Abstract
The hemagglutinin (HA) of influenza virus organizes the virus bud zone, a domain of the plasma membrane enriched in raft lipids. Using fluorescence lifetime imaging microscopy-fluorescence resonance energy transfer (FLIM-FRET), a technique that detects close colocalization of fluorescent proteins in transfected cells, we show that the viral proton channel M2 clusters with HA but not with a marker for inner leaflet rafts. The FRET signal between M2 and HA depends on the raft-targeting signals in HA and on an intact actin cytoskeleton. We conclude that M2 contains an intrinsic signal that targets the protein to the viral bud zone, which is organized by raft-associated HA and by cortical actin.
The assembly of influenza virus takes place at rafts in the host cell's plasma membrane (15, 20), microdomains rich in cholesterol and sphingolipids (24). Hemagglutinin (HA) is supposed to organize the virus bud zone, since it is intrinsically targeted to rafts where it causes budding of virus-like particles (2, 19). However, in the viral envelope, ordered and disordered lipid domains coexist, suggesting that the virus bud zone is not entirely composed of rafts (16). In addition, the diameters of the HA clusters observed by analytical immunogold electron microscopy and high resolution fluorescence microscopy are much larger than those commonly associated with rafts, and their irregular size boundaries indicate that others factors, such as the cytoskeleton, are involved in organizing the viral bud zone (8, 9, 13).
The viral neuraminidase also contains raft-targeting features and coclusters with HA at the plasma membrane (13). The proton channel M2, the third envelope protein, is seemingly excluded from membrane rafts (as judged by controversial cold-detergent extraction) and partially from nascent virus particles (31), although it contains a typical raft-targeting feature, palmitoylation (27), and has been proposed to bind cholesterol (18, 21). An interesting model suggests that M2 associates with the edge of rafts, where it catalyzes pinching-off of virus particles (21). Indeed, the cytoplasmic tail of M2 is required for efficient virus budding. It contains an M1 binding site recruiting the viral coat protein from internal membranes to the plasma membrane (3, 14, 29). In addition, it carries a palmitoylated amphipathic helix with the cholesterol binding site that might cause membrane curvature (3, 14, 29). Thus, to fulfill its function during virus budding, M2 must be targeted to the viral bud zone.
Fluorescence resonance energy transfer (FRET) is well suited to assess very close (smaller than 10 nm) colocalization of two proteins. Matching donor and acceptor fluorophores are fused to the proteins under study, and the constructs are coexpressed in a single cell. FRET can then be assessed by fluorescence lifetime imaging microscopy (FLIM), making use of the fact that the lifetime of the excited state of the donor will be shortened in the presence of the acceptor in close proximity. The FRET efficiency (E) can be calculated as follows:
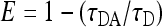 |
(1) |
with τDA being the lifetime of the donor in the presence and τD being the lifetime of the donor in the absence of the acceptor (determined from 10 cells expressing the donor only). For membrane proteins, however, it is not sufficient to calculate the FRET efficiency to assess clustering since energy transfer can simply occur by the random collision of two mobile molecules in the membrane. In this case, the FRET efficiency increases linearly with increasing acceptor fluorophore concentration at the membrane; in contrast, if FRET is due to the clustering of both proteins, the FRET efficiency is largely independent of the acceptor concentration. For quantitative evaluation, we used the equation
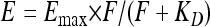 |
(2) |
which describes the FRET efficiency E as a hyperbolic function of the acceptor protein concentration assessed by its fluorescence intensity F. The associative properties of the donor and acceptor can be judged from the KD (equilibrium dissociation constant) value, which is very small compared to the intensity range of the acceptor in the case of clustering and in the same range as or larger than the acceptor intensity range in the event of FRET being due to the random collision of both molecules (30).
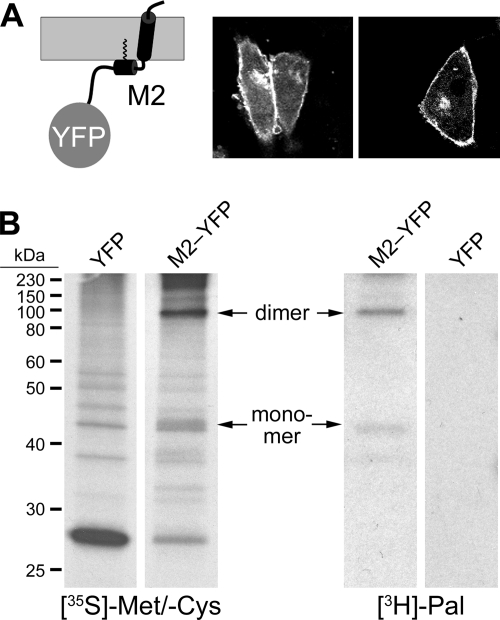
With FLIM-FRET, we have recently shown clustering of HA with markers for outer and inner leaflet rafts (6, 22). Here, we have used the same approach to analyze whether M2 associates with rafts and whether it coclusters with HA. We fused M2 via the linker PPVAT to monomeric yellow fluorescent protein (YFP). M2-YFP was efficiently transported to the plasma membrane of transfected cells (Fig. 1A) and palmitoylated as shown by metabolic labeling by using the previously described methodology (28) (Fig. 1B). Note that M2-YFP mostly ran as a disulfide-linked dimer, indicating that oligomerization had occurred (10). Since oligomerization and palmitoylation are known to occur in the endoplasmic reticulum (ER) and the early Golgi, respectively (11), attachment of YFP did not prevent transport of M2 along the exocytic pathway.
FIG. 1.
Generation and characterization of M2-YFP. (A) Scheme of M2-YFP in the membrane (gray) and expression of M2-YFP on the surface of transfected CHO cells. (B) Metabolic labeling of cells expressing YFP or M2-YFP with [35S]methionine/[35S]cysteine and [3H]palmitate for 6 h as indicated, immunoprecipitation with anti-green fluorescent protein (GFP) antibody, SDS-PAGE under nonreducing conditions, and fluorography. Positions of the M2-YFP monomer and dimer are marked with arrows. The M2 is from influenza A/Duck/Ukraine/1/63 (H3N8).
To assess oligomerization of the M2 probes by FLIM-FRET, we then coexpressed M2-YFP with M2 fused via the same linker to cerulean (Cer) (17). Fluorescence was recorded in cells expressing M2-Cer and M2-YFP at the plasma membrane by sequential excitation with 458- and 515-nm lasers using a confocal microscope, and fluorescence lifetime images of cerulean (excited at 440 nm with a pulsed laser diode) were acquired.
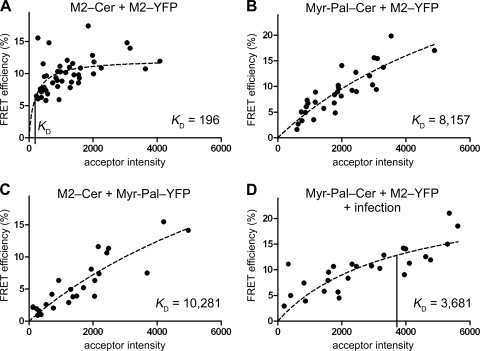
From these data, the mean FRET efficiency was calculated as 9.9%, lower than one would intuitively expect for a stable and close association of donor (M2-Cer) and acceptor (M2-YFP) fluorophores. However, FRET depends not only on the donor-acceptor distance but also on the relative orientation of the dipole moments of donor emission and acceptor absorption and cannot be compared between different FRET pairs. The FRET efficiency of each cell was plotted against the measured intensity of the acceptor in that cell (Fig. 2A, black dots). Fitting equation 1 to the data (Fig. 2A, dashed line) showed that the FRET efficiencies are independent of the intensity of the acceptor fluorescence and hence the expression level of M2-YFP. In addition, the calculated KD of 196 is lower than the acceptor intensities (ranging between 279 and 4,087), indicating stable association of M2-Cer with M2-YFP.
FIG. 2.
M2 associates with itself but not with membrane rafts. (A) Self-association of M2-Cer and M2-YFP evaluated by FLIM-FRET. The FRET efficiency E in the plasma membrane of each analyzed cell is plotted versus the acceptor intensity F; a hyperbolic function (equation 1, dashed line) is fitted to the data. The KD value to assess clustering is indicated as a vertical line on the x axis. Number of cells (n), 44; mean FRET efficiency ± standard error of the mean (SEM), 9.9% ± 0.4%; KD ± SEM, 196 ± 78. (B) FLIM-FRET of the raft marker Myr-Pal-Cer as the FRET donor with M2-YFP as the FRET acceptor. n, 31; E ± SEM, 9.0% ± 0.8%; KD ± SEM, 8,157 ± 4,679. (C) FLIM-FRET of M2-Cer as the FRET donor with the raft marker Myr-Pal-YFP as the FRET acceptor. n, 25; E ± SEM, 5.7% ± 0.8%; KD ± SEM, 10,281 ± 7,935. (D) FLIM-FRET of M2-YFP with Myr-Pal-Cer in virus-infected cells. Cells were transfected and 20 h later infected with fowl plague virus (H7N1) at a multiplicity of infection (MOI) of 40. FLIM-FRET measurements were performed 4 to 7 h postinfection. n, 26; E ± SEM, 10.2% ± 0.8%; KD ± SEM, 3,681 ± 1,964. Fluorescence was recorded with an Olympus FluoView 1000 confocal microscope, and FLIM was performed with the LSM upgrade kit and SymPhoTime software (PicoQuant).
We then assessed raft association of M2 using a double-acylated (myristoylated and palmitoylated) peptide fused to cerulean (Myr-Pal-Cer) as an established marker for inner leaflet rafts (6, 30) and M2-YFP as the FRET acceptor. A mean FRET efficiency of 9.0% was calculated, and the clustering analysis outlined above showed a clear dependence of FRET on the expression level of M2-YFP (Fig. 2B). Furthermore, a KD of 8,157 was calculated, which is higher than the highest acceptor intensity in the evaluation. The exchange of the fluorophores, i.e., using M2-Cer as the donor and Myr-Pal-YFP as the acceptor, yielded similar results (KD, 10,281; FRET efficiency, 5.7%) (Fig. 2C). Thus, in contrast to HA (6), M2 does not cluster with a marker for inner leaflet rafts in transfected cells. We then tested a possible association of M2-YFP with the raft marker in virus-infected cells. Only a small increase in the FRET efficiency (10.2%) and a marginal decrease in the KD (3,681) were measured, indicating minimal, if any, increase in clustering (compare Fig. 2D with B). However, we do not want to exclude the possibility that the association of M2 with raft lipids, such as cholesterol, increases during the course of a virus infection (18). It is likely that the assembly of viral proteins excludes the raft marker, a 28-kDa nonviral protein, from the budding site so that Myr-Pal-Cer is no longer available to interact with M2-YFP.
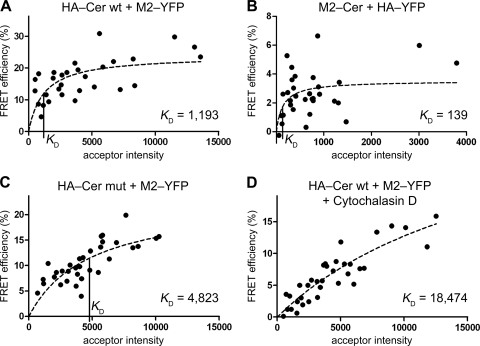
Next, we assessed clustering of M2-YFP with HA-Cer (Fig. 3A). We obtained a high degree of clustering, evidenced by a FRET efficiency considerably independent of the acceptor intensity and a low KD value of 1,193. Exchanging the fluorophores reduced the FRET efficiency, but the cluster analysis clearly revealed association of HA-YFP with M2-Cer (FRET efficiency, 2.6%; KD, 139) (Fig. 3B). To prove the specificity of HA-M2 clustering, we used an HA mutant in which the raft-targeting features—S acylation at three cysteine residues (12) and the sequence VIL in the exoplasmic half of the transmembrane domain (19, 26)—had been replaced by serines and alanines, respectively (6). A similar mutant of HA has been shown by electron microscopy to be uniformly distributed on the plasma membrane, in contrast to the patched appearance of wild-type HA (26). FLIM-FRET between the HA-Cer mutant (HA-Cer mut) and M2-YFP yielded a drop in the FRET efficiency from 16.3 to 10.6% and an increase in the KD from 1,193 to 4,823 (Fig. 3C) compared to the wild-type situation, indicating that clustering was moderately diminished.
FIG. 3.
Clustering of M2 with HA. (A) FLIM-FRET analysis (as described in the legend to Fig. 2) with the wild-type HA-Cer (HA-Cer wt) as the FRET donor and M2-YFP as the FRET acceptor. n, 30; E ± SEM, 16.3% ± 1.2%; KD ± SEM, 1,193 ± 509. (B) FLIM-FRET analysis with M2-Cer as the FRET donor and HA-YFP as the FRET acceptor. n, 32; E ± SEM, 2.6% ± 0.3%; KD ± SEM, 139 ± 112. (C) FLIM-FRET of HA-Cer mut with M2-YFP. n, 33; E ± SEM, 10.6% ± 0.7%; KD ± SEM, 4,823 ± 1,882. (D) FLIM-FRET of HA-Cer wt with M2-YFP in the presence of cytochalasin D (1 μM, added 20 h prior to the measurements). n, 35; E ± SEM, 6.4% ± 0.7%; KD ± SEM, 18,474 ± 8,647. The KD value to assess clustering is indicated as a vertical line on the x axis where applicable. The HA of fowl plague virus (H7N1) was used in the analysis. Relative donor and acceptor intensity values did not differ significantly (P > 0.01, unpaired, two-sided Student's t test) between the experiments involving HA-Cer wt (panel A; 100%), HA-Cer mut (panel C; 73%), and HA-Cer wt plus cytochalasin D (panel D; 122%) as the FRET donor and M2-YFP as the acceptor (100, 106, and 100%, respectively), showing that the use of the HA mutant and cytochalasin D treatment did not significantly alter surface expression of the probes.
The cytoskeleton has been assumed to organize raft domains (5) and to play a role in influenza virus assembly and budding (23, 25). We therefore performed FLIM-FRET measurements in the presence of the cytoskeleton-disrupting drug cytochalasin D. This treatment led to a further reduction in the FRET efficiency to a value of 6.4% and to a sharp increase in KD by more than one order of magnitude to a value of 18,474, which is much higher than the highest acceptor intensity in the evaluation and hence a clear indication of nonclustering (Fig. 3D).
In summary, we have shown by using FLIM-FRET that M2 clusters with HA but not with a marker for inner leaflet rafts. Colocalization of M2 with HA at the base of budding filamentous particles has very recently been shown by fluorescence microscopy (18). Studies of virus-infected cells with immunoelectron microscopy (immuno-EM), i.e., at a resolution comparable to the distance measured in FRET experiments, have provided mixed results on the colocalization of HA and M2 (3, 13). In addition, our work shows an intrinsic clustering of M2 and HA in the absence of other viral proteins, such as M1, which is assumed to bridge between the two proteins (3). This is in line with the observation that the clustering of HA with M2 persists even when the M1 binding site in M2 is deleted (3). Clustering might be due to a direct, previously unrecognized molecular interaction between HA and M2, which is disrupted by the described mutations at the C terminus of HA. However, since FLIM-FRET cannot distinguish between molecular interaction and very close proximity, it is also possible that M2 is targeted to the viral bud zone without the help of HA. Why then does M2 cluster with raft-associated HA but not with the raft marker? HA is supposed to organize a unique domain in the plasma membrane, the viral bud zone (4), in which it might be surrounded by a shell of specific lipids (1), which form ordered and disordered domains (16). Individual HA molecules inside the viral bud zone are mobile (8) and do not diffuse together with the raft marker in a stable complex for minutes (6). M2(-YFP) integrates into or localizes very near the HA(-Cer) clusters due to an intrinsic targeting signal, which might overlap with the unidentified signal for its delivery to the apical membrane (15). In contrast, HA lacking acylation sites and/or having mutations in the outer leaflet of its transmembrane region is dispersed over the cell surface, indicating that it is not able to organize the viral bud zone properly (3, 26), and thus HA-Cer mut interacts only poorly with M2-YFP. Likewise, in the absence of HA, the viral bud zone does not form and thus M2-YFP does not cluster with a raft marker, which is probably contained in small, dynamic rafts that do not integrate M2. Clustering of M2 with HA is furthermore dependent on an intact cytoskeleton. Evidence is accumulating that cortical actin drives the formation of protein nanoclusters and/or organizes the maintenance of raft domains (7). This might explain the involvement of actin in virus budding and its presence in purified virus particles (23, 25).
Acknowledgments
We gratefully acknowledge the Deutsche Forschungsgemeinschaft (DFG) for funding (grants SFB 740 and SPP 1175).
Footnotes
Published ahead of print on 29 September 2010.
REFERENCES
- 1.Anderson, R. G., and K. Jacobson. 2002. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science 296:1821-1825. [DOI] [PubMed] [Google Scholar]
- 2.Chen, B. J., and R. A. Lamb. 2008. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372:221-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, B. J., G. P. Leser, D. Jackson, and R. A. Lamb. 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82:10059-10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, B. J., G. P. Leser, E. Morita, and R. A. Lamb. 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 81:7111-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chichili, G. R., and W. Rodgers. 2009. Cytoskeleton-membrane interactions in membrane raft structure. Cell. Mol. Life Sci. 66:2319-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engel, S., S. Scolari, B. Thaa, N. Krebs, T. Korte, A. Herrmann, and M. Veit. 2010. FLIM-FRET and FRAP reveal association of influenza virus haemagglutinin with membrane rafts. Biochem. J. 425:567-573. [DOI] [PubMed] [Google Scholar]
- 7.Goswami, D., K. Gowrishankar, S. Bilgrami, S. Ghosh, R. Raghupathy, R. Chadda, R. Vishwakarma, M. Rao, and S. Mayor. 2008. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell 135:1085-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hess, S. T., T. J. Gould, M. V. Gudheti, S. A. Maas, K. D. Mills, and J. Zimmerberg. 2007. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl. Acad. Sci. U. S. A. 104:17370-17375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hess, S. T., M. Kumar, A. Verma, J. Farrington, A. Kenworthy, and J. Zimmerberg. 2005. Quantitative electron microscopy and fluorescence spectroscopy of the membrane distribution of influenza hemagglutinin. J. Cell Biol. 169:965-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holsinger, L. J., and R. A. Lamb. 1991. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology 183:32-43. [DOI] [PubMed] [Google Scholar]
- 11.Holsinger, L. J., M. A. Shaughnessy, A. Micko, L. H. Pinto, and R. A. Lamb. 1995. Analysis of the posttranslational modifications of the influenza virus M2 protein. J. Virol. 69:1219-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kordyukova, L. V., M. V. Serebryakova, L. A. Baratova, and M. Veit. 2008. S acylation of the hemagglutinin of influenza viruses: mass spectrometry reveals site-specific attachment of stearic acid to a transmembrane cysteine. J. Virol. 82:9288-9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leser, G. P., and R. A. Lamb. 2005. Influenza virus assembly and budding in raft-derived microdomains: a quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 342:215-227. [DOI] [PubMed] [Google Scholar]
- 14.McCown, M. F., and A. Pekosz. 2006. Distinct domains of the influenza A virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 80:8178-8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nayak, D. P., R. A. Balogun, H. Yamada, Z. H. Zhou, and S. Barman. 2009. Influenza virus morphogenesis and budding. Virus Res. 143:147-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polozov, I. V., L. Bezrukov, K. Gawrisch, and J. Zimmerberg. 2008. Progressive ordering with decreasing temperature of the phospholipids of influenza virus. Nat. Chem. Biol. 4:248-255. [DOI] [PubMed] [Google Scholar]
- 17.Rizzo, M. A., G. H. Springer, B. Granada, and D. W. Piston. 2004. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22:445-449. [DOI] [PubMed] [Google Scholar]
- 18.Rossman, J. S., X. Jing, G. P. Leser, V. Balannik, L. H. Pinto, and R. A. Lamb. 2010. Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J. Virol. 84:5078-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheiffele, P., M. G. Roth, and K. Simons. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitt, A. P., and R. A. Lamb. 2005. Influenza virus assembly and budding at the viral budozone. Adv. Virus Res. 64:383-416. [DOI] [PubMed] [Google Scholar]
- 21.Schroeder, C., H. Heider, E. Möncke-Buchner, and T. I. Lin. 2005. The influenza virus ion channel and maturation cofactor M2 is a cholesterol-binding protein. Eur. Biophys. J. 34:52-66. [DOI] [PubMed] [Google Scholar]
- 22.Scolari, S., S. Engel, N. Krebs, A. P. Plazzo, R. F. De Almeida, M. Prieto, M. Veit, and A. Herrmann. 2009. Lateral distribution of the transmembrane domain of influenza virus hemagglutinin revealed by time-resolved fluorescence imaging. J. Biol. Chem. 284:15708-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaw, M. L., K. L. Stone, C. M. Colangelo, E. E. Gulcicek, and P. Palese. 2008. Cellular proteins in influenza virus particles. PLoS Pathog. 4:e1000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature 387:569-572. [DOI] [PubMed] [Google Scholar]
- 25.Simpson-Holley, M., D. Ellis, D. Fisher, D. Elton, J. McCauley, and P. Digard. 2002. A functional link between the actin cytoskeleton and lipid rafts during budding of filamentous influenza virions. Virology 301:212-225. [DOI] [PubMed] [Google Scholar]
- 26.Takeda, M., G. P. Leser, C. J. Russell, and R. A. Lamb. 2003. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. U. S. A. 100:14610-14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veit, M., H. D. Klenk, A. Kendal, and R. Rott. 1991. The M2 protein of influenza A virus is acylated. J. Gen. Virol. 72:1461-1465. [DOI] [PubMed] [Google Scholar]
- 28.Veit, M., E. Ponimaskin, and M. F. Schmidt. 2008. Analysis of S-acylation of proteins. Methods Mol. Biol. 446:163-182. [DOI] [PubMed] [Google Scholar]
- 29.Wang, D., A. Harmon, J. Jin, D. H. Francis, J. Christopher-Hennings, E. Nelson, R. C. Montelaro, and F. Li. 2010. The lack of an inherent membrane targeting signal is responsible for the failure of the matrix (M1) protein of influenza A virus to bud into virus-like particles. J. Virol. 84:4673-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zacharias, D. A., J. D. Violin, A. C. Newton, and R. Y. Tsien. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296:913-916. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]