W. R. Elkins, and D. C. Montefiori. 1997. A molecularly cloned, pathogenic, neutralization-resistant simian immunodeficiency virus, SIVsmE543-3. J. Virol. 71:1608-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsch, V. M., G. Dapolito, P. R. Johnson, W. R. Elkins, W. T. London, R. J. Montali, S. Goldstein, and C. Brown. 1995. Induction of AIDS by simian immunodeficiency virus from an African green monkey: species-specific variation in pathogenicity correlates with the extent of in vivo replication. J. Virol. 69:955-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holzammer, S., E. Holznagel, A. Kaul, R. Kurth, and S. Norley. 2001. High virus loads in naturally and experimentally SIVagm-infected African green monkeys. Virology 283:324-331. [DOI] [PubMed] [Google Scholar]
- 21.Jacquelin, B., V. Mayau, B. Targat, A. S. Liovat, D. Kunkel, G. Petitjean, M. A. Dillies, P. Roques, C. Butor, G. Silvestri, L. D. Giavedoni, P. Lebon, F. Barre-Sinoussi, A. Benecke, and M. C. Muller-Trutwin. 2009. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J. Clin. Invest. 119:3544-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin, M. J., H. Hui, D. L. Robertson, M. C. Muller, F. Barre-Sinoussi, V. M. Hirsch, J. S. Allan, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J. 13:2935-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, P. R., S. Goldstein, W. T. London, A. Fomsgaard, and V. M. Hirsch. 1990. Molecular clones of SIVsm and SIVagm: experimental infection of macaques and African green monkeys. J. Med. Primatol. 19:279-286. [PubMed] [Google Scholar]
- 24.Keele, B. F., E. E. Giorgi, J. F. Salazar-Gonzalez, J. M. Decker, K. T. Pham, M. G. Salazar, C. Sun, T. Grayson, S. Wang, H. Li, X. Wei, C. Jiang, J. L. Kirchherr, F. Gao, J. A. Anderson, L. H. Ping, R. Swanstrom, G. D. Tomaras, W. A. Blattner, P. A. Goepfert, J. M. Kilby, M. S. Saag, E. L. Delwart, M. P. Busch, M. S. Cohen, D. C. Montefiori, B. F. Haynes, B. Gaschen, G. S. Athreya, H. Y. Lee, N. Wood, C. Seoighe, A. S. Perelson, T. Bhattacharya, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keele, B. F., H. Li, G. H. Learn, P. Hraber, E. E. Giorgi, T. Grayson, C. Sun, Y. Chen, W. W. Yeh, N. L. Letvin, J. R. Mascola, G. J. Nabel, B. F. Haynes, T. Bhattacharya, A. S. Perelson, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keele, B. F., J. H. Jones, K. A. Terio, J. D. Estes, R. S. Rudicell, M. L. Wilson, Y. Li, G. H. Learn, T. M. Beasley, J. Schumacher-Stankey, E. Wroblewski, A. Mosser, J. Raphael, S. Kamenya, E. V. Lonsdorf, D. A. Travis, T. Mlengeya, M. J. Kinsel, J. G. Else, G. Silvestri, J. Goodall, P. M. Sharp, G. M. Shaw, A. E. Pusey, and B. H. Hahn. 2009. Increased mortality and AIDS-like immunopathology in wild chimpanzees infected with SIVcpz. Nature 460:515-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirchhoff, F. 2009. Is the high virulence of HIV-1 an unfortunate coincidence of primate lentiviral evolution? Nat. Rev. Microbiol. 7:467-476. [DOI] [PubMed] [Google Scholar]
- 28.Kornfeld, C., M. J. Ploquin, I. Pandrea, A. Faye, R. Onanga, C. Apetrei, V. Poaty-Mavoungou, P. Rouquet, J. Estaquier, L. Mortara, J. F. Desoutter, C. Butor, R. Le Grand, P. Roques, F. Simon, F. Barre-Sinoussi, O. M. Diop, and M. C. Muller-Trutwin. 2005. Antiinflammatory profiles during primary SIV infection in African green monkeys are associated with protection against AIDS. J. Clin. Invest. 115:1082-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuiken, C., T. Leitner, B. Foley, B. Hahn, P. Marx, F. McCutchan, S. Wolinsky, and B. Korber (ed.). 2009. HIV sequence compendium 2009. Los Alamos National Laboratory, Los Alamos, NM.
- 30.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 31.Lee, H. Y., E. E. Giorgi, B. F. Keele, B. Gaschen, G. S. Athreya, J. F. Salazar-Gonzalez, K. T. Pham, P. A. Goepfert, J. M. Kilby, M. S. Saag, E. L. Delwart, M. P. Busch, B. H. Hahn, G. M. Shaw, B. T. Korber, T. Bhattacharya, and A. S. Perelson. 2009. Modeling sequence evolution in acute HIV-1 infection. J. Theor. Biol. 261:341-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, H., K. J. Bar, S. Wang, J. M. Decker, Y. Chen, C. Sun, J. F. Salazar-Gonzalez, M. G. Salazar, G. H. Learn, C. J. Morgan, J. E. Schumacher, P. Hraber, E. E. Giorgi, T. Bhattacharya, B. T. Korber, A. S. Perelson, J. J. Eron, M. S. Cohen, C. B. Hicks, B. F. Haynes, M. Markowitz, B. F. Keele, B. H. Hahn, and G. M. Shaw. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 13:e1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milush, J. M., J. D. Reeves, S. N. Gordon, D. Zhou, A. Muthukumar, D. A. Kosub, E. Chacko, L. D. Giavedoni, C. C. Ibegbu, K. S. Cole, J. L. Miamidian, M. Paiardini, A. P. Barry, S. I. Staprans, G. Silvestri, and D. L. Sodora. 2007. Virally induced CD4+ T-cell depletion is not sufficient to induce AIDS in a natural host. J. Immunol. 179:3047-3056. [DOI] [PubMed] [Google Scholar]
- 34.Morner, A., A. Bjorndal, V. KewalRamani, D. R. Littman, R. Inoue, R. Thorstensson, E. M. Fenyo, and E. Bjorling. 1999. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 73:2343-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Münch, J., D. Rajan, M. Schindler, A. Specht, E. Rücker, F. J. Novembre, E. Nerrienet, M. C. Müller-Trutwin, M. Peeters, B. H. Hahn, and F. Kirchhoff. 2007. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J. Virol. 81:13852-13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 37.Onanga, R., C. Kornfeld, I. Pandrea, J. Estaquier, S. Souquiere, P. Rouquet, V. P. Mavoungou, O. Bourry, S. M'Boup, F. Barre-Sinoussi, F. Simon, C. Apetrei, P. Roques, and M. C. Muller-Trutwin. 2002. High levels of viral replication contrast with only transient changes in CD4+ and CD8+ cell numbers during the early phase of experimental infection with simian immunodeficiency virus SIVmnd-1 in Mandrillus sphinx. J. Virol. 76:10256-10263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otsyula, M. G., A. Gettie, M. Suleman, R. Tarara, I. Mohamed, and P. Marx. 1995. Apparent lack of vertical transmission of simian immunodeficiency virus (SIV) in naturally infected African green monkeys, Cercopithecus aethiops. Ann. Trop. Med. Parasitol. 89:573-576. [DOI] [PubMed] [Google Scholar]
- 39.Paiardini, M., I. Pandrea, C. Apetrei, and G. Silvestri. 2009. Lessons learned from the natural hosts of HIV-related viruses. Annu. Rev. Med. 60:485-495. [DOI] [PubMed] [Google Scholar]
- 40.Pandrea, I., and C. Apetrei. 2010. Where the wild things are: pathogenesis of SIV infection in African nonhuman primate hosts. Curr. HIV/AIDS Rep. 7:28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandrea, I., C. Apetrei, J. Dufour, N. Dillon, J. Barbercheck, M. Metzger, B. Jacquelin, R. Bohm, P. A. Marx, F. Barre-Sinoussi, V. M. Hirsch, M. C. Muller-Trutwin, A. A. Lackner, and R. Veazey. 2006. Simian immunodeficiency virus (SIV) SIVagmSab infection of Caribbean African green monkeys: new model of the study of SIV pathogenesis in natural hosts. J. Virol. 80:4858-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pandrea, I., T. Gaufin, J. M. Brenchley, R. Gautam, C. Monjure, A. Gautam, C. Coleman, A. A. Lackner, R. M. Ribeiro, D. C. Douek, and C. Apetrei. 2008. Experimentally induced immune activation in natural hosts of SIV results in significant increases in viral replication. J. Immunol. 181:6687-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandrea, I., R. Onanga, S. Souquiere, A. Mouinga-Ondéme, O. Bourry, M. Makuwa, P. Rouquet, G. Silvestri, F. Simon, P. Roques, and C. Apetrei. 2008. Paucity of CD4+ CCR5+ T cells may prevent transmission of simian immunodeficiency virus in natural nonhuman primate hosts by breast-feeding. J. Virol. 82:5501-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandrea, I., C. Kornfeld, M. J.-Y. Ploquin, C. Apetrei, A. Faye, P. Rouquet, P. Roques, F. Simon, F. Barré-Sinoussi, M. C. Müller-Trutwin, and O. M. Diop. 2005. Impact of viral factors on very early in vivo replication profiles in simian immunodeficiency virus SIVagm-infected African green monkeys. J. Virol. 79:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pandrea, I., R. M. Ribeiro, R. Gautam, T. Gaufin, M. Pattison, M. Barnes, C. Monjure, C. Stoulig, G. Silvestri, M. Miller, A. S. Perelson, and C. Apetrei. 2008. Simian immunodeficiency virus SIVagm dynamics in African green monkeys. J. Virol. 82:3713-3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Papkalla, A., J. Munch, C. Otto, and F. Kirchhoff. 2002. Nef enhances human immunodeficiency virus type 1 infectivity and replication independently of viral coreceptor tropism. J. Virol. 76:8455-8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infection by macrophage-tropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pöhlmann, S., B. Lee, S. Meister, M. Krumbiegel, G. Leslie, R. W. Doms, and F. Kirchhoff. 2000. Simian immunodeficiency virus utilizes human and sooty mangabey but not rhesus macaque STRL33 for efficient entry. J. Virol. 74:5075-5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Proudfoot, A. E., C. A. Power, and M. K. Schwarz. 2010. Anti-chemokine small molecule drugs: a promising future? Expert Opin. Investig. Drugs 19:345-355. [DOI] [PubMed] [Google Scholar]
- 50.Rey-Cuillé, M. A., J. L. Berthier, M. C. Bomsel-Demontoy, Y. Chaduc, L. Montagnier, A. G. Hovanessian, and L. A. Chakrabarti. 1998. Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease. J. Virol. 72:3872-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 52.Salazar-Gonzalez, J. F., E. Bailes, K. T. Pham, M. G. Salazar, M. B. Guffey, B. F. Keele, C. A. Derdeyn, P. Farmer, E. Hunter, S. Allen, O. Manigart, J. Mulenga, J. A. Anderson, R. Swanstrom, B. F. Haynes, G. S. Athreya, B. T. Korber, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2008. Deciphering human immunodeficiency virus type 1 transmission and early envelope diversification by single-genome amplification and sequencing. J. Virol. 82:3952-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salazar-Gonzalez, J. F., M. G. Salazar, B. F. Keele, G. H. Learn, E. E. Giorgi, H. Li, J. M. Decker, S. Wang, J. Baalwa, M. H. Kraus, N. F. Parrish, K. S. Shaw, M. B. Guffey, K. J. Bar, K. L. Davis, C. Ochsenbauer-Jambor, J. C. Kappes, M. S. Saag, M. S. Cohen, J. Mulenga, C. A. Derdeyn, S. Allen, E. Hunter, M. Markowitz, P. Hraber, A. S. Perelson, T. Bhattacharya, B. F. Haynes, B. T. Korber, B. H. Hahn, and G. M. Shaw. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sauter, D., M. Schindler, A. Specht, W. N. Landford, J. Münch, K.-A. Kim, J. Votteler, U. Schubert, F. Bibollet-Ruche, B. F. Keele, J. Takehisa, Y. Ogando, C. Ochsenbauer, J. C. Kappes, A. Ayouba, M. Peeters, G. H. Learn, G. Shaw, P. M. Sharp, P. Bieniasz, B. H. Hahn, T. Hatziioannou, and F. Kirchhoff. 2009. Tetherin-driven evolution of Vpu and Nef function and the emergence of pandemic and non-pandemic HIV-1 strains. Cell Host Microbe 6:409-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schindler, M., J. Munch, O. Kutsch, H. Li, M. L. Santiago, F. Bibollet-Ruche, M. C. Muller-Trutwin, F. J. Novembre, M. Peeters, V. Courgnaud, E. Bailes, P. Roques, D. L. Sodora, G. Silvestri, P. M. Sharp, B. H. Hahn, and F. Kirchhoff. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055-1067. [DOI] [PubMed] [Google Scholar]
- 56.Schindler, M., J. Schmokel, A. Specht, H. Li, J. Munch, M. Khalid, D. L. Sodora, B. H. Hahn, G. Silvestri, and F. Kirchhoff. 2008. Inefficient Nef-mediated downmodulation of CD3 and MHC-I correlates with loss of CD4+ T cells in natural SIV infection. PLoS Pathog. 4:e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharron, M., S. Pohlmann, K. Price, E. Lolis, M. Tsang, F. Kirchhoff, R. W. Doms, and B. Lee. 2000. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood 96:41-49. [PubMed] [Google Scholar]
- 58.Silvestri, G., A. Fedanov, S. Germon, N. Kozyr, W. J. Kaiser, D. A. Garber, H. McClure, M. B. Feinberg, and S. I. Staprans. 2005. Divergent host responses during primary simian immunodeficiency virus SIVsm infection of natural sooty mangabey and nonnatural rhesus macaque hosts. J. Virol. 79:4043-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Silvestri, G., D. L. Sodora, R. A. Koup, M. Paiardini, S. P. O'Neil, H. M. McClure, S. I. Staprans, and M. B. Feinberg. 2003. Nonpathogenic SIV infection of sooty mangabeys is characterized by limited bystander immunopathology despite chronic high-level viremia. Immunity 18:441-452. [DOI] [PubMed] [Google Scholar]
- 60.Soares, M. A., D. L. Robertson, H. Hui, J. S. Allan, G. M. Shaw, and B. H. Hahn. 1997. A full-length and replication-competent proviral clone of SIVAGM from tantalus monkeys. Virology 228:394-399. [DOI] [PubMed] [Google Scholar]
- 61.Sodora, D. L., J. S. Allan, C. Apetrei, J. M. Brenchley, D. C. Douek, J. G. Else, J. D. Estes, B. H. Hahn, V. M. Hirsch, A. Kaur, F. Kirchhoff, M. Muller-Trutwin, I. Pandrea, J. E. Schmitz, and G. Silvestri. 2009. Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nat. Med. 15:861-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takehisa, J., M. H. Kraus, J. M. Decker, Y. Li, B. F. Keele, F. Bibollet-Ruche, K. P. Zammit, Z. Weng, M. L. Santiago, S. Kamenya, M. L. Wilson, A. E. Pusey, E. Bailes, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2007. Generation of infectious molecular clones of simian immunodeficiency virus from fecal consensus sequences of wild chimpanzees. J. Virol. 81:7463-7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tao, B., and P. N. Fultz. 1995. Molecular and biological analyses of quasispecies during evolution of a virulent simian immunodeficiency virus, SIVsmmPBj. J. Virol. 69:2031-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.VandeWoude, S., and C. Apetrei. 2006. Going wild: lessons from T-lymphotropic naturally occurring lentiviruses. Clin. Microbiol. Rev. 19:728-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang, S. J., L. A. Lopez, H. Hauser, C. M. Exline, K. G. Haworth, and P. M. Cannon. 2010. Anti-tetherin activities in Vpu-expressing primate lentiviruses. Retrovirology 18:7-13. [DOI] [PMC free article] [PubMed] [Google Scholar]