Abstract
The chloroviruses (family Phycodnaviridae), unlike most viruses, encode some, if not most, of the enzymes involved in the glycosylation of their structural proteins. Annotation of the gene product B736L from chlorovirus NY-2A suggests that it is a glycosyltransferase. The structure of the recombinantly expressed B736L protein was determined by X-ray crystallography to 2.3-Å resolution, and the protein was shown to have two nucleotide-binding folds like other glycosyltransferase type B enzymes. This is the second structure of a chlorovirus-encoded glycosyltransferase and the first structure of a chlorovirus type B enzyme to be determined. B736L is a retaining enzyme and belongs to glycosyltransferase family 4. The donor substrate was identified as GDP-mannose by isothermal titration calorimetry and was shown to bind into the cleft between the two domains in the protein. The active form of the enzyme is probably a dimer in which the active centers are separated by about 40 Å.
Glycosyltransferases constitute a large family of enzymes that catalyze the transfer of sugar moieties from donor molecules to specific acceptor molecules. Unlike other enzyme families that usually share conserved features in their primary sequences, glycosyltransferases can have highly diversified sequences that have been grouped into more than 90 families (designated GTn, where n = 1, 2, …) (http://www.CAZy.org) (1, 15). However, two families, GT2 and GT4, account for about half of the total number of glycosyltransferases. Despite the large variation in the primary sequences of glycosyltransferases, their three-dimensional structures are usually conserved. There are two major glycosyltransferase structural types, named GT-A and GT-B. The GT-A members contain a single nucleotide-binding domain consisting of six parallel β-strands flanked by connecting α-helices (referred to as a “Rossmann fold” in most of the literature on these enzymes and herein). GT-A enzyme activities are usually metal ion dependent. The GT-B type glycosyltransferases have two Rossmann folds separated by a cleft that forms the substrate-binding site. Metal ions are normally not required for GT-B function. Based on their catalytic mechanism, glycosyltransferases are also classified as either retaining or inverting enzymes depending on the geometry between the sugar donor and the receptor in the product molecule (e.g., depending on whether the anomeric carbon atom is linked to the acceptor via its α or β position). If the anomeric carbon atom has the same configuration in the donor and in the product, the enzyme is classified as a retaining enzyme; if the configurations are different, the enzyme is considered to be an inverting enzyme (2).
Many viruses, especially those that infect eukaryotic cells, have extensively glycosylated structural proteins. Glycans coating viral structural proteins serve multiple biological roles, e.g., they mimic host glycans to evade host cell immune reactions, aid in folding or assembly of viral structural proteins, function as a receptor recognized by cell surface proteins, or aid in stabilizing viral particles (see, e.g., reference 36).
Typically, viruses use host-encoded glycosyltransferases and glycosidases located in the endoplasmic reticulum (ER) and Golgi apparatus to add and remove N-linked sugar residues from virus glycoproteins either during or shortly after translation of the protein. This posttranslational processing aids in protein folding and requires other host-encoded enzymes. After folding and assembly, virus glycoproteins are transported by host-sorting and membrane transport functions to virus-specified regions in host membranes, where they displace host glycoproteins. Progeny viruses then bud through these virus-specific target membranes, in what is usually the final step in the assembly of infectious virions (3, 14, 21, 36). Thus, nascent viruses become infectious only by budding through the target membrane, usually the plasma membrane, as they are released from the cell. Consequently, the glycan portion of virus glycoproteins is host specific. The theme that emerges is that virus glycoproteins are synthesized and glycosylated by the same mechanisms as host glycoproteins. Therefore, the only way to alter glycosylation of virus proteins is to either grow the virus in a different host or have a mutation in the virus protein that alters the protein glycosylation site.
One explanation for this scenario is that, in general, viruses lack genes encoding glycosyltransferases. However, a few virus-encoded glycosyltransferases have been reported in recent years (see reference 17 for a review). Often these virus-encoded glycosyltransferases add sugars to compounds other than proteins. For instance, some phage-encoded glycosyltransferases modify virus DNA to protect it from host restriction endonucleases (see, e.g., reference 10), and a glycosyltransferase encoded by baculoviruses modifies a host insect ecdysteroid hormone, leading to its inactivation (22). Bovine herpesvirus 4 encodes a β-1,6-N-acetyl-glucosaminyltransferase that is localized in the Golgi apparatus and is probably involved in posttranslational modification of the virus structural proteins (32).
One group of viruses differs from the scenario that viruses use the host machinery located in the ER and the Golgi apparatus to glycosylate their glycoproteins. These viruses are the large, plaque-forming, double-stranded DNA (dsDNA)-containing chloroviruses (family Phycodnaviridae) that infect eukaryotic algae (4, 34, 39, 40). The chloroviruses have up to 400 protein-encoding genes (or coding sequences [CDSs]). Annotation of six chlorovirus genomes showed that each virus encodes 3 to 6 putative glycosyltransferases (7-9, 16, 33). Three of these viruses, NY-2A, AR158, and the prototype chlorovirus Paramecium bursaria chlorella virus 1 (PBCV-1), infect Chlorella strain NC64A. Two of the viruses, MT325 and FR483, infect Chlorella Pbi, and one of them, Acanthocystis turfacea chlorella virus (ATCV-1), infects Chlorella SAG 3.83.
Glycosylation of the PBCV-1 major capsid protein, Vp54, is at least partially performed by the viral glycosyltransferases (11, 20, 33, 38, 41). PBCV-1 encodes 5 putative glycosyltransferases. A previous structural study established that the N-terminal 211 amino acids of the A64R protein from PBCV-1 form a GT-A group glycosyltransferase that is a retaining enzyme belonging to the GT34 family and that UDP-glucose possibly serves as the donor sugar (41).
Among the four additional PBCV-1 glycosyltransferase-encoding genes, gene a546l encodes a 396-amino-acid protein that resembles members in the GT4 family of glycosyltransferases, based on amino acid sequence comparison of members in the CAZy classification (1, 15). Homologs of this protein, A546L, are encoded by 3 other chloroviruses, NY-2A, AR158, and ATCV-1. Here, we report the crystal structure of one of these homologs, B736L, at 2.3-Å resolution.
MATERIALS AND METHODS
Phylogenetic analyses.
Phylogenetic analyses began with a BLASTP search using the amino acid sequence of NY-2A B736L (RefSeq accession no. YP_001497932) against the NCBI nonredundant protein sequence database with default settings, as well as against the specific animal (NCBI taxonomy identification no. [taxid] 33208) and plant (taxid 3193) databases. A Bayesian inference tree of 27 characterized and hypothetical glycosyltransferase amino acid sequences was generated based on sequence alignment using MUSCLE and MrBayes programs within the Geneious Pro 4.8.5 software program (http://www.geneious.com). The phylogenetic tree was produced by utilization of a Poisson fixed amino acid rate matrix with the gamma rate variation and an evaluation category of four. The Markov chain Monte Carlo (MCMC) algorithm approach was used with a chain length setting of 1,100,000; the number of chains was 4, the temperature of heated chains was 0.2, and the subsampling frequency was 200, with a burn in length of 100,000 and a random seed number of 24,870. A zebrafish (Danio rerio) glycosyltransferase sequence (RefSeq accession no. XP_693602) served as the out-group.
Protein production and crystallization.
The B736L gene was amplified by PCR from virus NY-2A genomic DNA. Purified PCR products were treated with restriction endonucleases AseI and XhoI and cloned into pET30b (Novagen) by using the NdeI-XhoI sites that introduce a C-terminal His6 tag into the recombinant protein. Recombinant B736L was expressed in Escherichia coli strain BL21(DE3) CodonPlus-RIL cells at 20°C and was affinity purified to homogeneity by using cobalt-charged BD TALON resins. The eluted protein from the cobalt beads was concentrated and further purified on a Superdex 200 column (GE Healthcare). Based on the elution volume and appropriate calibration of the Superdex 200 column (see Fig. S1 in the supplemental material), the molecular mass was estimated to be about 84 kDa, approximately twice the calculated molecular mass of B736L (45.5 kDa). The selenomethionine derivative of B736L was produced and purified using procedures similar to those for the native protein. B736L crystals were obtained in 100 mM Tris-HCl (pH 7.0)-20% (wt/vol) polyethylene glycol 3350. B736L crystals in a complex with GDP-mannose were obtained by soaking apo-B736L crystals in the same well solution supplied with 10 mM GDP-mannose for 5 h. The selenomethionine derivative of B736L was crystallized by using conditions similar to those used for the native form but with 10 mM dithiothreitol (DTT).
ITC.
Isothermal titration calorimetry (ITC) was performed using a VP-ITC system (MicroCal Inc.). The enzyme and the different ligands were diluted into the same batch of buffer containing 50 mM Tris-HCl, pH 8.0, and 100 mM NaCl and were degassed under a vacuum. Titrations were performed by injecting 23 consecutive aliquots (5 μl each for the first 6 injections, 10 μl each for the following 13 injections, and 20 μl each for the last 4 injections) of CMP, UDP, ADP, GDP, GDP-mannose, GDP-fucose, or GDP-glucose solutions into the ITC cell (volume, 1.4 ml) containing B736L at 25°C. The substrate concentration was about 0.6 mM, whereas the protein concentration was about 1.2 mg/ml (∼25 μΜ). ITC data were corrected for heats of dilution of the substrate and product solutions. Binding stoichiometries, enthalpy values, and equilibrium dissociation constants were determined by fitting corrected data to a biomolecular interaction model using Origin7 software (MicroCal Inc.).
X-ray data collection, processing, structure determination, refinement, and analysis.
X-ray diffraction data were collected using synchrotron radiation on General Medicine and Cancer Institutes Collaborative Access Team (GM/CA-CAT) beamline 23ID at the Advanced Photon Source (Table 1). Crystals of B736L diffracted to 2.3-Å resolution, belonged to space group C2221 with two molecules in the asymmetric unit, and had the following cell parameters: a, 153.6 Å; b, 246.9 Å; and c, 67.2 Å. A single-wavelength anomalous diffraction (SAD) data set (λ = 0.9794 Å) was collected to 3.0-Å resolution from a single crystal of selenomethionine-labeled protein. All data were processed with the HKL2000 suite (Table 1) (23).
TABLE 1.
Data collection and refinement statistics
| Parameter | Value(s)a for: |
||
|---|---|---|---|
| SeMetb derivative | Native enzyme | GDP-mannose complex | |
| Data collection statistics | |||
| Wavelength (Å)/beamline | 0.9794/23ID | 1.03/23ID | 0.98/23ID |
| Space group | C2221 | C2221 | C2221 |
| Unit cell dimensions (Å) | |||
| a | 152.72 | 153.62 | 154.25 |
| b | 247.38 | 246.94 | 243.19 |
| c | 67.36 | 67.17 | 67.40 |
| No. of molecules in asymmetric unit | 2 | 2 | 2 |
| Resolution range (Å) | 50.0-3.0 (3.11-3.00) | 50.0-2.3 (2.38-2.30) | 50.0-2.7 (2.84-2.74) |
| Rsym (%) | 12.3 (54.8) | 7.4 (49.1) | 9.1 (56.6) |
| I/σ〈I〉 | 31.6 (7.7) | 27.1 (4.2) | 19.1 (2.3) |
| Completeness (%) | 99.2 (99.9) | 99.8 (99.6) | 98.4 (86.7) |
| No. of unique reflections | 26,599 | 56,045 | 33,711 |
| Refinement statistics | |||
| Rcryst (%) | 23.5 | 25.9 | |
| Rfree (%) | 26.8 | 29.0 | |
| RMSD | |||
| Bond length (Å) | 0.016 | 0.007 | |
| Bond angle (°) | 1.78 | 1.06 | |
| No. of: | |||
| Atoms in protein | 6,204 | 6,204 | |
| GDP-mannose molecules | 0 | 2 | |
| Water molecules | 107 | 9 | |
| Average B factor (Å2) | |||
| Protein | 62.2 | 91.4 | |
| Molecule A: domain N, C | 42.3, 48.5 | 67.2, 76.7 | |
| Molecule B: domain N, C | 107.0, 51.59 | 151.4, 74.4 | |
| GDP-mannose | 94.2 | ||
| Water | 52.0 | 46.9 | |
The values in parentheses represent the highest-resolution shell.
SeMet, selenomethionine.
The program SOLVE (31) was used for locating the Se atoms. The structure was then resolved using SAD phasing from the anomalous signals of the Se atoms with the program SOLVE (31). Density modification was performed with the program RESOLVE (30).
Model building and adjustments were carried out with the program Coot (5). The program REFMAC was used to refine all the structures (Table 1) (19). The resultant refined model consists of residues 1 to 392 and gave Rworking and Rfree factors of 23.5 and 26.8%, respectively. The 13 C-terminal residues and the C-terminal His6 tag were not visible in the electron density maps. The programs Chimera (25) and Ligplot (37) were used for producing figures.
Protein structure accession numbers.
The atomic coordinates have been deposited in the Protein Data Bank under accession codes 3OY7 and 3OY2.
RESULTS AND DISCUSSION
Sequence analysis.
As mentioned in the introduction, the six sequenced chloroviruses contain 3 to 6 putative glycosyltransferase-encoding genes (7-9, 16, 33). Typically, glycosyltransferases account for about 1 to 2% of the gene products of an organism, whether archaeal, bacterial, or eukaryotic (15). Interestingly, this proportion holds true for the chloroviruses. Single glycosyltransferases from viruses PBCV-1 (A546L), NY-2A (B736L), and AR158 (C661L), all of which infect the same host, are homologs. They are predicted to have a GT-B fold and belong to the GT4 family. B736L and C661L have 98% amino acid identity to each other and 81% amino acid identity to A546L. Virus ATCV-1, which infects Chlorella SAG.3.83, also encodes a protein (Z667L) that is predicted to be a homolog of B736L. Z667L has 26% amino acid identity to B736L.
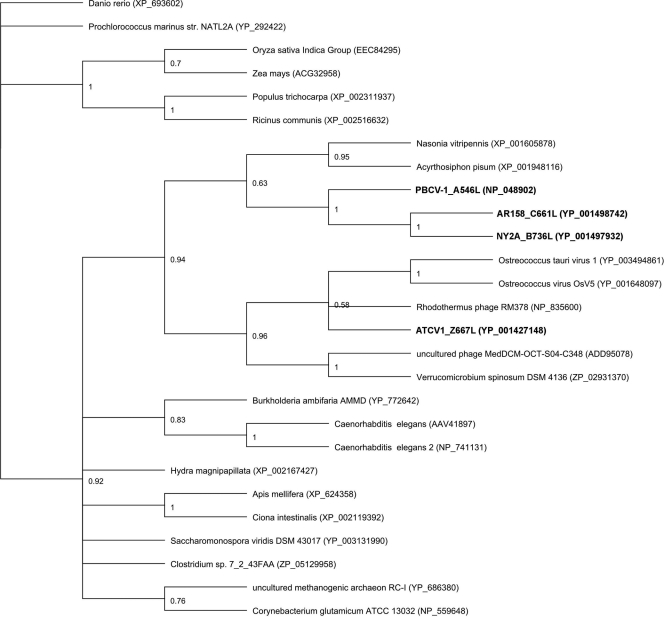
Molecular phylogenetic analyses of A546L, B736L, and C661L indicate that these homologous glycosyltransferases from the above-mentioned three viruses, all of which infect Chlorella NC64A, are in the same clade (Fig. 1). The ATCV-1 glycosyltransferase, Z667L, is in a related clade that also includes proteins from two other algal viruses (Ostreococcus tauri viruses 1 and OsV5), two bacteriophages, and a rare bacterium, Verrucomicrobium spinosum. These results are interesting for two reasons: first, the two sequenced chloroviruses, MT325 and FR483, that infect Chlorella Pbi lack a homolog of B736L (7), and second, B736L does not resemble the other chlorovirus-encoded glycosyltransferases. These findings suggest that the chloroviruses independently acquired the genes for the glycosyltransferases.
FIG. 1.
Bayesian inference tree of 27 glycosyltransferase protein sequences (designations of the four chlorella virus-encoded enzymes are in bold). The phylogenetic tree was generated using the MUSCLE alignment program with a maximum number of iterations of 8 and the MrBayes tree-building program within the Geneious Pro 4.8.5 software program. The Poisson amino acid rate matrix (gamma rate variation and an evaluation category of four) and the MCMC algorithm (chain length setting of 1,100,000; the number of chains was 4, the temperature of heated chains was 0.2, and the subsampling frequency was 200, with a burn in length of 100,000 and a random seed number of 24,870) were utilized with the Danio rerio sequence as the out-group (bootstrap values are shown).
The 405-amino-acid B736L protein from virus NY-2A was chosen as a representative of this group of glycosyltransferases and subjected to structural analysis.
Structure of B736L.
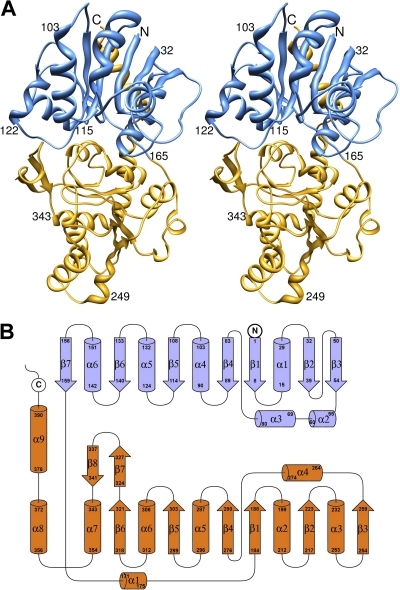
Two molecules related by a noncrystallographic 2-fold axis were in the asymmetrical unit. The B736L structure has two similar Rossmann folds (residues 1 to 163 and 164 to 355) separated by a deep cleft (Fig. 2), displaying a typical GT-B type structure. In addition, residues 356 to 392 form a long α-helix that spans over both the N-terminal and the C-terminal domains. A long antiparallel β hairpin insertion (residues 322 to 341) occurs between the end of the 6th β-strand of the C-terminal domain and the following α-helix. Structural homology searches using Dali show that the B736L structure resembles (with Z scores of ∼30) many GT-B type glycosyltransferases belonging to different glycosyltransferase families. The closest structural neighbor is a glycosyltransferase named MshA from Corynebacterium glutamicum (Protein Data Bank [PDB] no. 3c4v), with a Dali Z score of 31.5, a root mean square deviation [RMSD] of 3.0 Å, and 15% amino acid sequence identity (35). MshA is a member of the GT4 family of glycosyltransferases. Like in other GT-B type glycosyltransferases, there is a deep cleft between the two domains, which is where the donor and acceptor substrates bind. No metal ion was detected in the cleft of B736L, although Mg2+ ions were included in the crystallization process. Structural comparisons of B736L with other GT-B type glycosyltransferases show that the main structural differences are in the several loops near the substrate-binding cleft (Fig. 3).
FIG. 2.
Crystal structure of glycosyltransferase expressed from virus NY-2A gene b736l. (A) Stereoribbon diagram of the B736L structure. The N- and C-terminal domains are colored blue and yellow, respectively. (B) Topology diagram of the B736L structure. The N- and C-terminal domains are colored blue and orange, respectively.
FIG. 3.
Sequences and structural alignment of NY-2A glycosyltransferase B736L, MshA from Corynebacterium glutamicum (3C4V), BA1558 from Bacillus anthracis (2JJM), phosphatidylinositol mannosyltransferase PimA from Mycobacterium smegmatis (2GEK), and WaaG from Escherichia coli (2IV7). Secondary structural elements of each protein are shown above the alignments (arrows indicate β-strands, and coils indicate α-helices). Completely conserved residues are shown in white on a black background. Partially conserved residues are boxed. Residues involved in substrate binding to B736L are indicated by black and gray triangles. Residues critical for catalyzing the transferring reaction of B736L are indicated by black triangles. Residues involved in the dimer interface contacts are indicated by filled black circles.
The biological functional unit.
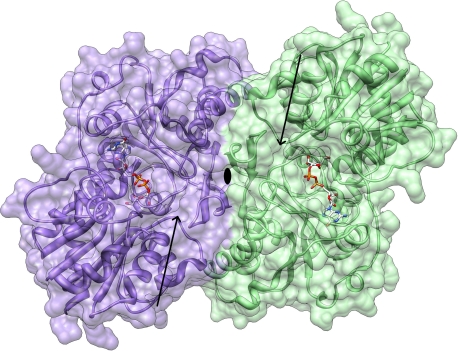
Size exclusion chromatography purification of B736L suggested that B736L exists as a dimer in solution (see Materials and Methods), which suggests that the crystallographic B736L dimer is probably the biological functional unit (Fig. 4). The B736L dimer is formed mainly through the C-terminal domain and especially by the β hairpin (residues 322 to 341). Interactions between the two molecules of the dimer include hydrogen bond formation, hydrophobic interactions, and salt bridge formation. The buried surface area between the two molecules is determined to be 2,577 Å2 when a probe radius of 1.4 Å is used. In the dimer structure, the N-terminal domains of B736L protrude from the dimeric interface formed mainly by the C-terminal domains. One of the N-terminal domains has close contacts with neighboring symmetry-related molecules, while the other N-terminal domain has only loose contacts with neighboring molecules. The electron densities of the N-terminal domain that has loose contacts with neighboring molecules indicate that it is highly flexible. This is also reflected by main-chain temperature factor comparisons of the N-terminal domain with its C-terminal domain in the more flexible monomer (average B factors, ∼107 and ∼50 Å2 for the N- and C-terminal domains, respectively) (Table 1).
FIG. 4.
Dimer structure of B736L. The two molecules are colored purple and green and are shown in ribbon and surface representations. The bound GDP-mannose substrates are depicted by a ball-and-stick model. Nitrogen, carbon, oxygen, and phosphorus atoms are colored blue, white, red, and yellow, respectively. The predicted acceptor-binding sites are indicated by arrows.
Translation/libration/screw refinement and analysis show movements of the flexible N-terminal domain relative to the rest of the dimer molecule that open and close the substrate-binding site in the cleft of the more flexible molecule. The less flexible monomer is held more tightly to other monomers in the crystal lattice than the more flexible monomer (Table 1). The N-terminal domain motion may be required for substrate binding or release. Similar domain motions have been discussed in analyses of other glycosyltransferase structures (18).
There are many reports of homodimeric glycosyltransferases (6, 13, 24, 29). The dimeric structures have increased thermostability (27). Some of these dimeric molecules, for instance, α1,3-fucosyltransferase from Helicobacter pylori, have been suggested to be biological functional units that target oligomeric substrates (6, 29). Among glycosyltransferases, including B736L, that form dimers and also have had their structure determined, the two active sites are often on the same side of the molecule. The approximate separation of the active centers is nevertheless quite large, being about 35Å for B736L. Notwithstanding this similarity, there is little similarity of residue types on the surface of contact between monomers that form the dimers. Thus, it is probable that the crystallographic dimer described herein has no functional significance except to increase the thermal stability of the molecule.
The donor substrate.
ITC is an efficient physical technique used to study binding of small molecules to large macromolecules by directly measuring the binding affinity, enthalpy changes, and binding stoichiometry of the interaction between the molecules in solution. UDP, GDP, CMP, and ADP are the most common nucleotides used by monosaccharide donors of glycosyltransferases. Usually, a glycosyltransferase recognizes a specific monosaccharide donor substrate that is derived from one of the four nucleotides. Although previous studies show that glycosyltransferases have only weak interactions with the nucleotides themselves, it is still possible to differentiate among the four possibilities (28). Thus, the ITC technique was used to screen the four nucleotides for their abilities to bind to B736L. Binding of GDP to B736L was significantly better than that of the other nucleotides (see Fig. S2 in the supplemental material), suggesting that GDP is the monosaccharide carrier used by B736L. Further ITC experiments with GDP-mannose, GDP-fucose, and GDP-glucose, the most common GDP sugars, indicated that GDP-mannose binds to the enzyme significantly better than the other two GDP sugars (Fig. 5). The disassociation constant of GDP-mannose for the enzyme is about 10 μM (change in enthalpy [ΔH] = −11.3 kcal·mol−1) (Fig. 5), which resembles that of monosaccharide donors for other glycosyltransferases (28). These results suggest that GDP-mannose is the specific monosaccharide donor for B736L.
FIG. 5.
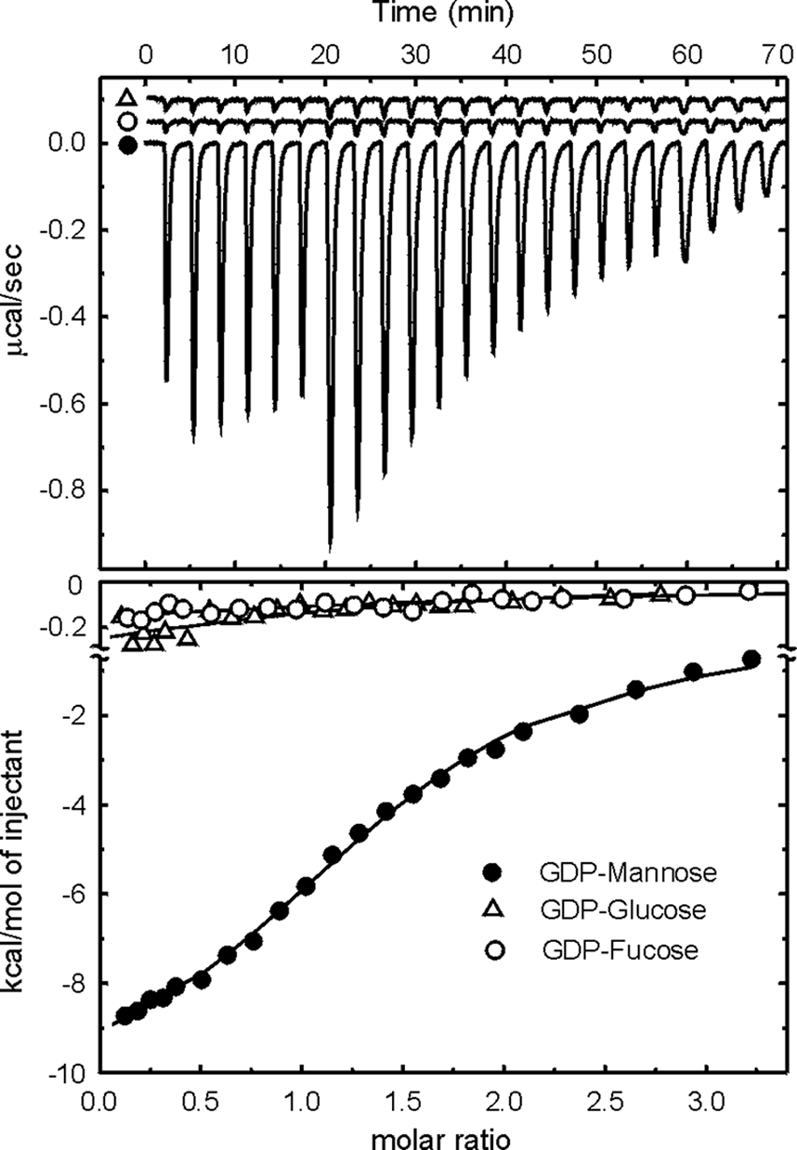
ITC measurements of enzyme-substrate interactions. The upper panel shows the titrations of B736L with GDP-mannose, GDP-glucose, or GDP-fucose. The lower panel shows the heat released when a substrate binds to the enzyme as a function of relative concentrations. Corrections have been made for the heat release due to dilution. Solid lines correspond to the best fit of data using a bimolecular interaction model.
Substrate binding.
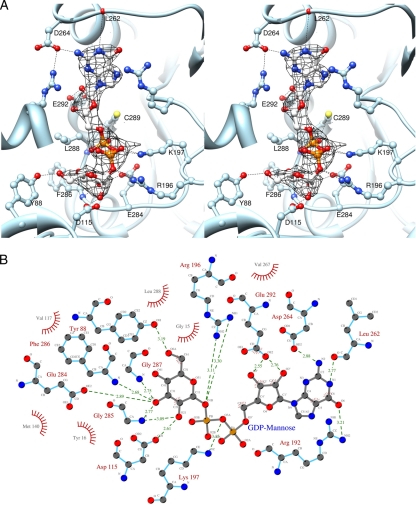
Based on the ITC results, B736L crystals in a complex with GDP-mannose were prepared by soaking the nucleotide-free enzyme crystals with GDP-mannose. The difference map (Fig. 6A) for a GDP-mannose-soaked crystal and a crystal of the native enzyme showed strong positive density in the pocket in the cleft of each monomer associated primarily with the C-terminal domain. This increased density is about the right volume for accommodating one GDP-mannose molecule. Model building and refinement for the bound GDP-mannose molecules (Fig. 6) show that they are coordinated by the side chains of residues Tyr88, Asp115, Arg196, Lys197, Asp264, Glu284, Cys289, and Glu292 through hydrogen bonds.
FIG. 6.
Structure of B736L in a complex with the donor substrate GDP-mannose. (A) Stereodiagram showing the interaction between GDP-mannose and protein residues. The GDP-mannose and the protein residue side chains are shown as ball-and-stick representations. Carbon, nitrogen, and oxygen atoms in the substrate are colored gray, blue, and red, respectively. The (Fo − Fc) electron density map is contoured at 2.6 sigma. (B) Schematic diagram showing the interactions between B736L and GDP-mannose. Distances are given in angstroms. Carbon, nitrogen, and oxygen atoms are labeled Cx, Nx, and Ox, respectively.
The structure of the refined complex shows that the overall fold of the protein is not significantly perturbed by GDP-mannose binding, as indicated by an RMSD between equivalent Cα atoms of the apoenzyme and the substrate-bound enzyme of only 0.56 Å. However, there are local conformational changes for the side chains of a few residues, such as Leu262 and Leu288.
Structural comparisons of B736L with other GDP-mannose-binding glycosyltransferases, such as the phosphatidylinositol mannosyltransferase PimA (PDB no. 2gej) (12), indicate that Glu284 is conserved among the residues that form hydrogen bonds with the mannose sugar ring of the donor. The two PimA residues that correspond to residues Glu284 and Asp115 of B736L have been proposed to be critical for catalysis (12). Although the acceptor for B736L is unknown, the rough position of the acceptor-binding site was established by homology modeling. A structural superposition of B736L with the structure of the glycosyltransferase MshA in a complex with its acceptor shows that residue Asp115 of B736L may have direct contacts with both the donor and the acceptor substrates (Fig. 6B). Thus, Asp115 should be a critical residue for catalyzing the transferring reaction.
Implications for viral glycosylation.
The structure of B736L provides additional support for the concept that the chlorella viruses encode most, if not all, of the machinery to glycosylate their major capsid proteins. The structure studies indicate that B736L binds GDP-mannose and possibly utilizes it as the donor substrate. However, definitive proof will require more complete biochemical analyses. The finding that GDP-mannose is the donor substrate is reasonable because mannose is one of the carbohydrate components of the PBCV-1 major capsid protein (38). However, sugar analysis of the NY-2A major capsid protein indicates that glucose, galactose, rhamnose, fucose, and xylose are the primary sugars (M. P. Chothi et al., unpublished results), i.e., no mannose was detected. There are at least three explanations for this apparent contradiction: (i) some sugar other than GDP-mannose might be the donor for B736L, (ii) B736L might not be a functional enzyme for the NY-2A virus, and/or (iii) the acceptor substrate might be not the virus major capsid protein but some other virus-encoded protein(s) that contains mannose. In addition to the major capsid protein, two proteins associated with PBCV-1 virions are glycosylated (26), and the same situation probably occurs in virus NY-2A. If B736L uses some sugar other than GDP-mannose as a donor substrate, a likely candidate might be GDP-rhamnose because mannose and rhamnose have the same oxydryl configuration.
Glycosylation of the major capsid protein in the chloroviruses probably occurs in the cytoplasm, not in the ER or Golgi apparatus of the host cell (32). Consistent with this hypothesis, B736L, like the other chlorovirus-encoded glycosyltransferases, lacks an N-terminal signal sequence that would target it to the ER. Thus, the chloroviruses apparently use their own glycosyltransferases, such as B736L and the previously described A64R protein from PBCV-1 (11, 41), to glycosylate their major capsid proteins. However, the major capsid protein from NY-2A contains 5 sugars but the virus encodes only 3 predicted glycosyltransferases. Likewise, the major capsid protein from PBCV-1 contains 7 sugars and the virus is predicted to encode 5 glycosyltransferases (32). Thus, at least two of the chlorovirus major capsid proteins have more sugars in their glycans than there are virus-encoded glycosyltransferases. The extra sugars may be added by host glycosyltransferases.
Supplementary Material
Acknowledgments
We are grateful to Baerbel Kaufmann and Siyang Sun for helpful discussions and to Sheryl Kelly for help in the preparation of the manuscript. We thank the staff of Advanced Photon Source (APS) beamline 23 for help in data collection. We thank Michele Tonetti and Madhu Parakkottil Chothi for conducting the sugar analyses on virus NY-2A.
The work was supported by NIH grant AI11219 to M.G.R., in part by Public Health Service grant GM32441 (to J.L.V.E.) and NIH grant P20RR15635 from the COBRE program of the National Center for Research Resources (to J.L.V.E.), and in part by the NIAMS Intramural Research Program.
Footnotes
Published ahead of print on 22 September 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Coutinho, P. M., E. Deleury, G. J. Davies, and B. Henrissat. 2003. An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328:307-317. [DOI] [PubMed] [Google Scholar]
- 2.Davies, G. T. 2001. Sweet secrets of synthesis. Nat. Struct. Biol. 8:98-100. [DOI] [PubMed] [Google Scholar]
- 3.Doms, R. W., R. A. Lamb, J. K. Rose, and A. Helenius. 1993. Folding and assembly of viral membrane proteins. Virology 193:545-562. [DOI] [PubMed] [Google Scholar]
- 4.Dunigan, D. D., L. A. Fitzgerald, and J. L. Van Etten. 2006. Phycodnaviruses: a peek at genetic diversity. Virus Res. 117:119-132. [DOI] [PubMed] [Google Scholar]
- 5.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 6.Finel, M., and M. Kurkela. 2008. The UDP-glucuronosyltransferases as oligomeric enzymes. Curr. Drug Metab. 9:70-76. [DOI] [PubMed] [Google Scholar]
- 7.Fitzgerald, L. A., M. V. Graves, X. Li, T. Feldblyum, J. Hartigan, and J. L. Van Etten. 2007. Sequence and annotation of the 314-kb MT325 and the 321-kb FR483 viruses that infect Chlorella Pbi. Virology 358:459-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fitzgerald, L. A., M. V. Graves, X. Li, T. Feldblyum, W. C. Neirman, and J. L. Van Etten. 2007. Sequence and annotation of the 369-kb NY-2A and the 345-kb AR158 viruses that infect Chlorella NC64A. Virology 358:472-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, L. A., M. V. Graves, X. Li, J. Hartican, A. P. J. Pfitzner, E. Hoffart, and J. L. Van Etten. 2007. Sequence and annotation of the 288-kb ATCV-1 virus that infects an endosymbiotic chlorella strain of the heliozoon Acanthocystis turfacea. Virology 362:350-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gram, H., and W. Ruger. 1986. The α-glucosyltransferases of bacteriophages T2, T4 and T6. A comparison of their primary structures. Mol. Gen. Genet. 202:467-470. [DOI] [PubMed] [Google Scholar]
- 11.Graves, M. V., C. T. Bernadt, R. Cerny, and J. L. Van Etten. 2001. Molecular and genetic evidence for a virus-encoded glycosyltransferase involved in protein glycosylation. Virology 285:332-345. [DOI] [PubMed] [Google Scholar]
- 12.Guerin, M. E., J. Kordulakova, F. Schaeffer, Z. Svetlikova, A. Buschiazzo, D. Giganti, B. Gicquel, K. Mikusova, M. Jackson, and P. M. Alzari. 2007. Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria. J. Biol. Chem. 282:20705-20714. [DOI] [PubMed] [Google Scholar]
- 13.Hung, M.-N., E. Rangarajan, C. Munger, G. Nadeau, T. Sulea, and A. Matte. 2006. Crystal structure of TDP-fucosamine acetyltrasnferase (WecD) from Escherichia coli, an enzyme required for enterobacterial common antigen synthesis. J. Bacteriol. 188:5606-5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter, E. 2007. Virus assembly, p. 141-168. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Walters Kluwer/Lippincott Williams & Wilkins, Philadelphia, PA.
- 15.Lairson, L. L., B. Henrissat, G. J. Davies, and S. G. Withers. 2008. Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77:521-555. [DOI] [PubMed] [Google Scholar]
- 16.Li, Y., Z. Lu, L. Sun, S. Ropp, G. F. Kutish, D. L. Rock, and J. L. Van Etten. 1997. Analysis of 74 kb of DNA located at the right end of the 330-kb chlorella virus PBCV-1 genome. Virology 237:360-377. [DOI] [PubMed] [Google Scholar]
- 17.Markine-Goriaynoff, N., L. Gillet, J. L. Van Etten, H. Korres, N. Verma, and A. Vanderplasschen. 2004. Glycosyltransferases encoded by viruses. J. Gen. Virol. 85:2741-2754. [DOI] [PubMed] [Google Scholar]
- 18.McCoy, J. G., E. Bitto, C. A. Bingman, G. E. Wesenberg, R. M. Bannen, D. A. Kondrashov, and G. N. Phillips, Jr. 2007. Structure and dynamics of UDP-glucose pyrophosphorylase from Arabidopsis thaliana with bound UDP-glucose and UTP. J. Mol. Biol. 366:830-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 20.Nandhagopal, N., A. A. Simpson, J. R. Gurnon, X. Yan, T. S. Baker, M. V. Graves, J. L. Van Etten, and M. G. Rossmann. 2002. The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. U. S. A. 99:14758-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olofsson, S., and J. E. S. Hansen. 1998. Host cell glycosylation of viral glycoproteins. A battlefield for host defence and viral resistance. Scand. J. Infect. Dis. 30:435-440. [DOI] [PubMed] [Google Scholar]
- 22.O'Reilly, D. R., and L. K. Miller. 1989. A baculovirus blocks insect molting by producing ecdysteroid UDP-glucosyltransferase. Science 245:1110-1112. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 24.Pereira, P. J., N. Empadinhas, L. Albuquerque, B. Sá-Moura, M. S. da Costa, and S. Macedo-Ribeiro. 2008. Mycobacterium tuberculosis glucosyl-3-phosphoglycerate synthase: structure of a key enzyme in methylglucose lipopolysaccharide biosynthesis. PLoS One 3:e3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 26.Que, Q., Y. Li, I.-N. Wang, L. C. Lane, W. G. Chaney, and J. L. Van Etten. 1994. Protein glycosylation and myristylation in Chlorella virus PBCV-1 and its antigenic variants. Virology 203:320-327. [DOI] [PubMed] [Google Scholar]
- 27.Schwab, T., D. Skegro, O. Mayans, and R. Sterner. 2008. A rationally designed monomeric variant of anthranilate phosphoribosyltransferase from Sulfolobus solfataricus is as active as the dimeric wild-type enzyme but less thermostable. J. Mol. Biol. 376:506-516. [DOI] [PubMed] [Google Scholar]
- 28.Sobhany, M., and M. Negishi. 2006. Characterization of specific donor binding to α1,4-N-acetylhexosaminyltransferase EXTL2 using isothermal titration calorimetry. Methods Enzymol. 416:3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun, H. Y., S. W. Lin, T. P. Ko, J. F. Pan, C. L. Liu, C. N. Lin, A. H. Wang, and C. H. Lin. 2007. Structure and mechanism of Helicobacter pylori fucosyltransferase. A basis for lipopolysaccharide variation and inhibitor design. J. Biol. Chem. 282:9973-9982. [DOI] [PubMed] [Google Scholar]
- 30.Terwilliger, T. C. 2000. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terwilliger, T. C., and J. Berendzen. 1999. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanderplasschen, A., N. Markine-Goriaynoff, P. Lomonte, M. Suzuki, N. Hiraoka, J.-C. Yeh, F. Bureau, L. Willems, E. Thiry, M. Fukuda, and P.-P. Pastoret. 2000. A multipotential β-1,6-N-acetylglucosaminyl-transferase is encoded by bovine herpesvirus type 4. Proc. Natl. Acad. Sci. U. S. A. 97:5756-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Etten, J. L., J. R. Gurnon, G. M. Yanai-Balser, D. D. Dunigan, and M. V. Graves. 2010. Chlorella viruses encode most, if not all, of the machinery to glycosylate their glycoproteins independent of the endoplasmic reticulum and Golgi. Biochim. Biophys. Acta 1800:152-159. [DOI] [PubMed] [Google Scholar]
- 34.Van Etten, J. L., L. C. Lane, and R. H. Meints. 1991. Viruses and viruslike particles of eukaryotic algae. Microbiol. Rev. 55:586-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vetting, M. W., P. A. Frantom, and J. S. Blanchard. 2008. Structural and enzymatic analysis of MshA from Corynebacterium glutamicum. Substrate-assisted catalysis. J. Biol. Chem. 283:15834-15844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vigerust, D. J., and V. L. Shepherd. 2007. Virus glycosylation: role in virulence and immune interactions. Trends Microbiol. 15:211-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallace, A. C., R. A. Laskowski, and J. M. Thornton. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127-134. [DOI] [PubMed] [Google Scholar]
- 38.Wang, I., Y. Li, Q. Que, M. Bhattacharya, L. C. Lane, W. G. Chaney, and J. L. Van Etten. 1993. Evidence for virus-encoded glycosylation specificity. Proc. Natl. Acad. Sci. U. S. A. 90:3840-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson, W. H., J. L. Van Etten, and M. J. Allen. 2009. The Phycodnaviridae: the story of how tiny giants rule the world, p. 1-42. In J. Van Etten (ed.), Lesser known large dsDNA viruses. Springer, Berlin, Germany. [DOI] [PMC free article] [PubMed]
- 40.Yamada, T., H. Onimatsu, and J. L. Van Etten. 2006. Chlorella viruses. Adv. Virus Res. 66:293-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Y., Y. Xiang, J. L. Van Etten, and M. G. Rossmann. 2007. Structure and function of a chlorella virus-encoded glycosyltransferase. Structure 15:1031-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.