Abstract
An immunocompetent, permissive, small-animal model would be valuable for the study of human immunodeficiency virus type 1 (HIV-1) pathogenesis and for the testing of drug and vaccine candidates. However, the development of such a model has been hampered by the inability of primary rodent cells to efficiently support several steps of the HIV-1 replication cycle. Although transgenesis of the HIV receptor complex and human cyclin T1 have been beneficial, additional late-phase blocks prevent robust replication of HIV-1 in rodents and limit the range of in vivo applications. In this study, we explored the HIV-1 susceptibility of rabbit primary T cells and macrophages. Envelope-specific and coreceptor-dependent entry of HIV-1 was achieved by expressing human CD4 and CCR5. A block of HIV-1 DNA synthesis, likely mediated by TRIM5, was overcome by limited changes to the HIV-1 gag gene. Unlike with mice and rats, primary cells from rabbits supported the functions of the regulatory viral proteins Tat and Rev, Gag processing, and the release of HIV-1 particles at levels comparable to those in human cells. While HIV-1 produced by rabbit T cells was highly infectious, a macrophage-specific infectivity defect became manifest by a complex pattern of mutations in the viral genome, only part of which were deamination dependent. These results demonstrate a considerable natural HIV-1 permissivity of the rabbit species and suggest that receptor complex transgenesis combined with modifications in gag and possibly vif of HIV-1 to evade species-specific restriction factors might render lagomorphs fully permissive to infection by this pathogenic human lentivirus.
New, infectable, small-animal models of human immunodeficiency virus type 1 (HIV-1) infection are needed to complement existing models for the study of viral pathogenesis, the in vivo evaluation of new antiviral compounds, and the testing of vaccine candidates. While existing animal models have made significant contributions, they have serious limitations. For example, the rhesus macaque model was instrumental for elucidating lentiviral pathogenesis and for evaluating fundamental concepts in vaccine development and drug intervention (38, 56, 70, 73). However, these nonhuman primate studies are limited by ethical concerns, small animal group sizes, and high cost. In addition, the animals can be infected only with simian immunodeficiency virus (SIV) or SIV/HIV chimeric viruses.
Over the past few years, considerable progress has been made in the development of xenotransplant models. Some of these allow transmission of HIV via the vaginal or rectal mucosa and display high-level viremia and CD4 T cell depletion (4, 21, 65). Recently, a feasible antiviral gene therapy approach was reported (55). Unfortunately, these models are technically challenging, time-consuming, and not amenable to widespread use. Moreover, HIV-1-infected, xenotransplant mice mount low or dysfunctional adaptive immune responses to HIV infection (5, 15), limiting studies of natural immune control and vaccine testing.
An alternative approach for developing HIV animal models in native rodents has been to identify and then surmount species-specific barriers that HIV encounters during its replication. The ultimate goal is to use this knowledge to generate immunocompetent transgenic small animals that are fully permissive for HIV replication. We and others have characterized such barriers in the early phase of HIV replication in mouse and rat cells (8, 32, 43, 50, 51, 72) and have successfully overcome them by transgenic expression of appropriate human cofactors (CD4, CCR5, CXCR4, and cyclin T1) in laboratory rodents (16, 42, 51, 76).
However, cells from triple-transgene mice are still resistant to infection (76). There remain additional undefined blocks to chromosomal integration into mouse T cells and within the late phase of HIV replication (6, 34, 67, 76). Our parallel efforts in the rat species have markedly increased in vivo susceptibility to systemic HIV-1 challenge. The human-CD4 (hCD4)/hCCR5-transgenic rat model has facilitated preclinical evaluation of antiviral drugs targeting entry, reverse transcriptase, and integrase (29, 30) and has contributed to aspects of viral pathogenesis (53, 59) and testing of a vaccine candidate (14). Despite a high proviral load in lymphatic organs, the current transgenic rat model still has limitations, including low and transient viremia and lack of HIV disease. As in the case of mice, these problems are due to ill-defined late-phase barriers that limit HIV production, particularly in primary T cells (42, 43, 51).
Species-specific barriers to HIV-1 infection might result from an inability of certain host factors to support specific steps of the replication cycle or from antiviral factors that specifically target viruses to restrict their propagation. Regarding the latter, members of the tripartite motif (TRIM)-bearing family of proteins have species- and retrovirus-specific antiviral properties. Their mechanisms of restriction are still incompletely understood, but targeting of incoming virions for proteasomal degradation and accelerated uncoating or steric hindrance by the capsid-binding TRIM proteins have been proposed (3, 58, 64, 68).
Previous work by several laboratories suggested that cell lines from rabbits impose early barriers to HIV-1 infection at the level of virus entry and reverse transcription (RT) (7, 20, 33, 36, 62), while only a few and, in part, conflicting reports on the efficiency of later steps in the viral life cycle have been published (33, 63). In the present study, we characterized the efficiency of critical steps throughout the HIV replication cycle in primary T cells and macrophages from New Zealand White rabbits.
MATERIALS AND METHODS
Animals.
New Zealand White rabbits (6- to 7-week-old females), female BALB/c mice, and Sprague-Dawley rats were obtained from Charles River Laboratories (Sulzfeld, Germany), housed in the animal facility of the University of Heidelberg, and sacrificed as reported previously (35, 43, 67). Human cyclin (hcyclin) T1-transgenic rats were recently described (51). Animal studies were conducted according to the German Animal Welfare Act and with authorization of the Regierungspräsidium Karlsruhe, Germany. Experiments were supervised by animal welfare officers of Heidelberg University.
Cells.
293T, TZM-bl, and HeLa cells (human cell lines), NIH 3T3 (mouse fibroblast) cells, and Rat2 (rat fibroblast-like) cells were cultivated as reported previously (40, 42, 43, 67). SIRC cells (rabbit cornea-derived fibroblasts) were maintained in minimal essential medium (MEM) supplemented with 10% fetal calf serum, 1% penicillin-streptomycin, and 1% l-glutamine (all from Invitrogen). SIRC hCD4/hCCR5 cells were generated by lentiviral transduction, fluorescence-activated cell sorting (FACS), and expansion of single-cell clones. Macrophages and activated T cell cultures were established from single-cell suspensions of rabbit, rat, and mouse spleen essentially as reported previously (29, 43, 66, 67). Human T cell and macrophage cultures were derived from single-donor peripheral blood mononuclear cells (PBMCs) and cultivated as described previously (43).
Viruses.
The molecular HIV-1 clone of pNL4-3 and its derivative pNL4-3 E-GFP, the latter of which carries an enhanced green fluorescent protein (gfp) gene within the nef locus driven by the 5′ long terminal repeat (5′-LTR), were from Malcom Martin and Nathaniel Landau, respectively, via the NIH AIDS Research and Reference Reagent Program. The molecular HIV-1 clone of p49.5 was a gift from Bruce Chesebro (17). The HIV-2 molecular clone pROD-A, carrying a gfp gene within the nef locus, was provided by Matthias Dittmar (60). The HIV-1 H/SCA packaging vector (which contains the first 150 amino acids of capsid from SIVmac239) (57) and the generation and characterization of vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped BlaM-Vpr-containing lentiviruses and lentiviral vectors were as described previously (29, 43, 67).
Virus entry and gene expression.
Fusion and entry of BlaM-Vpr-containing lentiviral vectors and vector-driven gene expression were measured as reported previously (27, 29, 67).
Reverse transcription and nuclear import.
The TaqMan-based quantification of total HIV-1 cDNA and 2-LTR circles in DNA extracts from infected cells was as described previously (29, 30, 67).
HIV-1 receptor complex-dependent entry.
Parental SIRC or SIRC hCD4/hCCR5 cells were infected with HIV-1 or HIV-1(H/SCA) GFP vectors, which had been pseudotyped with either VSV-G, strain NL4-3 Env (X4), or strain JR-FL Env (R5), with or without the coreceptor antagonist maraviroc (1 μM; Pfizer) or AMD3100 (2 μM; gift of Jose Este) or the peptidic fusion inhibitor T20 (50 μM; Roche). The percentage of infected (GFP-positive) cells was determined 2 days postinfection by flow cytometry. Primary rabbit macrophages were cotransfected with pCD4neo and pCCR5hygro using TurboFect (Fermentas). Surface expression of the human receptors was assessed using allophycocyanin (APC)-conjugated mouse anti-hCD4 and fluorescein isothiocyanate (FITC)-conjugated anti-hCCR5 antibodies (BD Bioscience). Subsequently, transfected rabbit macrophages were challenged with JR-FL Env-pseudotyped HIV-1 or HIV-1 (H/SCA) GFP vectors (5,000 infectious units [IU] determined on TZM-bl cells per 12 well), and reporter gene expression was examined 3 days later by microscopy (Olympus IX70) and flow cytometry (FACSCalibur with BDCell QuestPro4.0.2 software; BD Pharmingen) as described previously (41).
HIV LTR transactivation assays.
pBC12/CMV HIV-2 Tat (9) (pHIV-2 Tat) was a gift from Brian Cullen. pCMV4-Tat2ex carries the second exon of the tat gene from HIV-1NL4-3 (pHIV-1 Tat). The generation of minimal HIV LTR GFP reporter plasmids has been described previously (51). Cell lines were transfected with the pHIV-1 LTR GFP construct (1 ng) with the pHIV-1 Tat vector (10 ng) or an empty control vector (10 ng) using Lipofectamine 2000 (Invitrogen). Primary T cell cultures were cotransfected with pHIV-1 LTR GFP (4 ng) and pHIV-1 Tat (40 ng) or with pHIV-2 LTR GFP (20 ng) and pHIV-2 Tat (200 ng) by nucleofection (31, 41, 66), and GFP expression was quantified 1 day later. Primary rabbit macrophages were cotransfected using TurboFect with pHIV-1 LTR GFP and pHIV-1 Tat or with pHIV-2 LTR GFP and pHIV-2 Tat (in the same molar ratios for plasmids as for T cells), and GFP expression was quantified 3 days later.
HIV-1 Tat and Rev assays.
The Tat- and Rev-dependent pLRed(INS)2R expression construct (47) was a gift of Ruth Brack-Werner. Adherent cell lines or primary T cells were transfected with pLRed(INS)2R (1 μg) in the presence or absence of pHIV-1 Tat (200 ng) and/or pcDNA3.1 HIV-1 Rev (300 ng) together with peGFP-N1 (10 ng). Cells were treated with leptomycin B (LMB; 9.25 pmol; Sigma-Aldrich) where indicated below. One day posttransfection, the percentage of red fluorescent protein (RFP)- and GFP-positive cells and their mean fluorescence intensity (MFI) values were determined by flow cytometry. For each species and cell type, this product in the absence of Tat/Rev coexpression was arbitrarily set to 1. Primary T cells were nucleofected with either pNL4-3 GFP, carrying gfp in the nef locus (Rev-independent early gene expression), or pNL4-3 Gag-GFP, encoding an MA-GFP fusion protein (Rev-dependent late gene expression) driven by the HIV-1 LTR, constructed based on a cytomegalovirus (CMV)-driven construct (52). One day later, the MFI of GFP expression was determined in viable cells by flow cytometry, and the ratio of late MFI to early MFI (MFIlate/MFIearly) was calculated.
HIV-1 late-phase studies.
Activated primary human and rabbit T cells were transfected with wild-type (wt) pHIV-1NL4-3 or pNL4-3 E-GFP by electroporation (66). Rabbit macrophages were transfected with either pHIV-1NL4-3 or pNL4-3 E-GFP using TurboFect. As a reference control, human-monocyte-derived macrophages were infected overnight with VSV-G HIV-1NL4-3, followed by extensive washing and continued cultivation. Supernatants from T cell or macrophage cultures were harvested 36 h or 5 days posttransfection, respectively, and centrifuged through a 20% sucrose cushion. The virion-associated p24CA concentration and infectivity were determined by antigen enzyme-linked immunosorbent assay (ELISA) and TZM-bl blue-cell assay, respectively, and cell pellets were harvested for immunoblotting or for p24CA ELISA quantification as reported previously (40).
Immunoblotting.
Cell pellets were lysed in SDS lysis buffer. Proteins were separated by 12.5% SDS-PAGE and plotted onto nitrocellulose membranes. Blocked membranes were probed with the following primary antibodies: mouse monoclonal anti-HIV-1 p24CA antibody 183 (from Hans-Georg Kräusslich), sheep anti-HIV-1 p24CA antiserum (from Barbara Müller), rabbit polyclonal anti-Vpu antiserum (from Klaus Strebel), sheep anti-Nef antiserum (from Oliver T. Fackler), and rabbit anti-mitogen-activated protein kinase (anti-MAPK antiserum) (Santa Cruz Biotechnology). Washed membranes were subsequently probed with secondary antibodies conjugated to horseradish peroxidase and developed by enhanced chemiluminescence.
Mutation analysis.
DNA was extracted (DNeasy kit; Qiagen) from human TZM-bl cells 24 h after infection with HIV-1NL4-3 produced by either rabbit macrophages or T cells. DNA was DpnI digested and subjected to high-fidelity PCR using Pfu polymerase (Promega) and HIV-1-specific primers to amplify a nef/3′-LTR fragment, as reported by others (11). PCR products were gel purified, EcoRI/BamHI digested, and ligated into pcDNA3.1. All amplicon-positive clones were sequenced and the resulting sequences aligned with the proviral reference sequence.
RESULTS
Rabbit cells impose a postentry barrier to infection by HIV-1 and HIV-2 but not by SIVmac.
We first sought to confirm a species-specific postentry restriction for HIV-1 in a rabbit cell line (7, 20, 36, 62). VSV-G-pseudotyped HIV-1 GFP vectors, in which reporter gene expression is driven from a CMV promoter, were titrated on rabbit SIRC and human HeLa cells. Although entry efficiencies of VSV-G pseudotypes are similar in these two cell lines (data not shown), SIRC cells were only ∼12% as permissive as HeLa cells (Fig. 1A).
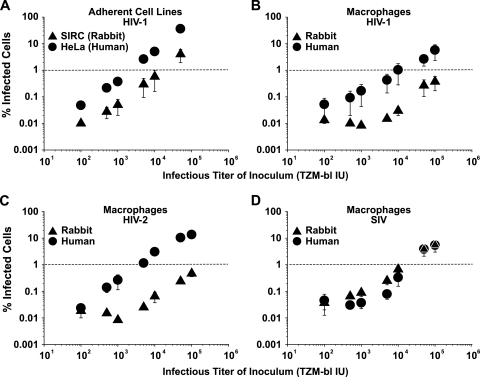
FIG. 1.
A rabbit-derived cell line and primary macrophages impose a barrier to infection by HIV but not by SIVmac. (A) Titration of VSV-G HIV-1 GFP vectors on SIRC and HeLa cells. (B to D) Titration of VSV-G pseudotyped HIV-1 (B)-, HIV-2 (C)-, or SIVmac (D)-based GFP vectors on spleen-derived rabbit macrophages and monocyte-derived human macrophages. Three days postchallenge, the percentage of infected (GFP-positive) cells was scored by flow cytometry. The percentages of infected cells are plotted as a function of the titer of the VSV-G GFP reporter virus inoculum (expressed in TZM-bl IU). Shown are the arithmetic means ± standard errors of the means (SEM) of results from two independent experiments.
To extend these studies to physiologically more relevant cells, we compared primary macrophages, derived from rabbit spleens, to human monocyte-derived macrophages for their ability to support lentiviral infection. The permissivity of primary rabbit macrophages to infection by VSV-G HIV-1 and HIV-2 GFP vectors was 9- to 47-fold lower than that of human macrophages (Fig. 1B and C). In contrast, infection levels after challenges with a VSV-G SIVmac GFP vector were comparable or slightly higher in macrophages from rabbits (Fig. 1D). In the rabbit macrophages, infection levels for the SIV vector were ∼10-fold higher than those of HIV-1 and HIV-2 vectors (Fig. 1B to D). These results demonstrate an HIV-specific postentry restriction in primary rabbit cells.
The postentry restriction in rabbit cells prevents HIV-1 DNA synthesis and can be overcome by limited changes of the gag gene.
To identify the step that is blocked in rabbit cells, we examined each step of the replication cycle, including virus entry, reverse transcription, nuclear import, and viral gene expression. To this end, we used a single challenge with a dual HIV-1 reporter vector and established methods (67). Based on the superior infectivity of the SIVmac vector in primary macrophages (Fig. 1D) and SIRC cells (data not shown) and recent characterizations of rabbit TRIM5 by Schaller and colleagues (62), we included a chimeric HIV-1 vector that contains the first 150 amino acids of capsid from SIVmac239 [HIV-1 (H/SCA) vector] (57).
SIRC cells were highly permissive for the HIV-1 (H/SCA) vector. Infection levels were 19- to 84-fold greater than those of the parental HIV-1 vector (Fig. 2A). This confirmed that the viral target of the postentry restriction of HIV-1 in rabbit cells is the capsid protein (62). Notably, this chimeric vector achieved higher infection levels (8-fold) in SIRC than in HeLa cells (Fig. 2B). The rabbit-specific restriction mapped to the level of reverse transcription. While the entry levels were similar for both vectors in SIRC cells (Fig. 2C, entry), HIV-1 cDNA synthesis was severely blocked for the BlaM-Vpr HIV-1 vector but not the BlaM-Vpr HIV-1 (H/SCA) chimera (Fig. 2C, reverse transcription). As a consequence of the lower HIV-1 cDNA synthesis, levels of HIV-1 2-LTR circles (Fig. 2C, nuclear import), used as a surrogate marker for nuclear import of the preintegration complex, and viral gene expression (Fig. 2C, gene expression) were reduced to similar degrees. Importantly, the HIV-1 (H/SCA) vector also displayed much greater infectivity (up to 195-fold) in rabbit macrophages than the HIV-1 vector (Fig. 3A). We also confirmed experimentally that, in this primary cell type, an early postentry event preceding or at nuclear import was affected (Fig. 3B), consistent with the restrictive activity of rabbit TRIM5.
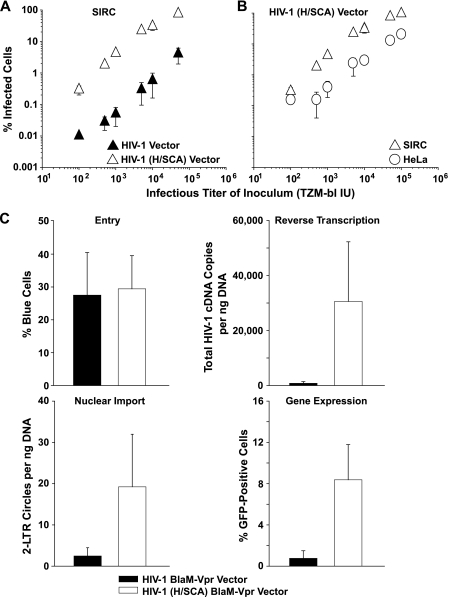
FIG. 2.
The postentry restriction in SIRC cells prevents HIV-1 cDNA synthesis and can be overcome by an HIV-1 (H/SCA) chimera. (A, B) Titration of VSV-G HIV-1 and HIV-1 (H/SCA) GFP vectors. The percentages of infected SIRC (rabbit) or HeLa (human) cells 3 days postinfection are plotted as a function of the titer of the viral inoculum. The cumulative data for the HIV-1 vector in panel A are identical to those shown in Fig. 1A. (C) The consecutive quantification of virus entry (flow cytometry-based virion fusion assay), reverse transcription (quantitative PCR for late RT products), nuclear import (quantitative PCR for 2-LTR circles), and gene expression (flow cytometry for GFP-positive cells) of SIRC cells infected with VSV-G HIV-1 or HIV-1 (H/SCA) GFP vectors carrying BlaM-Vpr was carried out in principle as reported previously (67). Shown are the arithmetic means ± SEM of results from four independent experiments.
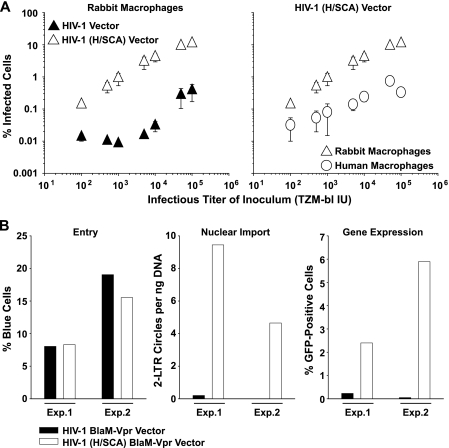
FIG. 3.
Primary rabbit macrophages are highly permissive for infection by the HIV-1 (H/SCA) chimera. (A) Titration of VSV-G HIV-1 or HIV-1 (H/SCA) GFP vectors on macrophages, carried out, in principle, as described in the legend of Fig. 2 (cumulative data for the HIV-1 vector are identical to those shown in Fig. 1B). Shown are the arithmetic means ± SEM of results from two independent experiments. (B) Consecutive quantification of virus entry, nuclear import, and gene expression from primary rabbit macrophages infected with either VSV-G HIV-1 or HIV-1 (H/SCA) GFP viruses carrying BlaM-Vpr, carried out, in principle, as reported in the legend to Fig. 2C. As a control of specificity for the entry signal, preincubation of virus stocks with a neutralizing anti-VSV-G antibody (13) reduced the entry signal from 8 to 19% down to 0.08 to 0.91% blue cells. Results from two independent experiments (Exp.1 and Exp.2) are shown. A high PCR background due to residual proviral plasmid contaminations in DNase-treated virus inocula precluded a reliable quantification of RT efficiency in infected macrophages (data not shown).
SIRC cells and primary rabbit macrophages are rendered permissive to R5 HIV-1 (H/SCA) infection by coexpression of human CD4 and CCR5.
Nonhuman cells generally do not support HIV-1 entry due to the absence of a functional CD4 receptor and chemokine coreceptors. A previous study by Speck and colleagues suggested that expression of the HIV receptor complex on SIRC cells supports HIV envelope-mediated cell-cell fusion (63). To determine if coexpression of hCD4 and hCCR5 could overcome the entry block in rabbit cells, we first generated a SIRC cell line-derived clone that stably expressed the human receptor complex. High coexpression of these human receptors on the surfaces of sorted and subcloned SIRC hCD4/hCCR5 cells was confirmed by flow cytometry (Fig. 4A). Importantly, in these cells, infections by HIV-1 (H/SCA) pseudotypes depended on the viral envelope and the coreceptor (Fig. 4B). Specifically, SIRC hCD4/hCCR5 cells were susceptible to infection by R5 Env (strain JR-FL) pseudotypes but nonpermissive for X4 Env (strain NL4-3) pseudotypes of HIV-1 (H/SCA) (Fig. 4B). As expected, both parental SIRC and SIRC hCD4/hCCR5 cells were permissive for VSV-G pseudotypes of the HIV-1 (H/SCA) vector but not of the TRIM5-restricted parental HIV-1 vector. The apparently lower degree of restriction of SIRC hCD4/hCCR5 cells than of the parental cells to infection by VSV-G HIV-1 wt pseudotypes is likely a clonal phenomenon unrelated to expression of the HIV receptor complex. As additional controls of specificity, SIRC hCD4/hCCR5 cells were pretreated with the CCR5 antagonist maraviroc or the fusion inhibitor T20, which resulted in a reduction of infection levels of the R5 Env HIV-1 (H/SCA) vector by 90% or 99.9%, respectively, while the CXCR4 antagonist AMD3100 had no effect (Fig. 4B). The incomplete inhibition by maraviroc is probably due to the high expression levels of the coreceptor on SIRC hCD4/hCCR5 cells (Fig. 4A) and the requirement for only very low cell surface concentrations of the coreceptor for HIV-1 entry, as reported previously (76).
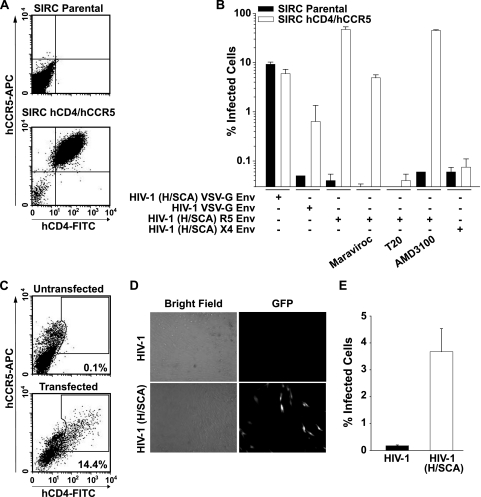
FIG. 4.
SIRC cells and primary rabbit macrophages are rendered permissive to R5 HIV-1 viruses by coexpression of human CD4 and CCR5. (A) FACS dot plots of parental SIRC cells and a SIRC clone, stably expressing the HIV receptor complex (SIRC hCD4/hCCR5). (B) Coreceptor-specific and entry inhibitor-sensitive R5 HIV-1 infection of SIRC hCD4/hCCR5 cells. Parental SIRC and SIRC hCD4/hCCR5 cells were challenged with the indicated HIV-1 or HIV-1 (H/SCA) GFP vectors pseudotyped with either VSV-G, an R5 (JR-FL) Env, or X4 (NL4-3) Env. Where indicated, cells were pretreated with the entry inhibitor maraviroc or AMD3100 or the fusion inhibitor T20. The percentage of infected (GFP-positive) cells was determined 2 days postchallenge. Shown are arithmetic means ± standard deviations (SD). (C) FACS dot plots of hCD4 and hCCR5 expression on rabbit macrophages, which had been transfected with corresponding expression constructs. The FACS gate indicates the receptor-positive cell population. (D) Microscopic images of transfected macrophages from panel C, which were subsequently challenged with JR-FL Env-pseudotyped HIV-1 or HIV-1 (H/SCA) GFP vectors. (E) Percentages of infected (GFP-positive) rabbit macrophages from panel D as determined by flow cytometry 3 days postinfection.
Seeking to extend these entry analyses to primary cells, we identified TurboFect as fairly well suited for the delivery of plasmid DNA into rabbit macrophages (data not shown). Cotransfection of expression plasmids for hCD4 and hCCR5 into spleen-derived rabbit macrophages resulted in a significant population of human-receptor-positive cells (Fig. 4C). Importantly, challenge of these cells with the R5 Env-pseudotyped HIV-1 (H/SCA) GFP vector, but not with the HIV-1 GFP counterpart, allowed for infection and reporter gene expression, as visualized by microscopy (Fig. 4D) and quantified by flow cytometry (Fig. 4E). Macrophage cultures, which were transfected with an empty plasmid or on which expression of hCD4 and hCCR5 was very low or absent (or only one of them was expressed), remained resistant to infection with the JR-FL Env HIV-1 (H/SCA) GFP vector (data not shown). Collectively, these results demonstrate that coexpression of hCD4 and hCCR5 on rabbit cells, including primary macrophages, is functional and confers coreceptor-specific virus entry, providing a strong basis for HIV receptor complex transgenesis in this species.
HIV-1 LTR transactivation is species and cell type specific and occurs efficiently in primary rabbit cells.
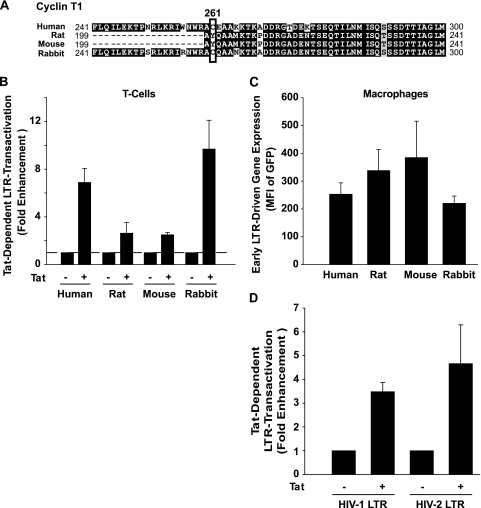
We (32, 43, 51) and others (1, 48, 72) have described a quantitative impairment of Tat-dependent HIV-1 LTR transactivation in native rat and mouse cells, which is due primarily to an inefficient transactivating responsive sequence (TAR)-Tat-cyclin T1 interaction. This species-specific limitation was mapped to a single amino acid in cyclin T1 (tyrosine-261 in rodents; cysteine-261 in humans) (10, 26).
Rabbit cyclin T1 is 91% identical at the amino acid level to human cyclin T1 and, notably, shares cysteine-261 with the human ortholog (Fig. 5A). It was previously suggested that Tat and Rev functions are at least partly preserved in SIRC cells (63). To assess HIV LTR-driven transcription in primary rabbit T cells, minimal reporter plasmids, consisting of the complete, PCR-amplified LTR and Gag leader sequences from either HIV-1NL4-3 or HIV-2ROD-A, driving the expression of GFP (pHIV LTR GFP) (51), and Tat expression constructs were cotransfected. The basal levels of Tat-independent transcription were low and comparable for T cells from all four species (data not shown). Importantly, coexpression of Tat increased levels of HIV LTR-driven gene expression in primary T cells from rabbits ∼10-fold (for HIV-1, Fig. 5B; data not shown for HIV-2), which was comparable to the enhancement in human T cells (7-fold for HIV-1 [Fig. 5B] and 9-fold for HIV-2 [data not shown]). In contrast, T cells from both rodents showed a minor, 1.2- to 2-fold Tat-mediated induction, in agreement with the results of previous studies conducted in the absence of hcyclin T1 coexpression (8, 43, 50).
FIG. 5.
Tat-dependent HIV-1-LTR transactivation is species and cell type specific and occurs efficiently in primary rabbit cells. (A) Partial amino acid sequence alignment of cyclin T1s of human, rabbit, rat, and mouse origins (ClustalW2 method and BOXSHADE). Identical amino acids are shaded in black, conserved or similar residues are in gray, and unrelated amino acids are in white. Dashes indicate gaps introduced to optimize the alignment. Amino acid 261 is boxed. (B) Primary T cells were nucleofected with pHIV-1 LTR GFP in the presence or absence of an HIV-1 Tat expression vector. The MFI of GFP expression was quantified in viable cells 24 h later by flow cytometry. Shown are the arithmetic means ± SEM of results of duplicates or triplicates of two to four independent experiments. (C) Macrophages were infected with VSV-G HIV-1NL4-3 GFP and analyzed for Tat- and LTR-dependent reporter gene expression by flow cytometry 3 days later. Infectious titers were chosen such that the percentage of GFP-positive macrophages for all species was in the single-cell infection range (human, 1.0%; rat, 3.6%; mouse, 7.8%; rabbit, 0.4%). Shown are the arithmetic means ± SEM of results of duplicates or quadruplicates of two to four independent experiments. (D) Primary rabbit macrophages were transfected with pHIV-1 or pHIV-2 LTR GFP vectors in the presence or absence of an HIV-1 or HIV-2 Tat expression vector, respectively. The MFI of GFP expression was quantified 3 days later by flow cytometry. Shown are the arithmetic means ± SEM of results from two independent experiments.
To assess HIV transcriptional activity in macrophages from all of these species, primary cultures were infected with the nearly full-length HIV-1NL4-3 GFP virus pseudotyped with VSV-G. Here, the 5′-LTR drives GFP expression from the nef locus. GFP expression from integrated proviruses under single-cell infection conditions is a quantitative surrogate of early, LTR-driven gene expression. Viral titers were chosen such that macrophages from all four species had relatively low numbers of infected, GFP-positive cells (0.4 to 7.8%). In contrast to T cells, macrophages from mice, rats, rabbits and humans supported LTR-driven gene expression from the nef locus to similar levels (Fig. 5C), indicating a less stringent requirement for cyclin T1 in this cell type in rodents. Tat's dependence on HIV LTR transactivation in primary rabbit macrophages was demonstrated using the HIV LTR GFP reporter/Tat cotransfection assay (Fig. 5D). In summary, these results reveal pronounced species- and cell type-specific differences in requirements for efficient HIV-1 transcription. Remarkably, primary T cells and macrophages from rabbits support Tat-dependent HIV LTR transactivation at levels similar to those from humans.
Rabbit cells support robust Rev/CRM1-dependent HIV-1 gene expression.
In rodent cells, the expression of late HIV gene products is low and represents a major posttranscriptional barrier. Inefficient Rev function has been suggested to limit the nuclear export of Rev-responsive element (RRE)-containing unspliced and singly spliced HIV mRNAs or to result in oversplicing of viral RNAs (8, 19, 50, 74, 77).
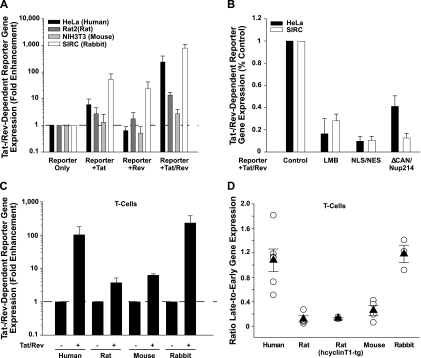
To simultaneously assess HIV Tat and Rev functions, we used a reporter plasmid that contains HIV-1 LTRs, the 5′ section of gag, containing inhibitory sequences (INS), and the RRE to direct Tat- and Rev-dependent expression of a red fluorescent protein (RFP) reporter [pLRed(INS)2R] (47, 75). First, adherent cell lines were transfected with the pLRed(INS)2R reporter with or without Tat and/or Rev expression constructs and peGFP-N1, and 1 day later, RFP expression was quantified in transfected (GFP-positive) cells by flow cytometry. Expression of either Tat or Rev alone was insufficient to induce RFP expression in HeLa, Rat2, and NIH 3T3 cells and, interestingly, already resulted in a marked reporter gene expression in SIRC cells (Fig. 6A). Coexpression of Tat and Rev triggered a drastic, 239- and 791-fold enhancement of RFP expression in HeLa and SIRC cells, respectively, while responses in Rat2 and NIH 3T3 cells were low or nearly absent (Fig. 6A). As controls for specificity, the Tat/Rev-dependent reporter gene expression in rabbit and human cells was markedly reduced by LMB treatment (23), coexpression of a dominant negative form of nucleoporin 214/CAN (ΔCAN/Nup214) (12), or saturating levels of short nuclear localization signal (NLS)/nuclear export signal (NES) sequences (22, 39) (Fig. 6B); all of these have been reported to interfere with CRM1-dependent nuclear export of viral RNAs.
FIG. 6.
Tat- and Rev/CRM1-dependent HIV-1 gene expression is robust in rabbit cells, including primary T cells. (A) The indicated cell lines were cotransfected with expression constructs encoding the Tat- and/or Rev-dependent RFP reporter, HIV-1 Tat and/or HIV-1 Rev, and GFP (to identify transfected cells). The product of the percentage of GFP/RFP-positive cells and the MFI of RFP, measured in the absence of Tat and Rev coexpression, was set to 1 for each cell line. Shown are arithmetic means ± SEM of results from triplicates of two to six experiments. (B) RFP reporter expression is CRM1 dependent. SIRC and HeLa cells were transfected as for panel A, with the addition of LMB and expression plasmids encoding either short NLS/NES sequences or the dominant negative nucleoporin ΔCAN/Nup214. (C) Primary T cells were nucleofected with the expression constructs described for panel A and analyzed 24 h later. Shown are arithmetic means ± SEM of results from duplicates of two to four independent experiments. (D) Primary T cells were nucleofected with the nearly full-length HIV-1 reporter construct pNL4-3 GFP (early gene expression) or pNL4-3 Gag-GFP (late gene expression). The MFI of GFP expression in viable cells was determined 24 h later by flow cytometry, and MFIlate/MFIearly ratios are depicted. Results for T cells from individual donors or animals (open circles) and the arithmetic means ± SEM (filled triangles) are depicted.
We next extended this reporter-based strategy to primary T cells. In agreement with the findings in cell lines, Tat/Rev coexpression resulted in 102- and 232-fold enhancements of RFP expression in transfected T cells from humans and rabbits, respectively (Fig. 6C), while T cells from rats and mice responded poorly. Finally, we used a second, independent experimental approach to assess the functionality of the viral Rev protein in primary T cells. We quantified the expression of a late, Rev-dependent gene product (MA-GFP) (52) relative to that of an early, Rev-independent gene product (Nef) in a nearly full-length proviral context. T cells from both rodents showed strong Rev-independent GFP reporter expression from the nef locus, whereas Rev-dependent MA-GFP expression was fairly weak, resulting in a “late-to-early” ratio of below 0.3 (Fig. 6D). Expectedly, transgenic expression of hcyclin T1 in rat T cells (51) enhanced GFP expression from both proviruses but did not significantly affect this ratio (Fig. 6D). T cells from rabbits and humans displayed a stronger expression of MA-GFP than did T cells from the rodent species, and GFP expression levels from both proviral loci were similar in these species (Fig. 6D). These results demonstrate that the two major barriers to HIV-1 replication in rodents, at the level of Tat-dependent viral transcription and Rev-dependent late viral gene expression, are absent in the rabbit species.
Primary rabbit T cells efficiently support the late phase of the HIV-1 replication cycle and release large numbers of infectious particles.
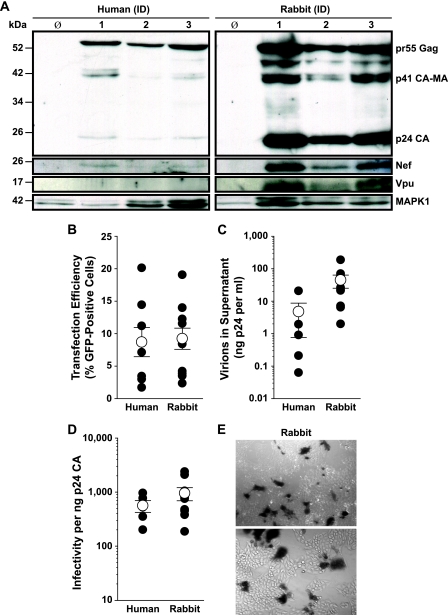
We next sought to quantitatively assess the efficiency of a series of steps in the late phase of the HIV-1 replication cycle in primary T cells from rabbits and humans. Speck and colleagues previously reported that mature virions can be released from provirus-transfected T cells (63). Here, we transfected activated T cells with HIV-1NL4-3 proviral DNA and analyzed them for the cell-associated expression of viral proteins, as well as the quantity and infectivity of virions released into the culture supernatant. In line with the above-described results for reporter system-based analyses of Tat and Rev function, cell lysates from provirus-transfected rabbit T cells contained abundant levels of early (Nef) and late (Gag, Vpu) HIV-1 proteins (Fig. 7A). At comparable transfection efficiencies (Fig. 7B), the cell-associated expression of these viral proteins in rabbit T cells was comparable to or higher than that in human reference T cells. Notably, processing of the p55Gag precursor to p24CA, which is generally believed to be impaired in rodent cells (8, 43, 50), was efficient in the rabbit species (Fig. 7A). Furthermore, the number of HIV virions released into the supernatant was comparable to or higher in T cells from rabbits than the number in T cells from humans (Fig. 7C). Under these experimental conditions, the average interspecies transfection efficiencies, despite considerable interindividual variability, were comparable (Fig. 7B). We can, however, not exclude the possibility that varied amounts of proviral DNA delivered per cell contributed to the somewhat higher levels of viral gene products in cell lysates (Fig. 7A) and released virions in T cells (Fig. 7C) from rabbits than from humans.
FIG. 7.
Robust HIV-1 protein expression, efficient Gag processing, and release of high levels of infectious HIV-1 from provirally transfected primary rabbit T cells. T cells from humans (left panels) or rabbits (right panels) were transfected with pHIV-1NL4-3 and, in parallel, with the proviral reporter pHIV-1NL4-3 GFP (transfection efficiency). After 36 h, supernatants and cells were harvested. (A) Western blots of cell lysates from three independent donors probed consecutively with anti-p24CA, anti-HIV-1 Nef, and anti-Vpu. MAPK expression served as a loading control. φ, untransfected control. Identification (ID) numbers indicate different donors. (B) Transfection efficiency determined by the percentage of viable, GFP-positive cells. (C, D) Concentrated, sucrose-pelleted virions were analyzed for p24CA concentration (C), and the infectious titer is depicted as the relative infectivity per ng p24CA (D). (B to D) Individual results from experiments with T cells from four human and eight rabbit donors (filled symbols) and the arithmetic means ± SEM of results (open symbols) are depicted. (E) Microscopic images of TZM-bl cells infected with HIV-1 virions derived from rabbit T cells and stained with a β-galactosidase substrate.
Finally, the infectivity of these transfection-derived HIV-1 particles was quantified in a standard TZM-bl reporter assay (Fig. 7E) (40). When infectivity values were normalized to the p24CA concentration of released virions, HIV-1 virions produced by primary T cells from rabbits and humans were equally infectious (Fig. 7D). Thus, the late phase of HIV-1 replication in primary rabbit T cells is efficient, revealing a degree of intrinsic susceptibility of lagomorph T cells to this human lentivirus that has not been reported for any other nonhuman species.
Rabbit macrophages efficiently release HIV-1 virions, but their infectivity is low.
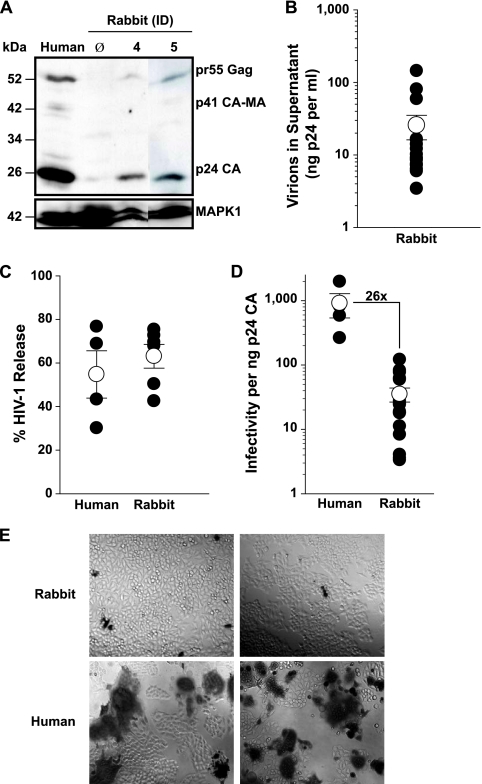
Next we examined the ability of primary rabbit macrophages to support the late phase of the HIV-1 replication cycle. Cells were transfected with proviral DNA. The transfection efficiency, assessed by parallel transfections with a nearly full-length proviral HIV-1 GFP reporter, ranged from 4 to 11% (data not shown). Despite this rather low transfection efficiency, Western blot analysis of macrophage lysates showed considerable expression and processing of Gag (Fig. 8A). Since human macrophages cannot be transfected efficiently with proviral DNA, VSV-G HIV-1NL4-3-infected cells served as references. Importantly, rabbit macrophage supernatants contained significant amounts of HIV-1, with virion-associated p24CA concentrations up to 144 ng per ml (mean, 26 ng p24CA per ml) (Fig. 8B). Levels of HIV-1 release, expressed as percentages of total p24CA (in cells and supernatants) that was secreted as virion-associated p24CA, were comparable for the HIV-1 provirus-transfected rabbit macrophages and VSV-G HIV-1-infected human macrophages (Fig. 8C), providing no evidence for a species-specific defect for wild-type HIV-1 at egress.
FIG. 8.
Primary rabbit macrophages efficiently release HIV-1 virions, but their infectivity is low. Rabbit macrophages were transfected with HIV-1 proviral DNA, and 5 days later, supernatants and cells were harvested. (A) Anti-p24CA Western blot of pHIV-1-transfected rabbit macrophages. MAPK expression served as a loading control. φ, untransfected control. ID numbers indicate different animals. VSV-G HIV-1NL4-3-infected human macrophages served as reference controls. (B) p24CA concentration of sucrose-pelleted supernatants from HIV-1NL4-3-transfected rabbit macrophages. (C) HIV-1 release from macrophages quantified as the percentage of total p24CA (in cells and supernatant) that was secreted as virion-associated p24CA, as reported previously (28). (D) The relative particle infectivity was determined as described for Fig. 7. (E) Microscopic images of TZM-bl cells infected with comparable amounts of HIV-1 virions derived from either provirus-transfected rabbit or HIV-1-infected human macrophages.
Unexpectedly, the infectivity of HIV-1 particles derived from rabbit macrophages was markedly less than those derived from human macrophages (26-fold) (Fig. 8D and E). Thus, rabbit macrophages apparently support all late steps of HIV-1 replication yet induce an infectivity defect in released particles.
Rabbit macrophage-derived HIV-1 carries a complex pattern of mutations.
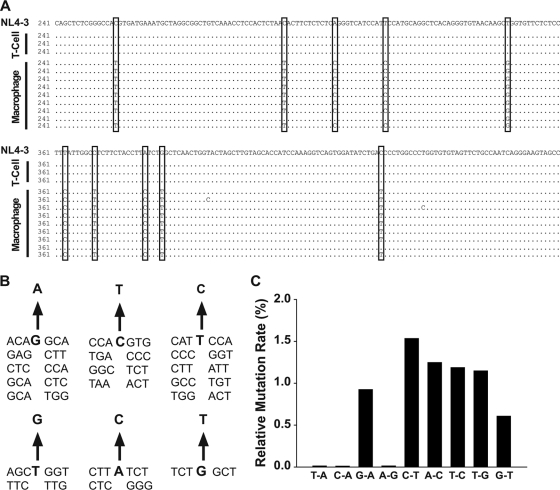
We hypothesized that the infectivity defect of HIV-1 released from rabbit macrophages might be due to the mutating activity of rabbit members of the APOBEC family. To characterize this, viral DNAs were recovered from TZM-bl cells infected with HIV-1NL4-3 produced by either macrophages or T cells from rabbits, a nef/3′-LTR fragment was then PCR amplified and cloned, and individual sequences were determined, in principle as reported previously (11). None of the sequences amplified from T cell-derived virus carried mutations (Fig. 9A), in line with the high infectivity of T cell-derived HIV-1 (Fig. 7D). In contrast, macrophage-derived HIV-1 cDNAs displayed multiple mutations in 51 out of 54 clones analyzed at an overall frequency of 0.74% (Fig. 9A and C). These clones were derived from 3 independent infection assays and clone analyses of TZM-bl cells, with HIV-1 virions derived from 4 different rabbit donor macrophages. Virtually all clones carried 6 different types of nucleotide changes (G→A, C→T, A→C, T→C, T→G, G→T) at identical nucleotide positions (boxed nucleotide changes in Fig. 9A). Besides this major pattern of mutations, additional single nucleotide changes were found in individual clones at multiple sites in the amplified region, resulting in a total of 8 different clones. The G→A and C→T mutations were detected at frequencies of 0.9 to 1.5%, respectively, consistent with both DNA editing and RNA editing by a cellular deaminase (11, 37). Remarkably, however, considerable levels of other, deamination-independent HIV-1 cDNA mutations (A→C, T→C, T→G, G→T), all of which occurred at frequencies of ∼1% (Fig. 9A and C), were also noted. The occurrence of preferential sites of mutations was analyzed (Fig. 9B). The G→A, C→T, and T→C changes showed no apparent consensus pattern. For T→G and A→C changes, two trinucleotide sequences (up- and downstream of the affected T and A, respectively) were noted. For the G→T mutation, TCTGGCT (where boldface indicates the mutated nucleotide) was the only sequence context for this mutation (Fig. 9B). Conceivably, these complex mutations and the resulting cDNA instability may combine to reduce the infectivity of HIV-1 derived from rabbit macrophages.
FIG. 9.
Mutation analysis of HIV-1 produced by primary rabbit cells. Viral cDNAs were recovered from TZM-bl cells infected with HIV-1NL4-3 produced by either rabbit T cells or macrophages, a nef/3′-LTR fragment of 650 bp was PCR amplified and cloned, and individual sequences were determined. (A) Shown are nucleotide positions 241 to 480 from 3 out of 4 sequences for T cell-derived HIV-1 and 9 out of 54 sequences for macrophage-derived HIV-1. Nucleotide changes relative to the unmodified HIV-1NL4-3 sequence (top) are indicated. Boxes indicate the most frequently found nucleotide changes. (B) Analysis of potential preferred sites of major mutations. Frequently found trinucleotides up- and downstream of the affected nucleotide are shown. (C) Relative mutation rates based on the total number of the indicated single nucleotide changes in all amplicons analyzed.
DISCUSSION
In the present study, we quantitatively characterized the ability of primary T cells and macrophages from rabbits to support critical steps in the HIV-1 replication cycle. We found that primary target cells in rabbits are permissive to HIV-1 infection and support the majority of replication steps as well as human cells do. Of the three replication barriers that HIV-1 encounters in lagomorphs, two can be readily overcome: expression of the HIV receptor complex and limited modifications in the N-terminal part of the HIV-1 capsid surmount blocks at entry and reverse transcription, respectively. In addition, we identified a virion infectivity defect in macrophages that is absent in T cells and that poses the final cellular barrier to full HIV-1 permissivity in this species.
The HIV-1 entry block is due to the general inability of CD4 and/or major chemokine receptors from nonhuman species to support virion binding and fusion (Fig. 10). Coexpression of hCD4 and hCCR5 in SIRC cells and primary macrophages mediated envelope-specific and coreceptor-dependent virion entry. These results agree with reports that surface expression of the human binding receptor and one of the major chemokine coreceptors is sufficient to overcome the entry block in cells from diverse primate and small-animal species (8, 16, 42, 43, 45, 49, 54, 76).
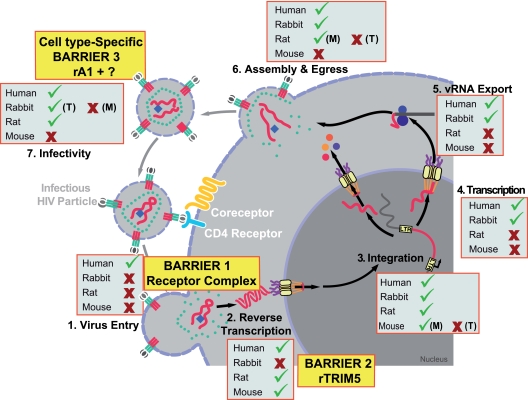
FIG. 10.
Summary of the efficiency of steps in HIV-1 replication in primary cells of human, rat, mouse, and rabbit origins. A schematic representation of consecutive steps in the HIV-1 replication cycle and the ability of primary cells from the respective species to support these steps (✓, efficient; ✖, inefficient or completely blocked [this study and references cited in the text]) are shown. (T), T cells; (M), macrophages; rTRIM5, rabbit TRIM5; rA1, rabbit APOBEC1. The three barriers to efficient HIV-1 replication in rabbit cells are highlighted.
In rabbit cells, unlike in mice and rats (32, 67, 76), HIV-1 encounters a barrier early postentry that leads to a strong block to viral DNA synthesis (7, 20, 36) (Fig. 10). While presaturation/infection studies indicative of a restriction factor were inconclusive in one study (20), Schaller and colleagues demonstrated that short hairpin RNA (shRNA)-mediated depletion of rabbit TRIM5 in SIRC cells enhanced the permissivity of restricted lentiviruses and that heterologous expression of rabbit TRIM5 in feline CRFK cells recapitulated the restrictive phenotype (62). Our mapping of the affected replication step and lentivirus-specific restriction pattern of the postentry block in primary rabbit macrophages (HIV-1 and HIV-2 restricted, SIVmac not restricted) is consistent with the reported antiviral activity of rabbit TRIM5 (62). Notably, the postentry restriction was independent of the route of HIV-1 entry. Most importantly, in the context of animal model development, introduction of the 150 N-terminal amino acids of capsid from SIVmac239 into HIV-1 allowed it to escape the postentry restriction in this primary cell type. The apparently lower levels of infectivity of the chimeric virus in human cells are in line with results from an earlier report (25). It will be interesting to determine if even smaller modifications in the HIV-1 capsid are sufficient for it to proceed unrestricted until integration and gene expression in rabbit cells. Preliminary data indicate that the stretch of 30 amino acids from positions 77 to 107 in capsid may be critical for the restriction.
Two major species-specific barriers to HIV-1 replication in rodents, one at the level of Tat-dependent LTR transactivation and the other affecting Rev- and RRE-dependent viral gene expression (8, 43, 46, 50, 69), are absent in the rabbit species (Fig. 10). This observation is supported by multiple lines of experimental evidence with replication stage-specific reporter assays and full-length infectious HIV-1. The absence of a transcriptional block can likely be explained at the genomic level. The Tat-interacting protein cyclin T1 is a key determinant for the inability of rodent cells to support efficient LTR transactivation, and a tyrosine instead of a cysteine at position 261 in the rodent orthologs plays a decisive role (10, 24, 26, 48). This residue is part of the Tat/TAR recognition motif of cyclin T1 (2). Critically, rabbit cyclin T1 shares cysteine-261 with the human ortholog. In light of the strong Tat-dependent LTR activity in both species, this finding underscores the functional importance of this amino acid in the cross-species context. Interestingly, rodent macrophages seem to pose an exception in their degree of dependence on cyclin T1 for LTR activity: irrespective of the amino acid residue at position 261, macrophages from rodents, humans, and rabbits supported comparable levels of provirus 5′-LTR-driven transactivation (this study and references 32 and 51). This may relate to the ability of HIV to exploit in macrophages a set of nuclear transcription factors and mechanisms of transcriptional regulation distinct from those used in other cell types (for reviews, see references 61 and 71).
Of great relevance for animal model development, the transcriptional impairment in primary T cells from rodents can be substantially ameliorated by hcyclin T1 transgenesis (51, 76). However, a molecular understanding for the deficit in Rev function in rodent cells or strategies to overcome it are entirely lacking (8, 18, 50, 69, 76). The severe defect in the Rev- and CRM1-dependent export of unspliced and singly spliced HIV-1 RNAs from the nucleus and oversplicing of the viral RNA result in grossly reduced levels of expression of structural proteins. Besides transcription and nuclear export of RRE-containing viral RNAs, the processing of Gag and particle assembly and release, as well as virion infectivity, pose additional postintegration defects in rodent cells. In primary mouse T cells, the cumulative impact of these late-phase deficits results in an up-to-500-fold reduction in the yield of infectious virus after a single cycle (76), and this has crippled efforts to advance a transgenic mouse model.
Importantly, we found a high natural HIV-1 permissivity in primary rabbit cells and, in particular, an ability of T cells to efficiently support the entire late phase of HIV-1 replication (Fig. 10). The number of infectious HIV-1 virions released after provirus transfection reflects the cumulative efficiency of late-phase steps from transcription over assembly, to release and maturation. At comparable transfection efficiencies, rabbit T cells released levels of infectious virions that were comparable to or higher than those of their human counterparts. We predict that an N-terminally CA-modified HIV-1 that can circumvent the postentry restriction could spread in primary rabbit T cells carrying the HIV receptor complex. However, neither the hCCR5 nor the hCXCR4 coreceptor could be transiently expressed in these cells (data not shown). Therefore, transgenic expression of the HIV receptor complex will be required to probe the full replication cycle in a dynamic infection context in primary T cells. Notably, the assembly and/or release of virions in provirus-transfected SIRC cells was rather inefficient (data not shown), underscoring the importance of performing studies into HIV-1 species susceptibility in the context of animal model development in physiologically relevant primary target cells. This finding unfortunately also precluded a spreading infection of CA-modified HIV-1 in SIRC hCD4/hCCR5 cells.
HIV-1 particles released from rabbit macrophages were 26-fold less infectious than those derived from human macrophages or from rabbit T cells. For macrophage-derived virus, a complex pattern of mutations in the HIV-1 cDNA that involved six different types of nucleotide changes was noted. Relevant in this context, the antiviral potency of rabbit APOBEC1 (rA1) expressed in 293T cells was recently reported (37). rA1 was efficiently incorporated into virions and reduced the infectivity of HIV-1 in a Vif-resistant manner by up to 300-fold. Catalytic-site mutants indicated a deaminase-dependent mechanism of restriction, with genomic RNA and reverse-transcribed proviral DNA serving as substrates. Of particular note, proviral mutation analysis by Ikeda et al. demonstrated an accumulation primarily of C→T and G→A changes (37), which in principle could account for approximately one-third of the mutations observed for rabbit macrophage-derived HIV-1 in our study. While Ikeda et al. noted a TTC* sequence as a preferred site for a C→T mutation in HIV-1 derived from rA1-expressing 293T cells, we found no apparent preference in the trinucleotides surrounding the affected C in macrophage-derived HIV-1. The mechanisms underlying the other specific mutations (A→C, T→C, T→G, G→T) are entirely unclear. T→C mutations can result from an amination reaction, while A→C, T→G, and G→T mutations require transversions of purine to pyrimidine or vice versa. Such transversions can apparently occur at higher frequency under oxidative stress (44). The contribution of rA1 expression to the macrophage infectivity phenotype should be analyzed, for example by RNA interference or by identification of a lentiviral vif allele which can counteract this deaminase. A complete understanding of this infectivity defect will be important to design strategies to overcome this final replication barrier. From a different perspective, the high permissivity of T cells alone may be sufficient to allow efficient HIV-1 (H/SCA) replication in receptor complex-transgenic rabbits in vivo. Important from a technical perspective, transgenesis is well established in this species, and more recently, knockout and knock-in technologies have also been accomplished in rabbits. We propose that receptor complex transgenesis combined with limited genetic modifications of HIV-1 gag and possibly vif to evade species-specific intrinsic restriction factors might render rabbits highly permissive to infection by HIV-1.
Acknowledgments
We are very grateful to Hans-Georg Kräusslich for support, discussion, and reagents. We thank Thomas Euler, Xavier Castell, and Sanjeev Kumar Kaushalya from the Department of Biomedical Optics, Max-Planck-Institute for Medical Research, Heidelberg, Germany, for providing access to rabbit carcasses. We thank Monika Langlotz and Viktor Sourjik for FACSAria analyses. We thank Christine Goffinet and Paul Burda for contributing unpublished data, reagents, and helpful discussions and Ina Ambiel for excellent technical support. We are grateful to Warner C. Greene and John C. W. Carroll for allowing us to use and modify their graphical representation of the HIV replication cycle. We thank Jens Bohne, Ruth Brack-Werner, Bruce Chesebro, Brian Cullen, Matthias Dittmar, Jose Este, Oliver Fackler, Nathaniel Landau, Malcom Martin, Barbara Müller, Jan Münch, Joseph Sodroski, Greg Towers, and Klaus Strebel for the gift of reagents. We are grateful to Gary Howard for expert editorial assistance. We thank members of the laboratory, Valerie Bosch, and Oliver T. Fackler for comments on the manuscript and Martin Löchelt for discussions.
O.T.K. is a member of the CellNetworks Cluster of Excellence EXC81.
Footnotes
Published ahead of print on 22 September 2010.
REFERENCES
- 1.Alonso, A., T. P. Cujec, and B. M. Peterlin. 1994. Effects of human chromosome 12 on interactions between Tat and TAR of human immunodeficiency virus type 1. J. Virol. 68:6505-6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anand, K., A. Schulte, K. Vogel-Bachmayr, K. Scheffzek, and M. Geyer. 2008. Structural insights into the cyclin T1-Tat-TAR RNA transcription activation complex from EIAV. Nat. Struct. Mol. Biol. 15:1287-1292. [DOI] [PubMed] [Google Scholar]
- 3.Anderson, J. L., E. M. Campbell, X. Wu, N. Vandegraaff, A. Engelman, and T. J. Hope. 2006. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J. Virol. 80:9754-9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baenziger, S., R. Tussiwand, E. Schlaepfer, L. Mazzucchelli, M. Heikenwalder, M. O. Kurrer, S. Behnke, J. Frey, A. Oxenius, H. Joller, A. Aguzzi, M. G. Manz, and R. F. Speck. 2006. Disseminated and sustained HIV infection in CD34+ cord blood cell-transplanted Rag2−/−gamma c−/− mice. Proc. Natl. Acad. Sci. U. S. A. 103:15951-15956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baenziger, S., P. Ziegler, L. Mazzucchelli, L. Bronz, R. F. Speck, and M. G. Manz. 2008. Human T cell development and HIV infection in human hemato-lymphoid system mice. Curr. Top. Microbiol. Immunol. 324:125-131. [DOI] [PubMed] [Google Scholar]
- 6.Baumann, J. G., D. Unutmaz, M. D. Miller, S. K. Breun, S. M. Grill, J. Mirro, D. R. Littman, A. Rein, and V. N. KewalRamani. 2004. Murine T cells potently restrict human immunodeficiency virus infection. J. Virol. 78:12537-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. U. S. A. 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bieniasz, P. D., and B. R. Cullen. 2000. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 74:9868-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1999. Highly divergent lentiviral Tat proteins activate viral gene expression by a common mechanism. Mol. Cell. Biol. 19:4592-4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bishop, K. N., R. K. Holmes, A. M. Sheehy, and M. H. Malim. 2004. APOBEC-mediated editing of viral RNA. Science 305:645. [DOI] [PubMed] [Google Scholar]
- 12.Bogerd, H. P., A. Echarri, T. M. Ross, and B. R. Cullen. 1998. Inhibition of human immunodeficiency virus Rev and human T cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J. Virol. 72:8627-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boritz, E., J. Gerlach, J. E. Johnson, and J. K. Rose. 1999. Replication-competent rhabdoviruses with human immunodeficiency virus type 1 coats and green fluorescent protein: entry by a pH-independent pathway. J. Virol. 73:6937-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosch, V., T. Pfeiffer, G. Devitt, I. Allespach, T. Ebensen, V. Emerson, C. A. Guzman, and O. T. Keppler. 2009. HIV pseudovirion vaccine exposing Env “fusion intermediates”—response to immunisation in human CD4/CCR5-transgenic rats. Vaccine 27:2202-2212. [DOI] [PubMed] [Google Scholar]
- 15.Brainard, D. M., E. Seung, N. Frahm, A. Cariappa, C. C. Bailey, W. K. Hart, H. S. Shin, S. F. Brooks, H. L. Knight, Q. Eichbaum, Y. G. Yang, M. Sykes, B. D. Walker, G. J. Freeman, S. Pillai, S. V. Westmoreland, C. Brander, A. D. Luster, and A. M. Tager. 2009. Induction of robust cellular and humoral virus-specific adaptive immune responses in human immunodeficiency virus-infected humanized BLT mice. J. Virol. 83:7305-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Browning, J., J. W. Horner, M. Pettoello-Mantovani, C. Raker, S. Yurasov, R. A. DePinho, and H. Goldstein. 1997. Mice transgenic for human CD4 and CCR5 are susceptible to HIV infection. Proc. Natl. Acad. Sci. U. S. A. 94:14637-14641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coskun, A. K., M. van Maanen, V. Nguyen, and R. E. Sutton. 2006. Human chromosome 2 carries a gene required for production of infectious human immunodeficiency virus type 1. J. Virol. 80:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullen, B. R. 2003. Nuclear mRNA export: insights from virology. Trends Biochem. Sci. 28:419-424. [DOI] [PubMed] [Google Scholar]
- 20.Cutino-Moguel, T., and A. Fassati. 2006. A phenotypic recessive, post-entry block in rabbit cells that results in aberrant trafficking of HIV-1. Traffic 7:978-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denton, P. W., J. D. Estes, Z. Sun, F. A. Othieno, B. L. Wei, A. K. Wege, D. A. Powell, D. Payne, A. T. Haase, and J. V. Garcia. 2008. Antiretroviral pre-exposure prophylaxis prevents vaginal transmission of HIV-1 in humanized BLT mice. PLoS Med. 5:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobbelstein, M., A. K. Arthur, S. Dehde, K. van Zee, A. Dickmanns, and E. Fanning. 1992. Intracistronic complementation reveals a new function of SV40 T antigen that co-operates with Rb and p53 binding to stimulate DNA synthesis in quiescent cells. Oncogene 7:837-847. [PubMed] [Google Scholar]
- 23.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 24.Fujinaga, K., T. P. Cujec, J. Peng, J. Garriga, D. H. Price, X. Grana, and B. M. Peterlin. 1998. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 72:7154-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujita, M., A. Yoshida, M. Miyaura, A. Sakurai, H. Akari, A. H. Koyama, and A. Adachi. 2001. Cyclophilin A-independent replication of a human immunodeficiency virus type 1 isolate carrying a small portion of the simian immunodeficiency virus SIVMAC gag capsid region. J. Virol. 75:10527-10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geuenich, S., C. Goffinet, S. Venzke, S. Nolkemper, I. Baumann, P. Plinkert, J. Reichling, and O. T. Keppler. 2008. Aqueous extracts from peppermint, sage and lemon balm leaves display potent anti-HIV-1 activity by increasing the virion density. Retrovirology 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goffinet, C., I. Allespach, S. Homann, H. M. Tervo, A. Habermann, D. Rupp, L. Oberbremer, C. Kern, N. Tibroni, S. Welsch, J. Krijnse-Locker, G. Banting, H. G. Krausslich, O. T. Fackler, and O. T. Keppler. 2009. HIV-1 antagonism of CD317 is species specific and involves Vpu-mediated proteasomal degradation of the restriction factor. Cell Host Microbe 5:285-297. [DOI] [PubMed] [Google Scholar]
- 29.Goffinet, C., I. Allespach, and O. T. Keppler. 2007. HIV-susceptible transgenic rats allow rapid preclinical testing of antiviral compounds targeting virus entry or reverse transcription. Proc. Natl. Acad. Sci. U. S. A. 104:1015-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goffinet, C., I. Allespach, L. Oberbremer, P. L. Golden, S. A. Foster, B. A. Johns, J. G. Weatherhead, S. J. Novick, K. E. Chiswell, E. P. Garvey, and O. T. Keppler. 2009. Pharmacovirological impact of an integrase inhibitor on human immunodeficiency virus type 1 cDNA species in vivo. J. Virol. 83:7706-7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goffinet, C., and O. T. Keppler. 2006. Efficient nonviral gene delivery into primary lymphocytes from rats and mice. FASEB J. 20:500-502. [DOI] [PubMed] [Google Scholar]
- 32.Goffinet, C., N. Michel, I. Allespach, H. M. Tervo, V. Hermann, H. G. Krausslich, W. C. Greene, and O. T. Keppler. 2007. Primary T-cells from human CD4/CCR5-transgenic rats support all early steps of HIV-1 replication including integration, but display impaired viral gene expression. Retrovirology 4:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hague, B. F., S. Sawasdikosol, T. J. Brown, K. Lee, D. P. Recker, and T. J. Kindt. 1992. CD4 and its role in infection of rabbit cell lines by human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 89:7963-7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatziioannou, T., S. Cowan, and P. D. Bieniasz. 2004. Capsid-dependent and -independent postentry restriction of primate lentivirus tropism in rodent cells. J. Virol. 78:1006-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hausselt, S. E., T. Euler, P. B. Detwiler, and W. Denk. 2007. A dendrite-autonomous mechanism for direction selectivity in retinal starburst amacrine cells. PLoS Biol. 5:e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hofmann, W., D. Schubert, J. LaBonte, L. Munson, S. Gibson, J. Scammell, P. Ferrigno, and J. Sodroski. 1999. Species-specific, postentry barriers to primate immunodeficiency virus infection. J. Virol. 73:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikeda, T., T. Ohsugi, T. Kimura, S. Matsushita, Y. Maeda, S. Harada, and A. Koito. 2008. The antiretroviral potency of APOBEC1 deaminase from small animal species. Nucleic Acids Res. 36:6859-6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson, P. R., B. C. Schnepp, J. Zhang, M. J. Connell, S. M. Greene, E. Yuste, R. C. Desrosiers, and K. R. Clark. 2009. Vector-mediated gene transfer engenders long-lived neutralizing activity and protection against SIV infection in monkeys. Nat. Med. 15:901-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katahira, J., T. Ishizaki, H. Sakai, A. Adachi, K. Yamamoto, and H. Shida. 1995. Effects of translation initiation factor eIF-5A on the functioning of human T cell leukemia virus type I Rex and human immunodeficiency virus Rev inhibited trans dominantly by a Rex mutant deficient in RNA binding. J. Virol. 69:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keppler, O. T., I. Allespach, L. Schuller, D. Fenard, W. C. Greene, and O. T. Fackler. 2005. Rodent cells support key functions of the human immunodeficiency virus type 1 pathogenicity factor Nef. J. Virol. 79:1655-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keppler, O. T., N. Tibroni, S. Venzke, S. Rauch, and O. T. Fackler. 2006. Modulation of specific surface receptors and activation sensitization in primary resting CD4+ T lymphocytes by the Nef protein of HIV-1. J. Leukoc. Biol. 79:616-627. [DOI] [PubMed] [Google Scholar]
- 42.Keppler, O. T., F. J. Welte, T. A. Ngo, P. S. Chin, K. S. Patton, C. L. Tsou, N. W. Abbey, M. E. Sharkey, R. M. Grant, Y. You, J. D. Scarborough, W. Ellmeier, D. R. Littman, M. Stevenson, I. F. Charo, B. G. Herndier, R. F. Speck, and M. A. Goldsmith. 2002. Progress toward a human CD4/CCR5 transgenic rat model for de novo infection by human immunodeficiency virus type 1. J. Exp. Med. 195:719-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keppler, O. T., W. Yonemoto, F. J. Welte, K. S. Patton, D. Iacovides, R. E. Atchison, T. Ngo, D. L. Hirschberg, R. F. Speck, and M. A. Goldsmith. 2001. Susceptibility of rat-derived cells to replication by human immunodeficiency virus type 1. J. Virol. 75:8063-8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kino, K., and H. Sugiyama. 2001. Possible cause of G-C→C-G transversion mutation by guanine oxidation product, imidazolone. Chem. Biol. 8:369-378. [DOI] [PubMed] [Google Scholar]
- 45.Koito, A., Y. Kameyama, C. Cheng-Mayer, and S. Matsushita. 2003. Susceptibility of mink (Mustera vision)-derived cells to replication by human immunodeficiency virus type 1. J. Virol. 77:5109-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Koito, A., H. Shigekane, and S. Matsushita. 2003. Ability of small animal cells to support the postintegration phase of human immunodeficiency virus type-1 replication. Virology 305:181-191. [DOI] [PubMed] [Google Scholar]
- 47.Kramer-Hammerle, S., F. Ceccherini-Silberstein, C. Bickel, H. Wolff, M. Vincendeau, T. Werner, V. Erfle, and R. Brack-Werner. 2005. Identification of a novel Rev-interacting cellular protein. BMC Cell Biol. 6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwak, Y. T., D. Ivanov, J. Guo, E. Nee, and R. B. Gaynor. 1999. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J. Mol. Biol. 288:57-69. [DOI] [PubMed] [Google Scholar]
- 49.LaBonte, J. A., G. J. Babcock, T. Patel, and J. Sodroski. 2002. Blockade of HIV-1 infection of New World monkey cells occurs primarily at the stage of virus entry. J. Exp. Med. 196:431-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mariani, R., G. Rutter, M. E. Harris, T. J. Hope, H. G. Krausslich, and N. R. Landau. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 74:3859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Michel, N., C. Goffinet, K. Ganter, I. Allespach, V. N. Kewalramani, M. Saifuddin, D. R. Littman, W. C. Greene, M. A. Goldsmith, and O. T. Keppler. 2009. Human cyclin T1 expression ameliorates a T cell-specific transcriptional limitation for HIV in transgenic rats, but is not sufficient for a spreading infection of prototypic R5 HIV-1 strains ex vivo. Retrovirology 6:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muller, B., J. Daecke, O. T. Fackler, M. T. Dittmar, H. Zentgraf, and H. G. Krausslich. 2004. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J. Virol. 78:10803-10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Munch, J., E. Rucker, L. Standker, K. Adermann, C. Goffinet, M. Schindler, S. Wildum, R. Chinnadurai, D. Rajan, A. Specht, G. Gimenez-Gallego, P. C. Sanchez, D. M. Fowler, A. Koulov, J. W. Kelly, W. Mothes, J. C. Grivel, L. Margolis, O. T. Keppler, W. G. Forssmann, and F. Kirchhoff. 2007. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell 131:1059-1071. [DOI] [PubMed] [Google Scholar]
- 54.Munk, C., J. Zielonka, H. Constabel, B. P. Kloke, B. Rengstl, M. Battenberg, F. Bonci, M. Pistello, M. Lochelt, and K. Cichutek. 2007. Multiple restrictions of human immunodeficiency virus type 1 in feline cells. J. Virol. 81:7048-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Neagu, M. R., P. Ziegler, T. Pertel, C. Strambio-De-Castillia, C. Grutter, G. Martinetti, L. Mazzucchelli, M. Grutter, M. G. Manz, and J. Luban. 2009. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J. Clin. Invest. 119:3035-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.North, T. W., K. K. Van Rompay, J. Higgins, T. B. Matthews, D. A. Wadford, N. C. Pedersen, and R. F. Schinazi. 2005. Suppression of virus load by highly active antiretroviral therapy in rhesus macaques infected with a recombinant simian immunodeficiency virus containing reverse transcriptase from human immunodeficiency virus type 1. J. Virol. 79:7349-7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Owens, C. M., B. Song, M. J. Perron, P. C. Yang, M. Stremlau, and J. Sodroski. 2004. Binding and susceptibility to postentry restriction factors in monkey cells are specified by distinct regions of the human immunodeficiency virus type 1 capsid. J. Virol. 78:5423-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perron, M. J., M. Stremlau, M. Lee, H. Javanbakht, B. Song, and J. Sodroski. 2007. The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J. Virol. 81:2138-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pettersen, J. A., G. Jones, C. Worthington, H. B. Krentz, O. T. Keppler, A. Hoke, M. J. Gill, and C. Power. 2006. Sensory neuropathy in human immunodeficiency virus/acquired immunodeficiency syndrome patients: protease inhibitor-mediated neurotoxicity. Ann. Neurol. 59:816-824. [DOI] [PubMed] [Google Scholar]
- 60.Reuter, S., P. Kaumanns, S. B. Buschhorn, and M. T. Dittmar. 2005. Role of HIV-2 envelope in Lv2-mediated restriction. Virology 332:347-358. [DOI] [PubMed] [Google Scholar]
- 61.Rohr, O., C. Marban, D. Aunis, and E. Schaeffer. 2003. Regulation of HIV-1 gene transcription: from lymphocytes to microglial cells. J. Leukoc. Biol. 74:736-749. [DOI] [PubMed] [Google Scholar]
- 62.Schaller, T., S. Hue, and G. J. Towers. 2007. An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J. Virol. 81:11713-11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Speck, R. F., M. L. Penn, J. Wimmer, U. Esser, B. F. Hague, T. J. Kindt, R. E. Atchison, and M. A. Goldsmith. 1998. Rabbit cells expressing human CD4 and human CCR5 are highly permissive for human immunodeficiency virus type 1 infection. J. Virol. 72:5728-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stremlau, M., M. Perron, M. Lee, Y. Li, B. Song, H. Javanbakht, F. Diaz-Griffero, D. J. Anderson, W. I. Sundquist, and J. Sodroski. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc. Natl. Acad. Sci. U. S. A. 103:5514-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun, Z., P. W. Denton, J. D. Estes, F. A. Othieno, B. L. Wei, A. K. Wege, M. W. Melkus, A. Padgett-Thomas, M. Zupancic, A. T. Haase, and J. V. Garcia. 2007. Intrarectal transmission, systemic infection, and CD4+ T cell depletion in humanized mice infected with HIV-1. J. Exp. Med. 204:705-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tervo, H. M., I. Allespach, and O. T. Keppler. 2008. High-level transfection of primary rabbit T lymphocytes. J. Immunol. Methods 336:85-89. [DOI] [PubMed] [Google Scholar]
- 67.Tervo, H. M., C. Goffinet, and O. T. Keppler. 2008. Mouse T-cells restrict replication of human immunodeficiency virus at the level of integration. Retrovirology 5:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Towers, G. J. 2007. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology 4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trono, D., and D. Baltimore. 1990. A human cell factor is essential for HIV-1 Rev action. EMBO J. 9:4155-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Veazey, R. S., P. J. Klasse, S. M. Schader, Q. Hu, T. J. Ketas, M. Lu, P. A. Marx, J. Dufour, R. J. Colonno, R. J. Shattock, M. S. Springer, and J. P. Moore. 2005. Protection of macaques from vaginal SHIV challenge by vaginally delivered inhibitors of virus-cell fusion. Nature 438:99-102. [DOI] [PubMed] [Google Scholar]
- 71.Venzke, S., and O. T. Keppler. 2006. Role of macrophages in HIV infection and persistence. Expert Rev. Clin. Immunol. 2:613-626. [DOI] [PubMed] [Google Scholar]
- 72.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 73.Williams, R., S. Bokhari, P. Silverstein, D. Pinson, A. Kumar, and S. Buch. 2008. Nonhuman primate models of NeuroAIDS. J. Neurovirol. 14:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winslow, B. J., and D. Trono. 1993. The blocks to human immunodeficiency virus type 1 Tat and Rev functions in mouse cell lines are independent. J. Virol. 67:2349-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wolff, H., R. Brack-Werner, M. Neumann, T. Werner, and R. Schneider. 2003. Integrated functional and bioinformatics approach for the identification and experimental verification of RNA signals: application to HIV-1 INS. Nucleic Acids Res. 31:2839-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, J. X., G. E. Diehl, and D. R. Littman. 2008. Relief of preintegration inhibition and characterization of additional blocks for HIV replication in primary mouse T cells. PLoS One 3:e2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zheng, Y. H., H. F. Yu, and B. M. Peterlin. 2003. Human p32 protein relieves a post-transcriptional block to HIV replication in murine cells. Nat. Cell Biol. 5:611-618. [DOI] [PubMed] [Google Scholar]