Abstract
In cells infected with the Kaposi's sarcoma-associated herpesvirus (KSHV), CSL/CBF1 signaling is essential for viral replication and promotes the survival of KSHV-infected cells. CSL/CBF1 is a DNA adaptor molecule which recruits coactivator and corepressor complexes to regulate viral and cellular gene transcription and which is a major downstream effector molecule of activated Notch. The interaction of KSHV RTA and LANA with CSL/CBF1 has been shown to balance the lytic and latent viral life cycle. Here we report that a third KSHV protein, viral interferon regulatory factor 4 (vIRF4/K10), but none of the three other KSHV-encoded vIRFs, interacts with CSL/CBF1. Two regions of vIRF4 with dissimilar affinities contribute to CSL/CBF1 binding. Similar to Notch, vIRF4 targets the hydrophobic pocket in the beta trefoil domain of CSL/CBF1 through a short peptide motif which closely resembles a motif found in Notch but does not strictly follow the ΦWΦP consensus conserved in human and mouse Notch proteins. Our results suggest that vIRF4 might compete with Notch for CSL/CBF1 binding and signaling.
The Kaposi's sarcoma-associated herpesvirus (KSHV) is the etiological agent of infectious Kaposi's sarcoma (KS) and is also closely associated with the pathogenesis of primary effusion lymphoma (PEL) and multicentric Castleman's disease (MCD). Like all herpesviruses, KSHV can switch between a lytic and latent life cycle. In KSHV-infected cells, CSL/Notch signaling has been shown to promote the survival of latently infected cells and to control the balance between establishment of latency and viral reactivation (12).
Notch receptors are transmembrane proteins. Upon ligand binding, Notch receptors are proteolytically cleaved in a multistep process involving γ-secretase activity at one stage. The intracellular Notch fragment (NICD) translocates into the cell nucleus and binds to the CSL/CBF1 protein. CSL [CBF1/RBP-Jκ, Su(H), and Lag1] is a sequence-specific DNA binding factor which recruits corepressor and coactivator complexes to cis-regulatory elements of its target genes. NICD is a transcription factor which, in complex with MAM (Mastermind), activates transcription. NICD consists of an unstructured N-terminal region called RAM (RBP-Jκ-associated molecule), followed by an Ankyrin (ANK) domain, a Q-rich region, and a PEST-rich region (reviewed in reference 16).
The crystal structure of the DNA/CBF1 complex, as well as that of the DNA/CBF1 complex associated with NICD fragments, has been solved. The CBF1 protein consists of an N-terminal domain (NTD), a modified β-trefoil domain (BTD), and a C-terminal domain (CTD). CBF1 binds DNA via extended regions in NTD and BTD. NICD interacts with two regions of CBF1, BTD and CTD. Twenty-five N-terminal residues of RAM bind with high affinity to a hydrophobic pocket built by the BTD (17, 30, 39, 42). A ΦWΦP motif within this stretch of 25 amino acids, where Φ represents any hydrophobic residue, is shared by all mouse and human Notch proteins, is highly conserved across the orthologs of diverse species, and is absolutely critical for the RAM/CBF1 interaction. In addition, the ANK domain interacts with the CTD of CBF1. This interaction is weak and is stabilized by association with MAM proteins (5, 33, 34).
Notch signaling is an evolutionary conserved signal transduction pathway which regulates multiple developmental processes, including stem cell renewal. Mammalian cells can express 4 Notch receptors. Dysregulated Notch signaling has been directly linked to multiple human disorders, including cancer (16). In KSHV-infected PEL B cells and KS cells of endothelial origin, Notch signaling is active and contributes to the cellular phenotype and survival in cell culture and in transplanted tumors in vivo (3, 4, 19, 22). KSHV infection can modulate Notch signaling at multiple levels. Viral genes regulate the transcription of components of the Notch signaling pathway or directly stabilize the intracellular Notch protein and thereby enhance or prolong Notch signaling (7, 23, 29). In addition, two viral proteins, RTA and LANA, have been shown to directly bind to CBF1, the central downstream component of Notch signaling. RTA (replication and transcription activator), the lytic switch protein, is a transactivator which either directly contacts DNA or uses transcription factors as adaptors to gain access to promoters. The interaction of RTA and CBF1 controls the activation of multiple viral target genes and is absolutely critical for virus reactivation (26-28, 38). LANA, the latency-associated nuclear antigen, is a multifunctional viral oncoprotein critical for the establishment of latency. The interaction of LANA with CBF1 antagonizes RTA functions and thereby stabilizes the status of latent infection in the cell (20, 21).
Since CBF1 appears to be a signaling molecule controlling cellular survival and the viral life cycle of KSHV-infected cells, we examined whether further viral CBF1 interaction partners could be identified. By means of a yeast two-hybrid screen, we discovered viral interferon regulatory factor 4 (vIRF4/K10) as a novel CBF1 interacting partner. The CBF1/vIRF4 interaction shares several characteristic features with the CBF1/Notch interaction. vIRF4 contacts CBF1 via two regions of differential affinity. The strong CBF1-interacting region of vIRF4 includes a peptide which closely resembles the conserved CBF1 binding motif described for NICD, which contacts BTD and can abolish CBF1/NICD complex formation. Our data suggest that vIRF4 is a potential antagonist of Notch/CBF1 signal transduction.
MATERIALS AND METHODS
Plasmids.
The Gateway-compatible destination vectors pGADT7, pHACR3, and pMycCR3 (41), as well as pHACS1, pMycCs1, the NICD expression vector (pED1), the CBF1 expression vector (AJ247), and the CBF1-dependent reporter gene construct (pGA981-6), have been described previously (6, 31, 32). The open reading frames of the EEF233AAA and KLV249AAA CBF1 mutants were inserted into the XbaI/XhoI restriction sites of pHACS1, generating the plasmids BS298 and BS299. F235I, K249M, and A258V (10) were inserted into the NotI/XbaI sites of pHACS1, generating the plasmids pKF202, pKF203, and pKF204. The designations of the amino acids throughout the manuscript refer to GenBank entry NM_005340 and Swiss-Prot entry Q06330. The vectors pGBKT7 and pGADT7 (Clontech), pcDNA3.1-lacZ (Invitrogen), and pGEX-6-P1 (GE-Healthcare) are commercially available. For bacterial expression of the His-tagged BTD CBF1 fragment, the region coding for amino acids (aa) 158 to 323 of CBF1 was amplified by PCR using forward (fw) and reverse (re) primers (fw, 5′ GGCGCCATGGAGAAGCAGTCATTGAAAAATGCTG; re, 5′ AGTGGTACCTTATGCCTTATCTGTGCTAATGAT) which carry restriction sites for NcoI or KpnI and ligated into the pETM11 vector, encoding an additional Z tag, provided by Arie Gerlof.
For expression of the glutathione S-transferase (GST)-tagged RAM domain, the region coding aa 1761 to 1890 of human Notch1 was amplified by PCR using primers containing restriction sites for EcoRI and NotI (fw, 5′ CGGAATTCATGCGGCGGCAGCATGGCC; re, 5′ TTAGCGGCCGCTTTAGCAGGAGGCGATCATGAG) and cloned into the vector pGEX-6-P1.
The pENTRY constructs encoding vIRF4, vIRF1, and vIRF3 have been published (40). For construction of vIRF4 deletion mutants, the fragments were amplified by PCR using the following forward and reverse primers: [KHe188.1], aa 315 to 911 (fw, 5′ GGAGATCTTCCTGTCACGG; re, 5′ ATGTAGACTATCCCAAATG); [KHe192.1], aa 731 to 911 (fw, 5′ CAGCGGGTGTCTTTTGTTG; re, 5′ ATGTAGACTATCCCAAATG); [KHe415], aa 1 to 314 (fw, 5′ CCTAAAGCCGGTGGC; re, 5′ CCATGGAAAACACGCAGG); [KHe416], aa 1 to 240 (fw, 5′ CCTAAAGCCGGTGGC; re, 5′ AGCAGGCGGATCTCCG); [KHe417], aa 1 to 160 (fw, 5′ CCTAAAGCCGGTGGC; re, 5′ GTAGCTTTGTTCTCTAGTG); [KHe370], aa 1 to 80 (fw, 5′ CCTAAAGCCGGTGGC; re, 5′ ACGTCCATACACCGTCTG); [KHe378], aa 161 to 240 (fw, 5′ GAGGAGGCAGGGGCTG; re, 5′ AGCAGGCGGATCTCCG); [KHe461], aa 140 to 240 (fw, 5′ CTGTTGTCCCCCCCAATG; re, 5′ AGCAGGCGGATCTCCG); [KHe462], aa 161 to 200 (fw, 5′ GAGGAGGCAGGGGCTG; re, 5′ GGCAGCCTTCTGTTCAGTC); [KHe463], aa 201 to 240 (fw, 5′ TGCTCCGTGTGGATACCAG; re, 5′ AGCAGGCGGATCTCCG).
All forward and reverse primers were flanked by attB sites in the 5′ flanking region AAAAAGCAGGCTCCGCCATG (fw) or AGAAAGCTGGGTCTA (re), respectively. The PCR products were cloned by BP clonase into pDONR207 (Invitrogen). The plasmid expressing [KHe142], aa 1 to 632, was created by restriction digestion with NdeI/HindIII of pENTRY/vIRF4, Klenow treatment, and subsequent blunt-end ligation.
For expression of the vIRF4 wild-type (wt) or deletion mutants, fragments were transferred into the destination vector pHACR3, pMycCR3, or pGADT7 by LR reactions (Gateway; Invitrogen).
Point mutations and deletions in wild-type pENTRY/vIRF4 were generated by using the Phusion site-directed mutagenesis kit from New England Biolabs with the following forward and reverse primers: [KHe785.1], aa 1 to 911, W471A/P473A (fw, 5′ ACGCCGTGGTGCCGGAAGTTCGCAC; re, 5′ GACCAACTGGGGCGTGGGCAACAGGCGCG), and [KHe815], aa 1 to 911, Δ201-240 (fw, 5′ CCATCAACGCTGACCGGCCCTTCAG; re, 5′ GGCAGCCTTCTGTTCAGTCGTTATATCGC).
vIRF4/K10 expression constructs were generated by hybridizing complementary single-stranded oligonucleotides encoding short stretches of open reading frames flanked by EcoRI/XhoI restriction sites and ligated into the corresponding sites of the vectors pHACR3 and pGADT7. The following oligonucleotides were used: [KHe568.1], aa 201 to 220 (fw, 5′ GATCGAATTCTGCTCCGTGTGGATACCAGTGAATGAGGGCGCATCTACCTCAGGTATGGGGTCCTCTGGGTGACTCGAGGATC; re, 5′ GATCCTCGAGTCACCCAGAGGACCCCATACCTGAGGTAGATGCGCCCTCATTCACTGGTATCCACACGGAG CAGAATTCGATC); [KHe568.2], aa 221 to 240 (fw, 5′ GATCGAATTCACGCGACAAGTTACGCAGGCCTCGTCCTTTACGTGGCGGGTTCCCGGAG ATCCGCCTGCTTGACTCGAGGATC; re, 5′ GATCCTCGAGTCAAGCAGGCGGATCTCCGGGAACCCGCCACGTAAAGGACGAGGCCTGCGTAA CTTGTCGCGTGAATTCGATC); [KHe812], aa 466 to 478, W471A/P473A (fw, 5′ GATCGAATTCGCAATACGCGCGCCTGTTGCCCACGCCCCAGTTGGTCACGCCGGATGACTCGAGGATC; re, 5′ GATCCTCGAGTCATCCGGCGTGACCAACTGGGGCGTGGGCAACAGGCGCGCGTATTGCGA ATTCGATC); and [KHe184], aa 466 to 478 (fw, 5′ GATCGAATTCGCAATACGCGCGCCTGTTTGGCACCCACCAGTTGGTCACGCCGGATGACTCGAGGATC; re, 5′ GATCCTCGAGTCATCCGGCGTGACCAACTGGTGGGTGCCAAACAGGCGCGCGTATTGCGAATTCGATC).
For construction of pENTRY/vIRF2 [KHe486], total RNA from 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced BCBL-1 cells was isolated and reverse transcribed, and a first-round PCR was done with the gene-specific forward (5′AGCTAGTTGTGGCGAGAAAG) and reverse (5′CCCACATGGGCCTACTATAAC) primers. This PCR was followed by a second-round PCR using forward (5′AAAAAGCAGGCTCCGCCATGCCTCGCTACACGGAG) and reverse (5′AGAAAGCTGGGTCTATTAGTCTCTGTGGTAAAATGG) primers which added attB sites.
Expression of proteins in bacteria and in vitro.
The recombinant His-tagged full-length CBF1 protein has been described previously (31). The expression vectors for His fusion proteins were transformed into BL21(DE3)pLysS (Promega) Escherichia coli, and bacteria were grown at 37°C until a cell optical density at 600 nm (OD600) of 0.6 was reached. The culture was then induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), grown for 2 h at 30°C, harvested by centrifugation, and frozen. Frozen bacterial pellets were resuspended in buffer containing 50 mM Tris (pH 8.0), 150 mM NaCl, 1 M urea, and protease inhibitors (Roche) and lysed by sonication. The resulting lysate was clarified by centrifugation (15,000 × g), loaded onto Perfecto Pro nickel-nitrilotriacetic acid agarose (5Prime), preequilibrated with buffer, and then applied to chromatography columns (Bio-Rad). The column was washed with 10 to 20 volumes of the buffer, and the protein was eluted by buffer with 50 mM reduced glutathione. Glutathione and urea were removed by dialysis against buffer containing 50 mM Tris (pH 8.0), 150 mM NaCl, 2 mM dithiothreitol (DTT), and 10 mM EDTA.
The expression vector for GST fusion proteins was transformed into Rosetta(DE3)pLysS (Novagen) E. coli, and bacteria were grown in LB medium at 37°C until a cell OD600 of 0.8 was reached. The culture was then induced with 1 mM IPTG, grown for 4 h at 30°C, and harvested by centrifugation, and cell pellets were frozen. Frozen bacterial pellets were resuspended in buffer containing 50 mM Tris (pH 8.0), 150 mM NaCl, 1 M urea, and complete protease inhibitors (Roche) and lysed by sonication. The resulting lysate was clarified by centrifugation (15,000 × g), loaded onto glutathione Sepharose 4B beads (GE health care), preequilibrated with buffer, and then applied to chromatography columns (Bio-Rad). The column was washed with 10 to 20 volumes of the buffer, and the protein was eluted with 250 mM Imidazole in lysis buffer. Imidazole and urea were removed by dialysis against buffer containing 50 mM Tris (pH 8.0), 150 mM NaCl, 2 mM DTT, and 10 mM EDTA.
In vitro-translated proteins were produced using T7-coupled reticulocyte lysates (Promega).
Peptide synthesis.
The solid-phase synthesis of the peptides, including the peptides for the plaque reduction assay, was carried out on a Pioneer automatic peptide synthesizer (Applied Biosystems, Foster City, CA) employing Fmoc chemistry with O-benzyltriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU)/diisopropylethyl amine activation. Side chain protections were as follows: Asp, Glu, Ser, Thr, and Tyr, t-Bu; Asn, Gln, and His, Trt; Arg, Pbf; Lys and Trp, Boc. Coupling time was 1 h. Double couplings were carried out if a difficult coupling was expected according to the program Peptide Companion (CoshiSoft/PeptiSearch, Tucson, AZ). All peptides were generated as carboxyl amides by synthesis on Rapp S RAM resin (Rapp Polymere, Tübingen, Germany). Biotin was incorporated at the C termini of indicated peptides with Fmoc-Lys(biotin)-OH (NovaBiochem/Merck, Nottingham, United Kingdom) and TBTU/diisopropylethyl amine activation for 18 h, followed by coupling of Fmoc-β-Ala-OH for 1 h. Peptides were cleaved from the resin and deprotected by a 3 h of treatment with trifluoroacetic acid (TFA) containing 3% triisobutylsilane and 2% water (10 ml/g resin). After precipitation with t-butylmethyl ether, the resulting crude peptides were purified by preparative high-pressure liquid chromatography (HPLC) (RP-18) with water-acetonitrile gradients containing 0.1% TFA and characterized by analytical HPLC. Sequences of the peptides were as follows: Notch, biotin-β-Ala-β-Ala-RRQHGQLWFPEGF-amide (wt) and biotin-β-Ala-β-Ala-RRQHGQLLAAEGF-amide (mutant [mt]); EBNA-2, biotin-β-Ala-β-Ala-HHLPSGPPWWPPV-amide (wt) and biotin-β-Ala-β-Ala-HHLPSGPPSRPPV-amide (mt); vIRF4, biotin-β-Ala-β-Ala-IRAPVWHPPVGHAVV-amide (wt) and biotin-β-Ala-β-Ala-IAAPVAHAPVGHA-amide.
Yeast two-hybrid screen.
The ORFeome library for KSHV and the screening procedure have been described previously (40). Yeast strains AH109 and Y187 were transformed with either the ORFeome library inserted into the vector pGADT7 DEST or CBF1 inserted into pGBKT7, respectively. Y187 transformed with pGBKT7 without an insert was processed in parallel. Yeast strains were cultured, transformed, and assayed for two-hybrid interactions according to the protocols provided by Invitrogen.
Yeast two-hybrid assay.
Yeast strain AH109 was transformed with 1 μg of either vIRF4 or vIRF4 mutants inserted in the Gal4 activation domain (AD) vector (pGADT7) together with 1 μg of CBF1 inserted in the Gal4 DNA binding domain vector (DBD) (pGBKT7). Cotransformations were also done with vectors containing no insert. Yeast cells were transformed using polyethylene glycol (PEG)/lithium acetate (LiAc) solution (40% PEG, 100 mM LiAc, 10 mM Tris, 1 mM EDTA, pH 7.5) together with salmon testes DNA. Transformed cells were plated onto synthetic complete medium lacking leucine and tryptophan. Representative colonies were replica plated onto medium lacking leucine, tryptophan, and adenine or histidine in the presence of different concentrations of 3-aminotriazole (3-AT).
Cell lines, culture conditions, and induction of the lytic cycle.
The cell lines BC-1 (2), BCBL-1 (35), HEK293 (11), and DG75 knockout (KO) (SM224.9) (31) have been described previously.
Cell lines were grown in 1640 RPMI supplemented with 10% fetal calf serum, 100 U penicillin per ml, 100 μg streptomycin per ml, and 4 mM glutamine and maintained at 37°C in a 5% CO2 atmosphere. BC-1 and BCBL-1 were supplemented with 20% fetal calf serum. For lytic cycle activation, cells were treated with 3 mM sodium butyrate (NaB) and/or 20 ng/ml TPA as appropriate.
Transfection of cells.
On the day of transfection, medium of HEK293 cells, plated on 10-cm dishes, was replaced by Optimem. Cells were transfected with 5 μg of each plasmid and 12 μg polyethylenimine (Sigma) in the presence of Optimem. After 4 h, the medium was replaced with RPMI 1640 cell culture medium containing 10% fetal bovine serum, and cells were further cultured for 24 h before harvesting. DG75 wild-type or KO (SM224.9) cells (1 × 107) were resuspended in 250 μl RPMI 1640 and transfected with 5 μg of each plasmid DNA by electroporation (220 V, 950 μF) using a Gene Pulser II system (Bio-Rad). Cells were harvested 48 h after transfection.
Antibodies.
The CBF1-specific rat monoclonal antibody RBJ-7A11 has been described previously (31). In order to produce rat monoclonal antibodies, a vIRF4 fragment consisting of the first 313 amino acids was expressed as a GST-tagged fusion protein, purified by glutathione Sepharose affinity chromatography, and used to immunize rats as described previously (18). The entire open reading frame of CBF1 was expressed as a His-GST tandem-tagged fusion protein and purified by Ni chelate affinity. The vIRF4-specific antibody vIRF-8F8 was used for immunoprecipitation and immunoblotting. The CBF1-specific antibody RBJ-6E7 was used for immunoprecipitation, the antibody RBP-7A11 was used for immunoblotting, and the antibody RBJ-1F1 was used for supershift experiments in electrophoretic mobility shift assays (EMSAs). The rat hemagglutinin (HA)-tag-specific antibody was purchased from Roche. The mouse Myc-tag-specific antibody was 9E10-derived supernatant. The His-specific mouse antibody was N-His 2F12 supernatant (E. Kremmer, unpublished).
Immunoprecipitation and immunoblotting.
Cells were lysed in NP-40 lysis buffer (1% NP-40, 140 mM NaCl, 5 mM MgCl2, 20 mM Tris, pH 7.6) supplemented with complete protease inhibitor cocktail (Roche) and sonicated. Total cell extracts were directly loaded onto the gel for immunoblotting or used for coimmunoprecipitation by adding 100 μl of hybridoma supernatant or 1 μg of purified antibody for 1 h on ice, followed by protein G-coupled Sepharose beads and incubation overnight. Immunoprecipitates were washed 3 times with NP-40 lysis buffer, Laemmli buffer was added to the beads, and the samples were boiled, submitted to electrophoresis by SDS-PAGE, and analyzed by immunoblotting on polyvinylidene difluoride (PVDF) membranes; proteins were detected by specific antibodies and the chemiluminescence system (Amersham Biosciences) according to the manufacturer's instructions.
Affinity precipitation of protein complexes by biotinylated oligonucleotides.
Five hundred micrograms cell extract of TPA and NaB-treated or untreated BC-1 cells was incubated for 10 min at 4°C with binding buffer (10 mM Tris-HCl, pH 8, 40 mM KCl, 1 mM dithiothreitol [DTT], 6% glycerol, 0.05% NP-40) and 20 μl of poly(dIdC) in a final volume of 1 ml. One μg of a biotin-labeled oligonucleotide carrying the CBF1 recognition motif of the Epstein-Barr virus (EBV) C-promoter (fw, 5′ AAGTTGGTGTAAACACGCCGTGGGAAAAAATTTATGGTT, re, 5′ GAACCATAAATTTTTTCCCACGGCGTGTTTACACCAACTT) was added and incubated for 4 h at 4°C. As a control, we used a nucleotide sequence carrying the C/EBP binding site (fw, 5′ TTATGTTAAGAATTTGTGCAAGTCTACCGTATT; re, 5′ AATACGGTAGACTTGCACAAATTCTTAACATAA).
Then, 50 μl of a 50% slurry of streptavidin-coated agarose beads (Invitrogen) was added, followed by 2 h of incubation at 4°C. Beads containing bound complexes were washed twice with Tris-EDTA (100 mM NaCl) and twice with binding buffer before adding Laemmli buffer. Samples were submitted to electrophoresis by SDS-PAGE and analyzed by immunoblotting.
Hybridization of peptide arrays with radiolabeled probes.
Peptides representing the entire open reading frame of vIRF4 as overlapping 15-mers were synthesized on a cellulose membrane as described previously (1). Nine μg plasmid DNA encoding CBF1 in pMycCS1 was in vitro transcribed and translated in the presence of [35S]methionine using the TNT-T7-coupled reticulocyte lysate (Promega). Membranes were activated with 100% ethanol, washed with TBS (138 mM NaCl, 2.7 mM KCl, 50 mM Tris base), blocked in blocking buffer (5 M NaCl, 1 M Tris base, pH 6.8, 2.5% skim milk powder, 0.05% Tween 20) overnight, and saturated with 100 mM cysteine in blocking buffer for 30 min. Membranes were incubated with the radiolabeled CBF1 probe in blocking buffer for 3 h. Membranes were washed for three times with TBS-T (TBS plus 0.05% Tween 20). Signals indicating protein binding were quantified on a phosporimager using the AIDA software program. The results are given as arbitrary units.
Reporter gene assays.
Transfections were done in triplicate, and 1 × 107 cells were transfected with a mixture of 5 μg luciferase reporter plasmid pGa981-6, 1 μg pcDNA3.1-lacZ, and 5 μg of each expression plasmid. The DNA amounts were adjusted by adding the corresponding empty vector. Cells were harvested 48 h after transfection, washed with phosphate-buffered saline (PBS), lysed in 100 μl LUC extraction buffer (10% [wt/vol] glycerin, 1% [wt/vol]) Triton X-100, 2 mM EDTA, 25 mM Tris-HCl, pH 7.8, 2 mM DTT). Cell debris was removed by centrifugation at 4°C and 16,000 × g for 15 min. Luciferase activity was determined using 10 μl of the clarified lysates in duplicate, followed by injection of 50 μl luciferase assay buffer [20 mM Tricine, 1.07 mM 4MgCO3· Mg(OH)2· 5H2O), 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM DTT, 270 μM coenzyme A, 470 μM luciferin, 530 mM ATP]. Luciferase activity was normalized for variations in transfection efficiencies by cotransfection of the cytomegalovirus (CMV) β-galactosidase (β-Gal) reporter plasmid (pcDNA3.1-lacZ). For the β-galactosidase assay, 10 μl of the lysates in duplicates were incubated for 20 min at room temperature with 100 μl β-Gal assay buffer (100 mM Na-P, pH 8.0, 1% Galacton [Tropix, Bedford, MA], 1 mM MgCl2). The reaction was started by adding 50 μl of β-Gal acceleration buffer (0.2 M NaOH, 10% Emerald enhancer) and measured in a luminometer (LB9501; Berthold).
EMSA.
The oligonucleotide Cp (SM316Cs, 5′-AAGTTGGTGTAAACACGCCGTGGGAAAAAA-3′; SM316Cas, 5′-GAACCATAAATTTTTTCCCACGG-3′) was annealed by mixing of equimolar ratios of sense and antisense oligonucleotides in annealing buffer (10 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 50 mM NaCl), incubation at 90°C for 10 min, and cooling down to 37°C. Twenty-five ng of the annealed oligonucleotides were blunt ended by Klenow polymerase in the presence of lsqb]α-32P]dCTP (3,000 Ci/mmol) and unlabeled dATP, dGTP, and dTTP at 37°C for 1 h. The labeled probe was separated from unincorporated nucleotides using Nick Sephadex G50 columns (Amersham Biosciences). A 0.5- to 1-ng amount of 32P-labeled oligonucleotide probe was incubated in a 20-μl reaction mixture containing 10 mM HEPES (pH 7.9), 1 mM EDTA, 200 mM KCl, 4% Ficoll, 2 μg bovine serum albumin (BSA), 2 μg poly(dI-dC), 4 mM DTT, and proteinase inhibitors, with or without competitive unlabeled oligonucleotide, and 5 pmol recombinant purified CBF1 protein or 40 pmol RAM protein protein-DNA complexes were resolved on 6% polyacrylamide gels in 1× Tris-borate-EDTA (TBE) buffer at room temperature for 3 h at 130 V. Gels were dried and exposed to X-ray films or Fujifilm imaging plates. After the Fujifilm imaging plates were scanned with the phosphorimager Fujifilm FLA-5100, the quantification of EMSA gels was performed using AIDA image analyzer software.
Peptide affinity precipitation.
Peptides were dissolved in PBS at a concentration of 5 mM. Streptavidin beads (Invitrogen) were washed with PBS-0.1% Triton X. Streptavidin beads were coated with 50 nM peptide per 1 ml beads for 3 h at 4°C, washed with PBS-0.1% Triton X, and subsequently dissolved as 50% slurry. For affinity precipitation, 500 pM of BTD-CBF1 was incubated with 40 μl streptavidin beads and preequilibrated with pulldown buffer containing 150 mM KCl, 20 mM HEPES, pH 7.9, 0.2 mM EDTA, 0.2% Triton X, and 25% (vol/vol) glycerol in a volume of 1 ml for 3 h at 4°C. Precipitates were washed with pulldown buffer, Laemmli buffer was added to the beads, and the samples were boiled, submitted to electrophoresis by SDS-PAGE, and analyzed by immunoblotting on PVDF membranes, and proteins were detected by specific antibodies with the chemiluminescence system (Amersham Biosciences) according to the manufacturer's instructions.
RESULTS
vIRF4 is a novel viral CBF1 binding partner.
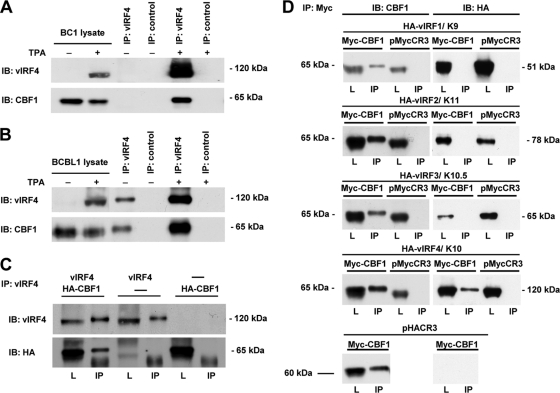
In order to screen for additional KSHV proteins, which bind to CBF1, we carried out a yeast two-hybrid screen using an ORFeome library of KSHV fused to the GAL4 activation domain (40, 8). The CBF1 sequence was fused to the GAL4 DNA binding domain and used as bait. Out of a group of several candidates, vIRF4 was consistently identified in different independent experiments (data not shown). To verify that this interaction can also be found in KSHV-infected cells, we performed coimmunoprecipitations using EBV plus KSHV-positive BC-1 cells and KSHV-positive BCBL-1 cells. Since vIRF4 is known to be expressed with early kinetics after reactivation of the virus, BC-1 and BCBL-1 cells were treated with sodium butyrate (NaB) and/or 12-O-tetradecanoylphorbol-13-acetate (TPA), and the cell lysates were examined for complex formation (Fig. 1A and B). Indeed, CBF1 coprecipitated with vIRF4-specific antibodies in lysates derived from both cell lines. The vIRF4 precipitates and CBF1 coprecipitates detected in untreated BCBL-1 cells are likely to be derived from spontaneously reactivating cells frequently seen in this cell line. We thus confirmed that vIRF4 is a novel viral interaction partner of CBF1 in KSHV-infected cells. Although the yeast two-hybrid system typically identifies binary interactions not mediated by additional bridging proteins, we next wished to confirm that the interaction can be formed by in vitro-translated proteins generated in reticulocyte lysates which are most likely devoid of any bridging factors critical for complex formation (Fig. 1C). In vitro translation products of both interaction partners, vIRF4 and HA-tagged CBF1, were examined for complex formation by coprecipitation. The CBF1 coprecipitation by the vIRF4-specific antibody was confirmed and shown to be strictly dependent on vIRF4 in the sample, thereby excluding a potential cross-reactivity of the vIRF4-specific antibody to CBF1.
FIG. 1.
CBF1 interacts directly and specifically with vIRF4. BC-1 (A) or BCBL-1 (B) cells (2 × 107) were treated with TPA or TPA plus NaB for 48 h to induce the lytic cycle or were left untreated. Total cellular protein extracts were used for immunoprecipitation (IP) and immunoblotting (IB) using the vIRF4-specific rat monoclonal antibody vIRF-8F8 or CBF1-specific rat monoclonal antibody RBJ-6E7 for IP and RBP-7A11 for IB. The protein amount loaded onto the lysate lanes corresponds to 2% of cell extracts analyzed by immunoprecipitation. (C) In vitro-translated vIRF4 and HA-CBF1 were submitted to immunoprecipitation and immunoblotting using antibodies specific for vIRF4 and HA. (D) HA-tagged vIRF1, vIRF2, vIRF3, and vIRF4 plus Myc-tagged CBF1 were coexpressed in HEK 293 cells by transfection of 5 μg of each expression construct. pMycCR3 and pHACR3 are the corresponding expression vectors carrying no insert. Complex formation was tested by coimmunoprecipitation using the Myc-specific antibody and immunoblotting by CBF1- and HA-specific antibodies. The amount of protein lysate (L) corresponds to 2% of the protein amount of the sample used for immunoprecipitation (IP).
We next wished to test whether, in addition to vIRF4, other viral IRFs interact with CBF1. We therefore coexpressed HA-tagged vIRF1/K9, vIRF2/K11, vIRF3/K10.5, or vIRF4/K10 in HEK293 cells and tested for complex formation with CBF1 by immunoprecipitation. While vIRF4 readily coprecipitated with CBF1, none of the other vIRFs could do so (Fig. 1D). Thus, vIRF4 appears to be the only vIRF interacting with CBF1.
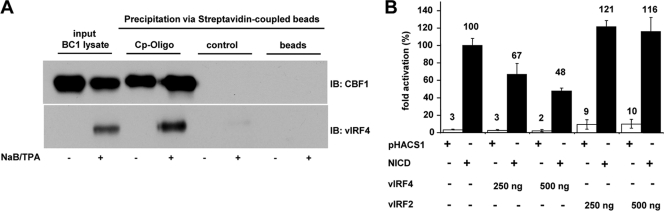
Since CBF1 is a DNA anchor protein, which can recruit further proteins to DNA, we tested whether vIRF4/CBF1 binding is compatible with CBF1/DNA complex formation by DNA affinity purification of vIRF4/DNA complexes. Our results showed that biotinylated oligonucleotides carrying the CBF1 binding site of the EBV C promoter (Cp) efficiently precipitated CBF1 and coprecipitated vIRF4 from BC-1 lysates induced for viral reactivation while an oligonucleotide carrying a C/EBP recognition site was used as a control (Fig. 2A).
FIG. 2.
vIRF4 can bind to CBF1/DNA complexes and interfere with Notch driven reporter gene activation. (A) BC-1 cells were treated with TPA and NaB for 24 h or left untreated. Protein extracts were harvested and a biotin labeled oligonucleotide derived from the EBV C-promoter carrying the CBF1 recognition motif was added. In order to control for a potential unspecific binding of vIRF4 to DNA a biotin labeled oligonucleotide carrying a C/EBP recognition site was analyzed in parallel. DNA-protein complexes were precipitated using streptavidin-coupled beads. CBF1 and vIRF4 were visualized by immunoblotting using the monoclonal antibodies (RBP-7A11) and (vIRF-8F8), respectively. (B) HEK 293 cells were cotransfected with 500 ng Notch fragment NICD expression constructs or control vectors (pHACS1) and a CBF1-dependent luciferase reporter gene (pGa981-6). Two hundred fifty or 500 ng of the HA-tagged vIRF4 or vIRF2 expression constructs were cotransfected when indicated. The results represent triplicates and are shown as relative transactivation compared to Notch transactivation, which is set to 100%. Standard deviations are shown as error bars. All results were normalized for β-galactosidase activity derived from the reporter construct pcDNA3.1-LacZ, included in each sample.
Since CBF1 is a central downstream component of Notch signaling, we tested if vIRF4 has the potential to interfere with Notch signal transduction. Taking advantage of the fact that the intracellular Notch fragment (NICD) can be used to mimic constitutively active Notch receptors, a CBF1-dependent reporter construct was coexpressed with vIRF4, NICD, or both proteins. While NICD activated the CBF1-dependent reporter gene, vIRF4 did not significantly modulate reporter gene expression. However, coexpression of NICD and vIRF4 but not vIRF2 significantly diminished reporter gene activity, indicating that vIRF4 might potentially interfere with Notch signaling (Fig. 2B). We thus speculated that vIRF4 might bind to CBF1 regions which are also targeted by Notch.
Two regions of vIRF4 contribute to CBF1 binding.
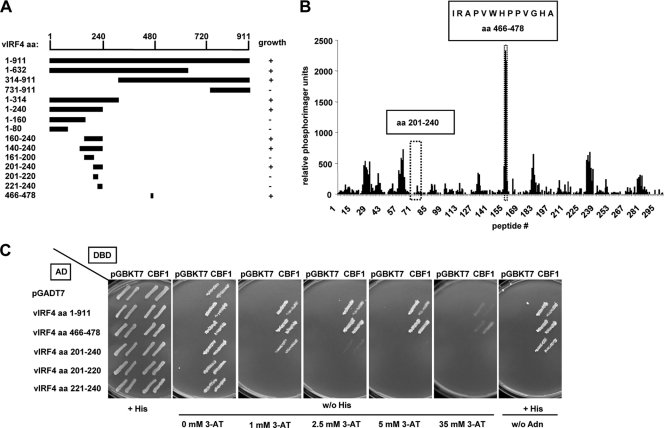
In order to map the CBF1 binding regions of vIRF4, two experimental approaches, yeast two-hybrid binding assays and hybridization of peptide arrays, were followed in parallel (Fig. 3).
FIG. 3.
CBF1 interacts with two regions of vIRF4. (A) Schematic overview on results obtained with yeast. A series of vIRF4 deletion constructs was generated, and the truncated protein was coexpressed as a GAL4 activation domain (AD) fusion with CBF1 GAL4 DNA binding domain fusion proteins (DBD) in the yeast strain AH109. Transformants were plated on semisolid synthetic medium lacking leucine and tryptophan. Representative colonies on these permissive plates were replica plated onto selective medium lacking histidine and supplemented with 2.5 mM 3-AT or lacking adenine and scored for growth. (B) Peptides representing the entire open reading frame of vIRF4 were synthesized and spotted onto membranes. The peptides were 15 amino acids in length and overlapped by 12 amino acids. CBF1 was in vitro transcribed and translated in the presence of [35S]methionine. The translation product was used as a probe. Signals indicating protein binding were quantified on a phosporimager using the AIDA software program, and the results are given as arbitrary units. The region corresponding to aa 466 to 478 (peptides 154 to 156) gave the best signals. Surprisingly, a second region, corresponding to aa 201 to 240 (peptides 64 to 80), which had scored for binding in the yeast two-hybrid experiment, did not score in this assay. (C) Two representative colonies of all transformants expressing the indicated vIRF4 fragments were grown on permissive synthetic medium and then replica plated onto selective plates lacking histidine supplemented with increasing amounts of 3-AT or lacking adenine and tested for interaction. pGADT7 and pGBKT7 are the yeast expression vectors carrying no insert.
For the yeast two-hybrid approach, a series of deletion constructs of vIRF4 encompassing N-terminal, C-terminal, and internal vIRF4 fragments was generated. These vIRF4 fragments were coexpressed in the yeast strain AH109 as GAL4 activation domain (AD) fusions together with CBF1 fused to the GAL4 DNA binding domain (DBD). These transformants and the respective controls were plated on permissive plates lacking leucine and tryptophan. Through replica plating, they were transferred to selective plates lacking adenine or histidine and supplemented with increasing amounts of 3-aminotriazole (3-AT). Interactions were scored as positive if they promoted colony growth on adenine-free plates and on histidine-free plates supplemented with at least 1 mM 3-AT (Fig. 3A). This analysis revealed that a fragment within the N terminus of vIRF4 consisting of aa 201 to 240 was sufficient to mediate weak CBF1 binding (Fig. 3C). In addition, a C-terminal fragment (aa 314 to 911) also promoted growth of CBF1-cotransformed colonies (data not shown). In order to map this secondary binding region further, an array of overlapping peptide fragments (15-mer peptides overlapping by 12 amino acids) encompassing the entire open reading frame of vIRF4 was synthesized on a cellulose membrane by SPOT synthesis (9) and probed with in vitro-translated 35S-labeled CBF1. The resulting signals were quantified by phosphorimaging (Fig. 3B). Strong signals were obtained for peptides no. 154 to 156, which share the 13-amino-acid sequence IRAPVWHPPVGHA, corresponding to aa 466 to 478 of vIRF4. Surprisingly, the region aa 201 to 240, which had shown weak interactions in the yeast two-hybrid system, did not score on the peptide array. Both regions were reexamined in yeast two-hybrid experiments (Fig. 3C). Region aa 466 to 478 coexpressed with CBF1 permitted yeast growth in histidine-free medium supplemented with up to 35 mM 3-AT and thus scored as strong as the complete open reading frame of vIRF4. In contrast, aa 201 to 240 could support yeast growth only in medium containing 2.5 mM 3-AT. Subfragments of aa 201 to 240 did not allow yeast growth. In summary, aa 466 to 478 appears to be a strong CBF1-interacting region, while aa 201 to 240 is sufficient to bind to CBF1 weakly and might have an auxiliary function.
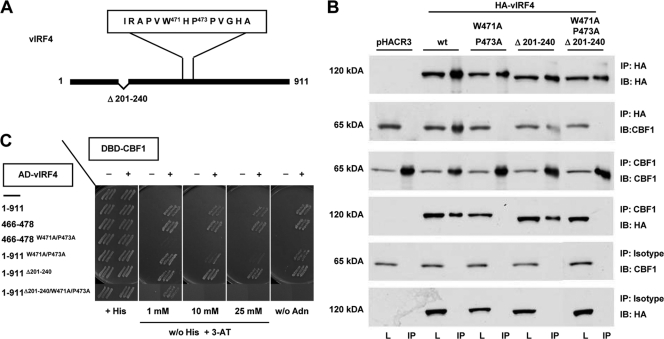
Next, we examined the molecular details of the interaction of aa 466 to 478 with CBF1. A library of mutant peptides replacing each single amino acid of the 13-residue peptide (aa 466 to 478) with an alanine was synthesized, arrayed on filter paper, and tested for interaction with 35S-labeled in vitro-translated CBF1. Alanine substitutions for tryptophan 471 (W471A) or proline (P473A) resulted in a significant loss of binding affinities (data not shown). A 13-residue peptide as depicted in Fig. 4A, carrying both substitutions (aa 466 to 478, W471A/P473A), was tested in yeast two-hybrid assays, and the loss of CBF1 binding was confirmed (Fig. 4B, row 4).
FIG. 4.
The high-affinity CBF1 binding motif in vIRF4 closely resembles the motif found in the Notch-RAM domain. (A) Schematic drawing of the vIRF4 regions targeted by mutagenesis. (B) HEK 293 cells were transfected with 10 μg expression plasmids encoding HA-vIRF4 and the respective HA-tagged mutants. CBF1 binding was tested by immunoprecipitation using HA-specific antibodies and immunoblotting by CBF1-specific antibody (RBP-7A11) or immunoprecipitation using the CBF1-specific RBJ-6E7 antibody and the HA-specific antibody for immunoblotting. Isotype controls, matching the HA-specific and RBJ-6E7 antibody, were performed in parallel. (C) AH209 yeast cells cotransformed with vectors expressing AD-vIRF4 or AD-vIRF4 mutants and DBD-CBF1 or the respective control vectors were tested for growth on selective medium supplemented with increasing amounts of 3-AT.
In order to test if the two binding regions are indeed relevant in the context of the complete open reading frame, a series of vIRF4 mutants was generated which carried either a deletion in position aa 201 to 240 (1-911Δ201-240), the alanine replacement mutations (1-911W471A/P473A), or both mutations (1-911Δ201-240/W471A/P473A). The CBF1 binding capacities of these mutants were tested in parallel in yeast two-hybrid assays and by coprecipitation in HEK293 cells (Fig. 4B and C). CBF1 binding of 1-911Δ201-240 was not severely impaired according to the yeast two-hybrid assay or coprecipitation experiments. In contrast, neither 1-911W471A/P473A nor 1-911Δ201-240/W471A/P473A coprecipitated CBF1 (Fig. 4B).
According to the yeast two-hybrid experiments, mutant 1-911W471A/P473A still retained a residual CBF1 binding activity, since yeast transformants grew in the presence of 10 mM 3-AT, as opposed to wild-type transformants, which grew on medium supplemented with 25 mM 3-AT (Fig. 4C). In contrast, 1-911Δ201-240/W471A/P471A yeast transformants did not grow on histidine-free medium supplemented with more than 1 mM 3-AT or on medium lacking adenine, indicating that aa 201 to 240 can contribute to the avidity of the vIRF4/CBF1 interaction. In summary, our data suggest that vIRF4 and CBF1 associate by a binary interaction but the WXP motif provides the dominant binding site.
vIRF4 contacts regions of CBF1 which are also critical for Notch binding.
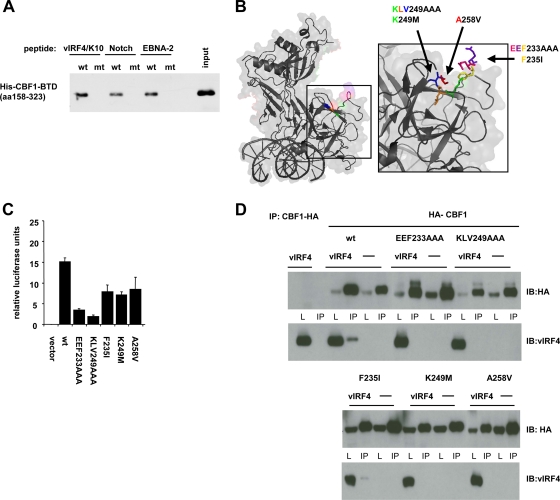
Interestingly, the sequence of the 13-residue vIRF4 peptide which appears to constitute the high-affinity binding site for CBF1 closely resembles peptides within the Notch RAM domain and the EBNA-2 conserved region 6 which are critical for CBF1 binding (Fig. 4A). Both peptides carry critical W and P residues in close proximity. However, the vIRF4 peptide (IRAPVWHPVGHA) does not perfectly match the consensus motif ΦWΦP of the Notch and EBNA-2, since the position of the second hydrophobic residue is occupied by a positively charged histidine. The ΦWΦP motifs of both Notch and EBNA-2 interact with a hydrophobic pocket on the surface of CBF1-BTD. To test whether the vIRF4 peptide also contacts the BTD, BTD (CBF1 fragment aa 158 to 323) was heterologously expressed in bacteria, and biotinylated vIRF4, Notch, or EBNA-2 peptides were used to affinity precipitate the BTD fragment of the CBF1 protein (Fig. 5A). These experiments demonstrated that all peptides which carry the conserved WXP residues readily precipitated the BTD while mutants with substitution of W and/or P were inactive.
FIG. 5.
vIRF4 and the Notch Ram domain target similar sites in CBF1. (A) The BTD of His-CBF1 (aa 158 to 323) was expressed in bacteria, and the purified protein was incubated with biotinylated peptides derived from vIRF4/K10, human Notch1, or EBNA-2. The conserved W and P residues were replaced in mutant peptides. Peptide/CBF1 complexes were affinity precipitated on streptavidin beads, and CBF1 was detected using a His-specific antibody in Western blots. (B) Illustration of the localization of the 3 mutations inserted into the CBF1 protein. (C) CBF1-negative DG75 cells (SM224.9) were transfected with 5 μg of expression plasmids for NICD and CBF1 (wt) or the respective CBF1 mutants and 3 μg CBF1-dependent luciferase reporter gene construct (pGa981-6). Results are shown as the means and standard deviations of triplicate experiments as relative luciferase units (RLU) normalized for β-galactosidase activity derived from the reporter construct (pcDNA3.1-lacZ) included in each sample. (D) HEK 293 cells were cotransfected with 5 μg of expression vectors for vIRF4 and HA-tagged CBF1 and the indicated CBF1 mutants (EEF233AAA, KLV249AAA, F235I, K249M, adn A258V). Immunoprecipitations were performed using an HA-specific antibody. For immunoblotting, HA-specific and vIRF4-specific antibodies were used.
Mutations located within the loop of the BTD or the hydrophobic pocket have been previously described to abolish the CBF1-Notch interaction (7, 11). These mutants include the CBF EEF233AAA, KLV249AAA, F235I, K249M, and A258V mutants (Fig. 5B). If vIRF4 and Notch target similar residues of CBF1, we expected that these loss-of-function mutants should be impaired for interaction with vIRF4 as well. We first confirmed the phenotype of the mutants we wanted to use. Since NICD/CBF1 coprecipitations work poorly in our hands, we performed luciferase assays using a CBF1-dependent reporter gene system. We have recently generated a CBF1-negative human DG75 B cell line by homologous recombination. NICD and CBF1 or CBF1 mutants were coexpressed in CBF1-negative DG75 cells. As expected, wild-type CBF1 reconstituted promoter activation, EEF233AAA and KLV249AAA mutants were strongly impaired in supporting reporter gene activation, and mutants carrying single amino acid substitutions were significantly impaired (Fig. 5C). Formation of coprecipitates was tested by coexpression of vIRF4 and the respective HA-tagged CBF1 mutants in HEK293 cells using an HA-specific antibody and immunoblotting (Fig. 5D). Indeed, both triple amino acid mutants, carrying EEF233AAA and KLV249AAA, known to abolish NICD binding, could not precipitate vIRF4, while wild-type CBF1 readily did so. The single-amino-acid substitution K249M and A258V mutants did not bind to vIRF4 either, while the F235I mutant retained residual vIRF4 binding activity. In summary, these results indicated that vIRF4 binds to CBF1 via the hydrophobic pocket in BTD and thus might compete for NICD/CBF1 complex formation.
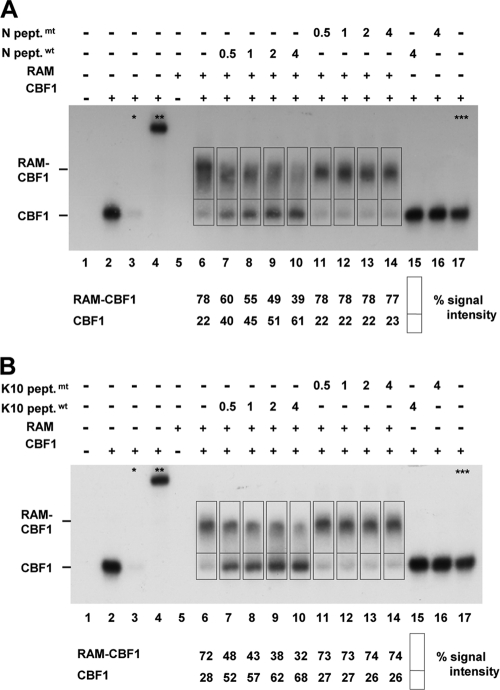
In order to directly prove that the vIRF4 peptide can interfere with NICD/CBF1 complex formation on DNA, we performed gel retardation experiments using bacterially expressed binding partners (Fig. 6). CBF1 and RAM complex formation on radiolabeled Cp oligonucleotides was tested in the presence of Notch- or vIRF4-derived peptides and mutant controls, which had also been used for affinity studies. All peptides efficiently and specifically abolished complex formation, confirming that the biological activities of all peptides are equivalent and histidine is tolerated in the position occupied by a nonpolar residue in the consensus sequence of most Notch proteins.
FIG. 6.
The vIRF4 peptide efficiently competes with RAM for CBF1 complex formation on DNA. Complex formation of full-length CBF1 and the Notch fragment RAM was tested in gel retardation assays. (A) The 32P-labeled Cp oligonucleotide was incubated with 5 pmol of purified CBF1 (lane 2). *, competition was performed with 100-fold excess of the unlabeled Cp oligonucleotide (lane 3); **, a CBF1-specific rat antibody was added to confirm CBF1/Cp complex formation (lane 4). Forty pmol purified RAM was added (lanes 5 to 14), and complex formation was analyzed in the presence of increasing amounts of a Notch peptide (N pept.wt; lanes 7 to 10 and 15; numbers indicate pmol) or a mutant Notch peptide (N pept.mt; lanes 11 to 14 and 16; numbers indicate pmol). ***, as a control, GST was added to lane 17. For the experiment shown in panel B, the Notch peptides were replaced by vIRF4/K10 peptides (K10 pept.wt, lanes 7 to 10 and 15) or mutant peptide (K10 pept.mt, lanes 11 to 14 and 16).
DISCUSSION
In this study, we describe the interaction of CBF1 with the early lytic vIRF4 protein. KSHV encodes a cluster of genes designated vIRFs since their open reading frames all share limited sequence homology with cellular IRFs (36). vIRF1, -2, and -3 have been shown to modulate interferon (IFN) induction and responsiveness and p53 and NF-κB activity (24). vIRF4 localizes predominantly to the nucleus in KS and PEL but can also be found in the cytoplasm cells in of MCD samples (15). Similarly, transient transfection of vIRF4 expression constructs into HEK293 cells generally results in a nuclear staining pattern. Interestingly, vIRF4 can interact with at least 15 KSHV proteins (40), and coexpression of vIRF4 with viral binding partners can influence its subcellular localization, revealing an intriguing plasticity of vIRF4 localization in response to binding partners (37). So far, vIRF4 is not assumed to be involved in IFN signaling. It interacts with the poly(A) binding protein (PABP) and could potentially interfere with host translation (15). In addition, vIRF4 stabilizes MDM2 by preventing its autoubiquitination and thereby indirectly promotes p53 degradation (25).
Although we do not provide direct structural data sets, our results strongly indicate that complexes of vIRF4 or Notch with CBF1 share important characteristics. The vIRF4/CBF1 interaction can be reconstituted in vitro and appears not to require additional adaptor proteins. However, the Notch/CBF1 interaction can also be reconstituted in vitro, but it is known that the mastermind protein is an integral component of Notch/CBF1 signaling (16). Thus, our data do not exclude the option that additional viral or cellular proteins support the vIRF4/CBF1 interaction. In particular, in EBV and KSHV doubly infected PEL cell lines, CBF1 might be a major hub which integrates viral and cellular signals by direct protein-protein interactions. Biophysical studies have shown that the ΦWΦP motif flanked by a stretch of basic amino acids within the RAM region of Notch is the primary site of high-affinity binding of NICD to CBF1 (14, 30). Parallel studies in yeast two-hybrid assays and coimmunoprecipitation showed that vIRF4 interacts with CBF1 via one high-affinity binding site consisting of not more than 13 amino acids and a secondary low-affinity binding site of 40 amino acids. The W and P residues within the high-affinity binding fragment are critical for the CBF1 interaction with BTD of the 13-residue vIRF4 peptide and of the entire vIRF4 protein. However, according to the results of the very sensitive semiquantitative yeast two-hybrid assay, a complete loss of CBF1 binding could be achieved only by alanine substitution for the W(471) and P(473) residues combined with the 201-to-240 deletion. Indeed, RTA also targets CBF1 via two nonconsecutive regions within its primary sequence (26).
In addition, our study also showed that vIRF4 cannot bind to CBF1 mutants which carry inactivating mutations located in the hydrophobic pocket of the BTD which prevent Notch binding. Thus, both proteins are likely to target the same region of the hydrophobic pocket, even if the ΦWΦP motif is not strictly conserved in vIRF4, where the second Φ residue is occupied by the positively charged histidine. Comparison of Notch orthologues reveals that Caenorhabditis remanei and C. briggsae Notch proteins do not obey the ΦWΦP rule since they carry an uncharged nonpolar threonine at the respective site (14). Indeed, a closer look at the published CBF1/RAM structure reveals that the respective hydrophobic residue is not oriented toward the hydrophobic pocket (42). Our gel retardation assays show that the vIRF4 peptide can specifically interfere with CBF1/RAM complex formation. These observations are further supported in reporter gene assays where vIRF4 interferes with CBF1-dependent Notch transactivation. In summary, it appears that vIRF4 and NICD binding to CBF1 will be mutually exclusive and vIRF4 can interfere with promoter activation by NICD via direct competition.
Both human gammaherpesviruses, Epstein-Barr virus (EBV) and KSHV, express viral factors which interact with CBF1, the central downstream element of Notch signaling. In EBV latently infected B cells, the Epstein-Barr virus nuclear antigens 2, 3A, 3B, and 3C interact with CBF1 to activate resting B cells and establish latent infection (13). In KSHV-infected cells, lytic cycle activation by RTA is CBF1 dependent and activation of multiple viral RTA target genes requires CBF1 response elements (38). In addition, LANA and RTA interactions with CBF1 might establish an autoregulatory loop which could stabilize the status of latency in infected cells (20). However, CBF1 is not required for the establishment of latency in KSHV-infected cells (27).
Up to now, there have been no X-ray structural data available describing CBF1 in complexes with viral proteins. However, good biochemical and biophysical evidence indicates that EBNA-2 also targets the CBF1 BTD through a small fragment carrying a ΦWΦP motif. Such detailed analysis is not available for RTA, LANA, or any of the EBNA-3 proteins. How vIRF4 modulates CBF1-dependent signaling in KSHV-infected cells, whether it directly modulates viral and cellular gene expression programs or rather interferes with the activity of other transcription factors targeting the hydrophobic pocket of CBF1, remains to be studied.
Acknowledgments
We thank Armin Baiker, Georg Malterer, Thomas Schulz, Arie Geerlof, and Diane Hayward for plasmids and cell lines and Susanne Daenicke for expert help in peptide array synthesis.
We thank the LifeScience Stiftung for financial support. B.A.S. and A.N. were supported by the Deutsche Jose Carreras Leukemie Stiftung (project DJCLS R 07/11), and J.H. was supported by Bayerisches Staatsministerium für Wissenschaft, Kultur und Kunst (Bayerisches Genomforschungsnetzwerk) and DFG SFB 576.
Footnotes
Published ahead of print on 22 September 2010.
REFERENCES
- 1.Beutling, U., K. Stading, T. Stradal, and R. Frank. 2008. Large-scale analysis of protein-protein interactions using cellulose-bound peptide arrays. Adv. Biochem. Eng. Biotechnol. 110:115-152. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 3.Curry, C. L., L. L. Reed, E. Broude, T. E. Golde, L. Miele, and K. E. Foreman. 2007. Notch inhibition in Kaposi's sarcoma tumor cells leads to mitotic catastrophe through nuclear factor-kappaB signaling. Mol. Cancer Ther. 6:1983-1992. [DOI] [PubMed] [Google Scholar]
- 4.Curry, C. L., L. L. Reed, T. E. Golde, L. Miele, B. J. Nickoloff, and K. E. Foreman. 2005. Gamma secretase inhibitor blocks Notch activation and induces apoptosis in Kaposi's sarcoma tumor cells. Oncogene 24:6333-6344. [DOI] [PubMed] [Google Scholar]
- 5.Del Bianco, C., J. C. Aster, and S. C. Blacklow. 2008. Mutational and energetic studies of Notch 1 transcription complexes. J. Mol. Biol. 376:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumont, E., K. P. Fuchs, G. Bommer, B. Christoph, E. Kremmer, and B. Kempkes. 2000. Neoplastic transformation by Notch is independent of transcriptional activation by RBP-J. signalling. Oncogene 19:556-561. [DOI] [PubMed] [Google Scholar]
- 7.Emuss, V., D. Lagos, A. Pizzey, F. Gratrix, S. R. Henderson, and C. Boshoff. 2009. KSHV manipulates Notch signaling by DLL4 and JAG1 to alter cell cycle genes in lymphatic endothelia. PLoS Pathog. 5:e1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fossum, E., C. C. Friedel, S. V. Rajagopala, B. Titz, A. Baiker, T. Schmidt, T. Kraus, T. Stellberger, C. Rutenberg, S. Suthram, S. Bandyopadhyay, D. Rose, A. von Brunn, M. Uhlmann, C. Zeretzke, Y. A. Dong, H. Boulet, M. Koegl, S. M. Bailer, U. Koszinowski, T. Ideker, P. Uetz, R. Zimmer, and J. Haas. 2009. Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5:e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frank, R. 1992. Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48:9217-9232. [Google Scholar]
- 10.Fuchs, K. P., G. Bommer, E. Dumont, B. Christoph, M. Vidal, E. Kremmer, and B. Kempkes. 2001. Mutational analysis of the J recombination signal sequence binding protein (RBP-J)/Epstein-Barr virus nuclear antigen 2 (EBNA2) and RBP-J/Notch interaction. Eur. J. Biochem. 268:4639-4646. [DOI] [PubMed] [Google Scholar]
- 11.Graham, F. L., J. Smiley, W. C. Russell, and R. Nairn. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59-74. [DOI] [PubMed] [Google Scholar]
- 12.Hayward, S. D. 2004. Viral interactions with the Notch pathway. Semin. Cancer Biol. 14:387-396. [DOI] [PubMed] [Google Scholar]
- 13.Hayward, S. D., J. Liu, and M. Fujimuro. 2006. Notch and Wnt signaling: mimicry and manipulation by gamma herpesviruses. Sci. STKE 2006:re4. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, S. E., M. X. Ilagan, R. Kopan, and D. Barrick. 2010. Thermodynamic analysis of the CSL × Notch interaction: distribution of binding energy of the Notch RAM region to the CSL beta-trefoil domain and the mode of competition with the viral transactivator EBNA2. J. Biol. Chem. 285:6681-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanno, T., Y. Sato, T. Sata, and H. Katano. 2006. Expression of Kaposi's sarcoma-associated herpesvirus-encoded K10/10.1 protein in tissues and its interaction with poly(A)-binding protein. Virology 352:100-109. [DOI] [PubMed] [Google Scholar]
- 16.Kopan, R., and M. X. Ilagan. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137:216-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovall, R. A., and W. A. Hendrickson. 2004. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 23:3441-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kremmer, E., B. R. Kranz, A. Hille, K. Klein, M. Eulitz, G. Hoffmann-Fezer, W. Feiden, K. Herrmann, H. J. Delecluse, G. Delsol, G. W. Bornkamm, N. Mueller-Lantzsch, and F. A. Grassert. 1995. Rat monoclonal antibodies differentiating between the Epstein-Barr virus nuclear antigens 2A (EBNA2A) and 2B (EBNA2B). Virology 208:336-342. [DOI] [PubMed] [Google Scholar]
- 19.Lan, K., T. Choudhuri, M. Murakami, D. A. Kuppers, and E. S. Robertson. 2006. Intracellular activated Notch1 is critical for proliferation of Kaposi's sarcoma-associated herpesvirus-associated B-lymphoma cell lines in vitro. J. Virol. 80:6411-6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lan, K., D. A. Kuppers, and E. S. Robertson. 2005. Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jkappa, the major downstream effector of the Notch signaling pathway. J. Virol. 79:3468-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan, K., D. A. Kuppers, S. C. Verma, and E. S. Robertson. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 78:6585-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan, K., M. Murakami, B. Bajaj, R. Kaul, Z. He, R. Gan, M. Feldman, and E. S. Robertson. 2009. Inhibition of KSHV-infected primary effusion lymphomas in NOD/SCID mice by gamma-secretase inhibitor. Cancer Biol. Ther. 8:2136-2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan, K., S. C. Verma, M. Murakami, B. Bajaj, R. Kaul, and E. S. Robertson. 2007. Kaposi's sarcoma herpesvirus-encoded latency-associated nuclear antigen stabilizes intracellular activated Notch by targeting the Sel10 protein. Proc. Natl. Acad. Sci. U. S. A. 104:16287-16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, H. R., M. H. Kim, J. S. Lee, C. Liang, and J. U. Jung. 2009. Viral interferon regulatory factors. J. Interferon Cytokine Res. 29:621-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, H. R., Z. Toth, Y. C. Shin, J. S. Lee, H. Chang, W. Gu, T. K. Oh, M. H. Kim, and J. U. Jung. 2009. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor 4 targets MDM2 to deregulate the p53 tumor suppressor pathway. J. Virol. 83:6739-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, Y., J. Chang, S. J. Lynch, D. M. Lukac, and D. Ganem. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang, Y., and D. Ganem. 2003. Lytic but not latent infection by Kaposi's sarcoma-associated herpesvirus requires host CSL protein, the mediator of Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 100:8490-8495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang, Y., and D. Ganem. 2004. RBP-J (CSL) is essential for activation of the K14/vGPCR promoter of Kaposi's sarcoma-associated herpesvirus by the lytic switch protein RTA. J. Virol. 78:6818-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, R., X. Li, A. Tulpule, Y. Zhou, J. S. Scehnet, S. Zhang, J. S. Lee, P. M. Chaudhary, J. Jung, and P. S. Gill. 2010. KSHV-induced notch components render endothelial and mural cell characteristics and cell survival. Blood 115:887-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubman, O. Y., M. X. Ilagan, R. Kopan, and D. Barrick. 2007. Quantitative dissection of the Notch:CSL interaction: insights into the Notch-mediated transcriptional switch. J. Mol. Biol. 365:577-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maier, S., M. Santak, A. Mantik, K. Grabusic, E. Kremmer, W. Hammerschmidt, and B. Kempkes. 2005. A somatic knockout of CBF1 in a human B-cell line reveals that induction of CD21 and CCR7 by EBNA-2 is strictly CBF1 dependent and that downregulation of immunoglobulin M is partially CBF1 independent. J. Virol. 79:8784-8792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minoguchi, S., Y. Taniguchi, H. Kato, T. Okazaki, L. J. Strobl, U. Zimber-Strobl, G. W. Bornkamm, and T. Honjo. 1997. RBP-L, a transcription factor related to RBP-Jkappa. Mol. Cell. Biol. 17:2679-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nam, Y., P. Sliz, L. Song, J. C. Aster, and S. C. Blacklow. 2006. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124:973-983. [DOI] [PubMed] [Google Scholar]
- 34.Nam, Y., A. P. Weng, J. C. Aster, and S. C. Blacklow. 2003. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J. Biol. Chem. 278:21232-21239. [DOI] [PubMed] [Google Scholar]
- 35.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 36.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. U. S. A. 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sander, G., A. Konrad, M. Thurau, E. Wies, R. Leubert, E. Kremmer, H. Dinkel, T. Schulz, F. Neipel, and M. Sturzl. 2008. Intracellular localization map of human herpesvirus 8 proteins. J. Virol. 82:1908-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staudt, M. R., and D. P. Dittmer. 2007. The Rta/Orf50 transactivator proteins of the gamma-herpesviridae. Curr. Top. Microbiol. Immunol. 312:71-100. [DOI] [PubMed] [Google Scholar]
- 39.Tamura, K., Y. Taniguchi, S. Minoguchi, T. Sakai, T. Tun, T. Furukawa, and T. Honjo. 1995. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr. Biol. 5:1416-1423. [DOI] [PubMed] [Google Scholar]
- 40.Uetz, P., Y. A. Dong, C. Zeretzke, C. Atzler, A. Baiker, B. Berger, S. V. Rajagopala, M. Roupelieva, D. Rose, E. Fossum, and J. Haas. 2006. Herpesviral protein networks and their interaction with the human proteome. Science 311:239-242. [DOI] [PubMed] [Google Scholar]
- 41.von Brunn, A., C. Teepe, J. C. Simpson, R. Pepperkok, C. C. Friedel, R. Zimmer, R. Roberts, R. Baric, and J. Haas. 2007. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One 2:e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson, J. J., and R. A. Kovall. 2006. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 124:985-996. [DOI] [PubMed] [Google Scholar]