Abstract
A novel database, under the acronym RISSC (Ribosomal Intergenic Spacer Sequence Collection), has been created. It compiles more than 1600 entries of edited DNA sequence data from the 16S–23S ribosomal spacers present in most prokaryotes and organelles (e.g. mitochondria and chloroplasts) and is accessible through the Internet (http://ulises.umh.es/RISSC), where systematic searches for specific words can be conducted, as well as BLAST-type sequence searches. Additionally, a characteristic feature of this region, the presence/absence and nature of tRNA genes within the spacer, is included in all the entries, even when not previously indicated in the original database. All these combined features could provide a useful documentation tool for studies on evolution, identification, typing and strain characterization, among others.
DESCRIPTION OF THE DATABASE
The development of free-access Molecular Biology databases via the Internet has increased dramatically over the past few years as the scientific community has become more familiar with this technological tool (1–3). Their utility is evident, even for the simplest databases, since they provide easy and fast access to a variety of relevant information that would otherwise be tedious and time-consuming to obtain.
One major issue in microbiology is the identification of microorganisms, even more so recently due to rekindling of prokaryotic biodiversity studies. Commonly, molecular identification techniques involve the sequencing of the prokaryotic 16S rRNA gene. This has proved to be useful in establishing coherent phylogenetic relationships at the taxonomic level of species or higher (genus, family, etc.) (4,5), but it often lacks accuracy in discriminating strains within the same species and sometimes even different species within the same genus (6). In contrast, the internal transcribed spacers (ITS), located between the 16S and 23S genes in most prokaryotic ribosomal RNA operons, are much more variable than the adjacent 16S and 23S ribosomal genes. Typically, this region consists of a series of conserved, alignable DNA stretches found in all strains of a single species, but rarely beyond the genus or family level, combined with hypervariable DNA segments (in the form of insertions, deletions and/or highly variable sequences of equal length) (7,8). The alignable stretches can be used for very precise species identification while the hypervariable stretches often allow strain characterization. These regions may vary even among the different operons within a single cell (intercistronic heterogeneity), particularly when several operons are present (9). The location of the ITS flanked by the highly conserved 16S and 23S rRNA genes allows for very easy PCR amplification using universal primers. In fact sequencing, or characterization by other means, of the ITS has become rather common over the past years for typing work in population genetics or molecular epidemiology (7,10–12).
From the previous argument it is easy to see the usefulness of a spacer database for fine species and/or strain characterization of Bacteria and Archaea. However, many of these sequences deposited in the EMBL/GenBank/DDBJ and other databases were not originally intended for these kinds of studies and they are very often submitted as part of a complete ribosomal operon sequence. Even when deposited as spacers, they may have partial 16S and/or 23S sequences attached, not even being characterized as such. Frequently, an ITS with flanking 16S/23S gene sequences may cause inconvenience when carrying out BLAST searches (since ribosomal homology often yields unwanted ‘best’ matches) or in experiments such as species or strain-specific primer design from aligned sequences. Moreover, one of the most conserved parts of the ITS, if present, the tRNA gene, also of great importance in typing and evolution experiments (13,14), is not always reported. Being highly conserved, the tRNA section can also bias BLAST searches. At RISSC we have carried out editing and tRNA searching of the 16S–23S spacers available at other databases, for better characterization.
USING RISSC
Upon entering the RISSC web page at http://ulises.umh.es/RISSC, visitors will find detailed instructions about how to proceed. Commands are presented in a similar way to other known databases to facilitate straightforward use. Among others, BLAST-type searches (15) can be conducted by downloading the appropriate program and obtaining the database of spacers cured from 16S and/or 23S tails (Fig. 1). The SIZE option is designed to delimitate a size range for a specific search. The ITS may vary greatly in length, from few bases to almost a kilobase and more (Fig. 2). The presence/absence of tRNA genes, as indicated by the tRNAscan-SE v1.11 program (http://www.genetics.wustl.edu/eddy/tRNAscan-SE) (16), is shown in the FEATURES field as well as their arrangement within the spacer. tRNA genes are important as phylogenetic markers, since their presence or absence in the ITS is characteristic of certain groups of prokaryotes (7,8,13). Nevertheless, almost 40% (302 sequences out of 790, August 30, 2000) of all these genes recorded at RISSC had previously gone unreported. Observations on the spacer sequence distribution according to the contributions of different phylogenetic groups also indicates that some groups are poorly represented (or not represented at all), with preference (in terms of number of entries) for those microorganism species of importance in clinical and applied microbiology (Table 1). This should be taken as an encouragement to scientists to expand their studies and shows how much work has yet to be done.
Figure 1.
BLAST search results from a chimerical sequence (100 bp of the 16S from Lactobacillus rhamnosus and 217 bp of the ITS from Lactobacillus zeae) using (A) the EMBL/GenBank/DDBJ database (the first 10 best matches) and (B) the RISSC database (all nine best matches). BLAST search in RISSC is unaffected by the presence of a 16S gene.
Figure 2.
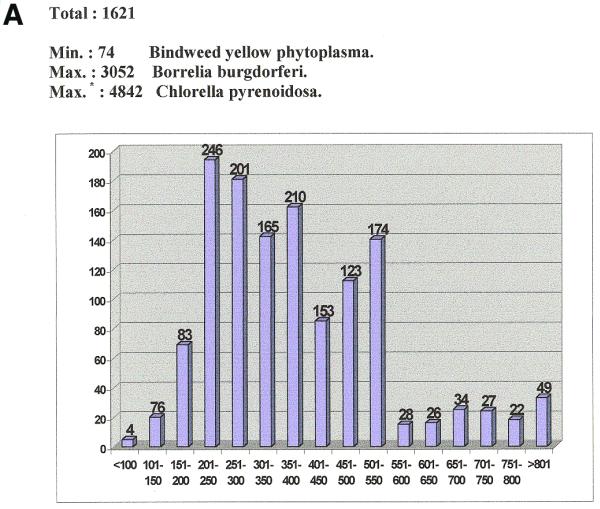
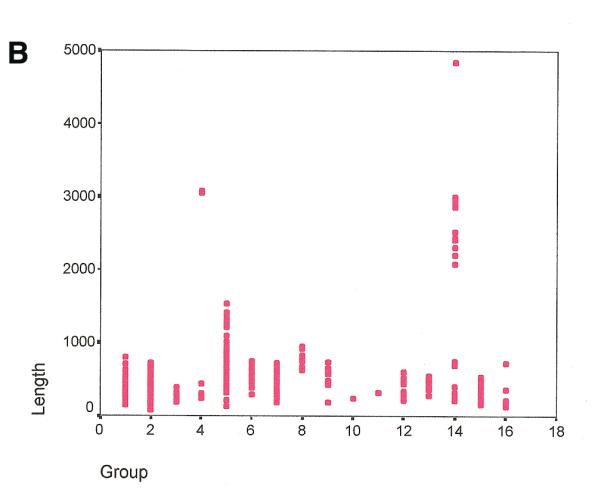
Size ranges for the sequences available at RISSC according to: (A) the whole database; (B) each phylogenetic group. Chloroplasts have the highest variability in size (August 30, 2000). Phylogenetic groups: 1, high GC Gram+ bacteria; 2, low GC Gram+ bacteria; 3, Planctomyces/Chlamydia; 4, spirochaetes; 5, α subdivision; 6, β subdivision; 7, γ subdivision; 8, δ/ɛ subdivision; 9, Cytophaga/Flexibacter/Bacteroides; 10, Thermotogales; 11, Aquificales; 12, unidentified; 13, cyanobacteria; 14, chloroplasts; 15, Euryarchaeota; 16, Crenarchaeota. *, plastids.
Table 1. Distribution of spacer sequences and tRNA genes according to their phylogeny (August 30, 2000).
| Sequences | Species | tRNA | ||||||||
| |
(size range) |
|
None |
Ile |
Ala |
Glu |
Ile–Ala |
Ala–Ile |
Glu–Ala |
Glu–Lys–Val |
| High GC Gram+ bacteria | 368 (144–802) | 136 | 368 | |||||||
| Low GC Gram+ bacteria | 457 (74–724) | 142 | 275 | 108 | 38 | 36 | ||||
| Planctomyces/Chlamydia | 50 (190–384) | 6 | 50 | |||||||
| Spirochaetes | 20 (245–3074) | 6 | 12 | 3 | 3 | 2 | ||||
| Proteobacteria | ||||||||||
| α subdivision | 159 (130–1529) | 31 | 10 | 7 | 1 | 140 | 1 | |||
| β subdivision | 47 (296–751) | 20 | 2 | 1 | 41 | 3 | ||||
| γ subdivision | 282 (185–725) | 53 | 16 | 3 | 6 | 97 | 86 | 67 | 2 | 5 |
| δ/ɛ subdivision | 15 (632–946) | 3 | 15 | |||||||
| Cytophaga/Flexibacter/Bacteroides | 15 (191–735) | 5 | 2 | 1 | 12 | |||||
| Thermotogales | 1 (241) | 1 | 1 | |||||||
| Aquificales | 2 (314) | |||||||||
| Unidentifieda | 20 (208–606) | 1 | 17 | 2 | ||||||
| Cyanobacteria | 56 (283–545) | 11 | 49 | 7 | ||||||
| Chloroplasts | 22 (216–4842) | 21 | 2 | 1 | 19 | |||||
| Euryarchaeota | 36 (162–528) | 23 | 2 | 34 | ||||||
| Crenarchaeota | 71 (129–724) | 17 | 71 |
Total sequences, 1621. Total tRNAs, 790; 302 not previously reported.
aPhylogeny not specified in the original query.
Acknowledgments
ACKNOWLEDGEMENTS
We thank I. Jarrín for her assistance in some of the statistical analyses. I.B. is holder of a doctoral grant from the Spanish Ministry of Culture, J.J.R.-S. is an IMPIVA grant holder, J.G.-M. is the recipient of a postdoctoral European Commission fellowship MAS3-CT-97-0154, UMH.DCET.DM.B, MIDAS project.
References
- 1.Burks C. (1999) Molecular Biology Database List. Nucleic Acids Res., 27, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baxevanis A.D. (2000) The molecular biology database collection: an online compilation of relevant database resources. Nucleic Acids Res., 28, 1–7. Updated article in this issue: Nucleic Acids Res. (2001), 29, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Discala C., Benigni,X., Barillot,E. and Vaysseix,G. (2000) DBcat: a catalog of 500 biological databases. Nucleic Acids Res., 28, 8–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hillis D.M. and Dixon,M.T. (1991) Ribosomal DNA: molecular evolution and phylogenetic inference. Q. Rev. Biol., 66, 411–453. [DOI] [PubMed] [Google Scholar]
- 5.Stackebrandt E. and Rainey,F.A. (1995) Partial and complete 16S rDNA sequences, their use in generation of 16S rDNA phylogenetic trees and their implications in molecular ecological studies. Molecular Microbial Ecology Manual, Vol. 3.1.1. Kluwer Academic Publishers, The Netherlands, pp. 1–17.
- 6.Normand P., Ponsonnet,C., Nesme,X., Neyra,M. and Simonet,P. (1996) ITS analysis of prokaryotes. Molecular Microbial Ecology Manual, Vol. 3.4.5. Kluwer Academic Publishers, The Netherlands, pp. 1–12.
- 7.García-Martínez, J., Acinas,S.G., Antón,A.I. and Rodríguez-Valera,F. (1999) Use of the 16S-23S ribosomal spacer region in studies of prokaryotic diversity. J. Microbiol. Methods, 36, 55–64. [DOI] [PubMed] [Google Scholar]
- 8.Gürtler V. and Stanisich,V.A. (1996) New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology, 142, 3–16. [DOI] [PubMed] [Google Scholar]
- 9.Condon C., Squires,C. and Squires,C.L. (1995) Control of rRNA transcription in Escherichia coli. Microbiol. Rev., 59, 623–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Antón A.I., Martínez-Murcia,A.J. and Rodríguez-Valera,F. (1998) Sequence diversity in the 16S-23S intergenic spacer region (ISR) of the rRNA operons in representatives of the Escherichia coli ECOR collection. J. Mol. Evol., 47, 62–72. [DOI] [PubMed] [Google Scholar]
- 11.Forsman P., Tilsala-Timisjärvi,A. and Alatossava,T. (1997) Identification of staphylococcal and streptococcal causes of bovine mastitis using 16S-23S rRNA spacer regions. Microbiology, 143, 3491–3500. [DOI] [PubMed] [Google Scholar]
- 12.Luz S.P., Rodríguez-Valera,F., Lan,R. and Reeves,P.R. (1998) Variation of the ribosomal operon 16S-23S gene spacer region in representatives of Salmonella enterica subspecies. J. Bacteriol ., 180, 2144–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Achenbach-Richter L. and Woese,C.R. (1988) ) The ribosomal gene spacer region in Archaebacteria. Syst. Appl. Microbiol., 10, 211–214. [DOI] [PubMed] [Google Scholar]
- 14.Daffonchio D., Borin,S., Frova,G., Manachini,P.L. and Sorlini,C. (1998) PCR fingerprinting of whole genomes: the spacers between the 16S and 23S rRNA genes and of intergenic tRNA gene regions reveal a different intraspecific genomic variability of Bacillus cereus and Bacillus licheniformis. Int. J. Syst. Bacteriol., 48, 107–116. [DOI] [PubMed] [Google Scholar]
- 15.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe T. and Eddy,S.R. (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed]



