Abstract
TRIM5α proteins recruit and restrict incoming cytoplasmic retroviruses. Primate TRIM5α sequence diversity underlies species-specific restriction and is likely caused by selective pressure from ancient pathogenic infections. Here we show that TRIM5α from the European brown hare restricts diverse retroviruses. Furthermore, it differs significantly in sequence from TRIM5α from the closely related rabbit, suggesting evolutionary changes in the last 12 million years since these species diverged. We propose that, like primates, lagomorphs have been subject to selective pressure from TRIM5-sensitive viruses, possibly related to the endogenous lentivirus RELIK found in both rabbits and hares.
TRIM5α proteins are cytoplasmic restriction factors (38) that are upregulated in response to viral infection (30, 32), bind to retroviral capsid protein (CA) (16, 25, 49), and inhibit retroviral infectivity in a species-specific way (31, 42, 48). TRIM5α belongs to the tripartite motif family of proteins, so named because of their RING-B-box-coiled-coil structure (RBCC), consisting of an N-terminal RING domain, often with E3 ubiquitin ligase activity (23), one or two B-box domains, and a coiled-coil (CC) domain.
The TRIM5α isoform contains a C-terminal PRYSPRY domain, a β-sheet fold (46) whose nonstructured interconnecting loops, the “variable loops” v1, v2, v3, and v4, form the surfaces that make contact with retroviral CA (11, 25, 37). Residues in these loops contribute directly to primate TRIM5α species specificity (12, 16, 22) and have undergone strong positive selection during primate evolution (21, 33, 36). The sequence identity in the RBCC of primate TRIM5α orthologues between Old World monkeys (OWM), New World monkeys (NWM) and hominoids is high, but the sequence identity in the PRYSPRY is low, particularly in the variable loops (33, 37, 39). It is thought that this is due to differential exposures to TRIM5α-sensitive pathogens since speciation (13, 21, 33, 34). The identity of the pathogens applying selection pressure on TRIM5α is unclear, but the primate phylogeny suggests selection over long periods of time that predate lentiviruses (33). However, the difficulty in ageing retroviruses, and particularly lentiviruses, which do not appear to endogenize readily, makes it difficult to gauge whether they are old enough to have provided selection pressure on TRIM5α. Recently, endogenous lentiviruses have been identified in rabbits, hares, and lemurs (5, 6, 14, 15). RELIK, the lentivirus found in rabbits and hares, is at least 12 million years old (15). We previously identified a TRIM5α orthologue in rabbits (35), and in order to consider whether there has been adaptive change in Lagomorpha TRIM5α over the last 12 million years, we here identify and characterize an active TRIM5α from hares. We show that there are striking differences in the PRYSPRY sequences of rabbit and hare TRIM5αs, suggesting adaptive change since these species split 12 million years ago.
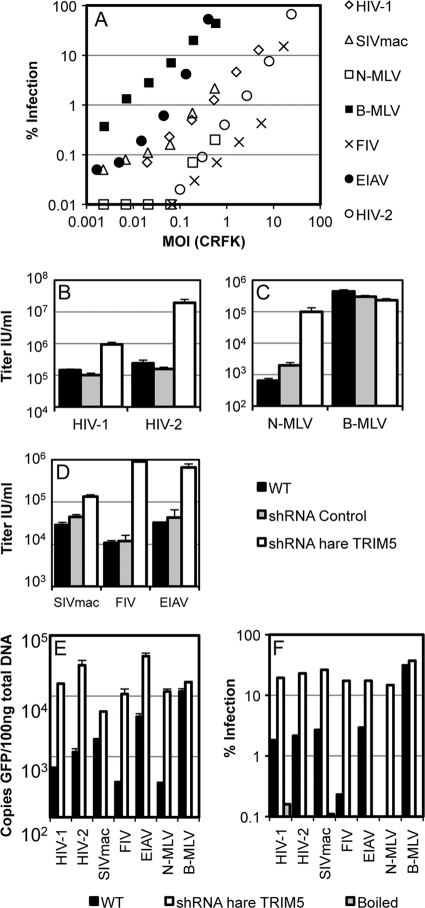
We first sought evidence for TRIM5α activity in the European brown hare, Lepus europaeus. We prepared vesicular stomatitis virus G protein (VSV-G)-pseudotyped, green fluorescent protein (GFP)-encoding retroviral vectors derived from human immunodeficiency virus type 1 (HIV-1) (2, 51), HIV-2 (7), simian immunodeficiency virus SIVmac (20), feline immunodeficiency virus (FIV) (28), equine infectious anemia virus (EIAV) (10), and N- and B-tropic murine leukemia virus (N-MLV and B-MLV) (4) as described previously (9) and titrated them on kidney fibroblasts from hares (Fig. 1A). In parallel, viruses were titrated on permissive feline kidney CrFK cells and the dose of virus plotted as the multiplicity of infection on CrFK cells. As such, B-MLV was equally infectious on CrFK and hare cells and, thus, infected 10% of the hare cells at a CrFK multiplicity of infection of 0.1. Most viruses were of significantly lower titer on hare cells, suggesting the existence of virus-specific blocks, as seen in rabbit cells (35). Of the seven retroviruses tested, HIV-1, HIV-2, SIVmac, N-MLV, EIAV, and FIV had reduced titers in hare cells. As is typical for restriction by TRIM5α, there was a difference of 3 orders of magnitude in titer between the most restricted (N-MLV) and unrestricted (B-MLV) (9, 27, 41). To examine whether a hare TRIM5α was responsible, we tested short hairpin RNAs (shRNAs) active against rabbit TRIM5α for their ability to restore the infectivity of a restricted virus (35). One shRNA, when expressed stably in drug-selected populations of hare cells, specifically restored the infectious titers of poorly infectious viruses (Fig. 1B to D). For example, N-MLV became as infectious as B-MLV. The expression of control shRNA had no impact on the infectivity of any virus.
FIG. 1.
Hare cells demonstrate TRIM5-like restriction. (A) GFP-encoding retroviral vectors were titrated on hare kidney cells. Viral doses were plotted as the multiplicity of infection (MOI) on CrFK cells. Data are representative of two independent experiments. (B to D) Transduction with TRIM5-specific shRNA rescues restricted infectivity of viruses as labeled. Titers are plotted as infectious units (IU)/ml. Data are representative of three independent experiments; error bars show standard errors of the means. (E) Hare cells were infected with retroviral vectors as shown, in triplicate. Two samples were extracted for DNA, and qPCR was performed for the GFP product of RT. (F) Infection in the third sample was determined by flow cytometry. Error bars show standard errors of the means for duplicate samples. Data are representative of two independent experiments.
TRIM5α in primates most often causes a block to reverse transcription (RT) (38). We infected unmodified hare cells or cells expressing TRIM5α shRNA with DNase-treated VSV-G-pseudotyped retroviruses and measured the early RT product by TaqMan quantitative PCR (qPCR) at 6 h postinfection as described previously (24) (Fig. 1E), as well as measuring infection 48 h later, as described above (Fig. 1F). Vector boiled at 95°C for 5 min served as a negative control for plasmid DNA contamination, and these samples gave values below the limit of reliable detection. This experiment showed that reduction of TRIM5α expression by shRNA rescued RT and the infectivity of restricted viruses. Together, these data suggest that hares express a TRIM5α orthologue that limits the infectivity of specific retroviruses before completion of RT.
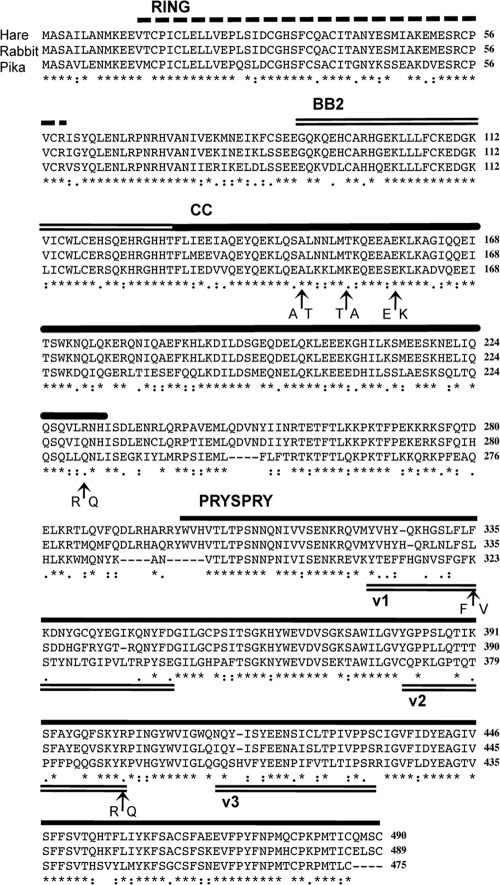
Next, we cloned a 1.5-kb putative TRIM5 cDNA from hare mRNA using forward primer TS40 (5′ ATC GGA ATT CCA CCA TGT ACC CAT ACG ACG TCC CAG ACT ACG CTG CTT CAG CAA TCT TAG CGA ATA TGA AGG AG 3′; containing an N-terminal HA tag, EcoRI site underlined), and reverse primer AF20 (5′ TAG CTT CGA ATC AAC AGC TCA TCT GGC AGA TTG TCA TGG 3′; BstBI site underlined) as described previously (50). Sequencing 20 hare TRIM5 PCR products revealed two alleles that differed at six positions (Fig. 2) (GenBank sequence accession numbers HM768824 and HM768825). The ratio of these alleles was 1:1, suggesting a single-copy heterozygous TRIM5 gene. Four polymorphisms were found in the coiled-coil domain, while two were found in PRYSPRY variable regions v1 and v2. No other TRIM sequences were amplified from either hare genomic DNA or cDNA using these primers. Sequence alignment with the rabbit TRIM5α protein showed the two to be highly conserved throughout, with 89% overall protein sequence identity (allele 1, 88.7%, and allele 2, 88.2%), and yet surprisingly divergent from rabbit in the PRYSPRY variable regions, with only 47% identity in v1 (both alleles, 14 out of 30 residues) (Fig. 2), a value comparable to that seen between human and rhesus macaque TRIM5α alleles (38). To extend the study from the Leporidae family to the Lagomorpha order as a whole, a pika TRIM5α sequence was reconstructed by searching the ongoing Ochotona princeps (American pika) whole-genome shotgun (WGS) project with the hare TRIM5α sequence using tBLASTn (1). Genome fragment cont2_132533, approximately 13 kb in length, was the most similar to the search sequence, allowing us to infer a pika TRIM5α sequence which was also aligned to the rabbit and hare TRIM5α sequences (Fig. 2). While the three lagomorph sequences show high similarity in the RBCC domains, there is almost no identity in the variable regions of the PRYSPRY, especially v1, where pika and hare TRIM5α share just 6 out of 30 residues (Fig. 2).
FIG. 2.
Alignment of TRIM5α protein sequences from hare, rabbit, and pika. RING, B-box type 2 (BB2), coiled-coil (CC), and PRYSPRY domains are shown. Hare allele one (GenBank sequence accession number HM768824) is shown; arrows indicate six polymorphisms from two hare TRIM5 alleles recovered from hare kidney fibroblasts. Overall identity between hare and rabbit proteins, 89%; identity in PRYSPRY v1, 47%. Inclusion of the pika sequence illustrates the variation in Lagomorpha TRIM5 PRYSPRYs that parallels the variation between primate TRIM5 sequences. Asterisk, identical residue; colon, conserved substitution; period, semiconserved substitution; gap, no conservation.
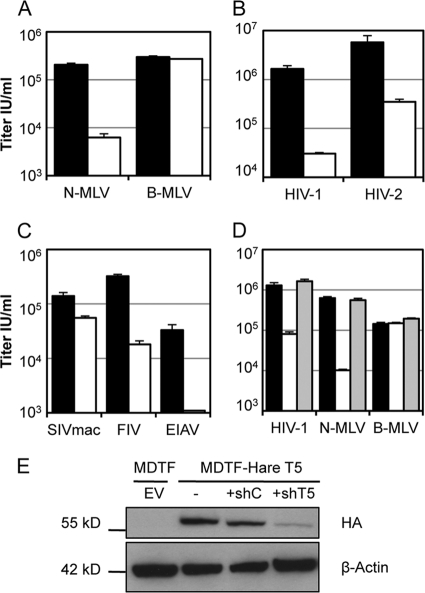
We cloned the hare TRIM5α cDNA (GenBank sequence accession number HM768824) into the MLV vector pCNCR as described previously (3) and transduced murine Mus dunni tail fibroblast (MDTF) cells. A clonal transduced cell line exhibited a restriction profile comparable in potency and specificity to that of hare cells (compare Fig. 3 to Fig. 1). This suggests that the allele cloned and expressed is responsible for the restriction profile of the hare cells. Importantly, transduction of the TRIM5-expressing MDTF cells with vector encoding TRIM5α-specific shRNA (35) rescued low-titer infection of HIV-1 and N-MLV, confirming that hare TRIM5α was responsible for restricted infection in these cells (Fig. 3D).
FIG. 3.
Expression of hare TRIM5α in permissive murine cells confers restriction properties of hare cells. (A to C) The retroviruses shown were titrated on MDTF cells expressing empty vector (black bars) or hare TRIM5 allele 1 (GenBank sequence accession number HM768824) (white bars), and infectious titers (IU/ml) were determined by flow cytometry. (D) Introduction of TRIM5-specific shRNA to MDTF-hare TRIM5 (shaded bars) rescued restricted infection by HIV-1 and N-MLV but not unrestricted infection by B-MLV. Error bars show standard errors of the means from two independent experiments. (E) Western blot detecting the hemagglutinin (HA) tag in MDTF cells expressing empty vector (EV) or HA-TRIM5α and hare control shRNA (shC) or TRIM5-specific shRNA (shT5). β-Actin was detected as a loading control.
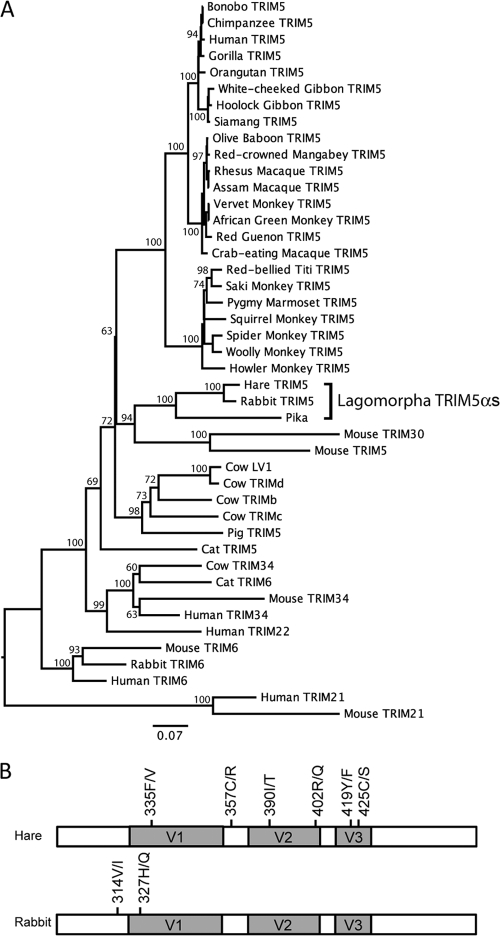
In order to confirm that the TRIM5α sequences we had identified were indeed TRIM5α orthologues, we aligned the nucleotide sequences with TRIM5α sequences from a variety of primates and nonprimates to construct a maximum-likelihood phylogenetic tree using PAUP* (40) (Fig. 4A). Percent bootstrap values (from 1,000 replicates) are shown on the branches. The two hare alleles and the reconstructed pika sequence clustered with the rabbit TRIM5α sequence, with pika TRIM5α basal to the leporid sequences, correlating with the order's taxonomy (18). Importantly the Lagomorpha TRIM5α genes do not cluster with the closely related TRIM5α paralogues TRIM6, TRIM22, and TRIM34 from humans, dogs, and cattle (34), confirming them as true TRIM5α orthologues (Fig. 4A).
FIG. 4.
A maximum-likelihood phylogenetic tree of full-length mammalian TRIM nucleotide sequences. (A) Hare and pika sequences fall within the TRIM5 cluster of the mammalian TRIM family, indicating that they are genuine TRIM5 orthologues. Branch lengths represent nucleotide substitutions per site. Percent bootstrap values (from 1,000 replicates) are shown on the branches. (B) Polymorphisms within the PRYSPRY sequences of four hares and six rabbits. Residues are numbered as in Fig. 2.
Balancing selection and maintenance of multiple TRIM5 alleles in the OWM rhesus macaques and sooty mangabeys has been described (21), and evidence suggests that individual alleles confer distinct restriction phenotypes both in vitro (21, 45) and in vivo (17, 44). We therefore performed selection analyses on the three full-length lagomorph TRIM5α sequences using hyphy (29) as implemented on the datamonkey website (http://www.datamonkey.org/) as described previously (8). We identified 11 codon positions with a statistically significant excess of nonsynonymous versus synonymous nucleotide substitutions, suggestive of adaptation. A striking 64% (7/11) of these lie in the v1 (positions 329, 332, 336, and 342), v2 (positions 391 and 403), and v3 (position 415) regions of the PRYSPRY domain (Table 1). Further analyses are warranted to determine the potential antiretroviral role of these positively selected residues; analysis of restriction of reconstructed RELIK viruses is likely to be informative. We also sought TRIM5 sequence variation within the Lagomorpha order. We sequenced the TRIM5α PRYSPRY domain of four hares (Lepus europaeus) and six rabbits (Oryctolagus cuniculus). Similar to the situation in primates, we found amino acid polymorphisms, six in hares and two in rabbits, in the variable loops within each TRIM5α (Fig. 4B). None of the polymorphisms were shared between hares and rabbits, implying that they have arisen and been fixed since divergence 12 million years ago (19, 43). The maintenance of polymorphisms in leporids in the variable loops known to control antiviral specificity in primates suggests that selection pressures may have been acting on TRIM5 genes in leporids as they have in primates.
TABLE 1.
Positively selected codons in the Lagomorpha TRIM5 genes
| Codona | Normalized E[dN − dS]b | Pr(dN > dS)c | Bayes factord | Regione | Amino acid in: |
||
|---|---|---|---|---|---|---|---|
| Hare | Rabbit | Pika | |||||
| 196 | 2.12 | 0.98 | 150 | CC | G | E | M |
| 288 | 2.12 | 0.99 | 166 | V | M | N | |
| 329 | 2.12 | 0.98 | 156 | v1 | K | Q | G |
| 332 | 2.13 | 0.99 | 212 | v1 | S | N | S |
| 336 | 2.16 | 1.00 | 4,743 | v1 | F/V | L | K |
| 342 | 2.14 | 0.99 | 387 | v1 | C | F | T |
| 391 | 2.13 | 0.99 | 204 | v2 | I | T | Q |
| 403 | 2.10 | 0.98 | 100 | v2 | R/Q | R | K |
| 415 | 2.12 | 0.98 | 143 | v3 | N | I | G |
| 457 | 2.13 | 0.99 | 240 | T | K | V | |
| 480 | 2.14 | 0.99 | 383 | Q | H | T | |
Consensus codon positions are as in Fig. 2.
Normalized posterior mean of the dN − dS difference. Codon-specific rates of synonymous (dS) and nonsynonymous (dN) nucleotide substitutions were estimated by random effect likelihood methods under the MG94 × HKY85 model of evolution. E, posterior mean.
Bayesian posterior probability (Pr) for positive selection (dN > dS) at each codon position.
A Bayes factor of more than 50 at a given site was considered to be strong support for positive selection.
CC, coiled-coil domain; v1, v2, and v3, variable loops in the PRYSPRY domain.
In conclusion, we show that TRIM5α sequences display striking diversity in the PRYSPRY variable loops between closely related species in the Lagomorpha order, suggesting adaptation to restrict different pathogens since these species diverged. We suggest that ancient retroviruses like RELIK, which infected the Lagomorpha germ line after the divergence of the Ochotonidae and Leporidae (14, 15, 43), may have driven the speciation of the hare and rabbit TRIM5α orthologues. It appears that despite the significant sequence differences between rabbit and hare TRIM5α, the panel of retroviruses we tested are unable to distinguish between them, at least for the alleles tested. The only possible exception is SIVmac, which appears to be more strongly restricted by hare than by rabbit TRIM5α (35). Of course other viruses, particularly the apparently older gamma retroviruses, may have contributed to the selection pressure, but it strikes us that MLVs are generally insensitive to TRIM5α, with only a single clone of MLV (N-MLV) having any sensitivity to any of the wild-type TRIM5α proteins thus far identified (26, 47). We further speculate that a similar situation may exist in primates, in which lentiviral infections may have provided selective pressure on primate TRIM5/TRIMCyp genes, and that a lack of endogenous lentiviruses in primates may reflect poor endogenization efficiencies rather than recent introduction of lentiviruses into primates.
Acknowledgments
This work was funded by a Medical Research Council Ph.D. studentship (A.J.F.), Wellcome Trust Senior Biomedical Research fellowship WT076608 (G.J.T.), the Medical Research Council, and the National Institute for Health Research UCL/UCLH Comprehensive Biomedical Research Centre.
We are grateful to Didier Trono, François-Loïc Cosset, Andrew Lever, Kyriacos Mitrophanos, and Eric Poeschla for plasmids, to Aris Katzourakis for helpful discussions, and to Jean-François Vautherot for the hare cell line.
Footnotes
Published ahead of print on 22 September 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bainbridge, J. W., C. Stephens, K. Parsley, C. Demaison, A. Halfyard, A. J. Thrasher, and R. R. Ali. 2001. In vivo gene transfer to the mouse eye using an HIV-based lentiviral vector; efficient long-term transduction of corneal endothelium and retinal pigment epithelium. Gene Ther. 8:1665-1668. [DOI] [PubMed] [Google Scholar]
- 3.Besnier, C., Y. Takeuchi, and G. Towers. 2002. Restriction of lentivirus in monkeys. Proc. Natl. Acad. Sci. U. S. A. 99:11920-11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock, M., K. N. Bishop, G. Towers, and J. P. Stoye. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74:7422-7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gifford, R. J., A. Katzourakis, M. Tristem, O. G. Pybus, M. Winters, and R. W. Shafer. 2008. A transitional endogenous lentivirus from the genome of a basal primate and implications for lentivirus evolution. Proc. Natl. Acad. Sci. U. S. A. 105:20362-20367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert, C., D. G. Maxfield, S. M. Goodman, and C. Feschotte. 2009. Parallel germline infiltration of a lentivirus in two Malagasy lemurs. PLoS Genet. 5:e1000425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffin, S. D., J. F. Allen, and A. M. Lever. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 75:12058-12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta, R. K., S. Hue, T. Schaller, E. Verschoor, D. Pillay, and G. J. Towers. 2009. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 5:e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatziioannou, T., S. Cowan, S. P. Goff, P. D. Bieniasz, and G. J. Towers. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J. 22:385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda, Y., M. K. Collins, P. A. Radcliffe, K. A. Mitrophanous, and Y. Takeuchi. 2002. Gene transduction efficiency in cells of different species by HIV and EIAV vectors. Gene Ther. 9:932-938. [DOI] [PubMed] [Google Scholar]
- 11.James, L. C., A. H. Keeble, Z. Khan, D. A. Rhodes, and J. Trowsdale. 2007. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proc. Natl. Acad. Sci. U. S. A. 104:6200-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Javanbakht, H., F. Diaz-Griffero, M. Stremlau, Z. Si, and J. Sodroski. 2005. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. J. Biol. Chem. 280:26933-26940. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, W. E., and S. L. Sawyer. 2009. Molecular evolution of the antiretroviral TRIM5 gene. Immunogenetics 61:163-176. [DOI] [PubMed] [Google Scholar]
- 14.Katzourakis, A., M. Tristem, O. G. Pybus, and R. J. Gifford. 2007. Discovery and analysis of the first endogenous lentivirus. Proc. Natl. Acad. Sci. U. S. A. 104:6261-6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keckesova, Z., L. M. Ylinen, G. J. Towers, R. J. Gifford, and A. Katzourakis. 2009. Identification of a RELIK orthologue in the European hare (Lepus europaeus) reveals a minimum age of 12 million years for the lagomorph lentiviruses. Virology 384:7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, Y., X. Li, M. Stremlau, M. Lee, and J. Sodroski. 2006. Removal of arginine 332 allows human TRIM5alpha to bind human immunodeficiency virus capsids and to restrict infection. J. Virol. 80:6738-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim, S. Y., T. Rogers, T. Chan, J. B. Whitney, J. Kim, J. Sodroski, and N. L. Letvin. 2010. TRIM5alpha modulates immunodeficiency virus control in rhesus monkeys. PLoS Pathog. 6:e1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matthee, C. A. 2009. Pikas, hares and rabbits (Lagomorpha), p. 487-489. In S. Blair Hedges and Sudhir Kumar (ed.), The timetree of life. Oxford University Press, New York, NY.
- 19.Matthee, C. A., B. J. van Vuuren, D. Bell, and T. J. Robinson. 2004. A molecular supermatrix of the rabbits and hares (Leporidae) allows for the identification of five intercontinental exchanges during the Miocene. Syst. Biol. 53:433-447. [DOI] [PubMed] [Google Scholar]
- 20.Negre, D., P. E. Mangeot, G. Duisit, S. Blanchard, P. O. Vidalain, P. Leissner, A. J. Winter, C. Rabourdin-Combe, M. Mehtali, P. Moullier, J. L. Darlix, and F. L. Cosset. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther. 7:1613-1623. [DOI] [PubMed] [Google Scholar]
- 21.Newman, R. M., L. Hall, M. Connole, G. L. Chen, S. Sato, E. Yuste, W. Diehl, E. Hunter, A. Kaur, G. M. Miller, and W. E. Johnson. 2006. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc. Natl. Acad. Sci. U. S. A. 103:19134-19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohkura, S., M. W. Yap, T. Sheldon, and J. P. Stoye. 2006. All three variable regions of the TRIM5alpha B30.2 domain can contribute to the specificity of retrovirus restriction. J. Virol. 80:8554-8565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozato, K., D. M. Shin, T. H. Chang, and H. C. Morse III. 2008. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8:849-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Passerini, L. D., Z. Keckesova, and G. J. Towers. 2006. Retroviral restriction factors Fv1 and TRIM5{alpha} act independently and can compete for incoming virus before reverse transcription. J. Virol. 80:2100-2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perez-Caballero, D., T. Hatziioannou, A. Yang, S. Cowan, and P. D. Bieniasz. 2005. Human tripartite motif 5alpha domains responsible for retrovirus restriction activity and specificity. J. Virol. 79:8969-8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez-Caballero, D., S. J. Soll, and P. D. Bieniasz. 2008. Evidence for restriction of ancient primate gammaretroviruses by APOBEC3 but not TRIM5alpha proteins. PLoS Pathog. 4:e1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perron, M. J., M. Stremlau, B. Song, W. Ulm, R. C. Mulligan, and J. Sodroski. 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proc. Natl. Acad. Sci. U. S. A. 101:11827-11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poeschla, E. M., F. Wong-Staal, and D. J. Looney. 1998. Efficient transduction of nondividing human cells by feline immunodeficiency virus lentiviral vectors. Nat. Med. 4:354-357. [DOI] [PubMed] [Google Scholar]
- 29.Pond, S. L., and S. D. Frost. 2005. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531-2533. [DOI] [PubMed] [Google Scholar]
- 30.Rajsbaum, R., J. P. Stoye, and A. O'Garra. 2008. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur. J. Immunol. 38:619-630. [DOI] [PubMed] [Google Scholar]
- 31.Rold, C. J., and C. Aiken. 2008. Proteasomal degradation of TRIM5alpha during retrovirus restriction. PLoS Pathog. 4:e1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadler, A. J., and B. R. Williams. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8:559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawyer, S. L., L. I. Wu, M. Emerman, and H. S. Malik. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc. Natl. Acad. Sci. U. S. A. 102:2832-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawyer, S. L., M. Emerman, and H. S. Malik. 2007. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 3:e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaller, T., S. Hue, and G. J. Towers. 2007. An active TRIM5 protein in rabbits indicates a common antiviral ancestor for mammalian TRIM5 proteins. J. Virol. 81:11713-11721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soares, E. A., A. N. Menezes, C. G. Schrago, M. A. Moreira, C. R. Bonvicino, M. A. Soares, and H. N. Seuanez. 2010. Evolution of TRIM5alpha B30.2 (SPRY) domain in New World primates. Infect. Genet. Evol. 10:246-253. [DOI] [PubMed] [Google Scholar]
- 37.Song, B., B. Gold, C. O'Huigin, H. Javanbakht, X. Li, M. Stremlau, C. Winkler, M. Dean, and J. Sodroski. 2005. The B30.2(SPRY) domain of the retroviral restriction factor TRIM5α exhibits lineage-specific length and sequence variation in primates. J. Virol. 79:6111-6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848-853. [DOI] [PubMed] [Google Scholar]
- 39.Stremlau, M., M. Perron, S. Welikala, and J. Sodroski. 2005. Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79:3139-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (* and other methods), 4 ed. Sinauer Associates, Sunderland, MA.
- 41.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. U. S. A. 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Towers, G. J. 2007. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology 4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Loo, W., J. Abrantes, and P. J. Esteves. 2009. Sharing of endogenous lentiviral gene fragments among leporid lineages separated for more than 12 million years. J. Virol. 83:2386-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Manen, D., M. A. Rits, C. Beugeling, K. van Dort, H. Schuitemaker, and N. A. Kootstra. 2008. The effect of Trim5 polymorphisms on the clinical course of HIV-1 infection. PLoS Pathog. 4:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson, S. J., B. L. Webb, C. Maplanka, R. M. Newman, E. J. Verschoor, J. L. Heeney, and G. J. Towers. 2008. Rhesus macaque TRIM5 alleles have divergent antiretroviral specificities. J. Virol. 82:7243-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo, J. S., J. H. Imm, C. K. Min, K. J. Kim, S. S. Cha, and B. H. Oh. 2006. Structural and functional insights into the B30.2/SPRY domain. EMBO J. 25:1353-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wood, A., B. L. Webb, B. Bartosch, T. Schaller, Y. Takeuchi, and G. J. Towers. 2009. Porcine endogenous retroviruses PERV A and A/C recombinant are insensitive to a range of divergent mammalian TRIM5alpha proteins including human TRIM5alpha. J. Gen. Virol. 90:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, X., J. L. Anderson, E. M. Campbell, A. M. Joseph, and T. J. Hope. 2006. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc. Natl. Acad. Sci. U. S. A. 103:7465-7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yap, M. W., S. Nisole, and J. P. Stoye. 2005. A single amino acid change in the SPRY domain of human Trim5alpha leads to HIV-1 restriction. Curr. Biol. 15:73-78. [DOI] [PubMed] [Google Scholar]
- 50.Ylinen, L. M., Z. Keckesova, B. L. Webb, R. J. Gifford, T. P. Smith, and G. J. Towers. 2006. Isolation of an active Lv1 gene from cattle indicates that tripartite motif protein-mediated innate immunity to retroviral infection is widespread among mammals. J. Virol. 80:7332-7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zufferey, R., D. Nagy, R. J. Mandel, L. Naldini, and D. Trono. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol. 15:871-875. [DOI] [PubMed] [Google Scholar]