Abstract
Although the herpes simplex virus type 1 (HSV-1) genome might be expected to induce a DNA damage response, the ATR kinase is not activated in infected cells. We previously proposed that spatial uncoupling of ATR from its interaction partner, ATRIP, could be the basis for inactivation of the ATR kinase in infected cells; however, we now show that ATR and ATRIP are in fact both recruited to HSV-1 replication compartments and can be coimmunoprecipitated from infected-cell lysates. ATRIP and replication protein A (RPA) are recruited to the earliest detectable prereplicative sites, stage II microfoci. In a normal cellular DNA damage response, ATR/ATRIP are recruited to stretches of RPA-coated single-stranded DNA in an RPA- and kinase-dependent manner, resulting in the phosphorylation of RPA by ATR in damage foci. In contrast, in HSV-1-infected cells, RPA is not phosphorylated, and endogenous phosphorylated RPA is excluded from stage II microfoci; in addition, the recruitment of ATR/ATRIP is independent of RPA and the kinase activity of ATR. Furthermore, we show that ATR/ATRIP play a beneficial role in viral gene expression and virus production. Although ICP0 has been shown to be important for partial inactivation of other cellular DNA repair pathways, we show that ICP0 is not responsible for the inactivation of ATR signaling and, furthermore, that neither ATR nor ATRIP is a target of ICP0 degradation. Thus, ATR and ATRIP may function outside the context of the canonical ATR damage signaling pathway during HSV-1 infection to participate in the viral life cycle.
Herpes simplex virus type 1 (HSV-1) is a large linear double-stranded DNA virus that replicates in the nucleus of the host cell. The incoming viral genome contains nicks and gaps (42), and cellular DNA repair machinery might be expected to recognize it as damaged, resulting in the activation of one or more cellular DNA damage pathways. Activation of DNA damage response pathways can result not only in repair of the damaged DNA but also in cell cycle arrest, gene silencing, and apoptosis (9). The later outcomes could result in suppression of viral gene expression and DNA replication and thus have negative consequences for lytic infection. Activation of a cellular DNA damage response during viral infection could, therefore, represent a form of intrinsic antiviral immunity (14, 15). On the other hand, HSV-1 and other DNA viruses which replicate in the nucleus have also been shown to utilize cellular DNA repair machinery to promote productive infection (28). Thus, HSV-1 has apparently evolved to manipulate the host DNA damage response by utilizing some components and inactivating others in an attempt to create an environment conducive to lytic viral infection.
The cellular DNA damage response is regulated by the three phosphoinositide 3-kinase-related kinases (PIKKs), DNA-PK (DNA-dependent protein kinase), ATM (ataxia-telangiectasia-mutated), and ATR (ATM and Rad3-related) (1, 9). DNA-PK and ATM respond predominantly to double-strand breaks, and ATR responds to stalled replication forks and long stretches of single-stranded DNA (ssDNA). DNA-PK is required for nonhomologous end joining (NHEJ), while ATM activation promotes homologous recombination. Interestingly, in some cell types, the catalytic subunit of DNA-PK (DNA-PKcs) is proteolytically degraded during infection by the immediate-early (IE) protein ICP0, a viral E3 ubiquitin ligase (25, 37), thereby resulting in the probable inactivation of the NHEJ pathway. ATM kinase activity, on the other hand, is activated during HSV-1 infection once viral DNA replication is initiated (26, 47, 56). Despite phosphorylation of several ATM targets, ATM signaling is also modulated by ICP0, which degrades the ubiquitin ligases RNF8 and RNF168. The function of these ubiquitin ligases is to promote the tethering of ATM pathway proteins at sites of cellular DNA damage (27). Thus, ICP0 functions to partially inactivate portions of both the DNA-PK- and ATM-mediated repair pathways.
During a cellular DNA damage response, ATM activation and processing of DNA ends generate ssDNA adjacent to double-stranded DNA (dsDNA), a structure that is known to activate ATR (9, 38). The ssDNA is coated by the cellular ssDNA binding protein, replication protein A (RPA), which then serves to recruit ATR through a direct interaction with ATR-interacting protein (ATRIP) (4, 12, 58). ATR signaling results in the phosphorylation of many substrates, including RPA and Chk1. During HSV-1 infection, the ATR substrates RPA and Chk1 are not phosphorylated (47, 54-56), indicating that ATR signaling may be disabled.
A hallmark of HSV-1 infection is the reorganization of the infected-cell nucleus, resulting in the formation of large globular replication compartments as well as the rearrangement of cellular proteins involved in several homeostatic pathways. In addition to cellular DNA repair proteins, HSV-1 infection also causes the reorganization of components of the cellular protein quality control pathways, resulting in the formation of virus-induced chaperone-enriched (VICE) domains, which act to maintain nuclear protein quality control during infection (31). Viral gene expression, DNA replication, and encapsidation of viral genomes occur in replication compartments (24, 39, 41). In this work we revisit the study of proteins recruited to and restricted from replication compartments in an attempt to better understand how HSV-1 manipulates components of the cellular DNA damage response for its own benefit.
MATERIALS AND METHODS
Cells and reagents.
Vero, HeLa, U2OS, HFF-1, 293T, and pEAK (293T derivative) cells were obtained from the American Type Culture Collection (ATCC). All cells except Vero cells were maintained in Dulbecco's modified Eagle medium with 10% fetal bovine serum; Vero cells were maintained in 5% fetal bovine serum. Vero-derived E11 cells were obtained from Neal Deluca (University of Pittsburgh School of Medicine, Pittsburgh, PA). These cells stably express ICP4 and ICP27 and were maintained in 400 μg/ml G418 (44). BAY 57-1293 [N-(5-(aminiosulfonyl)-4-methyl-1,3-thiazol-2-yl)-N-methyl-2-(4-(2-pyridinyl)phenyl)acetamide] was obtained from Gerald Kleymann (Bayer Pharmaceuticals, Wuppertal, Germany) (21) and used at a concentration of 100 μM as described previously (30).
Viruses.
The KOS strain was used as wild-type HSV-1. Viruses θβ and d106 are derived from KOS and were provided by Neal Deluca (University of Pittsburgh School of Medicine, Pittsburgh, PA). The ICP0-null virus, θβ, contains lacZ insertions in both copies of the ICP0 gene (45). The ICP0-expressing virus, d106, is deleted for all immediate-early genes except ICP0 and contains the green fluorescent protein (GFP) gene under the control of the human cytomegalovirus (HCMV) promoter inserted in the ICP27 gene (43). Viruses in1863 and dl1403/CMVlacZ are derived from strain 17+ and were obtained from Chris Preston (MRC Virology Unit, Glasgow, Scotland). These viruses contain the lacZ gene under the control of the HCMV promoter/enhancer inserted in the tk gene (18, 48). in1863 was used as the wild type in experiments with the ICP0-null virus, dl1403/CMVlacZ. KOS and in1863 were grown and titers were determined on Vero cells. θβ and dl1403/CMVlacZ were grown and titers were determined on U2OS cells. d106 was grown and titers were determined on E11 cells.
DNA constructs and transfection.
The Flag-ATR-wild type (pBJF-ATR-wt) and Flag-ATR-kinase-dead (pBJF-ATR-kd) constructs were obtained from Karlene Cimprich (Stanford University School of Medicine, Stanford, CA) (10, 11). Wild type ICP0 (pCI-110) was obtained from Roger Everett (MRC Virology Unit, Glasgow, Scotland) (17). pcDNA3 was purchased from Invitrogen, and pEGFP-N1 was purchased from Clontech. The following plasmids were previously described: wild-type Myc-ATRIP, wild-type Flag-ATRIP, Flag-ATRIP-1-107, and Flag-ATRIP-1-217 in the vector pUNI50 (12); wild-type hemagglutinin (HA)-ATRIP, HA-ATRIPΔ658-684, and HA-ATRIPΔ1-107 (4); and HA-ATRIPΔ112-225 (2). All HA-ATRIP plasmids are in pLPCX (Clontech) and contain wobble base mutations that make them resistant to shATRIP1, except for HA-ATRIPΔ1-107, which does not contain the target sequence for shATRIP1. Site-directed mutagenesis was used to introduce the checkpoint recruitment domain (CRD) point mutation D58K/D59K into wild-type HA-ATRIP to generate HA-ATRIP-crd. The forward CRD primer is 5′ CATGGGGACTTCACTGCCAAGAAGCTGGAGGAGCTTGACACC. Cells were transfected as indicated using Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's suggested protocol.
Immunoprecipitation.
Cells were lysed in 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.75% CHAPS, 1 mM NaF, 1 mM sodium vanadate, 1 mM dithiothreitol [DTT], and 1× protease inhibitor cocktail [Roche]) (2) and disrupted by passage through a 21-guage needle. Immunoprecipitation with the rATRIP Upstate antibody (2 μg) was performed in the presence of 30 μl of protein A-agarose overnight at 4°C. Immunoprecipitation with Myc antibody (2 μg) was performed in the presence of 30 μl of protein A/G-agarose for 3 h at 4°C.
IF analysis.
Immunofluorescence (IF) analysis was performed as described previously (30). Briefly, cells adhered to glass coverslips were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde, and permeabilized with 1% Triton X-100. Cells were blocked in 3% normal goat serum and reacted with antibodies as indicated. Primary antibodies included monoclonal mouse anti-ATRIP (mATRIP R&D) (1:400; R&D Systems), polyclonal rabbit anti-ATRIP (rATRIP Upstate) (1:200; Upstate), polyclonal rabbit anti-ATRIP 403 (rATRIP 403) (1:200) (12), monoclonal mouse anti-ICP8 (1:200; Abcam), polyclonal rabbit anti-ICP8 367 (1:400; provided by William Ruyechan, State University of New York, Buffalo, NY) (46), monoclonal rat anti-Hsc70 (1:200; Stressgen), polyclonal rabbit anti-Flag (1:200; Sigma), polyclonal rabbit anti-HA (1:200; Clontech), monoclonal mouse anti-RPA32 9H8 (1:200; GeneTex), and polyclonal rabbit anti-phospho-RPA32 S4/S8 (1:200; Bethyl Laboratories). TO-PRO-3 (1:1,000; Molecular Probes) was used as a nuclear counterstain. Alexa Fluor secondary antibodies (1:200; Molecular Probes) were used with fluorophores excitable at a wavelength of 488, 594, or 647. Images were captured using a Zeiss LSM 510 confocal NLO microscope equipped with argon and HeNe lasers and a Zeiss 63× objective lens (numerical aperture, 1.4). Images were processed and arranged using Adobe Photoshop CS3 and Illustrator CS3.
Western blot analysis.
Cells in 35-mm dishes were lysed in 2× SDS sample buffer (4% SDS, 20% glycerol, 100 mM Tris [pH 6.8], 100 mM DTT, 10% β-mercaptoethanol, 1× protease inhibitor cocktail [Roche], and 0.1% bromophenol blue) and boiled for 5 min. Proteins were resolved by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. Membranes were blocked for 1 h in 5% nonfat dry milk dissolved in Tris-buffered saline-Tween 20 (TBST). Primary antibodies were diluted in blocking solution and incubated overnight at 4°C. Primary antibodies used included monoclonal mouse anti-ATRIP (mATRIP R&D) (1:3,000; R&D Systems), polyclonal rabbit anti-ATRIP (rATRIP Upstate) (1:3,000; Upstate), polyclonal rabbit anti-ATRIP 403 (rATRIP 403) (1:3,000) (12), monoclonal mouse anti-ICP8 (1:1,000; Abcam), monoclonal mouse anti-ICP4 (1:1,000; Abcam), monoclonal mouse anti-ICP0 (1:1,000; East Coast Biotech), monoclonal mouse anti-β-actin (1:15,000; Sigma), monoclonal mouse anti-HA F7 (1:10,000; Santa Cruz), polyclonal goat anti-ATR N19 (1:1,000; Santa Cruz), monoclonal mouse anti-Chk1 (1:1,000; Santa Cruz), monoclonal mouse anti-Myc (1:5,000; Cell Signaling) polyclonal goat anti-PML N19 (1:1,000; Santa Cruz), monoclonal mouse anti-Ku70 Ab-4 (1:3,000; NeoMarkers), and rabbit polyclonal anti-phospho-Chk1 S345 (1:1,000; Cell Signaling). For phospho-specific antibodies, 2% bovine serum albumin (BSA) in TBST was used in place of milk.
Lentivirus generation and use.
The pLKO.1 system was used to package lentiviruses and deliver short hairpin RNA (shRNA) into target cells. The three-plasmid set containing the pLKO.1-TRC cloning vector (Addgene plasmid 10878) (35), psPAX2 (Addgene plasmid 12260), and pMD2.G (Addgene plasmid 12259) was purchased from Addgene, and the following shRNA target sequences were cloned into pLKO.1 according to the manufacturer's suggestions: shGFP, GCAAGCUGACCCUGAAGUUCA; shATRIP1, AAGGUCCACAGAUUAUUAGAU; shATRIP2, GGTCCACAGATTATTAGATTT; and shATR, AACCUCCGUGAUGUUGCUUGA. Lentivirus particles were produced by transient transfection of pEAK cells with pLKO.1, psPAX2, and pMD2.G at a ratio of 4:3:1. HFF-1 or Vero cells were transduced twice with the indicated lentiviruses 48 h apart and were used for experiments at 48 h after the second transduction.
Growth yield of KOS on shATRIP cells.
HFF-1 cells were transduced with lentiviruses expressing shGFP or shATRIP2 as described above. Cells were seeded into 35-mm dishes at a density of 4 × 105 cells per dish and infected with KOS at the indicated multiplicities of infection (MOIs). Virus was collected at 12 h postinfection, and titers were determined on Vero cells.
Probability of plaque formation.
Assays of the probability of plaque formation were performed as described by Everett et al. with the following modifications (18). HFF-1 cells were transduced with lentiviruses expressing shGFP or shATRIP2. Cells were seeded into 24-well dishes at a density of 1 × 105 cells per well and infected with 2-fold sequential dilutions of in1863 or dl1403/CMVlacZ. Cells were fixed at 24 h postinfection with 2% paraformaldehyde for 1 h and stained for β-galactosidase activity (5 mM potassium ferricyanide, 5 mM potassium ferricyanide, 2 mM MgCl2, and 1 mg/ml X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside]).
HU treatment of infected cells.
HeLa cells were infected as described in the figure legends. At the indicated times postinfection, cells were treated with 3 mM hydroxyurea (HU). Cells were harvested in 2× SDS sample buffer with phosphatase inhibitors at 2 h after the addition of HU.
RPA-ssDNA binding.
ATRIP binding to RPA-coated ssDNA (RPA-ssDNA) was done as previously described (4). Briefly, biotin-labeled ssDNA was bound to streptavidin beads and either left uncoated or coated with recombinant human RPA. RPA-ssDNA-streptavidin beads were then incubated with 293T cell lysates transiently expressing HA-ATRIP or HA-ATRIP-crd. The beads were washed, and proteins bound to the beads were eluted and separated by SDS-PAGE prior to Western blot analysis.
RESULTS
ATRIP colocalizes with ICP8 in HSV-1 replication compartments.
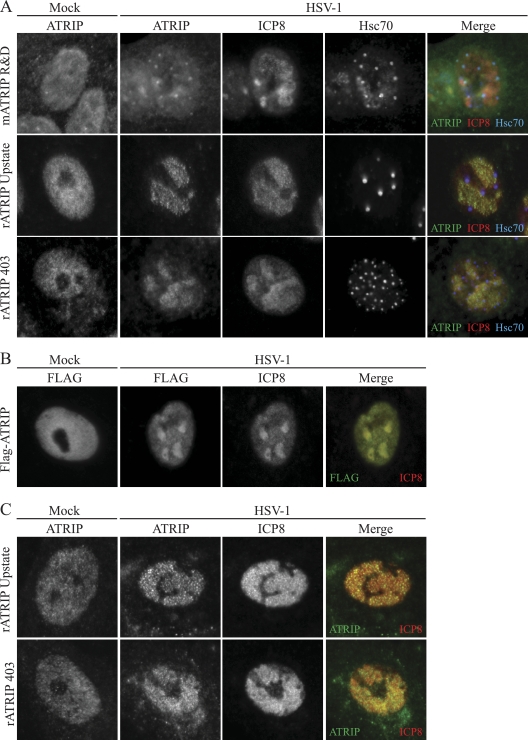
The obligate interacting partners ATR and ATRIP colocalize during a cellular DNA damage response (12). We have previously proposed that HSV-1 is able to disrupt the tight colocalization of ATR and ATRIP during infection by sequestering ATRIP away from ATR in virus-induced chaperone-enriched (VICE) domains (54). In that study a monoclonal antibody that was reported to react with ATRIP was used to probe the localization of ATRIP in HSV-1-infected cells. In this study we have confirmed that mATRIP R&D detects a protein that colocalizes with the VICE domain marker Hsc70 in infected cells (Fig. 1A). To confirm the localization of ATRIP in VICE domains, we used two rabbit polyclonal antibodies (rATRIP Upstate and rATRIP 403) and a Flag-tagged ATRIP expression construct. To our surprise, endogenous ATRIP detected with rATRIP Upstate and rATRIP 403 and overexpressed Flag-ATRIP colocalized with the viral single-stranded DNA binding protein ICP8 in replication compartments and not in VICE domains (Fig. 1A and B). It is possible that in Vero cells, mATRIP R&D detects a subpopulation of ATRIP distinct from that detected with the polyclonal antibodies and epitope tag. To test this, we next examined the staining pattern in limited-passage human diploid fibroblasts. HFF-1 cells were infected with HSV-1 and costained for ATRIP and ICP8. rATRIP Upstate and rATRIP 403 both detected endogenous ATRIP in replication compartments (Fig. 1C). Interestingly, mATRIP R&D did not detect ATRIP in HFF-1 cells. The two polyclonal ATRIP antibodies also detected ATRIP in replication compartments in HeLa cells (data not shown). Taken together, these data suggest that mATRIP R&D may not actually detect bona fide ATRIP.
FIG. 1.
ATRIP colocalizes with ICP8 in HSV-1 replication compartments. Vero cells were infected with HSV-1 at an MOI of 10 for 6 h. (A) Cells were fixed and stained with the indicated ATRIP antibody, ICP8 to indicate replication compartments, and rat Hsc70 to indicate VICE domains. Cells in the first row were stained with mATRIP R&D and rabbit ICP8 367. Cells in the second and third rows were stained with rATRIP Upstate or rATRIP 403 and monoclonal ICP8. (B) Vero cells were transfected with Flag-ATRIP and infected with HSV-1 at an MOI of 10 for 6 h. Cells were fixed and stained with rabbit Flag and monoclonal ICP8. (C) HFF-1 cells were infected with HSV-1 at an MOI of 10 for 6 h. Cells were fixed and stained with rATRIP Upstate or rATRIP 403 and monoclonal ICP8.
The mATRIP R&D antibody does not detect ATRIP.
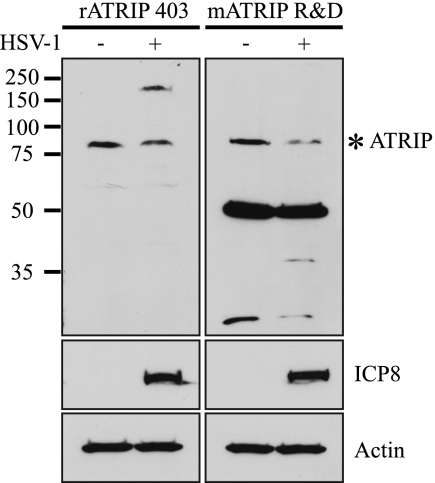
To directly address whether mATRIP R&D can detect ATRIP, mock- and HSV-1-infected Vero cell lysates were subjected to Western blot analysis using mATRIP R&D and the polyclonal antibody rATRIP 403 (Fig. 2). rATRIP 403 detected one band of approximately 80 kDa, the expected molecular mass of ATRIP, in uninfected Vero cell lysates. In HSV-1-infected cells we detected the 80-kDa band and another, much slower-migrating band. The slower-migrating species may represent a nonspecific interaction with a viral protein, as it is not present in uninfected cells and is not detected with an antibody directed against the tag in infected-cell lysates from cells expressing a tagged ATRIP (data not shown). mATRIP R&D detects an 80-kDa band and also detects a very prominent band at approximately 50 kDa in both mock- and HSV-1-infected lysates.
FIG. 2.
ATRIP antibodies recognize proteins of different sizes by Western blot analysis. Vero cells were mock infected or infected with HSV-1 at an MOI of 10 for 6 h. Cell lysates were prepared as described in Materials and Methods and separated by SDS-PAGE. Western blotting was performed with rATRIP 403 and mATRIP R&D. ICP8 served as an infection control, and actin served as a loading control.
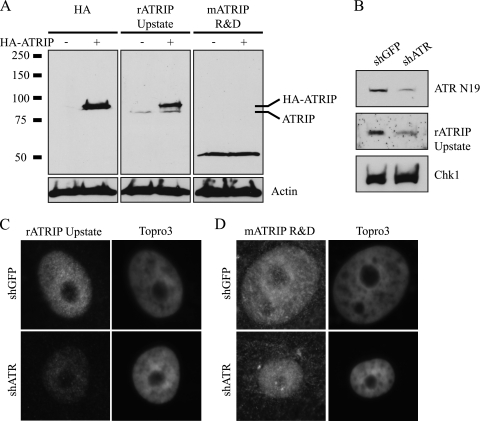
To confirm that the species detected by the polyclonal ATRIP antibodies correspond to bona fide ATRIP, Western blots of HeLa cell lysates transiently expressing HA-ATRIP were analyzed (Fig. 3A). HA antibody detected HA-ATRIP in the transfected samples but not in the mock-transfected sample. rATRIP Upstate detected an 80-kDa endogenous band in the mock-transfected cells and detected two bands, corresponding to endogenous and tagged HA-ATRIP, in the transfected sample. mATRIP R&D did not detect full-length endogenous ATRIP or tagged overexpressed ATRIP in HeLa cells (Fig. 3A, right panel). To confirm this result, we repeated the Western blot analysis with Vero cells depleted for ATRIP by infection with lentiviruses expressing short hairpin RNA targeting ATR (shATR). Since ATR and ATRIP are obligate interacting partners, depletion of one protein also reduces the protein levels of the other. As can be seen in Fig. 3B, shATR significantly reduced the levels of ATR and ATRIP in Vero cells. Parallel samples were processed for IF analysis (Fig. 3C). Cells depleted of ATRIP and stained with rATRIP Upstate showed a significant decrease in fluorescence intensity, suggesting that rATRIP Upstate does in fact detect ATRIP. On the other hand, cells depleted of ATRIP and stained with mATRIP R&D did not show any decrease in fluorescence intensity (Fig. 3C). Together, these lines of evidence demonstrate that although rATRIP Upstate and rATRIP 403 appear to recognize bona fide ATRIP, mATRIP R&D does not. From this point on, the mATRIP R&D antibody was excluded from further study. These data also support the conclusion that ATRIP is indeed a component of replication compartments.
FIG. 3.
mATRIP R&D does not detect ATRIP. (A) HeLa cells were mock transfected or transfected with HA-ATRIP. Western blotting was performed with HA, rATRIP Upstate, and mATRIP R&D. Actin served as a loading control. (B) Vero cells were infected twice with lentiviruses expressing shRNA to GFP or ATR. Knockdown was determined by Western blotting for ATR N19, rATRIP Upstate, and Chk1. (C and D) Knockdown was also determined by immunofluorescence with rATRIP Upstate (C) and mATRIP R&D (D).
ATR also colocalizes with ICP8 in HSV-1 replication compartments.
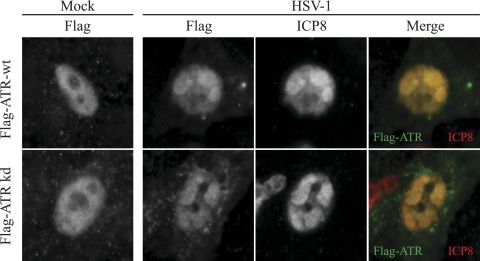
We previously reported that ATR is detected in the nucleoli of uninfected Vero cells and that this localization does not change upon infection with HSV-1 (54). Since we have now identified ATRIP as a component of replication compartments, we wished to revisit the question of ATR localization during infection. We confirmed our previous result that ATR detected by ATR antibody 138 (Bethyl Labs) is indeed in the nucleoli of infected Vero cells (data not shown). ATR is known to be alternatively spliced (13, 34), and these data suggest that ATR 138 may detect an isoform of ATR that is nucleolar and not reorganized during HSV-1 infection. We extended our analysis to include full-length epitope-tagged ATR cDNA, a reagent commonly used to study ATR localization. In uninfected cells Flag-ATR exhibited a nuclear diffuse staining pattern, and it colocalized with ICP8 to replication compartments in HSV-1-infected cells (Fig. 4).
FIG. 4.
ATR colocalizes with ICP8 in HSV-1 replication compartments. Vero cells were transfected with wild-type Flag-ATR (wt) or kinase-dead Flag-ATR (kd) and infected with HSV-1 at an MOI of 10 for 6 h. Cells were fixed and stained with rabbit Flag and monoclonal ICP8.
During a normal DNA damage response, the kinase activity of ATR is required for ATR recruitment to sites of DNA damage (5). We next tested whether ATR recruitment to replication compartments requires ATR kinase activity. Figure 4 shows that the kinase activity of ATR is not required for recruitment of ATR to replication compartments. This observation suggests that ATR and ATRIP are recruited to replication compartments by a mechanism that is independent of DNA damage.
ATR and ATRIP still interact in HSV-1-infected cells.
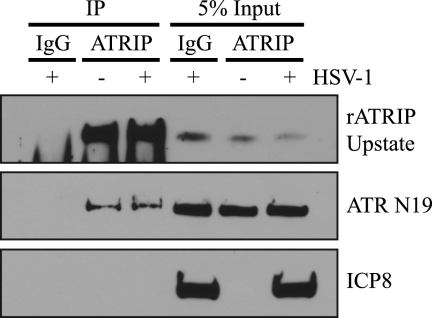
As obligate interacting proteins, ATR and ATRIP coimmunoprecipitate in uninfected and DNA-damaged cells (12). Based on our previous report that ATR and ATRIP appeared to be localized differently during infection, we originally proposed that HSV-1 infection resulted in their separation from one another (54). Based on the IF results reported above, however, it appears that ATR and ATRIP actually colocalize in replication compartments. To test directly whether ATR and ATRIP can interact with each other in HSV-1-infected cells, ATRIP was immunoprecipitated from mock- and HSV-1-infected Vero cell lysates using rATRIP Upstate. Figure 5 shows that ATR coimmunoprecipitated with ATRIP in mock- and HSV-1-infected cells. Levels of ATRIP and ATR in the coimmunoprecipitates were equivalent under both conditions. ICP8 was used as a marker for infection and did not coimmunoprecipitate with ATRIP (Fig. 5). Similar results were obtained using tagged overexpressed versions of ATR and ATRIP (data not shown). This experiment indicates not only that ATR and ATRIP are colocalized during HSV-1 infection but also that they still coimmunoprecipitate, confirming that ATR and ATRIP are not separated during infection as previously reported.
FIG. 5.
ATR and ATRIP can still interact in HSV-1-infected cells. Vero cells were infected with HSV-1 at an MOI of 10 for 6 h. ATRIP immunoprecipitation was performed from cell lysates with rATRIP Upstate, and products were separated by SDS-PAGE and blotted with rATRIP Upstate, ATR N19, and ICP8 antibodies.
ATRIP and RPA can be detected in the earliest detectable prereplicative sites, stage II microfoci.
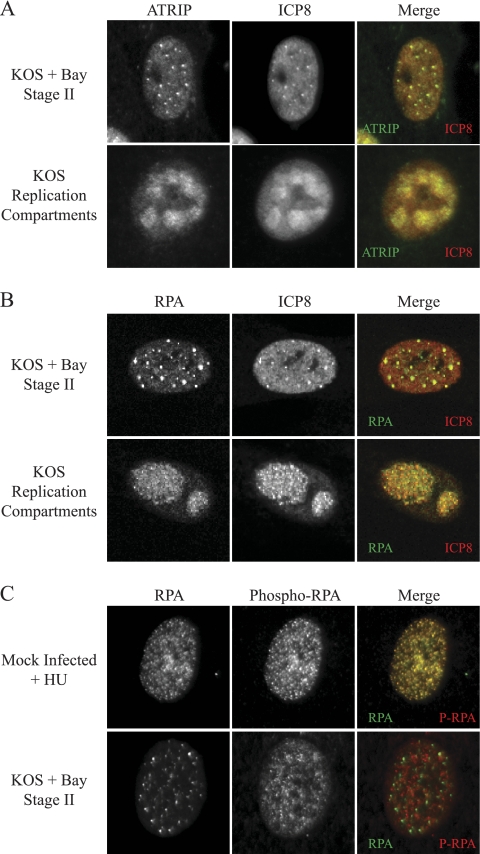
Our lab and others have described prereplicative sites that form as precursors to the formation of replication compartments (7, 29, 30, 32). It has also been reported that cellular proteins are recruited at various stages of replication compartment formation (40, 56), although it is not clear how they are recruited. The earliest detectable prereplicative sites are stage II microfoci, and they contain ICP8, the origin binding protein UL9, and the helicase-primase complex UL5/UL8/UL52 (30). Stage II microfoci can be detected in cells infected in the presence of the HSV-1 helicase-primase inhibitor BAY 57-1293 or mutants deleted for components of the helicase-primase complex (30). To determine whether ATRIP is recruited to stage II microfoci, we infected Vero cells in the presence of BAY 57-1293 and double labeled for ATRIP using rATRIP Upstate and ICP8. ATRIP was detected in stage II microfoci formed with BAY 57-1293 (Fig. 6A) and in stage II microfoci formed during infection with the primase-null mutant hr114 (data not shown). Figure 6B shows that RPA is also present in stage II foci.
FIG. 6.
ATRIP and RPA but not phospho-RPA are in stage II microfoci. Vero cells were infected with HSV-1 at an MOI of 2 for 6 h. Infected cells were treated with the helicase-primase inhibitor BAY 57-1293 to generate stage II microfoci. (A and B) Cells were fixed and stained with rATRIP Upstate and monoclonal ICP8 (A) or monoclonal RPA and rabbit ICP8 367 (B). (C) Vero cells were either mock infected and treated with 2 mM hydroxyurea for 6 h or infected with HSV-1 at an MOI of 10 for 6 h in the presence of BAY 57-1293. Cells were fixed and stained for RPA and phosphorylated RPA.
Previous reports have suggested that ATRIP is recruited to sites of DNA damage through a direct interaction with RPA (4, 58). We previously reported that RPA is not recruited to prereplicative sites until the formation of stage IIIb prereplicative sites (56). Now, using more sensitive detection methods, RPA could in fact be detected in stage II microfoci (Fig. 6B). During a normal ATR-mediated damage response, ATR/ATRIP and RPA colocalize in discrete foci that also include phosphorylated RPA (P-RPA). Therefore, we next asked whether stage II microfoci also contained P-RPA. Vero cells were infected with HSV-1 in the presence of BAY 57-1293 or were mock infected and treated with hydroxyurea (HU), a known activator of ATR signaling. Cells were then double labeled for RPA and P-RPA (Fig. 6C). Mock-infected cells treated with HU formed numerous punctate foci with colocalized RPA and P-RPA. On the other hand, cells with stage II microfoci contained punctate RPA foci, which did not colocalize with P-RPA. These data indicate that ATRIP and RPA are both recruited to stage II microfoci; however, as previously reported, there is no activation of ATR signaling. Furthermore, the endogenous phosphorylated RPA that is present prior to infection is excluded from stage II microfoci. These data taken together suggest that prereplicative sites do not represent areas of damaged DNA despite their superficial resemblance to damage foci.
ATRIP is recruited to replication compartments independently of RPA.
The RPA binding domain of ATRIP has been mapped to the N-terminal region of the protein and is adjacent to a predicted coiled-coil domain responsible for ATRIP oligomerization (2, 4) (Fig. 7). To test whether the RPA binding domain was sufficient to localize to replication compartments, we utilized wild-type Flag-ATRIP and Flag-ATRIP truncation mutants containing the RPA binding and the coiled-coil domains (Flag-ATRIP-1-217) or the RPA binding domain alone (Flag-ATRIP-1-107). Flag-ATRIP was able to localize to replication compartments in infected cells, as did the mutants Flag-ATRIP-1-217 and Flag-ATRIP-1-107 (Fig. 7). These observations suggest that the RPA binding domain is sufficient for localization to replication compartments.
FIG. 7.
Localization of ATRIP mutants to replication compartments. A schematic representation of ATRIP and the ATRIP mutants used in this study is shown, indicating the mutation, the ability to bind RPA, the ability to localize to DNA damage foci, and the ability to localize to replication compartments.
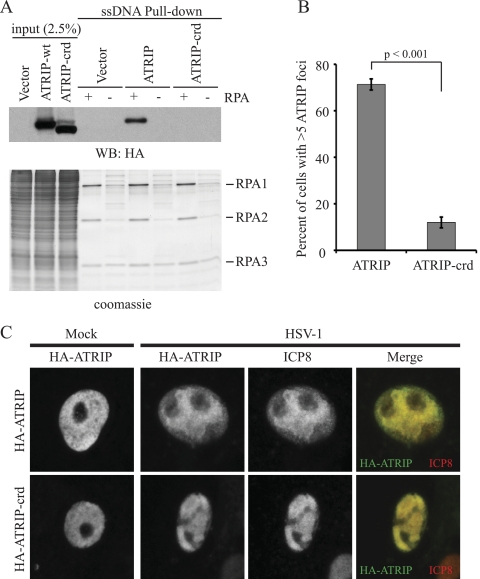
The ATRIP acidic residues D58 and D59 in the RPA binding domain have been reported to make direct contacts with a basic cleft on RPA70 (3). These acidic aspartic acid residues represent a conserved motif by which many DNA repair proteins are recruited to RPA, and this motif is called the checkpoint recruitment domain (CRD) (3, 57). Charge reversal mutations of aspartic acid to lysine are sufficient to prevent an ATRIP fragment (amino acids 1 to 217) from binding RPA-coated ssDNA (RPA-ssDNA) in vitro (57). To determine whether RPA binding was necessary for ATRIP recruitment to HSV-1 replication compartments more directly, we engineered the same charge reversal mutations into full-length HA-ATRIP to create HA-ATRIP-crd. Both HA-ATRIP and HA-ATRIP-crd were tested for their ability to bind RPA-ssDNA in vitro. ssDNA bound to streptavidin beads was coated with purified RPA and incubated with mammalian cell lysates from cells expressing either wild-type or mutant ATRIP. As previously reported, HA-ATRIP efficiently bound to the RPA-ssDNA but not to ssDNA alone (4) and HA-ATRIP-crd did not bind RPA-ssDNA or ssDNA alone (Fig. 8A), consistent with the observation that ATRIP-1-217-crd did not bind RPA-ssDNA (57). HA-ATRIP-crd migrated slightly faster than the wild-type protein, probably reflecting a change in isoelectric point (pI) due to the replacement of acidic residues with basic ones. This same mutation has also been shown to increase the mobility of another checkpoint protein, Rad9, which also binds RPA through conserved aspartic acid residues in its CRD (57).
FIG. 8.
ATRIP is recruited to replication compartments independently of RPA. Site-directed mutagenesis was used to introduce the checkpoint recruitment domain (crd) D58K,D59K mutation into full-length ATRIP. (A) Cells were transfected with HA-ATRIP or HA-ATRIP-crd, and cell lysates were incubated with beads bound to ssDNA that was either coated by recombinant RPA (+) or uncoated (−). Proteins bound to beads were eluted by boiling in SDS sample buffer, resolved by SDS-PAGE, and blotted with an HA antibody. (B) U2OS cells were infected with lentiviruses expressing shATRIP1 and then transfected with shRNA-resistant HA-ATRIP or HA-ATRIP-crd. Cells were then treated with 3 mM HU for 5.5 h and stained for HA and RPA. The percentage of cells with greater than five ATRIP foci per cell was determined for the HA-ATRIP-wt and the HA-ATRIP-crd mutation. The values represent the averages from three independent experiments, and the error bars represent the standard errors of the means. (C) Vero cells were transfected with HA-ATRIP or HA-ATRIP-crd and infected with HSV-1 at an MOI of 10 for 6 h. Cells were fixed and stained with HA and ICP8.
In a normal DNA damage response, the RPA binding domain of ATRIP is required for ATRIP localization to sites of DNA damage (4). Since HA-ATRIP-crd abrogates RPA binding, we hypothesized that it would also abrogate localization to DNA damage foci following replicative stress. To test this hypothesis, U2OS cells were infected with lentiviruses expressing shRNA to deplete endogenous ATRIP. Cells were then transfected with shRNA-resistant HA-ATRIP or HA-ATRIP-crd and treated with 3 mM HU for 5.5 h to induce a DNA damage response. Immunofluorescence analysis was performed to determine whether wild-type or mutant ATRIP could colocalize with RPA at sites of DNA damage. Cells expressing HA-ATRIP formed bright RPA foci that colocalized with ATRIP foci. On the other hand, cells expressing HA-ATRIP-crd also formed bright RPA foci, but these foci did not contain HA-ATRIP-crd. In fact, there was no HA-ATRIP-crd reorganization in response to HU. The percentage of cells with ATRIP foci was determined for both samples, and these data are displayed in Fig. 8B. A significant reduction was observed in the number of cells with ATRIP foci for the HA-ATRIP-crd mutant compared to the wild-type control. These data suggest that the HA-ATRIP-crd mutant cannot bind RPA and cannot localize to damage foci.
To test whether ATRIP is recruited to HSV-1 replication compartments through a direct interaction with RPA, we transiently overexpressed either HA-ATRIP or HA-ATRIP-crd in Vero cells and then infected them with HSV-1. Like Flag-ATRIP, HA-ATRIP also localized to replication compartments as determined by colocalization with ICP8 (Fig. 8C). Interestingly, HA-ATRIP-crd also colocalized with ICP8, indicating that RPA binding is not a requirement for ATRIP recruitment to replication compartments, as it is to sites of DNA damage. Consistent with the observation that the kinase activity of ATR is not required for localization to replication compartments (Fig. 4), these data also suggest that ATR/ATRIP are recruited to replication compartments by a mechanism different from that for recruitment to sites of DNA damage.
We next tested ATRIP mutants with deletions in the RPA binding domain, the coiled-coil domain, and the ATR binding domain in order to determine whether these regions are required for localization to replication compartments. None of these three mutants can localize to sites of DNA damage; however, all of them were able to localize to replication compartments (Fig. 7), again suggesting that ATRIP is recruited by a novel mechanism that does not represent normal DNA damage signaling.
ATR and ATRIP contribute to efficient HSV-1 infection.
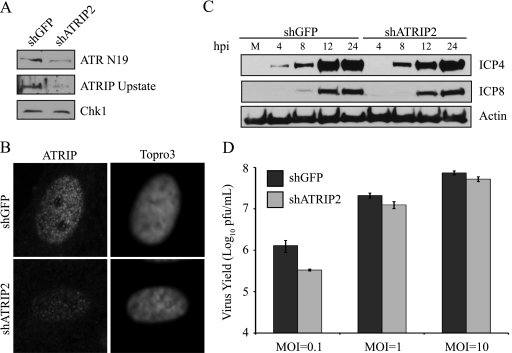
Since it does not appear that ATR and ATRIP were responding to DNA damage in the replication compartment, we next asked whether they were needed for viral infection. HFF-1 cells were infected with lentiviruses expressing shRNA to GFP or ATRIP, and virus growth was monitored. Knockdown of ATRIP was measured by both Western blot and immunofluorescence analyses to confirm that shRNA to ATRIP resulted in a significant depletion of both ATR and ATRIP (Fig. 9A and B). Viral gene expression in cells depleted of ATRIP was monitored by Western blot analysis for the viral proteins ICP4 and ICP8, which served as markers for immediate-early (IE) and early (E) genes, respectively. At an MOI of 0.1 PFU/cell, shATRIP cells exhibited a slight delay in the expression of both IE and E genes (Fig. 9C). For example, ICP4 could be detected as early as 4 h postinfection in shGFP cells but was not detected until 8 h postinfection in shATRIP cells. Likewise, ICP8 could be detected by 8 h in shGFP cells but was not detected until 12 h postinfection in shATRIP cells.
FIG. 9.
ATR and ATRIP are required for efficient HSV-1 replication and IE gene expression. HFF-1 cells were infected twice with lentiviruses expressing either shGFP or shATRIP2. (A) Knockdown was determined by Western blotting with rATRIP Upstate and ATR N19. Chk1 served as a loading control. (B) Knockdown of ATRIP was also assessed by immunofluorescence staining with rATRIP Upstate and Topro3. (C) Knockdown cells were infected with HSV-1 at an MOI of 0.1 and harvested at the indicated times postinfection. Western blotting were performed for representative immediate-early (ICP4) and early (ICP8) genes. Actin served as a loading control. (D) Knockdown cells were infected with HSV-1 at the indicated MOI. Progeny virus was collected at 12 h postinfection, and titers were determined on Vero cells. The values represent the averages from four independent experiments, and the error bars represent the standard errors of the means.
Since knockdown of ATRIP caused a delay in gene expression, we also asked whether viral yield was reduced in shATRIP cells. shGFP and shATRIP cells were infected at three different multiplicities of infection, viral progeny were collected at 12 h postinfection, and titers were determined on Vero cells. At an MOI of 0.1 PFU/cell, the growth of HSV-1 was reduced by 5-fold, or approximately 80%, on shATRIP cells compared to the shGFP controls (Fig. 9D). A smaller reduction in growth was also detected at MOIs of 1 and 10 PFU/cell. Similar delays in viral gene expression and reduction in virus yield were also obtained in HeLa cells (data not shown). Together these data indicate that ATR and ATRIP are not absolutely required for viral growth; however, they do contribute to an efficient infection, possibly by promoting gene expression of IE genes.
ATR and ATRIP are not targets of ICP0.
We previously suggested that ICP0 was able to uncouple ATRIP from ATR, as transfection of cells with an ICP0 expression plasmid resulted in a spatial uncoupling of ATRIP and ATR (54). Based on this result, we further suggested that ICP0 may be directly responsible for disabling the ATR signaling pathway in infected cells. These experiments, however, utilized antibodies which either do not react with bona fide ATRIP (mATRIP R&D) or interact with an alternate isoform of ATR (ATR 138). In order to further explore the possible role of ICP0 in disabling the ATR signaling pathway, we asked whether ATR and ATRIP are direct targets of ICP0 degradation.
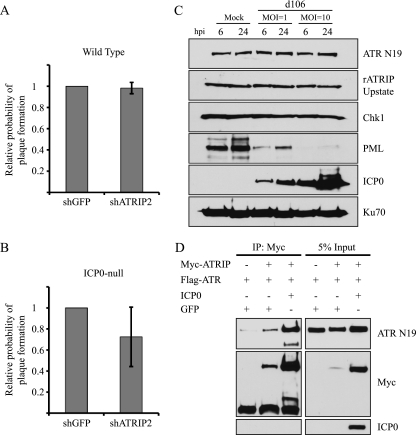
Cellular proteins that silence viral gene expression represent a form of intrinsic cellular resistance to viral infections, and ICP0 plays an important role during infection by degrading these cellular proteins (14-16). Consequently, the major phenotype of the ICP0-null virus is a reduced probability of initiating a productive infection due to one or more intrinsic antiviral defense mechanisms. Recent work from the Everett and Weitzman labs has shown that depletion of ICP0 targets from cells before infection improves the probability of plaque formation of the ICP0-null virus (18, 27). We therefore compared the probability of plaque formation of both wild-type HSV-1 and ICP0-null viruses on shATRIP cells compared to that on the shGFP controls. The number of plaques produced on shATRIP cells for a given dilution of virus was compared to the number of plaques produced by the same dilution of virus on shGFP cells. This method allowed us to determine a relative probability of plaque formation in ATRIP-depleted cells (18). We observed no difference in the probability of plaque formation of either wild-type HSV-1 or the ICP0-null virus when we depleted ATRIP (Fig. 10A and B). These data suggest that ATRIP does not appear to contribute to the repression of an ICP0-null virus.
FIG. 10.
ATR and ATRIP are not targets of ICP0. (A and B) HFF-1 cells were infected twice with lentiviruses expressing either shGFP or shATRIP2. Cells were then infected with wild-type HSV-1 (in1863) (A) or ICP0-null (dl1403/CMVlacZ) (B) at dilutions suitable for plaque assays. The relative probability of plaque formation on shATRIP2 cells was determined with respect to the same dilution of shGFP cells. The values represent the averages from three independent experiments, and the error bars represent the standard errors of the means. (C) HeLa cells were infected with d106, which expresses only ICP0, at an MOI of 1 or 10, and samples were harvested at 6 and 24 h postinfection as described in Materials and Methods and separated by SDS-PAGE. Western blotting was performed for ATR N19, rATRIP Upstate, Chk1, and PML. ICP0 served as an infection control, and Ku70 served as a loading control. (D) Vero cells were transfected with Myc-ATRIP, Flag-ATR, and ICP0 as indicated. ATRIP immunoprecipitation was performed from cell lysates with Myc antibody, and the products were separated by SDS-PAGE and blotted with Myc, ATR N19, and ICP0.
We next asked whether ATR and ATRIP are direct targets of ICP0 degradation, as has been previously reported for DNA-PKcs, PML, RNF8, RNF168, and several other cellular proteins potentially involved in repression of lytic infection (14, 16, 18, 27, 37). ICP0 was expressed by infecting cells with virus d106, which is deleted for all IE genes except ICP0 (43). HeLa cells were infected at MOIs of 1 and 10 PFU/cell and harvested at 6 and 24 h postinfection (Fig. 10C). Western blot analysis of d106-infected cell lysates shows that PML is efficiently degraded in the presence of ICP0, as has been previously reported (43). Despite the efficient degradation of PML, the levels of ATR, ATRIP, and Chk1 remained constant during infection, indicating that none of them are targets of ICP0.
Lastly, we wanted to confirm whether ICP0 could disrupt the interaction between ATR and ATRIP. Cells were transfected with Myc-ATRIP, Flag-ATR, and either GFP or ICP0. Myc-ATRIP was immunoprecipitated using a Myc antibody, and coprecipitating proteins were probed for ATR and ICP0 (Fig. 10D). Flag-ATR and Myc-ATRIP were still able to interact by coimmunoprecipitation analysis in cells coexpressing GFP or ICP0. Furthermore, ICP0 did not coimmunoprecipitate with ATRIP. This experiment demonstrates that ICP0 is not able to disrupt the interaction between ATR and ATRIP. Taken together, these data demonstrate the ATR and ATRIP are not involved in repression of the ICP0-null virus and that ATR, ATRIP, and Chk1 are not targets of ICP0-mediated degradation.
ATR signaling is disabled in HSV-1-infected cells.
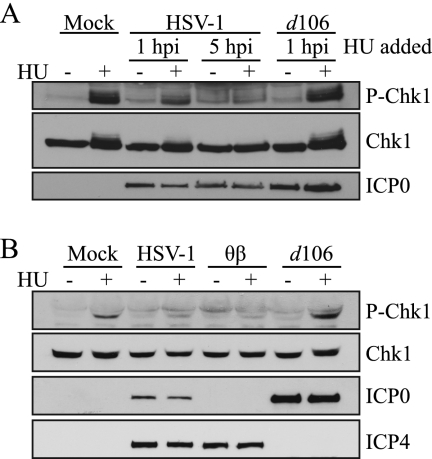
ATR and ATRIP still interact and are located in replication compartments of HSV-1-infected cells; however, ATR does not appear to be activated, suggesting that the ATR pathway may be disabled. We next asked whether the ATR pathway could be activated in HSV-1-infected cells by treating them with hydroxyurea, a known activator of ATR (9). HeLa cells were infected with HSV-1 for 1 h or 5 h and then treated with 3 mM HU for an additional 2 h. In uninfected cells, HU treatment results in robust phosphorylation of the ATR substrate Chk1 (Fig. 11A). In cells infected with HSV-1 for 1 h prior to treatment, Chk1 phosphorylation was reduced compared to that in the uninfected control. Interestingly, in cells infected with HSV-1 for 5 h prior to treatment, no Chk1 phosphorylation in response to HU could be detected. In addition, cells infected for 1 h with d106 and then treated with HU displayed no reduction in Chk1 phosphorylation. As noted above, d106 expresses only ICP0 and is deleted for all other IE genes (43); thus, ICP0 expression alone is not sufficient to prevent Chk1 phosphorylation in response to HU. Together, these results demonstrate that HSV-1 infection is sufficient to partially disable ATR signaling at 1 h postinfection and completely disable ATR signaling at 5 h postinfection.
FIG. 11.
HSV-1 blocks ATR signaling independently of ICP0. (A) HeLa cells were infected with HSV-1 or d106 at an MOI of 10, and 3 mM HU was added at the indicated times postinfection and incubated for another 2 h. Western blotting was performed for P-Chk1 S345, total Chk1, and ICP0. (B) HeLa cells were infected with KOS, θβ, or d106 at an MOI of 5, 3 mM HU was added at 6 h postinfection, and samples were collected 2 h later. Western blotting was performed as for panel A with the addition of ICP4.
To further explore the requirements of ICP0 in preventing the phosphorylation of Chk1 in response to HU during infection, we used the ICP0-null mutant θβ, which harbors lacZ insertions in both copies of the ICP0 gene (45). HeLa cells were infected with HSV-1, θβ, or d106 for 6 h and then treated with HU for an additional 2 h. We detected no inducible phosphorylation of Chk1 in cells infected with HSV-1 or θβ (Fig. 11B). However, we were able to detect Chk1 phosphorylation in d106-infected cells. Consistent with the observation that ATR and ATRIP are not targets of ICP0, these experiments show that ICP0 is not sufficient to block ATR signaling in response to replicative stress.
DISCUSSION
In this study several important observations were made. (i) ATRIP is recruited to the earliest reported prereplicative sites (stage II microfoci) as well as to mature replication compartments. (ii) ATRIP recruitment is independent of functional ATR signaling and RPA binding, and a number of ATRIP mutants defective in localization to sites of DNA damage can be recruited to replication compartments. (iii) Despite the presence of ATR, ATRIP, and unphosphorylated RPA in stage II microfoci in HSV-1-infected cells, endogenous phosphorylated RPA is excluded. (iv) ATR also appears to be recruited to replication compartments, and the ATR-ATRIP interaction remains intact in infected cells. (v) ATR and ATRIP appear to contribute to productive infection and are required for efficient expression of IE and E genes. (vi) Neither ATR nor ATRIP appears to be a target for ICP0 degradation, and ICP0 is apparently not required for HSV-1 to disable ATR signaling. Taken together, these results have important implications for the interaction between HSV-1 infection and the cellular damage response and add to a growing body of evidence suggesting that HSV-1 manipulates the host DNA damage response pathways. It now appears that HSV-1 can degrade or inactivate components of all three major DNA damage signaling pathways while potentially coopting other components for its own benefit.
ATR signaling may exert a negative influence on viral infection.
The activation of the ATM pathway in HSV-1-infected cells would generally be expected to activate ATR signaling (9). The observation that ATR signaling is disabled in infected cells is therefore of considerable interest, and we are interested in both the mechanism of inactivation and possible reasons for it. HSV-1 has previously been shown to interfere with signaling by two of the three major DNA damage signaling kinases, DNA-PK and ATM, through the action of ICP0, which specifically degrades DNA-PK, RNF8, and RNF168 (27, 37). The observation that ATR signaling is inactivated by HSV-1 infection led us to speculate that ATR signaling may also represent an intrinsically antiviral defense mechanism and that ICP0 might be responsible for counteracting negative consequences of ATR activation.
The observations that ATR and ATRIP contribute to a productive infection and are recruited to replication compartments suggest that ATR itself is not intrinsically antiviral; however, ATR signaling may still have negative outcomes for viral infection, including gene silencing and checkpoint signaling. For instance, during meiotic sex chromosome inactivation, the X and Y chromosomes are transcriptionally silenced by a mechanism that requires ATR (51, 52). Furthermore, ATR has been shown to colocalize with γH2AX on regions of heterochromatin or silenced chromosomes (19, 51, 52). It is also possible that phosphorylation of the downstream targets of ATR signaling, RPA and Chk1, may lead to checkpoint signaling that would be unfavorable to HSV-1 gene expression or DNA replication. It is also of interest that unphosphorylated RPA is recruited to viral prereplication sites while endogenous phosphorylated RPA is excluded, again suggesting that HSV-1 may have evolved to avoid downstream consequences of ATR signaling.
HSV-1 inactivates ATR signaling in an ICP0-independent manner.
The involvement of ICP0 in the degradation of PML, RNF8, and RNF168 is consistent with its known role in promoting lytic viral infection by counteracting intrinsically antiviral defense mechanisms. ICP0 is also known to interfere with the host interferon response (14, 20, 36). We were surprised to find that inactivation of ATR signaling in HSV-1-infected cells appears to occur by a mechanism that is independent of ICP0 and that neither ATR nor ATRIP is a target of ICP0 degradation. We conclude that unlike the mechanism by which the DNA-PK- and ATM-mediated damage responses are inactivated, another viral activity must be responsible for the disabling of ATR signaling in HSV-1-infected cells.
HSV-1 and adenovirus both disable ATR signaling.
Interestingly, ATR signaling is also disabled in cells infected with adenovirus (8) and in cells infected with HCMV at late times postinfection (33), consistent with the notion that ATR activation may exert a negative influence on infection by both herpesviruses and adenoviruses. Adenovirus type 5 (Ad5) apparently disables ATR activation by mislocalizing the MRN components of the ATM pathway and therefore prevents cross talk between signaling pathways (8). Ad12, on the other hand, disables ATR signaling by degrading the ATR activator TopBP1 (6). In HSV-1-infected cells, MRN is recruited to replication compartments (26, 47, 49, 56), and TopBP1 is not degraded (K. N. Mohni and S. K. Weller, unpublished observation), indicating that HSV-1 appears to disable ATR by a mechanism distinct from that of either Ad5 or Ad12. For example, ATR signaling generally occurs on cellular chromatin, and it is possible that ATR may not be able to signal damage on viral DNA because it adopts a different chromatin structure (22, 23). Alternatively, since ATR signaling requires the independent recruitment of cofactors such as the 9-1-1 complex and TopBP1 (9), it is possible that one or more of these cofactors are either degraded or sequestered to prevent ATR signaling in HSV-1-infected cells.
ATR/ATRIP are recruited to viral replication compartments and are important for efficient production of virus.
ATR/ATRIP are also recruited to adenovirus replication compartments in a manner that does not activate ATR signaling (8). This raises the possibility that ATR/ATRIP proteins themselves play a positive role in the viral life cycle. In a normal cellular DNA damage response, ATR/ATRIP are recruited to stretches of RPA-coated single-stranded DNA in an RPA- and kinase-dependent manner, resulting in the phosphorylation of RPA by ATR (4, 5, 9). In this paper we show that ATR and ATRIP recruitment is independent of functional ATR signaling and RPA binding and that ATRIP mutants defective in localization to sites of DNA damage can be recruited to replication compartments. These results suggest that ATR/ATRIP recruitment occurs by a mechanism entirely different from that used in a cellular DNA damage response.
It is possible that the recruitment of ATR and ATRIP to replication compartments is part of the mechanism by which HSV-1 disables ATR signaling, perhaps by sequestering ATR away from its required cofactors. It is also possible that ATR and ATRIP themselves play positive roles in viral infection during either immediate-early and early gene expression or DNA synthesis. For instance, they may function to recruit other cellular or viral factors that are important at the earliest stages of infection. Thus, although we have identified the seven major viral proteins required for HSV-1 DNA replication (53), a large number of cellular factors are also present in replication compartments, and the mechanism by which they are recruited and their precise functions remain a mystery. Work to identify the stage in the virus life cycle that utilizes ATR/ATRIP is under way.
In summary, the results presented in this paper augment a growing body of evidence demonstrating that HSV-1 has evolved to navigate an exquisitely complex web of cellular DNA repair pathways in order to promote lytic viral infection. We speculate that HSV-1 subverts intrinsically antiviral host defenses and engineers the recruitment of viral and cellular proteins that promote robust immediate-early and early gene expression and aid in viral DNA replication.
Acknowledgments
We thank Chris Preston, Neal Deluca, Roger Everett, William Ruyechan, Karlene Cimprich, Gerald Kleymann, David Root, and Didier Trono for generous gifts of reagents. We thank Samantha Marques for cloning the HA-ATRIP-crd plasmid. We thank members of the Weller laboratory for helpful comments and discussions.
This work was supported by Public Health Service grants AI21747 and AI69136 to S.K.W. and CA102729 from the National Cancer Institute to D.C.
Footnotes
Published ahead of print on 22 September 2010.
REFERENCES
- 1.Abraham, R. T. 2004. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst.) 3:883-887. [DOI] [PubMed] [Google Scholar]
- 2.Ball, H. L., and D. Cortez. 2005. ATRIP oligomerization is required for ATR-dependent checkpoint signaling. J. Biol. Chem. 280:31390-31396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, H. L., M. R. Ehrhardt, D. A. Mordes, G. G. Glick, W. J. Chazin, and D. Cortez. 2007. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol. Cell. Biol. 27:3367-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ball, H. L., J. S. Myers, and D. Cortez. 2005. ATRIP binding to replication protein A-single-stranded DNA promotes ATR-ATRIP localization but is dispensable for Chk1 phosphorylation. Mol. Biol. Cell 16:2372-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr, S. M., C. G. Leung, E. E. Chang, and K. A. Cimprich. 2003. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr. Biol. 13:1047-1051. [DOI] [PubMed] [Google Scholar]
- 6.Blackford, A. N., R. N. Patel, N. A. Forrester, K. Theil, P. Groitl, G. S. Stewart, A. M. Taylor, I. M. Morgan, T. Dobner, R. J. Grand, and A. S. Turnell. 2010. Adenovirus 12 E4orf6 inhibits ATR activation by promoting TOPBP1 degradation. Proc. Natl. Acad. Sci. U. S. A. 107:12251-12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkham, J., D. M. Coen, and S. K. Weller. 1998. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol. 72:10100-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carson, C. T., N. I. Orazio, D. V. Lee, J. Suh, S. Bekker-Jensen, F. D. Araujo, S. S. Lakdawala, C. E. Lilley, J. Bartek, J. Lukas, and M. D. Weitzman. 2009. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J. 28:652-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimprich, K. A., and D. Cortez. 2008. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 9:616-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cimprich, K. A., T. B. Shin, C. T. Keith, and S. L. Schreiber. 1996. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc. Natl. Acad. Sci. U. S. A. 93:2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cliby, W. A., C. J. Roberts, K. A. Cimprich, C. M. Stringer, J. R. Lamb, S. L. Schreiber, and S. H. Friend. 1998. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 17:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge. 2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 13.Durocher, F., Y. Labrie, P. Soucy, O. Sinilnikova, D. Labuda, P. Bessette, J. Chiquette, R. Laframboise, J. Lepine, B. Lesperance, G. Ouellette, R. Pichette, M. Plante, S. V. Tavtigian, and J. Simard. 2006. Mutation analysis and characterization of ATR sequence variants in breast cancer cases from high-risk French Canadian breast/ovarian cancer families. BMC Cancer 6:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 15.Everett, R. D. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 8:365-374. [DOI] [PubMed] [Google Scholar]
- 16.Everett, R. D., and M. K. Chelbi-Alix. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819-830. [DOI] [PubMed] [Google Scholar]
- 17.Everett, R. D., M. Meredith, and A. Orr. 1999. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol. 73:417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., C. Parada, P. Gripon, H. Sirma, and A. Orr. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 82:2661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Capetillo, O., S. K. Mahadevaiah, A. Celeste, P. J. Romanienko, R. D. Camerini-Otero, W. M. Bonner, K. Manova, P. Burgoyne, and A. Nussenzweig. 2003. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 4:497-508. [DOI] [PubMed] [Google Scholar]
- 20.Hagglund, R., and B. Roizman. 2004. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78:2169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleymann, G., R. Fischer, U. A. Betz, M. Hendrix, W. Bender, U. Schneider, G. Handke, P. Eckenberg, G. Hewlett, V. Pevzner, J. Baumeister, O. Weber, K. Henninger, J. Keldenich, A. Jensen, J. Kolb, U. Bach, A. Popp, J. Maben, I. Frappa, D. Haebich, O. Lockhoff, and H. Rubsamen-Waigmann. 2002. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat. Med. 8:392-398. [DOI] [PubMed] [Google Scholar]
- 22.Knipe, D. M., and A. Cliffe. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6:211-221. [DOI] [PubMed] [Google Scholar]
- 23.Kristie, T. M., Y. Liang, and J. L. Vogel. Control of alpha-herpesvirus IE gene expression by HCF-1 coupled chromatin modification activities. Biochim. Biophys. Acta 1799:257-265. [DOI] [PMC free article] [PubMed]
- 24.Lamberti, C., and S. K. Weller. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403-407. [DOI] [PubMed] [Google Scholar]
- 25.Lees-Miller, S. P., M. C. Long, M. A. Kilvert, V. Lam, S. A. Rice, and C. A. Spencer. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 70:7471-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lilley, C. E., C. T. Carson, A. R. Muotri, F. H. Gage, and M. D. Weitzman. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 102:5844-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lilley, C. E., M. S. Chaurushiya, C. Boutell, S. Landry, J. Suh, S. Panier, R. D. Everett, G. S. Stewart, D. Durocher, and M. D. Weitzman. 2010. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 29:943-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lilley, C. E., R. A. Schwartz, and M. D. Weitzman. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15:119-126. [DOI] [PubMed] [Google Scholar]
- 29.Liptak, L. M., S. L. Uprichard, and D. M. Knipe. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70:1759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livingston, C. M., N. A. DeLuca, D. E. Wilkinson, and S. K. Weller. 2008. Oligomerization of ICP4 and rearrangement of heat shock proteins may be important for herpes simplex virus type 1 prereplicative site formation. J. Virol. 82:6324-6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livingston, C. M., M. F. Ifrim, A. E. Cowan, and S. K. Weller. 2009. Virus-induced chaperone-enriched (VICE) domains function as nuclear protein quality control centers during HSV-1 infection. PLoS Pathog. 5:e1000619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 71:2390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo, M. H., K. Rosenke, K. Czornak, and E. A. Fortunato. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mannino, J. L., W. Kim, M. Wernick, S. V. Nguyen, R. Braquet, A. W. Adamson, Z. Den, M. A. Batzer, C. C. Collins, and K. D. Brown. 2001. Evidence for alternate splicing within the mRNA transcript encoding the DNA damage response kinase ATR. Gene 272:35-43. [DOI] [PubMed] [Google Scholar]
- 35.Moffat, J., D. A. Grueneberg, X. Yang, S. Y. Kim, A. M. Kloepfer, G. Hinkle, B. Piqani, T. M. Eisenhaure, B. Luo, J. K. Grenier, A. E. Carpenter, S. Y. Foo, S. A. Stewart, B. R. Stockwell, N. Hacohen, W. C. Hahn, E. S. Lander, D. M. Sabatini, and D. E. Root. 2006. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124:1283-1298. [DOI] [PubMed] [Google Scholar]
- 36.Paladino, P., and K. L. Mossman. 2009. Mechanisms employed by herpes simplex virus 1 to inhibit the interferon response. J. Interferon Cytokine Res. 29:599-607. [DOI] [PubMed] [Google Scholar]
- 37.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulsen, R. D., and K. A. Cimprich. 2007. The ATR pathway: fine-tuning the fork. DNA Repair (Amst.) 6:953-966. [DOI] [PubMed] [Google Scholar]
- 39.Phelan, A., J. Dunlop, A. H. Patel, N. D. Stow, and J. B. Clements. 1997. Nuclear sites of herpes simplex virus type 1 DNA replication and transcription colocalize at early times postinfection and are largely distinct from RNA processing factors. J. Virol. 71:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quadt, I., A. K. Gunther, D. Voss, M. Schelhaas, and D. Knebel-Morsdorf. 2006. TATA-binding protein and TBP-associated factors during herpes simplex virus type 1 infection: localization at viral DNA replication sites. Virus Res. 115:207-213. [DOI] [PubMed] [Google Scholar]
- 41.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 42.Roizman, B. 1979. The structure and isomerization of herpes simplex virus genomes. Cell 16:481-494. [DOI] [PubMed] [Google Scholar]
- 43.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Samaniego, L. A., A. L. Webb, and N. A. DeLuca. 1995. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J. Virol. 69:5705-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samaniego, L. A., N. Wu, and N. A. DeLuca. 1997. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J. Virol. 71:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shelton, L. S., A. G. Albright, W. T. Ruyechan, and F. J. Jenkins. 1994. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J. Virol. 68:521-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shirata, N., A. Kudoh, T. Daikoku, Y. Tatsumi, M. Fujita, T. Kiyono, Y. Sugaya, H. Isomura, K. Ishizaki, and T. Tsurumi. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 280:30336-30341. [DOI] [PubMed] [Google Scholar]
- 48.Strang, B. L., and N. D. Stow. 2007. Blocks to herpes simplex virus type 1 replication in a cell line, tsBN2, encoding a temperature-sensitive RCC1 protein. J. Gen. Virol. 88:376-383. [DOI] [PubMed] [Google Scholar]
- 49.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, T. J., E. E. McNamee, C. Day, and D. M. Knipe. 2003. Herpes simplex virus replication compartments can form by coalescence of smaller compartments. Virology 309:232-247. [DOI] [PubMed] [Google Scholar]
- 51.Turner, J. M. 2007. Meiotic sex chromosome inactivation. Development 134:1823-1831. [DOI] [PubMed] [Google Scholar]
- 52.Turner, J. M., S. K. Mahadevaiah, O. Fernandez-Capetillo, A. Nussenzweig, X. Xu, C. X. Deng, and P. S. Burgoyne. 2005. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat. Genet. 37:41-47. [DOI] [PubMed] [Google Scholar]
- 53.Weller, S. K. 2009. Herpesvirus genome replication, p. 249-265. In C. E. Cameron, M. Gotte, and K. D. Raney (ed.), Viral genome replication. Springer, New York, NY.
- 54.Wilkinson, D. E., and S. K. Weller. 2006. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J. Cell Sci. 119:2695-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilkinson, D. E., and S. K. Weller. 2005. Inhibition of the herpes simplex virus type 1 DNA polymerase induces hyperphosphorylation of replication protein A and its accumulation at S-phase-specific sites of DNA damage during infection. J. Virol. 79:7162-7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilkinson, D. E., and S. K. Weller. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, X., S. Vaithiyalingam, G. G. Glick, D. A. Mordes, W. J. Chazin, and D. Cortez. 2008. The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol. Cell. Biol. 28:7345-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]