Abstract
Herpes simplex virus (HSV) entry into cells is triggered by the binding of envelope glycoprotein D (gD) to a specific receptor, such as nectin-1 or herpesvirus entry mediator (HVEM), resulting in activation of the fusion effectors gB and gH and virus penetration. Here we report the identification of a hyperactive gB allele, D285N/A549T, selected by repeat passage of a gD mutant virus defective for nectin-1 binding through cells that express a gD-binding-impaired mutant nectin-1. The gB allele in a wild-type virus background enabled the use of other nectins as virus entry receptors. In addition, combination of the mutant allele with an epidermal growth factor receptor (EGFR)-retargeted gD gene yielded dramatically increased EGFR-specific virus entry compared to retargeted virus carrying wild-type gB. Entry of the gB mutant virus into nectin-1-bearing cells was markedly accelerated compared to that of wild-type virus, suggesting that the gB mutations affect a rate-limiting step in entry. Our observations indicate that ineffective gD activation can be complemented by hypersensitization of a downstream component of the entry cascade to gD signaling.
Entry of herpes simplex virus type 1 (HSV-1) into susceptible cells involves the coordinated activities of at least five viral envelope glycoproteins (9, 18, 33, 40). Virions initially bind to glycosaminoglycan (GAG) moieties of cell surface proteoglycans through glycoproteins B and C (gB and gC, respectively) (32, 51), facilitating the interaction of gD with one of its specific receptors, herpesvirus entry mediator (HVEM, or HveA), nectin-1 (HveC), or 3-O-sulfated heparan sulfate (24, 45, 50). Receptor binding is believed to result in a conformational change in gD, which in turn activates the fusion mechanism mediated by gB and the gH/gL heterodimer; fusion merges the virus envelope with the cell surface or endosomal membrane, resulting in capsid release into the cytoplasm (11, 23, 30, 37, 44, 47, 48). Prior to receptor binding, the N-terminal region of the gD ectodomain is folded back over the immunoglobulin (Ig)-like core domain in a position to engage the C-terminal effector region (pro-fusion domain), thereby keeping the effector domain in an inactive state (23, 37). Receptor binding disrupts this engagement and liberates the effector domain to activate gB and/or gH/gL. The crystal structure of the gB ectodomain shows unexpected homology to the postfusion form of glycoprotein G from vesicular stomatitis virus (VSV G), a well-characterized fusion protein (30), providing strong evidence that gB plays a major role in membrane fusion. In addition, gH displays structural hallmarks of fusion proteins (26, 27), and gB and gH each have fusogenic activity, as indicated by the finding that either alone is sufficient for membrane fusion during nuclear egress (20). However, gB and gH/gL are both required for complete fusion during virus entry, although gB is dispensable for hemifusion, an intermediate state (53).
Results from biochemical and bimolecular-complementation assays have shown that gD binds individually to both gB and gH/gL, regardless of the presence of gD receptors (4, 5, 25), while complexes of gB and gH/gL assemble only in the presence of receptor-bound gD (4, 5). These observations suggested that receptor-dependent gD activation brings gB and gH/gL together for execution of the fusion event. However, based on new evidence that gB and gH/gL can also interact in the absence of gD, an alternative model has been proposed in which activated gD signals to preformed gB-gH/gL complexes (6). While these models are not mutually exclusive, the functional significance of the detected complexes remains to be firmly established (15). However, there is broad consensus that the gD-receptor interaction triggers the initiation of fusion by direct interaction with either or both gB and gH/gL, indicating that the quality of the gD-receptor interaction is key to the efficiency of HSV infection.
Viruses have an intrinsic ability to evolve and adapt to changes in the environment, including the acquisition of an extended host range which can lead to epidemic infections (56). We previously described gain-of-function derivatives of a gD mutant virus, K26-gD:R222N/F223I, that was impaired in its ability to use nectin-1 as an entry receptor (54). Repeated passage of this virus through cells that express nectin-1 as the sole gD receptor yielded phenotypic revertants that had regained the ability to use nectin-1 for infection. This phenotype resulted from reversion or forward mutations at the parental mutant positions or from substitutions elsewhere in gD that likely affect the integrity of the discontinuous interface with nectin-1. Since these types of experiments can reveal novel factors or interactions that are important for virus entry, we performed a similar study at higher stringency in an attempt to avoid simple reversion mutations. The strategy was to use our previous gD:R222N/F223I mutant virus that is defective for entry via nectin-1 and ask if this virus could adapt to host cells expressing a mutant form of nectin-1 whose binding to wild-type gD is severely impaired. A specific goal of this effort was to find mutations in gD or other envelope glycoproteins that could enhance infection through atypical receptors, including cell-type-specific receptors that can be engaged by retargeted HSV vectors.
Here we report the identification of a hyperactive gB double mutation, gB:D285N/A549T, referred to herein as gB:N/T, that allows virus entry in the absence of authentic gD receptors, enhances virus entry through unconventional receptors, including a targeted receptor, and appears to act by sensitizing gB to activation by gD, directly or indirectly via gH/gL, and increasing the rate of virus entry into different host cells. Our observations demonstrate that hyperactive gB can compensate for ineffective gD-receptor interactions in the process of HSV entry into cells.
MATERIALS AND METHODS
Cells.
Baby hamster kidney J1.1-2, HVEM-transduced J/A, murine melanoma B78H1, HVEM-transduced B78/A, nectin-1-transduced B78/C (54), Chinese hamster ovary CHO-K1, nectin-1-transduced CHO/C (24), nectin-2-transduced CHO/Nec2 (55), epidermal growth factor receptor (EGFR)-transduced CHO/EGFR (46), GAG-deficient CHO derivative, pgsA-745 (19), African green monkey kidney Vero, gB-complementing D6 (10), and gD-complementing VD60 (40) cells were described previously. J/TMC and B78/TMC cells were established by transfection of J1.1-2 or B78H1 cells with plasmid pcDNA3TMC and selection for resistance to 0.4 mg/ml or 0.8 mg/ml G418, respectively. J/TMCΔC cells were established by transfection of J1.1-2 cells with plasmid pcDNA3TMCΔC and selection with 0.4 mg/ml G418. CHO/A and CHO/Nec4 cells were established by transfection of CHO-K1 cells with expression plasmids for HVEM (pBEC14) (45) or nectin-4 (p3×FLR4.C1) (49), respectively, and selection with 0.8 mg/ml G418. Clonal lines obtained by limiting dilution or cylinder cloning methods were confirmed for expression of the introduced receptor cDNAs in >95% of the cells by indirect immunofluorescence.
Viruses.
K26GFP, an HSV-1 KOS recombinant virus expressing VP26 as a fusion with green fluorescent protein (GFP) (17), K26-gD:2/3NI, an HVEM-restricted gD mutant derived from K26GFP (K26-gD:R222N/F223I in reference 54), and KΔT, a KOS derivative that does not incorporate gB into its envelope due to a deletion of 323 amino acids containing the membrane-spanning region of gB (10), were described previously. K-gB:wt and K-gB:N/T were established by cotransfection of Vero cells with KΔT viral DNA and plasmid pgB1:wt or pgB1:D285N/A549T, respectively, followed by plaque purification through three rounds of limiting dilution on Vero cells. The cloned viruses were sequenced through the entire gB and gD genes. In addition, PCR through the region deleted in KΔT was performed to rule out contamination with KΔT. The titers of K-gB:wt and K-gB:N/T stocks were determined on B78/C cells (PFU/ml) and by quantitative PCR (qPCR) to determine the number of genome copies (gc)/ml. K-gB:wtΔgD and K-gB:N/TΔgD were produced by cotransfection of VD60 cells with plasmid pΔgD-EGFP and viral DNAs of K-gB:wt or K-gB:N/T, respectively, with subsequent plaque purification on VD60 cells. The purified viruses were sequenced through the gD locus. In addition, two PCRs to amplify the entire gD coding region or an internal gD fragment, respectively, were performed to rule out contamination with the parent viruses. K-gB:wtΔgD and K-gB:N/TΔgD were passaged through Vero cells, and the numbers of gc/ml were determined by qPCR.
Plasmids.
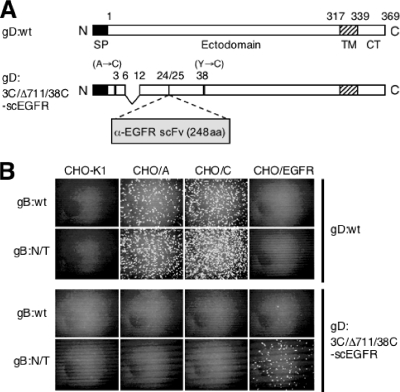
The TMC (triply mutated HveC) expression plasmid pcDNA3TMC was created by replacement of a V-domain-encoding fragment of pBG38 (24) with the corresponding fragment of pTMC153-his (38). Plasmid pcDNA3TMCΔC, encoding TMC with both C domains deleted, was created by deleting the coding sequences for nectin-1 codons 148 to 336 from pcDNA3TMC. Plasmid pgB1:wt contains the gB open reading frame (ORF) and flanking regulatory sequences from K26GFP (17); mutant counterparts, including pgB1:D285N/A549T, were created by replacement of appropriate fragments with the corresponding fragments of PCR products generated on DNA from selected virus isolates. The gD-null recombination plasmid pΔgD-EGFP was created by replacing the entire gD ORF of pgDSac with that of enhanced green fluorescent protein (EGFP) derived from pEGFP-C1 (Clontech). The EGFR-retargeting plasmid pgD:3C/Δ711/38C-scEGFR was generated by insertion of the 528 scFv sequence (3) into plasmid pgD:3C/Δ711/38C-NE, a pgDSac derivative that contains cloning sites between gD codons 24 and 25, a deletion of codons 7 to 11, and previously described mutations at positions 3 and 38 (A3C/Y38C) (14; unpublished results).
Selection of virus isolates.
Two separate selections were carried out. In the first, J/TMC cells were inoculated with K26-gD:2/3NI at 1,000 PFU/cell and rinsed after 8 h with acidic buffer. Progeny virus was harvested 2 days later from the cells and medium was expanded on J/A cells for infection of J/TMC cells at 100 PFU/cell, followed by acid treatment at 8 h. Progeny virus was harvested as described before and used for plaque purification by limiting dilution on J/TMC cells. The second selection was performed in a similar manner with the following modifications. B78/TMC cells were inoculated with the same initial virus at 100 PFU/cell, followed by expansion on J/A cells, and progeny virus was passaged twice more on B78/TMC cells and once on J/TMCΔC cells. Plaque purifications were performed on B78/TMC cells.
Primary infection and transient complementation assays.
Primary infection assays were performed as described previously (54). Briefly, cells were infected for 6 to 24 h and immunostained using monoclonal mouse anti-VP16 (1:400) or anti-ICP4 (1:300) as the primary antibodies (Santa Cruz) and Cy3-conjugated sheep anti-mouse IgG (1:400) (Sigma) as the secondary antibody. Transient complementation experiments with gB-null KΔT, gD-null K-gB:wtΔgD, or K-gB:N/TΔgD viruses were carried out essentially as described previously (1). Briefly, Vero cells were transfected with expression plasmids for wild-type or mutant gB or gD by lipofection, the cells were infected the next day with gB- or gD-null viruses at a multiplicity of infection (MOI) of 5, supernatants harvested the following day were used for infection of test cell lines, and infections were assessed as described above.
Immunofluorescence.
Indirect immunofluorescence was performed as described previously (54), using goat anti-mouse nectin-3 or nectin-4 polyclonal antibodies (R&D Systems) (1 μg/ml) as the primary antibodies and Cy3-conjugated rabbit anti-goat IgG (Sigma) (1:400) as the secondary antibody. The cell nuclei were stained by incubation with 1 μM Hoechst 33342 (Invitrogen) at room temperature for 10 min. Images were captured under a FluoView 1000 confocal microscope (Olympus).
Infection blocking assay.
CHO-K1 cells were incubated with rat anti-mouse nectin-3 (Cell Sciences) or rat anti-mouse nectin-4 (R&D Systems) monoclonal antibodies (both of the IgG2a isotype) or phosphate-buffered saline (PBS) for 1 h at room temperature and then infected with K-gB:N/T at 3,000 gc/cell for 2 h at 37°C, followed by acid treatment. Infections were assessed at 16 h postinfection (hpi) as described above.
Rate-of-entry assays.
Rate-of-entry assays were performed as described previously (32) with modifications. Cells were incubated with viruses at 4°C for 30 or 60 min and washed three times with cold PBS. The cells were then shifted to 37 or 30°C for various intervals followed by acid treatment. The cultures were incubated at 37°C and stained for VP16 expression at 8 h (primary infection) or 48 to 72 h (plaque formation).
RESULTS
Selection and classification of virus mutants.
Baby hamster kidney J1.1-2 and murine melanoma B78H1 cells are resistant to HSV infection due to the absence of gD receptors (13, 36, 43). Struyf and colleagues previously described a mutant version of nectin-1, QN76-77AA/M85F (referred to here as TMC, for triply mutated HveC), that is severely impaired for binding to gD and thus fails to support HSV entry (52). We created clonal TMC-expressing J and B78 cell lines, designated J/TMC and B78/TMC, by stable transfection. The virus mutant K26-gD:R222N/F223I, abbreviated here as K26-gD:2/3NI, has a highly diminished ability to use nectin-1 for infection due to a pair of mutations in gD but is largely unimpaired for infection through HVEM (54); it also expresses a VP26-GFP fusion protein (17) to facilitate the detection of virus infection and growth. We used this combination of defective virus and host cells to determine if HSV can evolve to complement these defects during reiterative high-MOI infection and progeny amplification on HVEM-expressing J (J/A) cells. Following 2 or 4 rounds of selection (see Materials and Methods), virus was isolated from a number of J/TMC and B78/TMC plaques and characterized. Direct sequencing was used to identify mutations in the gD gene of seven isolates derived from the first experiment and eight from the second. As shown in Table 1, 10 of the 15 isolates had one or two new missense mutations in the gD ORF in addition to the parental 2/3NI mutations. Distinct amino acid substitutions were found in isolates from the first (A185T; group 2) and second (Q178H; group 3) selections, and one isolate contained both substitutions (group 4). However, five of the seven isolates from the first experiment harbored no new gD mutations (group 1), suggesting that these viruses had undergone alterations outside the gD gene.
TABLE 1.
gD mutations in selected virus isolatesa
| Group | Substitution (parental + new)b | New base changeb | Frequency (no. with mutation/total)c | Isolate no.d |
|---|---|---|---|---|
| Expt 1 (2 passages) | ||||
| 1 | R222N/F223I | 5/7 | 1 | |
| 2 | R222N/F223I + A185T | GCC→ACC | 2/7 | 2 |
| Expt 2 (4 passages) | ||||
| 3 | R222N/F223I + Q178H | CAG→CAT | 7/8 | 3 |
| 4 | R222N/F223I + Q178H/A185T | CAG/GCC→CAT/ACC | 1/8 | 4 |
K26-gD:2/3NI was passaged twice or four times through TMC-expressing cells, and progeny viruses were cloned by limiting dilution on J/A cells.
Shown are amino acid or nucleotide changes (in boldface) in individual isolates.
Number of isolates with the indicated mutation/total number of analyzed isolates in each experiment.
Designation of a representative isolate from each group.
Tropism changes without new gD mutations.
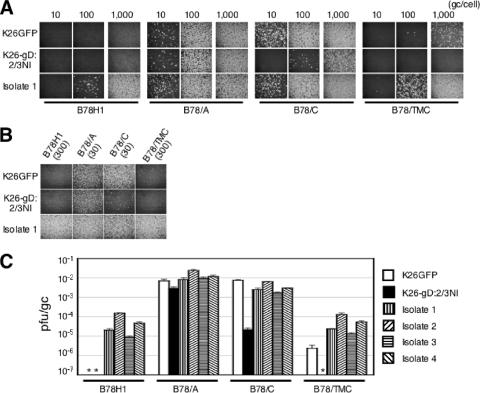
A representative isolate from group 1, referred to as isolate 1, was plaque purified by multiple rounds of limiting dilution on J/A cells and characterized for its ability to infect B78H1 cells and derivatives transduced with HVEM (B78/A), nectin-1 (B78/C), or TMC (B78/TMC). Control infections included the parental virus K26-gD:2/3NI and its wild-type gD counterpart, K26GFP (17). Virus input was standardized by using equal numbers of virus particles based on determinations of viral genome copy (gc) numbers by real-time quantitative PCR (34); in this manner, differences in infectious activities between viruses can be directly observed as differences in the number of transduced cells or plaques, whereas standardization to transducing units (tu) or PFU can obscure such differences. Cells were infected for 8 h, and expression of the tegument protein VP16 was visualized by indirect immunofluorescence (Fig. 1A). Remarkably, isolate 1 showed infection of receptor-deficient B78H1 cells, whereas no infection was observed for either of the control viruses. Furthermore, isolate 1 infected B78/TMC cells better than not only K26-gD:2/3NI (≥100-fold) but also K26GFP (∼10-fold). As expected, the three viruses showed similar infection efficiencies on B78/A cells, but isolate 1 yielded at least 10-fold more infected B78/C cells than K26-gD:2/3NI, similar to K26GFP. As shown in Fig. 1B, immunostaining for the immediate-early gene product ICP4 at 6 hpi showed essentially the same trend, indicating that the changes in isolate 1 principally facilitated virus entry rather than replication. Together, these results indicated that isolate 1 had acquired mutations outside the gD gene that (i) enable infection independent of known gD receptors, (ii) fully suppress the entry defect caused by the gD:2/3NI mutations, and (iii) compensate efficiently for the highly defective gD:2/3NI-TMC interaction. This profile was consistent with a general entry-enhancing effect, regardless of the particular gD-receptor interaction.
FIG. 1.
Tropisms of the selected virus isolates. (A) Cells were infected with isolate 1 or controls for 8 h at the various gc/cell indicated above the panels and immunostained for VP16. Infections at 1,000 gc/cell were performed separately. (B) Cells were infected with isolate 1 for 6 h at 30 or 300 gc/cell, as indicated in parentheses, and immunostained for ICP4. (C) Biological titers (PFU/ml) were divided by genome titers (gc/ml), and the mean values ± standard deviations (SD) from three determinations were plotted on a logarithmic scale. *, <10−7.
Identification and characterization of shared gB mutations.
Direct sequencing of the other essential glycoprotein genes of isolate 1 identified two missense mutations in the gB ORF, D285N and A549T. No changes were observed in the gH and gL ORFs, although we could not read through a highly GC-rich 20-nucleotide portion of gH (positions 1983 to 2102 in GenBank accession no. X03896). We plaque purified a representative isolate from each of the other groups listed in Table 1, denoted isolates 2 to 4, and determined the specific infectious activities of all four isolates, expressed as PFU/particle (gc) ratios, on the panel of B78 cell lines. The results (Fig. 1C) demonstrated that each of the isolates had a similar change in tropism compared to the parental K26:gD-2/3NI virus. We sequenced the gB genes of isolates 2 to 4 and found that each contained the same double mutation as isolate 1, referred to hereafter as N/T, without other changes. These results suggested that the N/T double mutation could be the primary cause of the altered tropism of the four isolates. It is noteworthy that the specific infectious activities of the four isolates were very similar on B78/TMC and B78H1 cells, indicating that the impaired gD receptor TMC did not play a key role in the selection of these isolates.
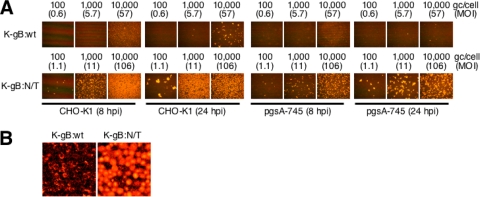
To separate the effects of the N/T mutations from those of other potential changes in these isolates, we transferred the gB:N/T allele into a wild-type virus background by homologous recombination with a gB-null virus, KΔT (10), a KOS derivative expressing an internally deleted gB that fails to be incorporated into the viral envelope due to the absence of the transmembrane domain. As a control, we also rescued KΔT with the wild-type gB gene. Isolates designated K-gB:N/T and K-gB:wt, respectively, were confirmed by DNA sequencing through the entire gB and gD ORFs. We tested these viruses for infection of CHO-K1 cells, another cell line that is resistant to HSV-1 due to the absence of gD receptors (45). We also used a GAG-deficient derivative of CHO-K1, pgsA-745 cells (19), to determine if the N/T mutations acted by facilitating virion attachment to GAGs, one of the known functions of gB (32, 51). At 8 hpi, K-gB:N/T produced readily detectable infection of CHO-K1 cells at a virus input of 100 gc/cell or more, whereas no infection by K-gB:wt was seen at a 10-fold-higher dose and limited infection was seen at a 100-fold-higher dose (Fig. 2A); the diffuse background at the highest K-gB:wt dose was due to extracellular virions attached to the cell surface (Fig. 2B). It is important to note that 100 gc/cell of K-gB:wt and K-gB:N/T corresponded to approximately 0.6 and 1.1 PFU/cell, respectively, on permissive B78/C cells (MOI of 0.6 to 1.1), such that the virus doses tested in this experiment were not exceptionally high. At 24 hpi, K-gB:N/T at 100 gc/cell produced foci of infected CHO-K1 cells, indicative of virus replication and lateral spread, while a 100-fold-higher dose of K-gB:wt yielded only individual infected cells and small foci (Fig. 2A). On GAG-deficient pgsA-745 cells, infection by K-gB:wt was barely detectable even at 24 hpi at the highest virus input (Fig. 2A). Consistent with the virion attachment function of GAGs, the diffuse background at 10,000 gc/cell at 8 hpi was reduced compared to that observed with CHO-K1 cells. In contrast, infected cells were distinguishable at a 100-fold-lower dose of K-gB:N/T at both 8 and 24 hpi. Together, these results demonstrated that gB:N/T enables infection of gD receptor-negative cells and indicated that this effect was not due to increased binding of gB to GAGs.
FIG. 2.
Effect of cellular glycosaminoglycans on K-gB:N/T infectivity. (A) Cells were infected for 8 or 24 h at the various gc/cell indicated above the panels and immunostained for VP16. MOIs (PFU/cell) based on virus titers on B78/C cells (PFU/ml) are included in parentheses. (B) Higher-magnification images of 10,000-gc/cell infections of CHO-K1 cells at 8 hpi.
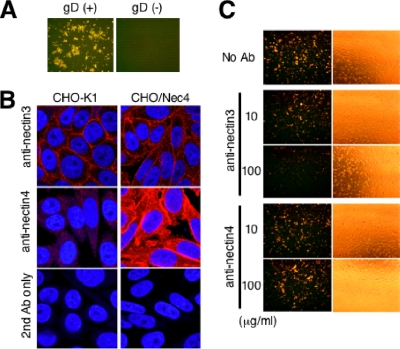
CHO-K1 infection by K-gB:N/T requires gD and is inhibited by anti-nectin-3 antibody.
Given the absence of known gD receptors on CHO-K1 cells (45), we asked whether gD itself is required for the enhanced infection of these cells by K-gB:N/T. We derived a gD-null virus from K-gB:N/T, designated K-gB:N/TΔgD, by replacement of the complete gD ORF with that of EGFP. CHO-K1 cells were infected with 1,000 gc/cell of K-gB:N/T or K-gB:N/TΔgD and stained for VP16 at 16 hpi. The results showed that gD is indispensable for infection of CHO-K1 cells by K-gB:N/T (Fig. 3A), raising the possibility that these cells express minor or cryptic gD receptors on their surface that can serve as HSV-1 entry receptors conditional on the presence of the gB:N/T double mutation. We considered nectin-3 as a candidate based on a previous report by Cocchi and colleagues that nectin-3 can mediate entry of HSV harboring a particular combination of gD mutations (12). Immunofluorescence analysis demonstrated the presence of nectin-3 on the surface of CHO-K1 cells (Fig. 3B, upper and lower left panels). Antibody blocking experiments showed that anti-nectin-3, but not isotype-matched anti-nectin-4, reduced infection by K-gB:N/T in a dose-dependent manner (Fig. 3C); the phase-contrast images indicate that this was not due to anti-nectin-3-mediated cell detachment. These results suggested that nectin-3 plays an essential role in gB:N/T mutant virus infection of CHO-K1 cells, most likely by functioning as a receptor for gD. Since nectin-3 is also expressed on J1.1-2 and B78H1 cells (12; unpublished results), it is possible that our original isolates were selected on the basis of weak gD:2/3NI binding to nectin-3.
FIG. 3.
Roles of gD and nectin-3 in infection of CHO-K1 cells by K-gB:N/T. (A) CHO-K1 cells were infected with K-gB:N/T (left panel) or K-gB:N/TΔgD (right panel) for 16 h at 1,000 gc/cell and immunostained for VP16. (B) CHO-K1 or CHO/Nec4 cells were immunostained for nectin-3 or nectin-4 and observed under a confocal microscope. Cells that reacted with the secondary antibody (Ab) alone are shown at the bottom. Nuclei were stained with Hoechst 33342. (C) CHO-K1 cells were preincubated with PBS or isotype-matched anti-nectin-3 or anti-nectin-4 monoclonal antibodies at 10 or 100 μg/ml; incubated with K-gB:N/T at 3,000 gc/cell for 2 h, followed by acidic buffer treatment; and immunostained for VP16 at 16 hpi. The panels to the right show phase-contrast images of the same fields.
Enhancement of K-gB:N/T virus infection by other nectins.
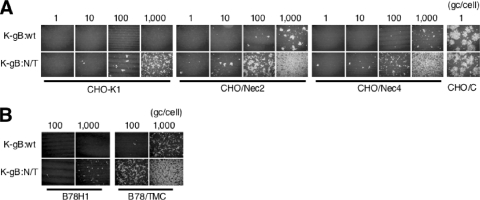
Since nectin-3 appeared to enable K-gB:N/T infection of CHO-K1 cells, we asked whether other nectin family members could function in a similar manner. At high virus input, CHO-K1 cells overexpressing human nectin-2 (CHO/Nec2) (55) or nectin-4 (CHO/Nec4) (Fig. 3B, right panels), but not regular CHO-K1 cells, showed infection by K-gB:wt (Fig. 4A), indicating that these nectins can act as inefficient entry receptors for wild-type virus; infection of CHO-K1 cells expressing human nectin-1 (CHO/C) at 1 gc/cell is shown for comparison. K-gB:N/T infection was observed at lower virus input on CHO-K1 cells and was also increased on the nectin lines (Fig. 4A). In addition, we found that K-gB:N/T was more efficient in infection of B78/TMC cells than K-gB:wt (Fig. 4B); the increased infection of the parental B78H1 cells detectable at high virus input is consistent with recognition of nectin-3 on these cells. Together, these data indicated that gB:N/T facilitates the use of multiple members of the nectin family as well as a defective version of nectin-1 for viral entry, raising the possibility that the gB mutations act in a general manner to enhance virus infection through unconventional receptors.
FIG. 4.
Effects of gB:N/T on infection through weak gD receptors. (A) Cells were infected for 24 h at the various gc/cell indicated above the panels and immunostained for VP16. (B) Cells were infected for 8 h at 100 or 1,000 gc/cell and immunostained for VP16.
gB:N/T enhances retargeted HSV infection.
To expand the observation that gB:N/T facilitates the use of atypical entry receptors, we used a transient complementation approach to determine whether the double mutation can enhance infection via an unrelated receptor. HSV can be retargeted by ablation of the native receptor recognition functions of gD and insertion of recognition elements for novel receptors (41, 42, 57), but the efficiency of retargeted infection tends to be lower than that of natural infection through authentic receptors. We generated a gD-null derivative of K-gB:wt, designated K-gB:wtΔgD, by replacement of the gD ORF with that of enhanced GFP (EGFP), as done earlier to produce K-gB:N/TΔgD. Using equal amounts of these two gD-null viruses (PFU determined on gD-complementing VD60 cells), we performed transient complementation assays with a retargeted gD construct, pgD:3C/Δ711/38C-scEGFR, containing mutations that severely impair virus infection through nectin-1 (A3C/Y38C) (14, 54) and HVEM (deletion of residues 7 to 11; unpublished results), and an insertion of the ORF for a single-chain antibody (scFv) directed against the EGF receptor (EGFR) (3) between residues 24 and 25 (Fig. 5A). Vero cells were transfected with pgD:3C/Δ711/38C-scEGFR or the parental gD:wt expression construct and infected the next day with K-gB:wtΔgD or K-gB:N/TΔgD, and supernatants harvested the following day were used to infect CHO-K1 cells or derivatives expressing HVEM (CHO/A), nectin-1 (CHO/C), or EGFR (CHO/EGFR). As shown in Fig. 5B, the retargeted gD construct enabled infection exclusively of CHO/EGFR cells and only by K-gB:N/TΔgD, whereas the gD:wt construct complemented both viruses for infection of CHO/A and CHO/C cells, but not CHO/EGFR cells. These results indicated that gB:N/T provided a function allowing virus entry by gD recognition of the EGFR that is not provided by gB:wt. We suggest that this reflects weakness in the interaction between retargeted gD and the EGFR that can be complemented by increased activity of a downstream component of the entry cascade, specifically gB. This interpretation is consistent with the observation that gB:N/T enhances infection through nectin-1-related surface antigens such as nectins 2 to 4 and TMC that may weakly interact with gD and indicates that this mechanism is not limited to nectin-related molecules.
FIG. 5.
Effects of gB:N/T on EGFR-specific infection by transiently retargeted HSV. (A) Schematic representation of gD:wt and retargeted gD:3C/Δ711/38C-scEGFR. SP, signal peptide; TM, transmembrane region; CT, cytoplasmic domain; α-EGFR scFV, anti-EGFR single-chain antibody; aa, amino acids. (B) Vero cells were transfected with expression plasmids for the gD proteins indicated to the right and then infected with K-gB:wtΔgD or K-gB:N/TΔgD (indicated to the left) at MOIs of 5, followed by treatment with acidic buffer. Equal volumes of supernatant collected the next day were used for infection of the cells indicated at the top. Cells were immunostained for ICP4 at 6 hpi.
Acceleration of virus entry by the gB:N/T mutations.
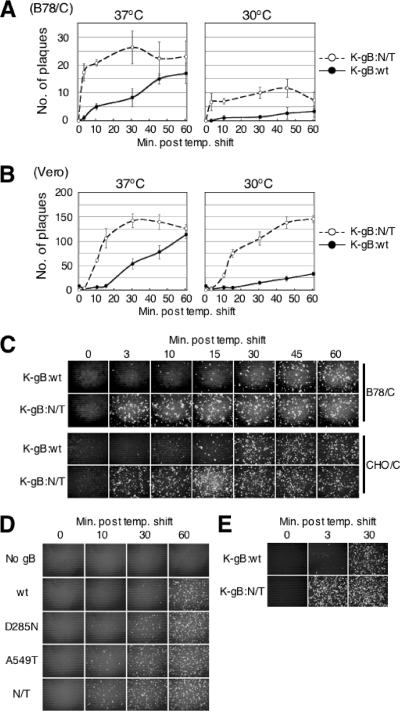
We performed rate-of-entry assays to determine whether the gB:N/T mutations might affect the kinetics of viral entry, as previously observed for other gB mutations (8, 16, 32). We used nectin-1-expressing cells for these experiments because they could be infected by the wild-type virus, unlike receptor-deficient cells (Fig. 4A), and thus could provide a baseline for comparison with the mutant virus. B78/C cells were incubated for 30 min at 4°C with K-gB:wt or K-gB:N/T at 200 PFU per well, washed thoroughly, and incubated at 37°C or 30°C for 0 to 60 min, and extracellular virus was inactivated by low-pH treatment. The cells were then overlaid with methylcellulose-containing medium and incubated at 37°C for 3 days to allow plaque formation. At 37°C, entry of gB:wt virus progressed steadily over time, whereas entry of the mutant virus during the first 3 min exceeded that of the wild-type virus at 60 min (Fig. 6A). However, following this initial phase of rapid entry by the mutant virus, additional entry was limited such that at 60 min nearly equal numbers of the two viruses had entered the cells. This observation suggested that the dramatic differences at the early time points reflected a difference in virus entry kinetics rather than in virus input. At 30°C, entry also proceeded more rapidly for K-gB:N/T than for K-gB:wt, although the extent of entry was lower overall than at 37°C. These results demonstrated that the gB:N/T mutations accelerate virus entry via the natural gD receptor nectin-1.
FIG. 6.
Effects of gB:N/T on virus entry kinetics. B78/C cells (A) or Vero cells (B) were incubated with 200 PFU of K-gB:wt or K-gB:N/T at 4°C for 30 min, washed thoroughly, incubated at 37 or 30°C for the indicated times, and treated with acidic buffer. Cells were then incubated at 37°C under methylcellulose-containing medium for 2 to 3 days to allow plaque formation. Plaques were counted, and the mean values ± SD from three determinations were plotted. temp., temperature. (C) Cells were incubated with K-gB:wt or K-gB:N/T at 4°C for 30 min at 0.3 PFU/cell (B78/C) or for 1 h at 1 PFU/cell (CHO/C), washed thoroughly, incubated at 37°C for the various times indicated above the panels, and treated with acidic buffer. Cells were then incubated at 37°C for 8 h and immunostained for VP16. (D) Vero cells were transfected with expression plasmids for the gB genes indicated to the left, infected with KΔT, and treated with acidic buffer. Fresh Vero monolayers were then incubated with equal volumes of the respective supernatants at 4°C for 30 min, and the kinetics of virus entry were examined as in panel C. (E) Entry kinetics on CHO/A cells. Virus input was 3 PFU/cell, and the experiment was performed as in panel C, with minor modifications.
HSV-1 entry into gD receptor-transduced B78 cells reportedly takes place by a low-pH-independent endocytic pathway (44), whereas entry into Vero cells occurs by membrane fusion at the cell surface (21, 22); entry into receptor-transduced CHO-K1 cells is mediated by a low-pH-dependent endocytic pathway (47, 48). To determine whether the gB:N/T mutations accelerate entry through each of these different pathways, we repeated the rate-of-entry assays on Vero and CHO/C cells. As shown in Fig. 6B, the results on Vero cells at 37°C were similar to those on B78/C cells, although both viruses showed a delay in early kinetics compared to B78/C cells. As on B78/C cells, the entry kinetics were slower at 30°C, but the 60-min entry level of K-gB:N/T was as high as the maximum reached at 37°C. Since CHO/C cells do not form well-defined plaques, we assessed the rates of entry on these cells by anti-VP16 staining at 8 h post-acidic wash to visualize infected cells; B78/C cells were included for comparison. The entry kinetics of both viruses were similar on the two cell lines (Fig. 6C) and consistent for B78/C cells with the results of Fig. 6A. K-gB:wt showed a gradual increase in infection, reaching a maximum at 30 to 60 min, while as little as 3 min sufficed for near-maximum infection by K-gB:N/T. These results demonstrated that the gB:N/T mutations accelerate virus entry into receptor-bearing cells regardless of the pathway used by wild-type virus. In addition, they showed that the N/T mutations not only affect entry via atypical receptors, but also affect entry into naturally (Vero) and artificially (B78/C and CHO/C) susceptible cells.
We used transiently complemented gB-null virus to determine whether the individual mutations of gB:N/T affected the rate of virus entry into Vero cells. The results in Fig. 6D show that both mutations accelerated entry compared to the wild-type gene. The A549T allele had a greater effect than the D285N allele, but the mutations appeared most effective in combination. These results indicated that each of the gB:N/T mutations contributed, albeit unequally, to the phenotype of the double mutant protein by accelerating virus entry.
Finally, we used HVEM-expressing CHO/A cells to exclude the possibility that the observed differences in entry kinetics between K-gB:wt and K-gB:N/T are specific for nectin-1-mediated entry. As shown in Fig. 6E, the gB:N/T allele accelerated virus entry into CHO/A cells as well, indicating that the effect of the N/T mutations on entry kinetics is not limited to a specific receptor.
DISCUSSION
In this report, we describe a double mutation in HSV-1 gB, D285N/A549T, that enhances virus infection through unconventional entry receptors and accelerates entry into susceptible cells. The double mutation was identified through in vitro evolution experiments involving reiterative passage of a nectin-1-detargeted gD-mutant virus, K26-gD:2/3NI (54), on cells that express a gD-binding-impaired receptor. Our data suggest that these conditions selected for viruses that could enter by recognition of nectin-3, present on the parent lines of the J/TMC and B78/TMC cells employed in the selection protocols (12; data not shown). First, the four characterized isolates had very similar particle/PFU ratios on B78H1 and B78/TMC cells, indicating that the defective TMC receptor was not essential for the acquired infection activity. Second, the gB mutations alone were sufficient to allow infection of CHO-K1 cells, and this infection was blocked by anti-nectin-3 antibodies. We showed that gD is required for CHO-K1 infection by K-gB:N/T, indicating that gD can interact with nectin-3 to produce a normally inconsequential entry signal that is enhanced by the gB mutations resulting in detectable entry. Using a fundamentally similar selection strategy, Cocchi and colleagues have previously described the isolation of a derivative of HSV-1 strain MP that had gained the ability to infect J cells in a nectin-3-dependent manner (12). The authors identified three amino acid substitutions in the strain MP gD gene that were all required for the new phenotype. Our observations are different in that they show complementation of deficient gD-receptor interactions not by compensatory mutations in gD itself, but by mutations that appear to hypersensitize a downstream component of the entry cascade to activating signals originating in gD. This mechanism allows the use of a range of atypical entry receptors and as such represents an attractive strategy for the virus to adapt to a changing environment or survive debilitating mutations in gD. We believe that the different outcome of the earlier study with strain MP can be ascribed to our use of a selection virus that was impaired for gD interaction with nectin-1 (K26-gD:2/3NI), whereas the selection virus used in the previous study contained wild-type gD.
The recently reported crystal structure of the gB ectodomain shows that amino acid 285 of HSV-1 gB is located in the base domain (domain I), which contains putative fusion loops believed to associate with membranes during fusion (28, 29). The core region of domain I has a fold that resembles a pleckstrin homology (PH) domain, indicative of involvement of this region in signal transmission (39). It has been reported that HSV-1 entry is blocked by monoclonal antibodies directed against this domain (31, 35). Furthermore, Avitabile and colleagues have reported, using pull-down assays, that replacement of the PH fold in domain I of HSV-1 gB (amino acids 271 to 361) with the homologous region of Kaposi's sarcoma-associated herpesvirus (KSHV) gB hampered the interaction of gB with gH/gL, implying that this region may be one of the contact sites between these molecules (6). Thus, it may be speculated that the D285N mutation identified in our study facilitates cooperation between gB and gH/gL during the fusion process.
Amino acid 549 of gB is located in the core domain (domain III), which contains a long, 44-residue α-helix (αC) followed by a short, 9-residue helix (αD) (30). Comparative analysis of the structures of Epstein-Barr virus (EBV) gB and VSV G by Backovic and colleagues has predicted that domain III undergoes significant refolding during the transition from the pre- to the postfusion conformation (7). A previous study identified a valine-to-alanine substitution at amino acid 553 as a rate-of-entry (roe) mutation accelerating virus entry (8). The effect of this mutation on virus entry via alternate receptors is unknown, and it is therefore unclear whether V553A is equivalent to our A549T mutation. However, residues 549 and 553 are both located in the short αD helix, suggesting that this region may play an important role in the predicted conformational change in gB that may be integral to the fusion event. It may be speculated that mutations in this region establish an intermediate state that no longer requires a strong signal from gD to complete the conformational change, but instead can readily progress toward the postfusion state, even in response to minimal gD interaction with a receptor, thus explaining both the requirement for gD and the ability of the gB:N/T virus to use unconventional receptors for entry. This effect may be referred to as hypersensitization of gB to activating signals from gD, which includes the possibility that the mechanism involves a change in the yet unknown region of gB-gD contact.
We demonstrated that gB:N/T accelerates virus uptake by each of three cell lines that reportedly undergo infection by distinct pathways. Entry of HSV-1 into Vero cells takes place by viral envelope fusion with the cell surface membrane, a process referred to as the “direct fusion pathway” (21, 22). In contrast, B78/C cells are infected via gD receptor-dependent, low-pH-independent endocytosis (44), while infection of CHO/C cells involves low-pH-dependent endocytosis (47, 48). Since fusion at the cell surface is believed to be executed by gB and gH/gL in response to gD-receptor interaction, the faster entry kinetics observed with gB:N/T may be attributed to more efficient activation of gB by gD and/or accelerated execution of the fusion reaction in cooperation with gH/gL. However, our rate-of-entry assays exclusively measured virus uptake, which on B78/C and CHO/C cells does not normally involve fusion. Thus, the effects of the N/T mutations on the entry kinetics into these cells can only be ascribed to acceleration of the fusion process itself if they also cause a switch from endocytic entry to cell surface fusion, perhaps as a direct result. There is precedent for such a switch involving gB (2), and this course of events would be consistent with the molecular mechanisms of the two mutations suggested above. However, additional studies are required to exclude the possibility that gB:N/T accelerates virus uptake into endosomes.
We examined the effects of the individual mutations of gB:N/T and found that both increased the virus entry kinetics on Vero cells. While the A549T mutation alone allowed almost as much entry as the double mutation in 10 min, it is possible that greater differences between these two can be observed at shorter infection times or with weaker target receptors such as TMC. This analysis awaits the creation of new recombinant viruses to enable consistent measurement over a range of time points, at different infection temperatures, and on multiple cell lines.
We demonstrated the potential utility of the gB:N/T mutations in enhancing retargeted virus entry, using a mutant recombinant gD gene that enables gD-null virus infection via the EGFR instead of the natural gD receptors. In view of the evidence presented in this study that gB:N/T facilitates the use of cryptic gD receptors for virus entry, such as nectin-3, it will be important to determine whether the gB mutations increase off-target infection of retargeted viruses. In our transient complementation assay, the gB:N/T, gD-null viral backbone complemented with EGFR-retargeted gD did not show increased entry into CHO-K1, CHO/A, or CHO/C cells compared to the gB:wt, gD-null backbone. It is possible that large targeting moieties inserted into gD, such as the scFv used in this study (248 residues), in combination with the HVEM- and nectin-1-detargeting mutations present in our retargeted gD construct abolish gD binding to cryptic receptors. We are in the process of constructing retargeted recombinant viruses to further validate the utility of gB:N/T.
In conclusion, this study describes a pair of mutations in glycoprotein B that appear to hypersensitize the HSV entry machinery to activation by gD, providing amplification of weak signals resulting from interaction of gD with unconventional receptors to accelerate the rate of virus entry and thereby achieve efficient infection. While the effects of these mutations on natural virus infection in vivo remain to be determined, they provide an immediate means to dramatically increase the infectivity of retargeted HSV vectors which has broad implications for the use of this virus for therapeutic purposes as well as fundamental research to distinguish and modify specific cell populations in vitro and in vivo.
Acknowledgments
We thank Gary Cohen, Patricia Spear, Gabriella Campadelli-Fiume, David Johnson, Stanley Person, Prashant Desai, Stephen Russell, and Marc Lopez for reagents and Simon Watkins for the use of his imaging facility. We also thank Zhanna Hakhverdyan for technical assistance and Arthur Frampton and Kenji Nakano for helpful discussions.
This work was supported by NIH grants CA119298, NS40923, and DK044935 to J.C.G.
Footnotes
Published ahead of print on 22 September 2010.
REFERENCES
- 1.Anderson, D. B., S. Laquerre, K. Ghosh, H. P. Ghosh, W. F. Goins, J. B. Cohen, and J. C. Glorioso. 2000. Pseudotyping of glycoprotein D-deficient herpes simplex virus type 1 with vesicular stomatitis virus glycoprotein G enables mutant virus attachment and entry. J. Virol. 74:2481-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arii, J., M. Uema, T. Morimoto, H. Sagara, H. Akashi, E. Ono, H. Arase, and Y. Kawaguchi. 2009. Entry of herpes simplex virus 1 and other alphaherpesviruses via the paired immunoglobulin-like type 2 receptor alpha. J. Virol. 83:4520-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asano, R., Y. Sone, K. Makabe, K. Tsumoto, H. Hayashi, Y. Katayose, M. Unno, T. Kudo, and I. Kumagai. 2006. Humanization of the bispecific epidermal growth factor receptor x CD3 diabody and its efficacy as a potential clinical reagent. Clin. Cancer Res. 12:4036-4042. [DOI] [PubMed] [Google Scholar]
- 4.Atanasiu, D., J. C. Whitbeck, T. M. Cairns, B. Reilly, G. H. Cohen, and R. J. Eisenberg. 2007. Bimolecular complementation reveals that glycoproteins gB and gH/gL of herpes simplex virus interact with each other during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 104:18718-18723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avitabile, E., C. Forghieri, and G. Campadelli-Fiume. 2007. Complexes between herpes simplex virus glycoproteins gD, gB, and gH detected in cells by complementation of split enhanced green fluorescent protein. J. Virol. 81:11532-11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avitabile, E., C. Forghieri, and G. Campadelli-Fiume. 2009. Cross talk among the glycoproteins involved in herpes simplex virus entry and fusion: the interaction between gB and gH/gL does not necessarily require gD. J. Virol. 83:10752-10760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Backovic, M., R. Longnecker, and T. S. Jardetzky. 2009. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc. Natl. Acad. Sci. U. S. A. 106:2880-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bzik, D. J., B. A. Fox, N. A. DeLuca, and S. Person. 1984. Nucleotide sequence of a region of the herpes simplex virus type 1 gB glycoprotein gene: mutations affecting rate of virus entry and cell fusion. Virology 137:185-190. [DOI] [PubMed] [Google Scholar]
- 9.Cai, W. H., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai, W. Z., S. Person, S. C. Warner, J. H. Zhou, and N. A. DeLuca. 1987. Linker-insertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J. Virol. 61:714-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 12.Cocchi, F., L. Menotti, V. Di Ninni, M. Lopez, and G. Campadelli-Fiume. 2004. The herpes simplex virus JMP mutant enters receptor-negative J cells through a novel pathway independent of the known receptors nectin1, HveA, and nectin2. J. Virol. 78:4720-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocchi, F., L. Menotti, P. Mirandola, M. Lopez, and G. Campadelli-Fiume. 1998. The ectodomain of a novel member of the immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 72:9992-10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connolly, S. A., D. J. Landsburg, A. Carfi, J. C. Whitbeck, Y. Zuo, D. C. Wiley, G. H. Cohen, and R. J. Eisenberg. 2005. Potential nectin-1 binding site on herpes simplex virus glycoprotein D. J. Virol. 79:1282-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connolly, S. A., G. P. Leser, T. S. Jardetzky, and R. A. Lamb. 2009. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J. Virol. 83:10857-10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeLuca, N., D. J. Bzik, V. C. Bond, S. Person, and W. Snipes. 1982. Nucleotide sequences of herpes simplex virus type 1 (HSV-1) affecting virus entry, cell fusion, and production of glycoprotein gb (VP7). Virology 122:411-423. [DOI] [PubMed] [Google Scholar]
- 17.Desai, P., and S. Person. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 72:7563-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desai, P. J., P. A. Schaffer, and A. C. Minson. 1988. Excretion of non-infectious virus particles lacking glycoprotein H by a temperature-sensitive mutant of herpes simplex virus type 1: evidence that gH is essential for virion infectivity. J. Gen. Virol. 69:1147-1156. [DOI] [PubMed] [Google Scholar]
- 19.Esko, J. D., T. E. Stewart, and W. H. Taylor. 1985. Animal cell mutants defective in glycosaminoglycan biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 82:3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farnsworth, A., T. W. Wisner, and D. C. Johnson. 2007. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81:319-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuller, A. O., R. E. Santos, and P. G. Spear. 1989. Neutralizing antibodies specific for glycoprotein H of herpes simplex virus permit viral attachment to cells but prevent penetration. J. Virol. 63:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuller, A. O., and P. G. Spear. 1987. Anti-glycoprotein D antibodies that permit adsorption but block infection by herpes simplex virus 1 prevent virion-cell fusion at the cell surface. Proc. Natl. Acad. Sci. U. S. A. 84:5454-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fusco, D., C. Forghieri, and G. Campadelli-Fiume. 2005. The pro-fusion domain of herpes simplex virus glycoprotein D (gD) interacts with the gD N terminus and is displaced by soluble forms of viral receptors. Proc. Natl. Acad. Sci. U. S. A. 102:9323-9328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geraghty, R. J., C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor-related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 25.Gianni, T., M. Amasio, and G. Campadelli-Fiume. 2009. Herpes simplex virus gD forms distinct complexes with fusion executors gB and gH/gL in part through the C-terminal profusion domain. J. Biol. Chem. 284:17370-17382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gianni, T., P. L. Martelli, R. Casadio, and G. Campadelli-Fiume. 2005. The ectodomain of herpes simplex virus glycoprotein H contains a membrane alpha-helix with attributes of an internal fusion peptide, positionally conserved in the Herpesviridae family. J. Virol. 79:2931-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gianni, T., L. Menotti, and G. Campadelli-Fiume. 2005. A heptad repeat in herpes simplex virus 1 gH, located downstream of the alpha-helix with attributes of a fusion peptide, is critical for virus entry and fusion. J. Virol. 79:7042-7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hannah, B. P., T. M. Cairns, F. C. Bender, J. C. Whitbeck, H. Lou, R. J. Eisenberg, and G. H. Cohen. 2009. Herpes simplex virus glycoprotein B associates with target membranes via its fusion loops. J. Virol. 83:6825-6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hannah, B. P., E. E. Heldwein, F. C. Bender, G. H. Cohen, and R. J. Eisenberg. 2007. Mutational evidence of internal fusion loops in herpes simplex virus glycoprotein B. J. Virol. 81:4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heldwein, E. E., H. Lou, F. C. Bender, G. H. Cohen, R. J. Eisenberg, and S. C. Harrison. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science 313:217-220. [DOI] [PubMed] [Google Scholar]
- 31.Highlander, S. L., W. H. Cai, S. Person, M. Levine, and J. C. Glorioso. 1988. Monoclonal antibodies define a domain on herpes simplex virus glycoprotein B involved in virus penetration. J. Virol. 62:1881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Highlander, S. L., D. J. Dorney, P. J. Gage, T. C. Holland, W. Cai, S. Person, M. Levine, and J. C. Glorioso. 1989. Identification of mar mutations in herpes simplex virus type 1 glycoprotein B which alter antigenic structure and function in virus penetration. J. Virol. 63:730-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hutchinson, L., H. Browne, V. Wargent, N. Davis-Poynter, S. Primorac, K. Goldsmith, A. C. Minson, and D. C. Johnson. 1992. A novel herpes simplex virus glycoprotein, gL, forms a complex with glycoprotein H (gH) and affects normal folding and surface expression of gH. J. Virol. 66:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang, C., M. Ataai, A. Ozuer, D. Krisky, J. Wechuck, S. Pornsuwan, F. Pourarian, and J. C. Glorioso. 2006. Inactivation of herpes simplex type 1 gene vector on immobilized metal affinity chromatography: oxidative damage by hydroxyl free radicals and its prevention. Biotechnol. Bioeng. 95:48-57. [DOI] [PubMed] [Google Scholar]
- 35.Kousoulas, K. G., P. E. Pellett, L. Pereira, and B. Roizman. 1984. Mutations affecting conformation or sequence of neutralizing epitopes identified by reactivity of viable plaques segregate from syn and ts domains of HSV-1(F) gB gene. Virology 135:379-394. [DOI] [PubMed] [Google Scholar]
- 36.Krummenacher, C., I. Baribaud, J. F. Sanzo, G. H. Cohen, and R. J. Eisenberg. 2002. Effects of herpes simplex virus on structure and function of nectin-1/HveC. J. Virol. 76:2424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krummenacher, C., V. M. Supekar, J. C. Whitbeck, E. Lazear, S. A. Connolly, R. J. Eisenberg, G. H. Cohen, D. C. Wiley, and A. Carfi. 2005. Structure of unliganded HSV gD reveals a mechanism for receptor-mediated activation of virus entry. EMBO J. 24:4144-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwon, H., Q. Bai, H. J. Baek, K. Felmet, E. A. Burton, W. F. Goins, J. B. Cohen, and J. C. Glorioso. 2006. Soluble V domain of nectin-1/HveC enables entry of herpes simplex virus type 1 (HSV-1) into HSV-resistant cells by binding to viral glycoprotein D. J. Virol. 80:138-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemmon, M. A. 2004. Pleckstrin homology domains: not just for phosphoinositides. Biochem. Soc. Trans. 32:707-711. [DOI] [PubMed] [Google Scholar]
- 40.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by beta-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menotti, L., A. Cerretani, H. Hengel, and G. Campadelli-Fiume. 2008. Construction of a fully retargeted herpes simplex virus 1 recombinant capable of entering cells solely via human epidermal growth factor receptor 2. J. Virol. 82:10153-10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Menotti, L., G. Nicoletti, V. Gatta, S. Croci, L. Landuzzi, C. De Giovanni, P. Nanni, P. L. Lollini, and G. Campadelli-Fiume. 2009. Inhibition of human tumor growth in mice by an oncolytic herpes simplex virus designed to target solely HER-2-positive cells. Proc. Natl. Acad. Sci. U. S. A. 106:9039-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, C. G., C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and N. W. Fraser. 2001. Development of a syngenic murine B16 cell line-derived melanoma susceptible to destruction by neuroattenuated HSV-1. Mol. Ther. 3:160-168. [DOI] [PubMed] [Google Scholar]
- 44.Milne, R. S., A. V. Nicola, J. C. Whitbeck, R. J. Eisenberg, and G. H. Cohen. 2005. Glycoprotein D receptor-dependent, low-pH-independent endocytic entry of herpes simplex virus type 1. J. Virol. 79:6655-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura, T., K. W. Peng, S. Vongpunsawad, M. Harvey, H. Mizuguchi, T. Hayakawa, R. Cattaneo, and S. J. Russell. 2004. Antibody-targeted cell fusion. Nat. Biotechnol. 22:331-336. [DOI] [PubMed] [Google Scholar]
- 47.Nicola, A. V., A. M. McEvoy, and S. E. Straus. 2003. Roles for endocytosis and low pH in herpes simplex virus entry into HeLa and Chinese hamster ovary cells. J. Virol. 77:5324-5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicola, A. V., and S. E. Straus. 2004. Cellular and viral requirements for rapid endocytic entry of herpes simplex virus. J. Virol. 78:7508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reymond, N., S. Fabre, E. Lecocq, J. Adelaide, P. Dubreuil, and M. Lopez. 2001. Nectin4/PRR4, a new afadin-associated member of the nectin family that trans-interacts with nectin1/PRR1 through V domain interaction. J. Biol. Chem. 276:43205-43215. [DOI] [PubMed] [Google Scholar]
- 50.Shukla, D., J. Liu, P. Blaiklock, N. W. Shworak, X. Bai, J. D. Esko, G. H. Cohen, R. J. Eisenberg, R. D. Rosenberg, and P. G. Spear. 1999. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 99:13-22. [DOI] [PubMed] [Google Scholar]
- 51.Shukla, D., and P. G. Spear. 2001. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J. Clin. Invest. 108:503-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Struyf, F., W. M. Martinez, and P. G. Spear. 2002. Mutations in the N-terminal domains of nectin-1 and nectin-2 reveal differences in requirements for entry of various alphaherpesviruses and for nectin-nectin interactions. J. Virol. 76:12940-12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian, R. P., and R. J. Geraghty. 2007. Herpes simplex virus type 1 mediates fusion through a hemifusion intermediate by sequential activity of glycoproteins D, H, L, and B. Proc. Natl. Acad. Sci. U. S. A. 104:2903-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uchida, H., W. A. Shah, A. Ozuer, A. R. Frampton, Jr., W. F. Goins, P. Grandi, J. B. Cohen, and J. C. Glorioso. 2009. Generation of herpesvirus entry mediator (HVEM)-restricted herpes simplex virus type 1 mutant viruses: resistance of HVEM-expressing cells and identification of mutations that rescue nectin-1 recognition. J. Virol. 83:2951-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Warner, M. S., R. J. Geraghty, W. M. Martinez, R. I. Montgomery, J. C. Whitbeck, R. Xu, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. A cell surface protein with herpesvirus entry activity (HveB) confers susceptibility to infection by mutants of herpes simplex virus type 1, herpes simplex virus type 2, and pseudorabies virus. Virology 246:179-189. [DOI] [PubMed] [Google Scholar]
- 56.Woolhouse, M. E., and S. Gowtage-Sequeria. 2005. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 11:1842-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou, G., and B. Roizman. 2006. Construction and properties of a herpes simplex virus 1 designed to enter cells solely via the IL-13alpha2 receptor. Proc. Natl. Acad. Sci. U. S. A. 103:5508-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]