Abstract
Ubiquitination is a critical regulator of the host immune response to viral infection, and many viruses, including coronaviruses, encode proteins that target the ubiquitination system. To explore the link between coronavirus infection and the ubiquitin system, we asked whether protein degradation by the 26S proteasome plays a role in severe coronavirus infections using a murine model of SARS-like pneumonitis induced by murine hepatitis virus strain 1 (MHV-1). In vitro, the pretreatment of peritoneal macrophages with inhibitors of the proteasome (pyrrolidine dithiocarbamate [PDTC], MG132, and PS-341) markedly inhibited MHV-1 replication at an early step in its replication cycle, as evidenced by inhibition of viral RNA production. Proteasome inhibition also blocked viral cytotoxicity in macrophages, as well as the induction of inflammatory mediators such as IP-10, gamma interferon (IFN-γ), and monocyte chemoattractant protein 1 (MCP-1). In vivo, intranasal inoculation of MHV-1 results in a lethal pneumonitis in A/J mice. Treatment of A/J mice with the proteasome inhibitor PDTC, MG132, or PS-341 led to 40% survival (P < 0.01), with a concomitant improvement of lung histology, reduced pulmonary viral replication, decreased pulmonary STAT phosphorylation, and reduced pulmonary inflammatory cytokine expression. These data demonstrate that inhibition of the cellular proteasome attenuates pneumonitis and cytokine gene expression in vivo by reducing MHV-1 replication and the resulting inflammatory response. The results further suggest that targeting the proteasome may be an effective new treatment for severe coronavirus infections.
Severe acute respiratory syndrome (SARS) was first introduced into the human population in the Guangdong Province in China and rapidly spread to several other countries (31). SARS is caused by infection with the SARS coronavirus (SARS-CoV) and is characterized by an atypical pneumonia and lymphopenia. Two-thirds of the SARS-infected patients developed a viral pneumonitis, of which 10% developed acute respiratory distress syndrome. During the outbreak in 2002 to 2003, 8,000 people were infected and 774 people died from respiratory failure (36; WHO, Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003 [http://www.who.int]). At present there are no effective treatments for SARS as well as other coronavirus infections. Finding an effective treatment for coronavirus infections could be protective in the event of a reemergent coronavirus outbreak (7).
We have recently reported that a rodent model of SARS mimics many of the features of severe clinical SARS pathology (11, 12). Intranasal infection of A/J mice with strain 1 of murine hepatitis virus (MHV-1) causes a lethal form of pneumonitis, characterized by marked innate immune inflammatory cytokine production and replication of the virus in pulmonary macrophages (11, 12). MHV-1 infection is uniformly fatal in infected A/J mice; the resultant disease serves as a model to understand the pathology of the most severe SARS cases. In mice, the pulmonary damage is histologically similar to that seen in human SARS and is similarly associated with a marked upregulation of inflammatory mediators, including monocyte chemoattractant protein 1 (MCP-1), IP-10, MIG, gamma interferon (IFN-γ), interleukin-8 (IL-8), and IL-6 (11, 12, 25). These innate immune mediators are likely to play roles in human SARS and MHV-1 SARS-like pathogenesis.
A critical aspect of the host innate immune response to viral illness is the upregulation of the antiviral type 1 IFN response. With respect to SARS, type 1 IFN responses have been reported to be suppressed by SARS-CoV in several models and in clinical cases (11, 39, 45, 52). In our model, MHV-1-infected A/J mice produce less type 1 IFN than resistant strains of mice and they respond poorly to IFN-β therapy (11). Type I IFN has been used clinically in the treatment of established SARS infections but has shown only limited efficacy (25). In the absence of an effective antiviral treatment, the innate immune pathways present a potential target for therapeutic intervention (7).
Ubiquitination, the process by which cellular proteins are conjugated to the 7.5-kDa ubiquitin (Ub) protein, is a critical regulator of innate and adaptive immune pathways (40). There are several possible fates for ubiquitinated proteins: degradation by the 26S proteasome, trafficking to various subcellular sites, altered interactions with other proteins, and altered signal transduction functions (28). The fates of the ubiquitinated proteins, many of which overlap, can play a role in innate immunity. Since the first discovery that papillomavirus encodes an E3 ubiquitin ligase that targets p53, it has become widely appreciated that many viruses encode proteins that target or exploit ubiquitination pathways (37, 43). For example, Epstein-Barr virus and herpes simplex virus proteins interact with the host deubiquitinating (DUB) protein USP7 (13, 17). Ubiquitination of IRF3 has been implicated in the viral control of the innate immune system (22, 48, 49). DUB may also be important for viral functions, such as the assembly of viral replicase proteins with double-membrane vesicles at the site of replication, a process that parasitizes autophagy (39).
All coronaviruses, including MHV (A59 and JHM), infectious bronchitis virus, and human CoV229E SARS coronavirus, encode one or more papain-like proteases (PLpros) (PL1pro and PL2pro) (3, 5, 19, 23, 50). One role for the PL2pro proteases is to cleave the coronavirus polyprotein into its component parts. This enzyme, isolated from the SARS-CoV, has also been shown to have DUB activity both in vitro and in HeLa cells (23), suggesting that it might also play a role in modulating the host ubiquitination pathways. PLpro proteases harbor an N-terminal Ub-like domain reported to mediate interactions between PLpro DUB activity and the cellular proteasome (35). Although there is no direct link between the proteasome and SARS-CoV DUB activity, the presence of the Ub1 domain and of SARS-CoV DUB activity suggests that the proteasome may be being exploited by the virus either to evade the immune response or to promote viral replication. These interactions also suggest that the ubiquitination system might be a target for antiviral therapeutic intervention.
We explored the role of the cellular proteasome in MHV-1 replication and in the innate immune response to the virus by testing the effects of small-molecule proteasome inhibitors in both cell-based and murine models of SARS pneumonitis. We compared the results in the SARS model to a well-described model of lymphocytic choriomeningitis virus (LCMV) hepatitis in order to test for virus-specific effects. To control for nonspecific effects of the inhibitors, we used three different agents: pyrrolidine dithiocarbamate (PDTC), MG132, and PS-341 (bortezomib, Velcade). PDTC is a chelating agent that reversibly inhibits the proteasome complex, MG132 is a peptide aldehyde protease inhibitor, and PS-341 is a peptide boronic acid inhibitor (1, 20, 38). PS-341 is a clinically approved drug currently being used in the treatment of multiple myeloma.
MATERIALS AND METHODS
Animals, buffers, and reagents.
Pathogen-free female A/J mice 6 to 7 weeks old were purchased from Jackson, chow fed, and allowed to acclimatize for at least 1 week prior to experiments. Thioglycolate (3%; Life Technologies, Inc.) was prepared in accordance with the manufacturer's instructions. Endotoxin-free H21 and Hanks balanced salt solution (HBSS; Sigma) were obtained from Life Technologies, Inc. Fetal bovine serum (FBS) was obtained from HyClone. A 5 mM pyrrolidine dithiocarbamate (PDTC; Sigma) stock solution was prepared in saline (pH 7.4), and a 20 μM MG132 (Biomol) stock solution was prepared in dimethyl sulfoxide (DMSO). PS-341 was synthesized by American Custom Chemicals Corp., San Diego, CA, and prepared in DMSO at a concentration of 40 mM.
Cell and MHV-1 preparation.
Peritoneal exudate macrophages (PEM) were harvested in ice-cold HBSS 3 days following a 2-ml intraperitoneal (i.p.) injection of 3% sterile thioglycolate. Cells were washed twice in cold HBSS and resuspended in Dulbecco's modified Eagle medium (DMEM; Gibco), 2% fetal calf serum (FCS), and l-Gln at 1 × 106 to 10 × 106 cells/ml. This procedure consistently yields a >96% macrophage cell population with Wright's stain, with >97% viability by trypan blue exclusion (27). For most experiments 1 × 106 cells were plated on 6-well polystyrene plates (Sarstedt) and allowed to incubate overnight at 37°C and 5% CO2. Nonadherent cells were washed away with RPMI 1640 (Gibco) and replaced with RPMI 1640-2% FCS-l-Gln. MHV-1 was obtained and purified as described previously (9). Virus was grown to titers of 10 × 106 to 50 × 106 PFU/ml H21 on confluent 17CL1 cells.
Measuring PEM viability and MHV-1 viral replication.
For studies of viral replication in PEM, cells were pretreated for 60 min at 37°C and 5% CO2 in the presence or absence of PDTC (50 μM), MG132 (2 μM), or PS-341 (0.1 μM). One to 18 h after infection by MHV-1 (multiplicity of infection [MOI] = 1), cells and culture media were harvested and freeze-thawed at −20°C and virus titers on L2 cells were determined as previously described (22). Viability was measured by trypan blue exclusion on the Vi-CELL series cell viability analyzer (Beckman Coulter, Mississauga, ON, Canada).
LCMV viral titers.
PEM (1 × 106) were allowed to adhere to a 24-well plate for 4 h. The cells were then treated with either vehicle alone, PS341 at a final concentration of 0.1 μM, MG-132 at 2 μM, or PDTC at 50 μM for 60 min. After being washed, the cells were treated with LCMV strain WE (gift from Pamela Ohashi, Princess Margaret Hospital) at an MOI of 1 for 1 h, followed by another wash. At this point supernatant containing the proteasome inhibitor was added back to the PEM. Cell culture supernatants were collected 18 h postinfection (p.i.) and assayed for viral titers using a plaque assay adapted from Battegay et al. (4).
In vivo LCMV WE infectious model.
C57BL/6 mice were injected with LCMV WE intravenously (i.v.) at 2 ×106 PFU (n = 5 per treatment group) or with vehicle alone (n = 10). Animals were treated immediately postinfection with vehicle or with one of the proteasome inhibitors and every day following until sacrifice at day 8 p.i. (10 μg MG132/dose, 100 μg PDTC/dose, or 5 μg PS-341/dose and 0.5 ml saline subcutaneously [s.c.]). Liver tissue samples were collected, and viral titers were assessed as described above (4).
SARS pneumonitis model.
As previously described (11), A/J mice were inoculated with 5,000 PFU MHV-1 via an intranasal route. Mice were treated with PDTC (5 mg/kg of body weight daily s.c.), MG132 (0.5 mg/kg daily s.c.), or PS-341 (0.25 mg/kg daily s.c.) and 0.5 ml saline s.c. daily. All mice were monitored for signs of suffering and were euthanized at humane endpoints according to protocols approved by the hospital animal care committee. Data were analyzed using Prism software (Graphpad Software). At various times after infection plasma and serum were collected by cardiac puncture and lung tissue was harvested and immersed in 10% formalin for hematoxylin and eosin (H&E) histology or prepared for real-time PCR.
RNA isolation and real-time PCR (qPCR).
RNA was isolated using the TRIzol method in accordance with the manufacturer's specifications (Invitrogen). RNA was reverse transcribed with the First-Strand cDNA synthesis kit (Amersham Biosciences) using the manufacturer's protocol and the Amp 2400 PCR system (Perkin Elmer). Quantitative PCR (qPCR) was performed with SYBR green (Roche, Montreal, QC, Canada) on the LightCycler 480 system (Roche) using a standard thermal cycling protocol. Plates (384 wells/plate) and optical covers were purchased from Roche. Analysis was performed using LightCycler 480 software with a standard curve relative quantification method. Samples were normalized to hypoxanthine phosphoribosyltransferase (HPRT) (sense primer, 5′-AGAGGGAAATCGTGCGTGAC-3′; antisense primer, 5′-GATTCAACTTGCGCTCATCTTAGGC-3′) and β-actin (sense primer, 5′-ACTCCAAAGTTTTTATGGCTGGT-3′; antisense primer, 5′-CAATAGTGATGACCTGGCCGT) housekeeping genes. PCR products were amplified with the following primer sequences: IFN-α sense, 5′-ACTCCAAAGTTTTTATGGCTGGT-3′; IFN-α antisense, 5′-GTACTGCCCAGAAGTTTCACATT; IFN-γ sense, 5′-ATGAACGCTACACACTGCATC-3′; IFN-γ antisense, 5′-CCATCCTTTTGCCAGTTCCTC-3′; MCP-1 sense, 5′-TTAAAAACCTGGATCGGAACCAA-3′; MCP-1 antisense, 5′-GCATTAGCTTCAGATTTACGGGT-3′; MIG sense, 5′-TCCTTTTGGGCATCATCTTCC-3′; MIG antisense, 5′-TTTGTAGTGGATCGTGCCTCG-3′; IFN-β sense, 5′-TGAATGGAAAGATCAACCTCACCTA-3′; IFN-β antisense, 5′-CTCTTCTGCATCTTCTCCGTCA-3′; IP-10 sense, GCCGTCATTTTCTGCCTCAT-3′; IP-10 antisense, 5′-GCTTCCCTATGGCCCTCAT-3′; tumor necrosis factor alpha (TNF-α) sense, 5′-CATCTTCTCAAAATTCGAGTGACAA-3′; TNF-α antisense, 5′-TGGGAGTAGACAAGGTACAACCC-3′.
Western blot analysis.
At various times after exposure to MHV-1, PEM were pelleted or lung tissue was homogenized and lysed in ice-cold cell lysis buffer. Whole-cell lysates were prepared with 2× Laemmli buffer and 0.1 M dithiothreitol (DTT) buffer, immediately followed by incubation at 100°C for 5 min. Cytosolic fractions were isolated with 1% Triton X-100, 150 mM NaCl, 10 mM Tris-HCl (pH 7.4), 2 mM sodium orthovanadate, 10 μg/ml leupeptin, 50 mM NaF, 5 mM EDTA, 1 mM EGTA, and 1 mM phenylmethylsulfonyl fluoride. Postnuclear supernatants were collected following centrifugation at 10,000 × g for 5 min and diluted with 2× Laemmli buffer and 0.1 mM DTT. Lysates prepared from 1 × 106 cells were separated by 12.5% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore). Blots were probed with mouse monoclonal anti-MHV nucleocapsid (kind gift from Julian Leibowitz, Texas A&M University, College Station, TX), mouse monoclonal antibody to ubiquitinated protein clone FK2 (Biomol), rabbit anti-mouse pStat1 (Tyr 701)-R (Santa Cruz), rabbit anti-mouse Stat1 p84/p91 (M-22) (Santa Cruz), and monoclonal anti-β-actin clone AC-15 (Sigma). Subsequently, membranes were incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody: sheep anti-mouse IgG-HRP (Amersham) or donkey anti-rabbit IgG-HRP (Santa Cruz Biotechnology). Blots were developed using an enhanced chemiluminescence (ECL)-based system (Amersham Pharmacia Biotech).
Northern blot analysis.
Six hours after infection with MHV-1, PEM were harvested and RNA was isolated by the TRIzol method. MHV-1 RNA was separated on a 1.2% agarose-formaldehyde denatured gel and transferred onto Hybond-N+ (Amersham Biosciences) nylon membranes. Northern blotting of RNA was performed according to the ExpressHyb manual (Clontech). The hybridization probes was amplified from the C terminus of the untranslated region of MHV-1 by reverse transcription-PCR (RT-PCR) using the following primers: sense, MHV_UTR_B5′ (5′-GAT GAA GTA GAT AAT GTA AGC GT-3′); antisense, MHV_UTR_3′ (5′-TGC CAC AAC CTT CTC TAT CTG TTA T-3′). DNA probes (268 bp) were labeled by random priming using the Rediprime II random prime labeling system (Amersham Biosciences) according to the manufacturer's directions. Results were analyzed using the GS-800 calibrated densitometer (Bio-Rad) and Quantity One software (Bio-Rad).
Histology.
Samples for histological analysis were fixed in 10% formalin and were processed by standard methods as described previously (11). Histological assessment for pulmonary disease (pneumonitis, consolidation, and peribronchial inflammation) was performed by a pathologist (M.J.P.) in a blind, random manner.
RESULTS
MHV-1 replication and cytotoxicity are blocked by inhibition of the cellular proteasome.
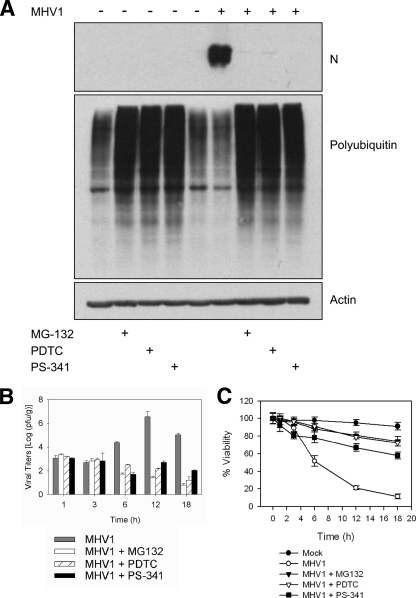
MHV-1 infects and replicates in A/J mouse peritoneal PEM in culture, causing cellular necrosis and the formation of large syncytia (data not shown). Necrotic cell death plays a role in the tissue damage of severe coronavirus infections such as SARS (7, 47). Since coronaviruses express the PLP2 DUB enzyme, which has been implicated in coronavirus-induced pathogenesis (48), we tested the effect of inhibiting the function of the cellular proteasome on viral cytotoxicity and replication of coronavirus. Therefore, we pretreated PEM isolated from A/J mice with PDTC, MG132, or PS-341 for 1 h prior to infection with MHV-1. When PEM infected with MHV-1 were left untreated, the level of polyubiquitination was decreased compared to that in the control PEM (uninfected and untreated) and PEM expressed high levels of viral nucleocapsid protein (N protein), an index of MHV-1 replication (Fig. 1A). However, in the presence of both MHV-1 infection and proteasome inhibition, N protein expression was abrogated and cellular polyubiquitination levels were similar to those for control groups treated with the inhibitor alone.
FIG. 1.
MHV-1 replication in PEM is inhibited by PS-341. PEM isolated from A/J mice were infected with MHV-1 (MOI = 1) and treated with one of three proteasome inhibitors: PS-341 (0.1 μM), MG132 (2 μM), or PDTC (50 μM). (A) PS-341 treatment of MHV-1-infected PEM inhibited N protein expression and increased levels of ubiquitinated proteins, as determined by Western blotting. Data are representative of two independent experiments. Each lane contains pooled triplicate wells. (B) PS-341 treatment decreased viral replication in MHV-1-infected PEM. Error bars indicate standard deviations (SD). (C) PEM viability was increased in the presence of PS-341, MG132, and PDTC. Error bars indicate SD. Panels B and C are representative of three independent experiments. Each sample was done in triplicate.
MHV-1 replication was also inhibited in treated cells, as determined by measuring viral titers (Fig. 1B). Decreased replication was associated with improved cellular viability (Fig. 1C) and improved cell morphology (data not shown). These data suggest that proteasome inhibition negatively regulates viral replication and decreases the cytotoxic effects of MHV-1 infection in PEM.
Proteasome inhibition has an effect on early MHV-1 replication.
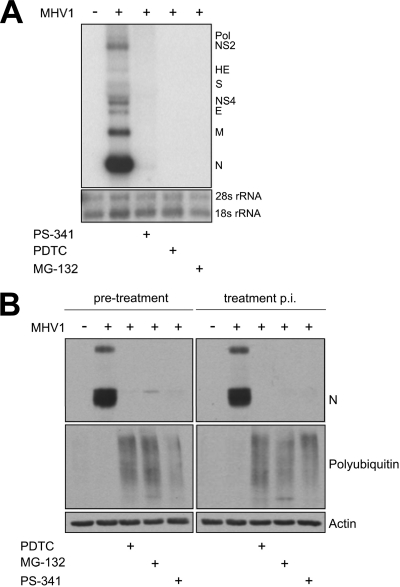
In order to determine whether proteasome inhibition affects early or late stages of the MHV-1 life cycle, we examined the time course of expression of viral RNA and protein in the presence and absence of proteasome inhibition. By Northern blot analysis, infection of PEM with MHV-1 in the presence of PDTC, MG132, or PS-341 decreased viral replication, as indicated by the absence of subgenomic mRNA (Fig. 2A). The marked decrease in subgenomic viral RNA could be explained by an inhibition of viral entry into the cell or an inhibition of viral replication. To distinguish between these possibilities, we treated PEM with PS-341 either 1 h prior to infection or 1 h after infection with MHV-1 and measured viral replication at several time points. The effect of proteasome inhibition was observed 6 h p.i. but not at earlier time points (data not shown), indicating that viable virus was present in both treatment groups. As determined by measuring N protein expression, viral replication was decreased at 6 h postinfection regardless of whether cells were treated with proteasome inhibitors before or 1 h after infection with MHV-1, indicating that viable virus was present in both the proteasome inhibitor pretreatment and the treatment p.i. groups (Fig. 2B). These findings suggest that the effect of proteasome inhibition is not mediated at the level of viral binding to or viral entry into the PEM.
FIG. 2.
PS-341 inhibits transcription of genes encoding viral proteins. (A) PS-341 inhibited the expression of MHV-1 subgenomic mRNA, as indicated by Northern blotting. PEM were harvested 6 h p.i. Each lane contains six pooled replicate wells. (B) PS-341 effectively inhibits N protein expression when PEM were treated with the proteasome inhibitor prior to and after infection with MHV-1. Each lane contains pooled triplicate wells. Data are representative of two independent experiments.
Proteasome inhibition suppresses MHV-1-induced inflammatory cell activation.
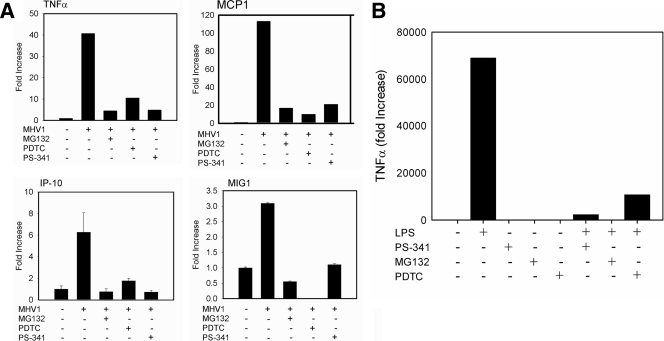
Similar to SARS-mediated disease in humans, MHV-1 infection can induce a massive and uncontrolled immune response in mice, initiated and driven by the induction of proinflammatory mediators (7, 11). Pneumonitis, a characteristic symptom of MHV-1-induced disease, is driven by a pronounced innate immune response partly initiated and amplified by proinflammatory cytokines (7). Therefore, we tested whether proteasome inhibition has an effect on virally induced cellular activation as a potential mechanism of limiting disease pathogenesis. We measured the transcription levels of genes encoding the following inflammatory mediators, which have been found to be relevant to SARS and which are relevant to inflammatory responses: IP-10, MCP-1, MIG-1, and TNF-α (11). The mRNA levels for the four cytokines were markedly increased following MHV-1 infection but suppressed when proteasome activity was inhibited (Fig. 3A). The effect on cytokine expression might be due either to decreased viral replication (Fig. 1B) or to the acknowledged effect of proteasome inhibitors on cytokine production (10, 44). To confirm that the proteasome inhibitors can have a direct effect on cytokine expression in our system, we stimulated PEM with a bacterial endotoxin, lipopolysaccharide (LPS; 50 ng/ml), in the presence or absence of proteasome inhibition (Fig. 3B). Cytokine expression was determined by measuring TNF-α mRNA expression levels as before. All proteasome inhibitors decreased TNF-α expression following LPS stimulation. Thus, the inhibition of the cellular proteasome affects MHV-1 replication, MHV-1 cytotoxicity, and inflammatory macrophage activation in vitro.
FIG. 3.
PS-341 decreases levels of inflammatory cytokines in response to MHV-1 infection. (A) mRNA expression levels of TNF-α, IP-10, MCP-1, and MIG-1 in PEM decreased following treatment with proteasome inhibitors, as measured by real-time PCR. Of the cytokines tested, MCP-1 and TNF-α mRNA increased the most prominently. Data are representative of three independent experiments. Samples were run in triplicate. Error bars indicate SD. (B) Proteasome inhibitors downregulate TNF-α mRNA expression in PEM stimulated by LPS. PEM isolated from A/J mice were treated with 50 ng/ml LPS, and TNF-α mRNA levels were measured by real-time PCR after 6 h. PEM cultures were either preincubated for 1 h with PDTC, MG132, or PS-341 or left untreated. TNF-α mRNA expression was downregulated following treatment with any of the three proteasome inhibitors (n = 3/group). Data are representative of 2 independent experiments.
Proteasome inhibitor treatment improves survival of MHV-1-infected A/J mice.
The in vitro results mentioned in the previous sections suggest that inhibition of the cellular proteasome has two potential benefits for the host: a decrease in viral replication and protection from virally induced inflammatory mediators. To explore whether the effects of cellular proteasome inhibition might be translated to an in vivo system, we used a murine SARS-like MHV-1 model and treated the infected mice with one of three of the proteasome inhibitors PDTC, MG132, and PS-341, the last being the only proteasome inhibitor being used clinically. The intranasal inoculation of A/J mice with 5,000 PFU of MHV-1 has a 100% fatality rate (11). By a treatment regimen of PDTC, MG132, or PS-341, the mortality rate of MHV-1 disease was reduced, with 40% of mice surviving long-term (Fig. 4A).
FIG. 4.
In vivo treatment of mice with PS-341 increases survival and attenuates viral replication in vivo. A/J mice were infected with 5,000 PFU MHV-1 intranasally and then treated with PDTC (5 mg/kg daily s.c.), MG132 (0.5 mg/kg daily s.c.), or PS-341 (0.25 mg/kg daily s.c.). (A) Survival of MHV-1-infected A/J mice increased significantly following treatment with PS-341. Standard errors of the means (SEM) were calculated using the log rank test. *, P = 0.003; **, P = 0.0003 (n = 10 for untreated and PS-341-treated animals; n = 5 for MG132- and PDTC-treated animals). (B) Prolonged survival was associated with a 1.1-log decrease in viral titers in PS-341-treated mice. Error bars indicate SD; P values for PS-341-treated versus untreated mice (using a one-way analysis of variance) are shown (n = 10 for untreated group; n = 6 for PS-341-treated mice).
At day 7 after infection with MHV-1, lung histology of untreated A/J mice showed severe peribronchitis and interstitial pneumonia affecting the entire lung, which resulted in complete lung consolidation followed by death (Table 1). PS-341-treated mice also developed peribronchitis and interstitial pneumonia; however, at day 7, the percentage of the lung involved (25 to 49%) decreased, with a marked improvement in the area of the lung that was consolidated (<25% in the PS-341 group compared to 100% consolidation in untreated mice). Both PDTC- and MG132-treated groups showed improved lung histology compared to the untreated, MHV-1-infected control group. Overall, MG132 produced the most prominent improvement in peribronchitis, interstitial pneumonia, and lung consolidation. These results suggest that SARS-like coronavirus infections are amenable to treatment with agents that inhibit the proteasome in vivo.
TABLE 1.
Lung pathology is improved following PS-341 treatment of MHV-1-infected mice
| Group | % of lung affecteda at indicated day p.i. by: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Peribronchitis |
Interstitial pneumonia |
Lung consolidation |
|||||||
| 1.5 | 4 | 7 | 1.5 | 4 | 7 | 1.5 | 4 | 7 | |
| Uninfected | − | − | − | − | − | − | − | − | − |
| MHV-1 | + | ++ | ++++ | + | ++ | ++++ | − | + | ++++ |
| MHV-1 + MG132 | + | + | − | ++ | + | + | − | − | − |
| MHV-1 + PDTC | ++ | − | +++ | +++ | + | +++ | +++ | − | − |
| MHV-1 + PS-341 | +++ | ++ | ++ | +++ | ++ | ++ | +++ | + | + |
−, no abnormal pathology; +, <25% of lung affected; ++, 25 to 50% of lung affected; +++, 50 to 75% of lung affected; ++++, 75 to 100% of lung affected.
Proteasome inhibition in vivo is associated with decreased production of pulmonary inflammatory mediators.
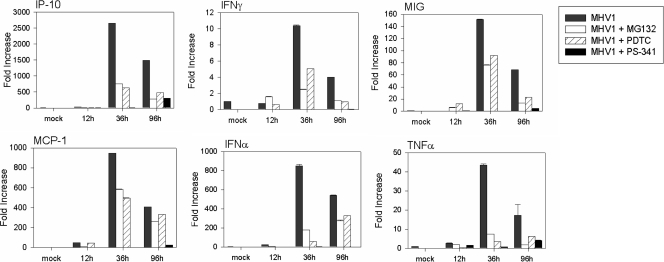
The increase in survival in the proteasome inhibitor treatment groups observed in vivo might have been influenced by decreased viral replication and/or decreased inflammatory cell activation, since both are key survival factors. Therefore, we studied the effects of cellular proteasome inhibition on viral replication and on activation markers of the innate immune response induced by MHV-1 in vivo. To this end, mice were treated daily subcutaneously with a regimen of 5 mg/kg PDTC, 0.5 mg/kg MG132, or 0.25 mg/kg PS-341. Lung tissue was harvested at days 1.5, 4, and 7 days after inoculation of MHV-1 intranasally and tested for viral titers. All three proteasome inhibitors decreased pulmonary MHV-1 replication at all times studied (data not shown); the most marked effect was seen following PS-341 treatment, where MHV-1 replication at day 7 postinfection decreased by 1.1 logs (P < 0.01) (Fig. 4B). In addition, mRNA was extracted from the lung tissue to measure by real-time PCR the expression levels of IP-10, MCP-1, MIG, IFN-γ, and TNF-α, as well as type 1 IFN. PS-341 showed a consistent and marked inhibition of inflammatory mediator gene expression, particularly IFN-α (Fig. 5). Taken together, these results suggest that a modest effect on viral replication, coupled with a more marked effect on inflammatory gene expression, contributes to the improved histology and outcomes seen in the in vivo SARS model. This improvement was achieved despite a reduction in type 1 IFN gene expression.
FIG. 5.
PS-341 treatment attenuates cytokine induction by MHV-1 in vivo. MHV-1 infection upregulates the mRNA expression of IP-10, IFN-γ, TNF-α, MCP-1, IFN-β, and MIG in the lung. All the mentioned cytokines reach peak levels of expression 36 h postinfection. In the presence of PDTC, MG132, and PS-341, cytokine mRNA levels are markedly reduced. PS-341 attenuates the mRNA expression levels more dramatically than either PDTC or MG-132 (n = 3 for each group). Data are representative of two independent experiments.
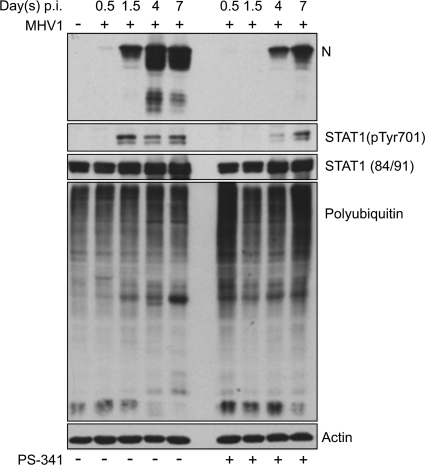
PS-341 delays expression of N protein.
In order to gain insight into the mechanisms underlying the inhibition of pulmonary inflammatory mediator gene expression in vivo, we determined the pulmonary expression of the N protein, one of the major mechanisms through which coronaviruses influence cellular activation (41, 42, 47). We also asked whether STAT1 phosphorylation was altered in treated animals, since activation of the JAK/STAT cascade is upstream of the induction of many inflammatory mediators and alterations in this signaling cascade have previously been associated with inhibition of the cellular proteasome (26). As shown in Fig. 6, treatment of A/J mice with PS-341 significantly delayed the expression of N protein in the lung and decreased the absolute amount of N protein. As well, STAT1 phosphorylation was delayed in treated mice. The ability of PS-341 to inhibit the mouse cellular proteasome in pulmonary tissue was confirmed by the increase in ubiquitinated proteins at days 0.5 and 7. The in vivo innate immune response to MHV-1 in the presence of PS-341 confirms our in vitro data that proteasome inhibition is an important factor both in activating the innate immune response and facilitating viral replication.
FIG. 6.
PS-341 alters expression and phosphorylation of viral and cellular proteins. Shown is a Western blot analysis of lung tissue lysate from MHV-1-infected mice. PS-341 treatment reduced levels of viral protein (N protein) production compared to levels in untreated controls. The protein levels of STAT-1 remained unchanged between the untreated and PS-341-treated groups; however, the phosphorylation of STAT-1 was markedly decreased in PS-341-treated mice. PS-341 treatment increased the levels of polyubiquitinated proteins. Data are representative of three independent experiments. Each well contains pooled triplicate wells.
Proteasome inhibition of viral replication is virus specific.
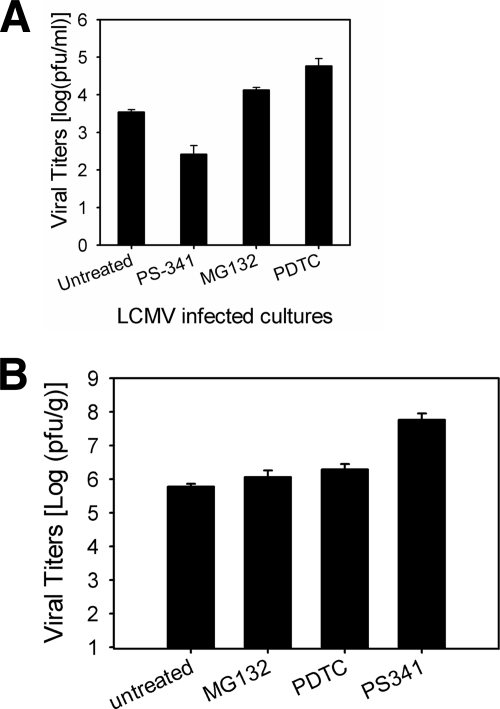
In order to test for virus-specific effects, we tested the efficacy of proteasome inhibition on a second infectious agent, LCMV WE, using a well-described model of LCMV hepatitis in vivo and PEM infection in vitro (3a, 51). PEM infected with LCMV in vitro did not show a consistent decrease in replication when treated with PS-341, MG132, or PDTC. PEM cultures that were treated with PS-341 did show slightly decreased MHV-1 production (Fig. 7A). However, it is unlikely that this effect is a direct result of proteasome inhibition per se since neither MG132 nor PDTC inhibited viral replication. The general lack of effect of proteasome inhibition on LCMV replication in vitro was mirrored in subsequent in vivo studies. C57BL/6 mice infected with LCMV WE and treated with proteasome inhibitors did not show a consistent decrease in viral titers derived from liver tissue harvested at day 8 p.i. (Fig. 7B). Interestingly, while PS-341 showed some degree of inhibition in vitro, the opposite effect was observed in vivo. Given the consistent lack of effect of PDTC and MG132, the effect of PS-341 on LCMV replication is not likely to be due to proteasome inhibition per se, which contrasts markedly with the inhibition of MHV-1 production (both in vitro and in vivo).
FIG. 7.
LCMV WE infection is unaffected by proteasome inhibition both in vitro and in vivo. We tested the efficacy of proteasome inhibition on a second infectious agent, LCMV WE, to assess the specificity of proteasome inhibition on coronavirus infections. (A) PEM (1 × 106) were pretreated with either vehicle alone or a final concentration of 0.1 μM PS341, 2 μM MG-132, or 50 μM PDTC for 60 min. The cells were washed and subsequently infected with LCMV WE at an MOI of 1. LCMV replication in vitro was not inhibited when PEM were treated with proteasome inhibitors. (B) C57BL/6 mice (n = 5 per treatment group, n = 10 for vehicle alone) were infected i.v. with 2.0 × 106 PFU LCMV WE. Viral titers in livers collected from mice at day 8 postinfection when the infection reaches peak levels were assessed (51). In contrast to those of MHV-1, LCMV WE titers were not inhibited by proteasome inhibition in vivo.
DISCUSSION
In this study we present evidence that inhibition of the cellular proteasome has important consequences for coronavirus replication and innate immune activation. In vitro, pretreatment of PEM with three proteasome inhibitors dramatically reduced viral replication and the production of inflammatory mediators. In vivo the inhibition of the proteasome had a clear beneficial effect in the murine model of SARS, an effect that seemed to be mediated by decreased inflammatory cell activation, as evidenced by a reduction in inflammatory cytokine gene expression. Taken together, these results suggest that inhibition of the cellular proteasome could be considered a therapy for SARS.
The fact that the SARS-CoV papain-like protease (PLpro) has deubiquitinating (DUB) activity both in in vitro studies and in HeLa cells emphasizes the link between severe coronavirus infections and ubiquitination-dependent pathways (3, 23, 48). PLpro cleaves the coronavirus polyprotein at the N terminus of the replicase at the sequence LXGG, which is the consensus deubiquitination sequence targeted by other DUB proteases such as USP14, HAUSP, and UCH-L1. The SARS-CoV and avian infectious bronchitis virus (aIBV) encode only one PLpro, whereas all other coronaviruses encode two PLpros (3), at least one of which has the potential for DUB activity. The SARS-CoV PLpro cleaves a common ubiquitin substrate, Ub-7-amino-4-methylcoumarin (Ub-AMC), and deconjugates ISG15 (isopeptide bond cleavage) both in cis and in trans (3, 23).
Although coronaviruses, including the SARS coronavirus, have the potential to target ubiquitination pathways through a DUB protein, the role of the viral protease in deubiquitination remains unclear. Even though the SARS PLpro recognizes the LXGG consensus deubiquitination sequence, the enzyme has much lower affinity for substrates than other cellular DUB enzymes and does not significantly contribute to cellular deubiquitination (3, 23). Inhibition of SARS-CoV DUB blocks coronavirus replication but less effectively than the 26S proteasome inhibitors described here (34).
The mechanism through which proteasome inhibition disrupts MHV-1 replication and activation of inflammatory cells is unclear but is not likely one that affects viral entry into the cell. PDTC, MG132, and PS-341 inhibited viral replication at the level of RNA transcription. In vitro, the effect of proteasome inhibition was observed only after 6 h of infection and persisted even when the inhibitor was introduced after infection of PEM (Fig. 1). In vivo, proteasome inhibition had relatively little effect on viral replication but did attenuate inflammatory cytokine expression (Fig. 5). Moreover, proteasome inhibition also led to decreased redox activation but did not inhibit coronavirus-induced tyrosine phosphorylation (data not shown), consistent with an effect focused more on the viral replication machinery than on early viral signaling. Taken together, these data suggest that inhibition of the cellular proteasome leads to inhibition of MHV-1 replication and cellular activation at steps after internalization of the virus.
Previous work has suggested that disrupting the cellular proteasome can also inhibit the release of some strains of coronaviruses into the cytoplasm from internalizing lysosomes. Yu and Lai found that the release of the MHV JHM strain into the cytoplasm was sensitive to inhibition of the cellular proteasome with MG132 and lactacystin (46). In this study, treatment of cells with MG132 and lactacystin resulted in decreased MHV JHM replication and accumulation of viral particles in late endosomes and lysosomes. Although these effects may have been due to inhibition of the proteasome, there was no detectable change in Ub-conjugated viral proteins or cellular proteins associated with MHV, suggesting an alternative mechanism. In this regard, the authors noted that MG132 and lactacystin can also inhibit lysosomal proteins cathepsin B and A, respectively (2, 20, 24).
The beneficial effects of proteasome inhibition in the murine SARS model correlate with an inhibition of cytokine production and improved histopathology more than with a marked inhibition of viral replication. The cellular proteasome plays an important role in macrophage inflammatory activation (18); indeed, in our model system proteasome inhibition markedly decreases PEM cytokine production after exposure to endotoxin (Fig. 3B). Based on these data one might expect some inhibition of virally induced macrophage activation, though this study is the first to our knowledge to demonstrate this for coronaviruses. The consequences of this attenuation of inflammatory cell activation are mixed. Inhibiting aspects of the innate immune response can ameliorate survival in models of coronavirus infection, even without an effect on viral replication. For example, inhibition of the FGL2 membrane prothrombinase, an important mediator of the innate immune response to MHV-3-induced fulminant hepatitis, improves survival without affecting early viral replication (21). The interaction between viral replication, cytokine effects, and disease pathogenesis can be complex: in the same model, inhibiting tyrosine kinase activation with tyrphostin A59 blocks some aspects of the innate immune response, e.g., hepatic expression of FGL2, but does not improve survival, possibly because viral replication is increased (I. D. McGilvray, unpublished observations). Tissue damage resulting from coronavirus infection is the result of both direct cell cytotoxicity and activation of inflammatory cells and cascades; both mechanisms are important targets for therapy but either may, in certain contexts, play more or less of a role.
In this study, the effect of the proteasome inhibitors in the murine model of SARS is a global suppression of cytokine expression. This sort of suppression likely has both positive and negative aspects. For example, many of the detrimental effects of SARS are likely due to an overwhelming production of cytokines. In this case, suppression of inflammatory cell activation would be expected to be beneficial. On the other hand, pulmonary IFN-α mRNA expression was decreased by all three proteasome inhibitors in the murine model. Since IFN-α is a key antiviral effector and one that is associated with a positive clinical outcome (25), its suppression by the proteasome inhibitors may be one reason that the effect of the proteasome inhibitors is not more marked. Other studies have shown that the levels of type I IFN are suppressed following SARS-CoV infection, both in a proportion of SARS patients and in several animal models (8, 39, 52). In the MHV-1 model of SARS, there is an induction of IFN-α, which is at first glance in contradiction to these studies. However, as noted earlier, while there is induction of type 1 IFN in mice susceptible to a SARS-like pneumonitis following MHV-1 infection, resistant mice express more type 1 IFN, a pattern more in keeping with that suggested in studies of SARS-CoV infection of peripheral blood monocytes and macrophages (45). In addition, cell- and model-specific differences are likely to underlie some of the conflicting results. We would expect that any intervention with the capacity to alleviate an otherwise uniformly fatal model of SARS-like coronavirus infection would have that much more effect in a less severe form of disease.
The effect of proteasome inhibition on viral infection and disease is likely to be specific to the virus involved. For example, proteasome inhibition with PS-341 promotes Epstein-Barr virus-related gene expression in cell culture (6, 15, 16, 53). We have found that proteasome inhibition has little to no effect on replication of LCMV either in vitro or in vivo (Fig. 7A and B). Similarly, PS-341 has no effect on J6/JFH hepatitis C virus (HCV) replication in Huh7.5 cells (McGilvray, unpublished). Moreover, treatment of clinical multiple myeloma with PS-341 may be associated with an increased rate of varicella-zoster virus reactivation (6). Interpreting the latter possibility is difficult, since it may reflect the role of the cellular proteasome in either the viral infection or the host response to the virus or both. Taken together, these examples illustrate the variability in infection routes and host responses and demonstrate that the role of the cellular proteasome will vary with the specific virus in question.
Recently, two articles published by Raaben et al. assessed the potential to use PS-341 as an anti-CoV agent against both SARS-CoV infection and MHV infection in mice (32, 33). Both the current study and those by Raaben et al. demonstrated an early decrease in viral replication with proteasome inhibition in vitro. However, our study demonstrates a clear benefit of treatment of coronavirus infection in vivo using a MHV-1 pneumonitis model in A/J mice, while those of Raaben et al. showed increased viral titers and an adverse effect of PS-341 treatment in an MHV-A59 hepatitis model in C57BL/6 mice. There are a number of factors that differ between the studies and likely contribute to the different outcomes. These factors include the mouse strain (A/J in our pneumonitis model, C57BL/6 in the Raaben et al. hepatitis model) and the dose (PS-341 at 0.25 mg/kg/dose daily versus 1 mg/kg given on day −1 and day 2 p.i.), and delivery route (s.c. versus i.p.). The viral strains differed as well: Raaben et al. used a recombinant MHV EFLM virus (derived from the A59 strain) and wild-type MHV-A59, leading to an acute hepatitis in C57BL/6 mice, while our study focused on lung-tropic MHV-1 (with no liver disease) in A/J mice. We have shown previously that different combinations of coronavirus and host strains result in distinct outcomes in a variety of in vivo models (11, 28, 29). For example, MHV-3, a coronavirus that causes an acute fulminant hepatitis, kills BALB/c mice within 3 to 4 days postinfection but is cleared by A/J mice and has intermediate effects in C57BL/6 mice (29, 30). Interestingly, with MHV-1 pneumonitis infection the opposite effect is seen: BALB/c and C57BL/6 mice are able to clear the virus, but A/J mice succumb to the virus within 7 to 8 days postinfection (11). De Albuquerque et al. (11) showed that MHV-1-induced disease in A/J mice resembles the pathology of SARS and therefore serves as an ideal model for studying SARS-like disease. In this model, our study demonstrates that PS-341 treatment increases survival, decreases viral load, and inhibits inflammatory cytokine expression.
In summary, this study presents evidence that MHV-1 replication and induction of inflammatory cell activation can be attenuated by inhibition of the cellular proteasome. The inhibition of coronavirus replication occurs at an early step, but not at the point of viral entry into the cell. Proteasome inhibition has consequences both at the cellular and whole-animal levels, with similar levels of inhibition of inflammatory cell activation in both settings. The suppression of inflammatory cell activation seems to be particularly important for the beneficial effect of proteasome inhibition in the murine SARS model. Taken together, these results suggest that proteasome inhibition is a novel therapeutic intervention that could be considered in cases of clinical coronavirus infection.
Acknowledgments
I.D.M. is supported in part through Canadian Institutes of Health Research operating grant 62488. G.L. is supported by Canadian Institutes of Health Research operating grant CIHR 79561. A.B. has a scholarship from the University of Toronto Training Program in Regenerative Medicine (STP-53882).
Footnotes
Published ahead of print on 22 September 2010.
REFERENCES
- 1.Adams, J., V. J. Palombella, E. A. Sausville, J. Johnson, A. Destree, D. D. Lazarus, J. Maas, C. S. Pien, S. Prakash, and P. J. Elliott. 1999. Proteasome inhibitors: a novel class of potent and effective antitumor agents. Cancer Res. 59:2615-2622. [PubMed] [Google Scholar]
- 2.Authier, F., M. Metioui, A. W. Bell, and J. S. Mort. 1999. Negative regulation of epidermal growth factor signaling by selective proteolytic mechanisms in the endosome mediated by cathepsin B. J. Biol. Chem. 274:33723-33731. [DOI] [PubMed] [Google Scholar]
- 3.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Barthold, S. W., and A. L. Smith. 2007. Lymphocytic choriomeningitis virus, p. 179-213. In J. G. Fox, S. W. Barthold, M. Y. Davisson, C. Newcomer, F. Quimby, and A. L. Smith (ed.), The mouse in biomedical research. Academic Press/Elsevier, San Diego, CA.
- 4.Battegay, M., S. Cooper, A. Althage, J. Banziger, H. Hengartner, and R. M. Zinkernagel. 1991. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J. Virol. Methods 33:191-198. [DOI] [PubMed] [Google Scholar]
- 5.Bonilla, P. J., S. A. Hughes, J. D. Pinon, and S. R. Weiss. 1995. Characterization of the leader papain-like proteinase of MHV-A59: identification of a new in vitro cleavage site. Virology 209:489-497. [DOI] [PubMed] [Google Scholar]
- 6.Chanan-Khan, A., P. Sonneveld, M. W. Schuster, E. A. Stadtmauer, T. Facon, J. L. Harousseau, D. Ben-Yehuda, S. Lonial, H. Goldschmidt, D. Reece, R. Neuwirth, K. C. Anderson, and P. G. Richardson. 2008. Analysis of herpes zoster events among bortezomib-treated patients in the phase III APEX study. J. Clin. Oncol. 26:4784-4790. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., and K. Subbarao. 2007. The immunobiology of SARS*. Annu. Rev. Immunol. 25:443-472. [DOI] [PubMed] [Google Scholar]
- 8.Cinatl, J., B. Morgenstern, G. Bauer, P. Chandra, H. Rabenau, and H. W. Doerr. 2003. Treatment of SARS with human interferons. Lancet 362:293-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dackiw, A. P., K. Zakrzewski, A. B. Nathens, P. Y. Cheung, R. Fingerote, G. A. Levy, and O. D. Rotstein. 1995. Induction of macrophage procoagulant activity by murine hepatitis virus strain 3: role of tyrosine phosphorylation. J. Virol. 69:5824-5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasanu, C. A., and D. T. Alexandrescu. 2009. Does bortezomib induce de facto Varicella Zoster virus reactivation in patients with multiple myeloma? J. Clin. Oncol. 27:2293-2294. [DOI] [PubMed] [Google Scholar]
- 11.De Albuquerque, N., E. Baig, X. Ma, J. Zhang, W. He, A. Rowe, M. Habal, M. Liu, I. Shalev, G. P. Downey, R. Gorczynski, J. Butany, J. Leibowitz, S. R. Weiss, I. D. McGilvray, M. J. Phillips, E. N. Fish, and G. A. Levy. 2006. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J. mice. J. Virol. 80:10382-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Albuquerque, N., E. Baig, M. Xuezhong, I. Shalev, M. J. Phillips, M. Habal, J. Leibowitz, I. McGilvray, J. Butany, E. Fish, and G. Levy. 2006. Murine hepatitis virus strain 1 as a model for severe acute respiratory distress syndrome (SARS). Adv. Exp. Med. Biol. 581:373-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reference deleted.
- 15.Fu, D. X., Y. Tanhehco, J. Chen, C. A. Foss, J. J. Fox, J. M. Chong, R. F. Hobbs, M. Fukayama, G. Sgouros, J. Kowalski, M. G. Pomper, and R. F. Ambinder. 2008. Bortezomib-induced enzyme-targeted radiation therapy in herpesvirus-associated tumors. Nat. Med. 14:1118-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao, G., J. Zhang, X. Si, J. Wong, C. Cheung, B. McManus, and H. Luo. 2008. Proteasome inhibition attenuates coxsackievirus-induced myocardial damage in mice. Am. J. Physiol. Heart Circ. Physiol. 295:H401-H408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holowaty, M. N., M. Zeghouf, H. Wu, J. Tellam, V. Athanasopoulos, J. Greenblatt, and L. Frappier. 2003. Protein profiling with Epstein-Barr nuclear antigen-1 reveals an interaction with the herpesvirus-associated ubiquitin-specific protease HAUSP/USP7. J. Biol. Chem. 278:29987-29994. [DOI] [PubMed] [Google Scholar]
- 18.Kalantari, P., O. F. Harandi, P. A. Hankey, and A. J. Henderson. 2008. HIV-1 Tat mediates degradation of RON receptor tyrosine kinase, a regulator of inflammation. J. Immunol. 181:1548-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanjanahaluethai, A., Z. Chen, D. Jukneliene, and S. C. Baker. 2007. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology 361:391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, D. H., and A. L. Goldberg. 1998. Proteasome inhibitors: valuable new tools for cell biologists. Trends Cell Biol. 8:397-403. [DOI] [PubMed] [Google Scholar]
- 21.Li, C., L. S. Fung, S. Chung, A. Crow, N. Myers-Mason, M. J. Phillips, J. L. Leibowitz, E. Cole, C. A. Ottaway, and G. Levy. 1992. Monoclonal antiprothrombinase (3D4.3) prevents mortality from murine hepatitis virus (MHV-3) infection. J. Exp. Med. 176:689-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, M., D. Chen, A. Shiloh, J. Luo, A. Y. Nikolaev, J. Qin, and W. Gu. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416:648-653. [DOI] [PubMed] [Google Scholar]
- 23.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Menard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longva, K. E., F. D. Blystad, E. Stang, A. M. Larsen, L. E. Johannessen, and I. H. Madshus. 2002. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J. Cell Biol. 156:843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loutfy, M. R., L. M. Blatt, K. A. Siminovitch, S. Ward, B. Wolff, H. Lho, D. H. Pham, H. Deif, E. A. LaMere, M. Chang, K. C. Kain, G. A. Farcas, P. Ferguson, M. Latchford, G. Levy, J. W. Dennis, E. K. Lai, and E. N. Fish. 2003. Interferon alfa con-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. JAMA 290:3222-3228. [DOI] [PubMed] [Google Scholar]
- 26.McConkey, D. J., and K. Zhu. 2008. Mechanisms of proteasome inhibitor action and resistance in cancer. Drug Resist. Updat. 11:164-179. [DOI] [PubMed] [Google Scholar]
- 27.McGilvray, I. D., Z. Lu, A. C. Wei, A. P. Dackiw, J. C. Marshall, A. Kapus, G. Levy, and O. D. Rotstein. 1998. Murine hepatitis virus strain 3 induces the macrophage prothrombinase fgl-2 through p38 mitogen-activated protein kinase activation. J. Biol. Chem. 273:32222-32229. [DOI] [PubMed] [Google Scholar]
- 28.Muratani, M., and W. P. Tansey. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4:192-201. [DOI] [PubMed] [Google Scholar]
- 29.Pope, M., S. W. Chung, T. Mosmann, J. L. Leibowitz, R. M. Gorczynski, and G. A. Levy. 1996. Resistance of naive mice to murine hepatitis virus strain 3 requires development of a Th1, but not a Th2, response, whereas pre-existing antibody partially protects against primary infection. J. Immunol. 156:3342-3349. [PubMed] [Google Scholar]
- 30.Pope, M., P. A. Marsden, E. Cole, S. Sloan, L. S. Fung, Q. Ning, J. W. Ding, J. L. Leibowitz, M. J. Phillips, and G. A. Levy. 1998. Resistance to murine hepatitis virus strain 3 is dependent on production of nitric oxide. J. Virol. 72:7084-7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Brunham, and A. J. McGeer. 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348:1995-2005. [DOI] [PubMed] [Google Scholar]
- 32.Raaben, M., G. C. Grinwis, P. J. Rottier, and C. A. de Haan. 2010. The proteasome inhibitor Velcade enhances rather than reduces disease in mouse hepatitis coronavirus-infected mice. J. Virol. 84:7880-7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raaben, M., C. C. Posthuma, M. H. Verheije, E. G. te Lintelo, M. Kikkert, J. W. Drijfhout, E. J. Snijder, P. J. Rottier, and C. A. de Haan. 2010. The ubiquitin-proteasome system plays an important role during various stages of the coronavirus infection cycle. J. Virol. 84:7869-7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ratia, K., S. Pegan, J. Takayama, K. Sleeman, M. Coughlin, S. Baliji, R. Chaudhuri, W. Fu, B. S. Prabhakar, M. E. Johnson, S. C. Baker, A. K. Ghosh, and A. D. Mesecar. 2008. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 105:16119-16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 103:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 37.Shackelford, J., and J. S. Pagano. 2005. Targeting of host-cell ubiquitin pathways by viruses. Essays Biochem. 41:139-156. [DOI] [PubMed] [Google Scholar]
- 38.Si, X., B. M. McManus, J. Zhang, J. Yuan, C. Cheung, M. Esfandiarei, A. Suarez, A. Morgan, and H. Luo. 2005. Pyrrolidine dithiocarbamate reduces coxsackievirus B3 replication through inhibition of the ubiquitin-proteasome pathway. J. Virol. 79:8014-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spiegel, M., A. Pichlmair, L. Martinez-Sobrido, J. Cros, A. Garcia-Sastre, O. Haller, and F. Weber. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 79:2079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun, S. C., and S. C. Ley. 2008. New insights into NF-κB regulation and function. Trends Immunol. 29:469-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surjit, M., and S. K. Lal. 2008. The SARS-CoV nucleocapsid protein: a protein with multifarious activities. Infect. Genet. Evol. 8:397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urbanowski, M. D., C. S. Ilkow, and T. C. Hobman. 2008. Modulation of signaling pathways by RNA virus capsid proteins. Cell Signal. 20:1227-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Waddell, S., and J. R. Jenkins. 1998. Defining the minimal requirements for papilloma viral E6-mediated inhibition of human p53 activity in fission yeast. Oncogene 16:1759-1765. [DOI] [PubMed] [Google Scholar]
- 44.Yannaki, E., A. Papadopoulou, E. Athanasiou, P. Kaloyannidis, A. Paraskeva, D. Bougiouklis, P. Palladas, M. Yiangou, and A. Anagnostopoulos. 18 August 2010, posting date. The proteasome inhibitor, bortezomib, drastically affects inflammation and bone disease in a rat model of adjuvant-induced arthritis. Arthritis Rheum. doi: 10.1002/art.27690. [DOI] [PubMed]
- 45.Yilla, M., B. H. Harcourt, C. J. Hickman, M. McGrew, A. Tamin, C. S. Goldsmith, W. J. Bellini, and L. J. Anderson. 2005. SARS-coronavirus replication in human peripheral monocytes/macrophages. Virus Res. 107:93-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu, G. Y., and M. M. Lai. 2005. The ubiquitin-proteasome system facilitates the transfer of murine coronavirus from endosome to cytoplasm during virus entry. J. Virol. 79:644-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuwaraj, S., M. Cattral, M. Pope, and G. Levy. 1996. Murine hepatitis virus: molecular biology and pathogenesis. Viral Hepatitis Rev. 2:125-142. [Google Scholar]
- 48.Zheng, D., G. Chen, B. Guo, G. Cheng, and H. Tang. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 18:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhong, B., L. Zhang, C. Lei, Y. Li, A. P. Mao, Y. Yang, Y. Y. Wang, X. L. Zhang, and H. B. Shu. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity. 30:397-407. [DOI] [PubMed] [Google Scholar]
- 50.Ziebuhr, J., B. Schelle, N. Karl, E. Minskaia, S. Bayer, S. G. Siddell, A. E. Gorbalenya, and V. Thiel. 2007. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 81:3922-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zinkernagel, R. M., E. Haenseler, T. Leist, A. Cerny, H. Hengartner, and A. Althage. 1986. T cell-mediated hepatitis in mice infected with lymphocytic choriomeningitis virus. Liver cell destruction by H-2 class I-restricted virus-specific cytotoxic T cells as a physiological correlate of the 51Cr-release assay? J. Exp. Med. 164:1075-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zorzitto, J., C. L. Galligan, J. J. Ueng, and E. N. Fish. 2006. Characterization of the antiviral effects of interferon-alpha against a SARS-like coronavirus infection in vitro. Cell Res. 16:220-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou, P., J. Kawada, L. Pesnicak, and J. I. Cohen. 2007. Bortezomib induces apoptosis of Epstein-Barr virus (EBV)-transformed B cells and prolongs survival of mice inoculated with EBV-transformed B cells. J. Virol. 81:10029-10036. [DOI] [PMC free article] [PubMed] [Google Scholar]