Abstract
The Lettuce infectious yellows virus (LIYV) RNA 2 mutant p1-5b was previously isolated from Bemisia tabaci-transmitted virus maintained in Chenopodium murale plants. p1-5b RNA 2 contains a single-nucleotide deletion in the minor coat protein (CPm) open reading frame (ORF) that is predicted to result in a frameshift and premature termination of the protein. Using the recently developed agroinoculation system for LIYV, we tested RNA 2 containing the p1-5b CPm mutant genotype (agro-pR6-5b) in Nicotiana benthamiana plants. We showed that plant infection triggered by agro-pR6-5b spread systemically and resulted in the formation of virions similar to those produced in p1-5b-inoculated protoplasts. However, virions derived from these mutant CPm genotypes were not transmitted by whiteflies, even though virion concentrations were above the typical transmission thresholds. In contrast, and as demonstrated for the first time, an engineered restoration mutant (agro-pR6-5bM1) was capable of both systemic movement in plants and whitefly transmission. These results provide strong molecular evidence that the full-length LIYV-encoded CPm is dispensable for systemic plant movement but is required for whitefly transmission.
Members of the genus Crinivirus are emerging plant viruses in many parts of the world. An important factor contributing to the increase in the incidence of these viruses is their association with and transmission by whitefly vectors that have increased in distribution in the last several decades. Lettuce infectious yellows virus (LIYV), the type member of the genus Crinivirus (family Closteroviridae), is specifically transmitted by the sweet potato whitefly, Bemisia tabaci biotype A, in a semipersistent, noncirculative manner (6). The virus is confined to phloem cells within infected plants and is not transmissible to plants by leaf rub inoculation. The bipartite single-stranded positive-sense LIYV genome components, consisting of RNA 1 (approximately 8.1 kb) and RNA 2 (approximately 7.2 kb), are separately encapsidated in flexuous filamentous particles that are characteristic of the family Closteroviridae (8, 11). These virions are comprised of four protein components: the major coat protein (CP), the minor coat protein (CPm), an Hsp70 homolog (Hsp70h), and a 59-kDa protein (P59). Like other viruses in the family Closteroviridae, LIYV has bipolar virions with a “body” composed mainly of the CP and a “head” that is formed by the assembly of CPm subunits (2, 4, 7, 22, 28). Hsp70h and P59 are detected in LIYV virions (22), but their locations have not been identified, as they are not readily detected by immunogold labeling and transmission electron microscopy (IGL-TEM). For two members of the family Closteroviridae, Citrus tristeza virus (CTV) and Beet yellows virus (BYV), the combination of Hsp70h, P61 (the homolog of LIYV P59 in CTV) or P64 (the homolog of LIYV P59 in BYV), and CPm encapsidates the 5′ end (∼630 to 650 nucleotides [nt]) of the RNA genome, demonstrating the complex interactions that exist among the capsid proteins and the genomic RNA (15, 21).
In our previous studies, we demonstrated the transmission of LIYV using an in vitro acquisition and whitefly transmission system (13, 22). Results from previous work implicated a role for LIYV CPm in whitefly transmission. Antibodies to CPm blocked the in vitro acquisition/transmission of LIYV virion preparations by B. tabaci biotype A, while antibodies to CP, Hsp70h, and P59 did not (22). The in vitro whitefly membrane-feeding system had also been used to demonstrate B. tabaci biotype A transmission of virions that were derived from cloned infectious cDNAs of LIYV RNA 1 and RNA 2 of several genotypes, including pR6 (the first cloned wild-type [WT] infectious cDNA of LIYV RNA 2 [10]), establishing for the first time that these cloned constructs contained all of the information necessary for protoplast infection, virion formation, whitefly transmission, and infection in plants (12). In that study, the mutant p1-5b was among the cloned LIYV RNA 2 cDNAs derived from B. tabaci biotype A-transmitted virus maintained in Chenopodium murale plants.
p1-5b contains a single-adenine-residue deletion in the CPm open reading frame (ORF) at nucleotide 592, a deletion that is predicted to result in a frameshift, 14 new amino acids, and premature termination of the protein (12). The predicted p1-5b CPm has 211 amino acids, compared to 453 amino acids in the wild-type (pR6 genotype) protein. The p1-5b genotype also contains three other nucleotide changes in the CPm ORF relative to the pR6 infectious clone sequence (27), all of which result in amino acid changes. In contrast, the p1-5b CP, Hsp70h, and P59 sequences are identical to that of pR6 (12). Possible polymorphisms throughout the rest of the p1-5b clone were not characterized. In a prior study, B. tabaci biotype A transmission of p1-5b virions was not observed, even though the mutation did not affect its infectivity in protoplasts (as determined by virion yields) and apparent particle morphology (12). However, those studies were disadvantaged by the necessity of propagation in protoplasts to obtain specific genotypes from infectious cloned cDNAs. Protoplasts yield low quantities of virion relative to plants, and virion concentration is a critical parameter in whitefly transmission (13). Although virion concentrations in those experiments were above typical thresholds for whitefly transmission (12, 13), low concentrations may still be limiting for transmission, making negative transmission results difficult to interpret. Obtaining adequate virion concentrations of specific genotypes for whitefly transmission to plants has therefore been a significant hurdle to LIYV transmission studies.
The recently developed agroinoculation method for LIYV (24) permits the study of systemic plant infection by distinct LIYV genotypes, including those that are whitefly transmission deficient, and the recovery of higher virion yields than were possible using protoplasts. The objective of this study was to further examine the function of the LIVY CPm by extending our observations of p1-5b. We constructed mutants with the CPm frameshift restored to determine if engineered mutations that either restored or disrupted the formation of an intact CPm also affected systemic plant infection, virion formation, and B. tabaci biotype A transmission. Our study revealed that a mutant engineered with the restored CPm ORF produced a WT infection profile characterized by systemic virus movement within agroinoculated plants and the generation of CPm-containing virions that were whitefly transmissible. Intriguingly, systemic virus movement was also observed for a mutant engineered to express the 1-5b CPm, but the virions lacked an identifiable CPm and were defective in whitefly transmission. These results represent a significant advance in addressing challenging questions and hypotheses about Crinivirus whitefly transmission properties not testable using earlier systems.
MATERIALS AND METHODS
Engineering of CPm mutants.
To restore the CPm ORF in p1-5b (Fig. 1C), an adenine residue was engineered at p1-5b nucleotide position 5555 by PCR-mediated mutagenesis, and the resulting construct was p1-5bM1 (Fig. 1C). Specifically, p1-5bM1 was derived by using complementary oligonucleotide primers LIYV 18 (5′-GTTTACACAAAAATGGGTAATCCTATATCTTATAAC-3′) and LIYV 17 (5′-GTTATAAGATATAGGATTACCCATTTTTGTGTAAAC-3′) (the adenine and its complementary thymine are indicated in boldface type). pR6-5b was constructed by replacing an 876-bp NheI-cut fragment in cloned WT pR6 (between nucleotide positions 4884 and 5759, which includes the adenine nucleotide at position 5555 and encompasses the 3′ end of the CP ORF as well as the 5′ end of the CPm ORF) with the corresponding fragment of p1-5b that is missing the adenine nucleotide at position 5555 (Fig. 1C). Subsequent procedures employed for transformation, colony analysis, and plasmid purification were performed according to methods described previously by Sambrook and Russell (20). All engineered constructs were sequenced to confirm the introduced nucleotide change and to ensure that no spurious substitutions or deletions were present.
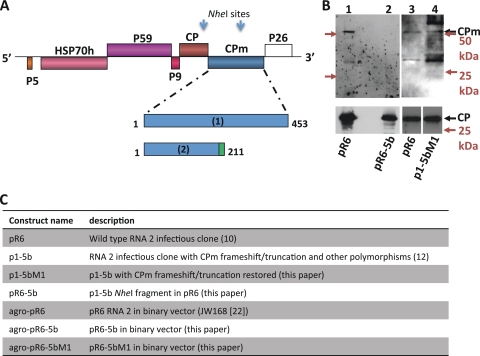
FIG. 1.
Predicted sizes of the CPm encoded by the WT and mutants of LIYV and immunoblot analysis of virions. (A) Genome organization of LIYV RNA 2 and amino acid positions of expected translation products encoded by the CPm gene. Expected translation products were encoded by (1) the full-length CPm gene of whitefly-transmissible cloned WT pR6 and the CPm restoration mutants p1-5bM1 and agro-pR6-5bM1 and (2) the CPm gene of the non-whitefly-transmissible mutants p1-5b, pR6-5b, and agro-pR6-5b, including 14 predicted novel amino acids (green) at the C terminus due to the frameshift-preceding termination. The complete CPm corresponds to an intact protein with 453 amino acids, whereas that from p1-5b, pR6-5b, and agro-pR6-5b corresponds to a truncated protein with 211 amino acids. The relative positions of the LIYV RNA 2 ORFs encoding P5, HSP70h, P59, P9, CP, CPm, and P26 are indicated. Arrows indicate NheI sites used for the construction of agroinfiltration plasmids. (B) Western blot analysis of virions. Shown are pR6 virions (lanes 1 and 3) purified from lettuce plants inoculated by whitefly transmission of virions from pR6-infected protoplasts and pR6-5b and p1-5bM1 virions (lanes 2 and 4) purified from inoculated protoplasts. Virion proteins were detected with antisera raised against the CPm (top) or whole virions (bottom). Positions of the CP (28 kDa) and CPm (52 kDa) are indicated. CPm antiserum frequently detected both the expected 52-kDa protein and a ca. 35-kDa protein, which may be a CPm degradation product (lanes 1, 3, and 4). Migrations of molecular mass markers are indicated with colored text and arrows. Separate arrows are shown for the 50-kDa and 25-kDa marker locations for each blot at the top. (C) Summary of constructs used for experiments described herein.
Construction of LIYV agroinoculation plasmids and agroinoculation.
RNA 2 CPm mutant binary vector constructs were based on agro-pR6 (pR6 genotype RNA 2 agroinoculation construct, also named JW168) (24) (Fig. 1C). NheI fragments from p1-5b and p1-5bM1 were swapped into agro-pR6 and screened for directionality (Fig. 1A). The resultant clones were named agro-pR6-5b (where the NheI fragment source was p1-5b) and agro-pR6-5bM1 (where the NheI fragment source was p1-5bM1) (Fig. 1C). Insert regions were sequenced to confirm the genotype.
The procedures for the transformation and preparation of Agrobacterium tumefaciens strain C58C1 were previously described (24). Constructs were coinfiltrated with the wild-type RNA 1 agroinoculation construct JW100 (24) and either a 35S Tomato bushy stunt virus (TBSV) P19 silencing suppressor construct or a 35S Tobacco etch virus P1/HC-Pro silencing suppressor construct (5, 23) into Nicotiana benthamiana plants at about the 5- to 6-leaf stage as previously described (23). Plants were assayed for infection by symptom scoring and reverse transcription (RT)-PCR using primers P26-F and P26-r2 (13) at 2 to 4 weeks postinfiltration. Symptomatic plants were assayed for the retention of engineered mutations by the sequencing of RT-PCR products obtained from infected plants at various time points between 2 and 12 weeks postinfiltration (see Results for further details on individual experiments).
Protoplast inoculation, virion purification, IGL-TEM, and whitefly transmission.
Procedures for the isolation of protoplasts derived from cultured suspension cells of Nicotiana tabacum var. Xanthi were previously described (14). The extraction of virion RNA and synthesis of capped transcripts of cloned constructs were described previously (9, 10, 26). Protoplasts were inoculated with capped transcripts of the cloned cDNA constructs of RNAs 1 and 2 as described previously (10). Specifically, protoplasts were inoculated with capped transcripts of p9/55, the full-length cDNA clone of RNA 1 (10), and one of the following full-length cDNA clones of RNA 2, using ∼2 μg (RNA 1 transcript) and ∼6 μg (RNA 2 transcript) per half-million protoplasts: pR6 (10), p1-5b (12), p1-5bM1, and pR6-5b. Procedures for the purification of virions from infected protoplasts and plants, estimation of virion concentrations, Western blot analysis, IGL-TEM, and whitefly transmission were performed according to previously described methods (13, 22). The nomenclature of virions obtained from the in vitro transcript-inoculated protoplasts is according to previous conventions (12). For example, p1-5b virions are those obtained from p9/55 (RNA 1)- and p1-5b (RNA 2)-inoculated protoplasts. Virion concentrations were estimated by densitometry with SDS-PAGE or Western immunoblots, as indicated in Table 2, using ImageJ 1.42q software (NIH). Diluted proteins of known concentration were used to generate a linear formula for the concentration, which was used to calculate virion (CP) concentrations.
RT-PCR, immunocapture RT-PCR, cloning, and sequencing.
For the analysis of CPm nucleotides following protoplast inoculation, total RNAs were extracted from protoplasts by using Tri reagent (Invitrogen) according to the manufacturer's recommendations. First-strand cDNA synthesis by Superscript II reverse transcriptase (Invitrogen), followed by PCR amplification with Pfu DNA polymerase (Stratagene) and the direct sequencing of PCR-amplified products, was performed by using primers LIYV 10, LIYV 11, LIYV 12, and LIYV 13 (12). Immunocapture RT-PCR was done according to a method described previously (18), with modifications. Following the precoating of PCR tubes with LIYV virion-specific IgG at ∼2.3 μg/ml for 30 min at 37°C and rinsing with phosphate-buffered saline plus 0.05% Tween 20 (PBST), tubes were incubated with a blocking buffer (PBST with 2% polyvinyl pyrrolidone and 2% bovine serum albumin) for 30 min at 37°C. After the second wash, tubes were incubated in blocking buffer with samples, including greenhouse-maintained wild-type virions (100 ng), pR6 virions (10 ng), p1-5b virions (5 ng), wild-type virion RNAs (100 ng), and a 1/20 dilution of a purified preparation of protoplast-derived virions inoculated with the p9/55 (RNA 1) in vitro-synthesized transcript. Following the final wash, RT-PCR was performed by using primers LIYV 38 (5′-TCACAATTACCATTGGGCGAAG-3′) and LIYV 39 (5′-TCCGCTCTTAGTATTCGCAG-3′), which amplify 273 nucleotides (from nucleotide positions 6 to 278) of the LIYV RNA 2 5′ region; P26-F and P26-r2 (13), which amplify a 303-bp fragment of the LIYV RNA 2 3′ region; LIYV 40 (5′-CCGTTGGACAAGGTAAGATT-3′) and LIYV 41 (5′-GTCACACTCCAACCATTACC-3′), which amplify 486 nucleotides (from nucleotide positions 19 to 504) of the LIYV RNA 1 5′ region; and LIYV 42 (5′-TGTCGCCGTTATACGCATTG-3′) and LIYV 43 (5′-CGTTGCACAACCTAAGACGA-3′), which amplify 255 nucleotides (from nucleotide positions 7085 to 7339) of the LIYV RNA 1 3′ region.
For the analysis of CPm nucleotides of LIYV with the 1-5bM1 genotype (agro-pR6-5bM1) after whitefly transmission, total RNAs were extracted from systemically infected lettuce leaves by using the RNeasy plant minikit (Qiagen) and subjected to RT-PCR with Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega) and Herculase II fusion DNA polymerase (Agilent Technologies). RT-PCR was performed by using primers LIYV 19 (5′-GAAATATCAGCAATCGGGCATTGTCT-3′) and LIYV 20 (5′-CACCACTGATGCTCAACTCACTTCG-3′), which amplify 1,063 nucleotides (from nucleotide positions 4847 to 5909) spanning the CPm ORF in which the mutation in 1-5bM1 was engineered. The resulting purified RT-PCR product was A tailed by incubation with Taq polymerase for 30 min at 72°C with dATP to facilitate subsequent cloning into the pGEM-T Easy vector (Promega). cDNA clones from four randomly selected infected plant samples were sequenced in both directions.
RESULTS
Western blot and IGL-TEM analyses of virions recovered from inoculated protoplasts.
Due to the presence of a premature stop codon in the CPm of the p1-5b mutant, only 211 encoded amino acids (<50% of the intact wild-type CPm), corresponding to a peptide of 21 kDa, were expected to be expressed from the p1-5b genome (Fig. 1A) (12). This mutant was competent for replication in protoplasts and yielded virions as determined by TEM analyses (12). However, Western blot analyses of purified p1-5b virions did not show the complete CPm or a fragment corresponding to the expected 21-kDa CPm truncation, suggesting that they were absent on mutant virions (12).
To test the hypothesis that a complete CPm is required for positive detection on virions, we analyzed p1-5b and the engineered mutants p1-5bM1 (p1-5b engineered to express a complete CPm) and pR6-5b (an engineered mutant in which the p1-5b CPm gene had been incorporated within the cloned WT [pR6] background [Fig. 1A and C]). As a comparison, LIYV virions purified from pR6-infected protoplasts and/or lettuce plants and greenhouse-maintained WT-infected lettuce plants were included in the study. Western blot analyses of pR6 virions identified proteins with sizes corresponding to those of the intact CP and CPm (Fig. 1B, lanes 1 and 3), consistent with our previously reported results (12). The complete CP and CPm were also identified for virions of p1-5bM1 in the same Western analyses (Fig. 1B, lane 4). Thus, the mutation engineered into the CPm gene of p1-5b removed the premature stop codon and resulted in the restoration of an intact CPm while retaining the other nucleotide polymorphisms of the p1-5b clone relative to the wild-type pR6 clone. However, Western blot analyses of purified pR6-5b virions positively identified only the CP but not the CPm (Fig. 1B, lane 2), indicating that the loss of detection of the CPm for the 1-5b CPm genotype was irrespective of the genetic background of the LIYV isolate.
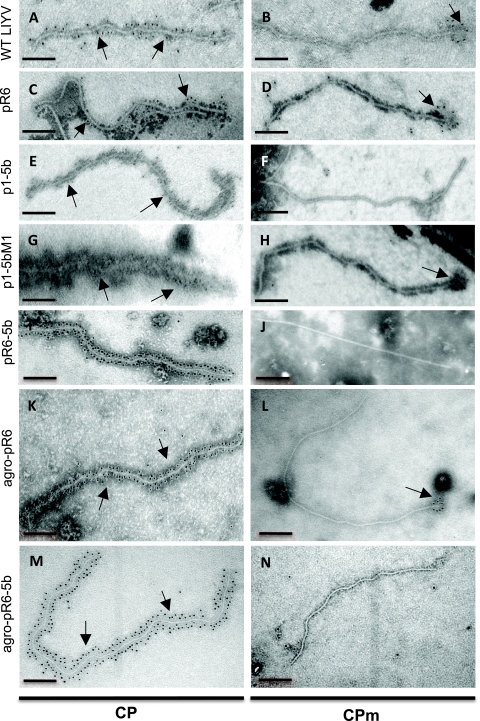
We also used antibodies specific to the CP and CPm in IGL-TEM analyses to obtain physical evidence of the presence of the CP and CPm on virions purified from protoplasts. These virions were derived from protoplasts inoculated with the in vitro-synthesized transcripts of LIYV RNA 1 (p9/55) and one of the following LIYV RNA 2 constructs: pR6, p1-5b, p1-5bM1, or pR6-5b. IGL-TEM observation of virions prepared from p1-5b-, p1-5bM1-, and pR6-5b-inoculated protoplasts revealed the presence of virions that were similar in length and morphology to greenhouse-maintained WT and pR6 virions, consistent with results from our previous observations (Fig. 2A to J) (12). Positively labeled particles were counted, and the percentage of labeling was expressed as the number of labeled particles over the total number of particles observed (Table 1 and Fig. 2). When antibodies to the CP were used, specific labeling was readily and abundantly (100%) observed along the entire extent of the “body” of the particles for greenhouse-maintained WT and pR6 virions regardless of whether the latter virions were purified from protoplasts or plants (Table 1 and Fig. 2A and C). Specific labeling (at one end of the particle) was also seen for greenhouse-maintained WT and pR6 virions when antibodies to CPm were used, although the percentage of labeling was lower (44 to 88%) than that of particles tested using antibodies to the CP (Table 1 and Fig. 2A to D). Specific labeling (92%) was seen when p1-5b virions were tested by using antibodies to the CP (Table 1 and Fig. 2E). In contrast, none of the 50 virions examined were specifically labeled when tested by using antibodies to the CPm (Table 1 and Fig. 2F). Similarly, IGL-TEM analyses positively identified the CP (100%) but not the CPm (0%) from purified pR6-5b virions (Table 1 and Fig. 2I and J). This finding was consistent with the Western blot results for pR6-5b virions and was expected, since p1-5b lacked a detectable full-length CPm. Therefore, we used the virions of p1-5bM1 to determine whether the restoration of an intact CPm resulted in the restoration of detectable specific labeling on p1-5b virions. Abundant (100%) specific labeling was observed when p1-5bM1 virions were tested by using antibodies to CP (Table 1 and Fig. 2G). With CPm antibodies, up to 82% of the virions examined were labeled (Table 1 and Fig. 2H).
FIG. 2.
IGL-TEM analyses of LIYV virions. Virions were tested by using antibodies against the LIYV CP and CPm followed by goat anti-rabbit IgG labeled with 5-nm gold particles. (A and B) Greenhouse-maintained wild-type virions purified from infected lettuce plants. (C and D) pR6 virions purified from protoplasts of N. tabacum var. Xanthi. (E and F) p1-5b virions purified from N. tabacum var. Xanthi protoplasts. (G and H) p1-5bM1 virions purified from N. tabacum var. Xanthi protoplasts. (I and J) pR6-5b virions purified from N. tabacum var. Xanthi protoplasts. (K and L) Agro-pR6 virions purified from agroinoculated N. benthamiana. (M and N) Agro-pR6-5b virions purified from agroinoculated N. benthamiana. Each bar represents 200 nm. Arrows indicate gold labeling sites.
TABLE 1.
IGL-TEM analysis of LIYV
| LIYV genotype | Major coat protein (CP) |
Minor coat protein (CPm) |
||||
|---|---|---|---|---|---|---|
| No. of observed virions | No. of labeled virionsd | % labeling | No. of observed virions | No. of labeled virionsd | % labeling | |
| WTa | 50 | 50 | 100 | 44 | 38 | 88 |
| pR6a | 50 | 50 | 100 | 40 | 30 | 80 |
| pR6b | 8 | 8 | 100 | 25 | 11 | 44 |
| p1-5bb | 46 | 42 | 92 | 50 | 0 | 0 |
| p1-5bM1b | 50 | 50 | 100 | 21 | 17 | 82 |
| pR6-5bb | 50 | 50 | 100 | 48 | 0 | 0 |
| Agro-pR6c | 50 | 50 | 100 | 52 | 47 | 94 |
| Agro-pR6-5bc | 50 | 50 | 100 | 50 | 0 | 0 |
Virions purified from plants (WT, greenhouse-maintained wild-type virus; pR6, cloned infectious wild-type virus inoculated by whitefly transmission of virions purified from infected protoplasts).
Virions purified from inoculated N. tabacum var. Xanthi protoplasts.
Virions purified from agroinoculated N. benthamiana plants.
Immunogold labeling of virions was performed as previously described (20), using primary antibodies specific to the major and minor coat proteins.
Immunocapture RT-PCR assay for mutant p1-5b virions purified from protoplasts.
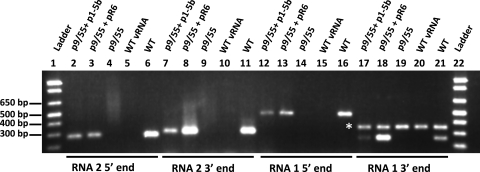
Closterovirus encapsidation studies have revealed that the polar ends of these virions in which CPm is located correspond to the 5′ ends of the encapsidated genomic RNAs (15, 21). Thus, it was of interest to determine if the 5′ ends of genomic RNAs 1 and 2 were present in the p1-5b virions since an intact p1-5b CPm was not detectable based on the above-described analyses. Immunocapture RT-PCR of p1-5b virions was performed by using antibodies raised against whole LIYV virions and primers specific to the 5′ and 3′ ends of both RNAs 1 and 2 (Fig. 3; see also Materials and Methods). Greenhouse-maintained WT and pR6 virions were included as positive controls, while virion RNAs and the in vitro transcript of LIYV RNA 1 (p9/55) were used as negative controls in the assay (Fig. 3). Our results indicated that 486 and 273 nucleotides at the 5′ ends of RNAs 1 and 2, respectively, as well as 255 and 303 nucleotides at the 3′ ends of RNAs 1 and 2, respectively, of wild-type controls and p1-5b (and the pR6 positive controls) were detected (Fig. 3), suggesting that these regions in the genomic RNAs were present in the virions.
FIG. 3.
Immunocapture RT-PCR assay for virions. Virion preparations were obtained from protoplasts inoculated with the in vitro transcripts derived from the cDNA of LIYV RNA 1 (p9/55) (lanes 4, 9, 14, and 19) alone or along with pR6 RNA 2 (lanes 3, 8, 13, and 18) or p1-5b RNA 2 (lanes 2, 7, 12, and 17). Greenhouse-maintained wild-type virions (lanes 6, 11, 16, and 21) and virion RNAs (lanes 5, 10, 15, and 20) purified from LIYV-infected plants were included as positive and negative controls, respectively. Immunocaptured samples were subjected to RT-PCR using primers specific to the 5′ region, nucleotide positions 6 to 278 and nucleotide positions 19 to 504 of RNA 2 (lanes 2 to 6) and RNA 1 (lanes 12 to 16), respectively, and primers specific to the 3′ region, nucleotide positions 6235 to 6537 and nucleotide positions 7085 to 7339 of RNA 2 (lanes 7 to 11) and RNA 1 (lanes 17 to 21), respectively. Sizes in base pairs for DNA standards (lanes 1 and 22) are indicated. The asterisk indicates the position of a nonspecifically amplified product present in samples in lanes 17 to 21. vRNA, viral RNA.
Systemic plant infection, sequence retention, and virion formation of CPm mutants.
LIYV agroinoculation constructs with the p1-5b and p1-5bM1 CPm genotypes (referred to here as agro-pR6-5b and agro-pR6-5bM1, respectively) were tested compared to a wild-type (pR6 genotype) RNA 2 agroinoculation construct, agro-pR6, in all cases coinfiltrated with an RNA 1 construct (24). In these experiments, we repeatedly observed a systemic infection of plants agroinfiltrated with agro-pR6-5b, agro-pR6-5bM1, or the positive control (agro-pR6). Infiltrated N. benthamiana plants showed typical LIYV symptoms, including interveinal chlorosis beginning at lower leaves and progressing upward and a quality of leaf brittleness. Western blotting and/or RT-PCR confirmed LIYV infection of noninoculated leaves (data not shown). Plants infected by each of the genotypes listed above showed indistinguishable symptoms and timing of symptom appearance, suggesting no noticeable delay or defect in infection for the CPm mutants.
To determine whether infected plants retained the expected CPm genotypes, including the CPm frameshift in agro-pR6-5b, we tested plants by RT-PCR and nucleotide sequence analysis. All plants tested within 6 to 8 weeks postinfiltration exhibited the expected genotypes for the entire CPm ORF (based on results from the testing of approximately 16 plants). When three symptomatic plants infiltrated with the agro-pR6-5b construct were tested by RT-PCR and sequencing at 3 months postinoculation (mpi), all consensus sequences had the expected genotype. However, when a set of eight plants, including the same three tested at 3 mpi, were tested again a month later, two of the eight plants (including one of the three plants sequenced previously) exhibited a compensatory frameshift mutation in the CPm gene: an adenine residue insertion in a 4-adenine tract seven nucleotides upstream of the mutant adenine deletion. This insertion was not a bona fide “reversion,” since the site of nucleotide insertion was not the same as the mutant deletion site. However, it was expected to restore the CPm frame for the translation of a full-length CPm with two additional amino acid changes: M to N at CPm amino acid 196 and N to Y at amino acid 198. Other polymorphisms of the CPm genotype in agro-pR6-5b relative to the wild-type infectious clone agro-pR6 were retained in these compensatory mutants. These late-appearing compensatory mutants were not further tested, as mixed infections arising from viruses with the revertant and the 5b genotypes were likely present in these plants, thereby possibly confounding virion concentration-dependent transmission results.
We recovered agro-pR6 and agro-pR6-5b virions from the systemically infected leaves of agroinfiltrated plants with the expected sequences and subjected them to IGL-TEM analysis using antibodies to the LIYV CP and CPm (Table 1 and Fig. 2). The results indicated that the CP was readily detected in 100% of these virions (Table 1 and Fig. 2K and M), consistent with results obtained by using virions purified from protoplasts (Table 1 and Fig. 2C and I). In contrast, although the CPm was readily detected in 94% of the agro-pR6 virions, it was not observed in any of 50 particles of agro-pR6-5b (Table 1 and Fig. 2L and N). These results were consistent with those obtained by using virions purified from protoplasts (Table 1 and Fig. 2D and J).
Whitefly transmission of CPm mutant virions.
In our previous study, the p1-5b virions were defective in transmission by B. tabaci biotype A, while transmission of the greenhouse-maintained WT and pR6 virions was observed (12). Here, we tested virions of the restoration mutant p1-5bM1, which expresses the complete CPm, for transmission by B. tabaci biotype A. Tobacco protoplasts were inoculated with the in vitro-synthesized transcripts of p9/55, the cloned cDNA construct of LIYV RNA 1, and that of p1-5bM1, and the resulting purified virions were subjected to in vitro acquisition and transmission by B. tabaci biotype A. In four transmission experiments in which the average concentration of p1-5bM1 virions was approximately 7.9 ng/μl and hundreds of whiteflies were used per transmission, no transmission was observed for a cumulative total of 30 test plants. However, it should be noted that although the average virion concentrations in these experiments were within the range that supports virion transmission (13), they were close to limiting concentrations for efficient transmission.
In order to confirm the above-described transmission data in a system where much higher virion yields could be obtained, N. benthamiana plants were systemically infected with LIYV agro-pR6-5b and agro-pR6-5bM1. It should be noted that N. benthamiana plants were used for agroinoculation because methods so far have not succeeded in delivering LIYV to lettuce or other preferred feeding hosts of the LIYV whitefly vector B. tabaci biotype A (24). Because these whiteflies feed very poorly on N. benthamiana plants, and this negatively affects virus transmission, virions were purified from agroinfected plants and used for membrane feeding in transmission experiments. In this way, virion concentrations used for feeding could also be measured and compared between experiments. Since the 1-5b mutation was not 100% retained following agroinoculation, particularly at time points after 3 mpi, the expected genotypes in all plants used for subsequent whitefly transmissions were confirmed by RT-PCR and sequencing within 1 week prior to transmission experiments, and plants were used for virion purification between 6 and 8 weeks postinfiltration (at which time points no mutations were found). Virion concentrations were estimated by SDS-PAGE and Western blotting or Coomassie staining and comparison with standards of known concentrations (data not shown). In three experiments, with concentrations ranging from 32 to 78 ng/μl, transmission of agro-pR6-5bM1 virions was observed (experiments 1 to 3) (Table 2), while the transmission of agro-pR6-5b virions, with concentrations ranging from 15 to 21 ng/μl, was not (experiments 1 and 2) (Table 2). pR6-5bM1 CPm genotypes in four randomly selected lettuce plants were verified after transmission by RNA isolation and sequencing (see Materials and Methods) (data not shown). The transmission of agro-pR6-5b virions was not observed even when the concentration exceeded that of agro-pR6-5bM1 by more than 9-fold (experiment 3) (Table 2). The transmission of virions from greenhouse-maintained WT plants or plants infiltrated with the agro-pR6 RNA 2 construct was also observed (experiments 1 and 2) (Table 2). Because of the higher virion concentrations obtained in these experiments, the transmission of agro-pR6-5bM1 virions demonstrated here (Table 2) may be taken with greater confidence over that of the protoplast-derived p1-5bM1 virions described above.
TABLE 2.
Whitefly transmission of virions purified from plants infected with three LIYV genotypes
| RNA 2 genotypec | Expt 1a |
Expt 2a |
Expt 3b |
Total no. of infected plants/total no. of plants inoculated | |||
|---|---|---|---|---|---|---|---|
| Virion concn (ng/μl) | No. of infected plants/total no. of plants inoculated | Virion concn (ng/μl) | No. of infected plants/total no. of plants inoculated | Virion concn (ng/μl) | No. of infected plants/total no. of plants inoculated | ||
| Agro-pR6-5b | 28 | 0/4 | 21 | 0/5 | 300 | 0/10 | 0/24 |
| 15 | 0/5 | ||||||
| Agro-pR6-5bM1 | 55 | 0/4 | 37 | 5/5 | 32 | 10/10 | 17/24 |
| 78 | 2/5 | ||||||
| Agro-pR6d | 4 | 1/4 | 1/4 | ||||
| WTe | 68 | 5/5 C. muralef | 5/10 | ||||
| 9 | 0/5 lettuce | ||||||
Virion concentrations estimated by densitometry relative to protein standards from Coomassie blue-stained SDS-PAGE gels.
Virion concentrations estimated by densitometry relative to virions of known concentrations from Western blots.
Virions derived from clones with the CPm genotypes agro-pR6, agro-pR6-5b, and agro-pR6-5bM1 were prepared from systemically infected agroinoculated Nicotiana benthamia plants after CPm genotypes in systemic tissue were confirmed by RT-PCR and sequencing.
Wild-type pR6 clone in a binary vector.
Greenhouse-maintained wild-type LIYV.
Greenhouse-maintained WT virions were prepared from the host plant Chenopodium murale or Lactuca sativa L. (lettuce), as indicated for experiment 2.
DISCUSSION
The current study extends our previous findings for the mutant p1-5b by comparing this LIYV genotype to an engineered restoration genotype, p1-5bM1, and assessing virion structure (encapsidation), systemic plant infection by agroinoculation, and transmissibility by B. tabaci biotype A. In immunoblotting and IGL-TEM analyses of p1-5b and p1-5bM1 virions, CPm was not detected in the truncation mutant virions, consistent with the notion that a complete CPm is likely the preferred form to be incorporated into the assembled virion. In order to not be biased by our own observations, it was necessary for the IGL-TEM analyses to be both qualitative and quantitative. Therefore, we counted every particle, labeled or otherwise (depending on the virions and the antibody used), in our analysis, and the data presented in Table 1 and Fig. 2 represent our observations of the similarity and dissimilarity in the specific labeling characteristics between the p1-5b and p1-5bM1 virions.
Studies of CTV and BYV have shown that genomic RNA encapsidation involves a complex interaction between Hsp70h, P61 (the homolog of P59 in CTV)/P64 (the homolog of P59 in BYV), and the CPm, and the interaction occurs near the 5′ end of the genomic RNA (16, 21). How and/or whether similar interactions occur for LIYV is not known; however, given the similarity in the structural compositions among these viruses, it may be that LIYV behaves similarly. However, our immunocapture/RT-PCR on LIYV virions indicates that the 5′ and 3′ ends of LIYV RNAs 1 and 2 are present even in virions derived from the 1-5b CPm truncation genotype. Similar results have been reported for BYV, for which CPm mutants still had complete RNA protection, likely due to compensation by the major CP (4). It is also possible that the CP or other capsid proteins (Hsp70h or P59) compensate in encapsidating the CPm mutant or that the truncated LIYV CPm is still incorporated into virions but lacks immunogenicity, thereby precluding detection by immunoblot or immunogold labeling assays.
Based on transmission experiments with plant- and protoplast-derived virions, it is clear that the p1-5b genotype, which lacks a complete CPm, is not transmissible to plants by B. tabaci biotype A. The restoration of the CPm mutation in p1-5b was performed by engineering p1-5bM1 so that it had an intact CPm but retained other sequence polymorphisms of the 1-5b CPm relative to wild-type pR6. The agroinoculated pR6 chimeras containing these CPm genotypes (agro-pR6-5b and agro-pR6-5bM1) both moved systemically in plants when coinoculated with an RNA 1 construct, but only agro-pR6-5bM1 virions were transmissible by whitefly (Table 2). These results provide strong evidence that the CPm is a whitefly transmission determinant for LIYV and are in agreement with previous work implicating the CPm in whitefly transmission (12, 22). We also have preliminary data indicating that one basis for the defect in whitefly transmission of virions derived from the mutant CPm genotype in p1-5b (agro-pR6-5b) is impaired virion retention in the whitefly's foregut (J. C. K. Ng, unpublished data). Continuing work will reveal the relationship of 1-5b LIYV virions with whiteflies, including the retention of p1-5b versus wild-type and p1-5bM1 virions. Further analyses will also be necessary to delimit the portions of the CPm, apparently in the C-terminal 31 kDa, that are required for whitefly transmission. A comparison of the CPm amino acid sequences among the criniviruses revealed a low to moderate level of identity among them (19), with more similarities being seen in the C termini (alignment not shown). Although we have not yet identified any distinct motifs that can be linked to the whitefly-specific transmission of these viruses, it is plausible to speculate that if all Criniviruses require the CPm for whitefly transmission, sequence variations in the CPm might play a role in differences in whitefly transmission specificity, as was suggested previously (25).
Using the agroinoculation method to bypass protoplast inoculation and whitefly transmission to deliver specific genotypes to plants, we have also been able to assess systemic plant movement for these mutants. Our results show that full-length CPm is dispensable for systemic LIYV movement in agroinoculated N. benthamiana plants, since systemic infection of the 1-5b genotype was repeatedly identified and retained in noninoculated tissues. We have also recently found systemic infection in plants agroinoculated with another engineered CPm genotype, R6M6 (27), containing a 5′ double stop codon insertion for an earlier truncation than in the 1-5b genotype, for a predicted CPm fragment of only 71 amino acids (data not shown). In contrast, in repeated experiments, we have not detected systemic infection or symptom development of Hsp70h, P59, or CP knockout mutants by agroinfiltration. However, confirmation of local replication is necessary to validate negative results for these mutants.
The systemic movement of the LIYV CPm mutants is intriguing and somewhat surprising, since all four conserved capsid components (CP, CPm, Hsp70h, and the P59 homolog) have been shown to be required for the cell-to-cell movement of another closterovirus, BYV, in a local-lesion host (1, 3). The requirements and nature of movement components for LIYV have not been accessible prior to the development of the agroinoculation system, and long-distance movement mechanisms for the phloem-limited LIYV are likely to have important distinctions versus the cell-to-cell movement mechanisms for the non-phloem-limited BYV, perhaps including the role of the CPm in movement. We must also consider the possibility that systemic movement without an intact CPm is host specific for N. benthamiana, as plant delivery methods without the use of whiteflies are not yet available for any other hosts. It is also noteworthy that a compensatory mutation predicted to restore full-length CPm production appeared over time in some of the agro-pR6-5b-infected plants after systemic infections had been established. This finding suggests that there may be selection for the full-length CPm for some as-yet-undefined role in plants. For example, it is tempting to speculate that the full-length CPm may be required to confer an increased, albeit subtle, competitive ability in some aspects of replication, movement, and/or tissue tropism beyond what our estimation of purified virion concentrations could discern. Similarly, predicted compensatory mutations without selection by aphid transmission has been reported for viruses carrying mutations in a transmission-determining structural protein (P3/P5) in the polerovirus Potato leafroll virus (PLRV) (17). The PLRV P5 readthrough protein is also associated with other roles in host plants, including the establishment of phloem limitation.
The results presented here clearly elucidate how the complex nature of the LIYV virions (and similarity with other viruses of the Closteroviridae) regulates their transmissibility by insect vectors. These results also provide insight into potentially novel movement requirements for LIYV relative to other studied members of the family Closteroviridae.
Acknowledgments
Funding for this work was provided by startup funds provided by the College of Natural and Agricultural Sciences of the University of California, Riverside, to J.C.K.N. and a USDA NRICGP grant to B.W.F.
We also acknowledge and thank Josilyn Endres and Chawin Mongkolsiriwattana for technical assistance.
Footnotes
Published ahead of print on 22 September 2010.
REFERENCES
- 1.Agranovsky, A. A., A. S. Folimonov, S. Folimonova, S. Morozov, J. Schiemann, D. Lesemann, and J. G. Atabekov. 1998. Beet yellows closterovirus HSP70-like protein mediates the cell-to-cell movement of a potexvirus transport-deficient mutant and a hordeivirus-based chimeric virus. J. Gen. Virol. 79:889-895. [DOI] [PubMed] [Google Scholar]
- 2.Agranovsky, A. A., D. E. Lesemann, E. Maiss, R. Hull, and J. G. Atabekov. 1995. “Rattlesnake” structure of a filamentous plant RNA virus built of two capsid proteins. Proc. Natl. Acad. Sci. U. S. A. 92:2470-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alzhanova, D. V., Y. Hagiwara, V. V. Peremyslov, and V. V. Dolja. 2000. Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology 268:192-200. [DOI] [PubMed] [Google Scholar]
- 4.Alzhanova, D. V., A. J. Napuli, R. Creamer, and V. V. Dolja. 2001. Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. EMBO J. 20:6997-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiba, M., J. C. Reed, A. I. Prokhnevsky, E. J. Chapman, M. Mawassi, E. V. Koonin, J. C. Carrington, and V. V. Dolja. 2006. Diverse suppressors of RNA silencing enhance agroinfection by a viral replicon. Virology 346:7-14. [DOI] [PubMed] [Google Scholar]
- 6.Duffus, J. E., R. C. Larsen, and H. Y. Liu. 1986. Lettuce infectious yellows virus—a new type of whitefly-transmitted virus. Phytopathology 76:97-100. [Google Scholar]
- 7.Febres, V. J., L. Ashoulin, M. Mawassi, A. Frank, M. Bar-Joseph, K. L. Manjunath, R. F. Lee, and C. L. Niblett. 1996. The p27 protein is present at one end of citrus tristeza virus particles. Phytopathology 86:1331-1335. [Google Scholar]
- 8.Karasev, A. V. 2000. Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 38:293-324. [DOI] [PubMed] [Google Scholar]
- 9.Klaassen, V. A., M. Boeshore, V. V. Dolja, and B. W. Falk. 1994. Partial characterization of the lettuce infectious yellows virus genomic RNAs, identification of the coat protein gene and comparison of its amino acid sequence with those of other filamentous RNA plant viruses. J. Gen. Virol. 75:1525-1533. [DOI] [PubMed] [Google Scholar]
- 10.Klaassen, V. A., D. Mayhew, D. Fisher, and B. W. Falk. 1996. In vitro transcripts from cloned cDNAs of the lettuce infectious yellows closterovirus bipartite genomic RNAs are competent for replication in Nicotiana benthamiana protoplasts. Virology 222:169-175. [DOI] [PubMed] [Google Scholar]
- 11.Martelli, G. P., A. A. Agranovsky, M. Bar-Joseph, D. Boscia, T. Candresse, R. H. Coutts, V. V. Dolja, B. W. Falk, D. Gonsalves, W. Jelkmann, A. V. Karasev, A. Minafra, S. Namba, H. J. Vetten, G. C. Wisler, and N. Yoshikawa. 2002. The family Closteroviridae revised. Arch. Virol. 147:2039-2044. [DOI] [PubMed] [Google Scholar]
- 12.Ng, J. C., and B. W. Falk. 2006. Bemisia tabaci transmission of specific Lettuce infectious yellows virus genotypes derived from in vitro synthesized transcript-inoculated protoplasts. Virology 352:209-215. [DOI] [PubMed] [Google Scholar]
- 13.Ng, J. C., T. Tian, and B. W. Falk. 2004. Quantitative parameters determining whitefly (Bemisia tabaci) transmission of Lettuce infectious yellows virus and an engineered defective RNA. J. Gen. Virol. 85:2697-2707. [DOI] [PubMed] [Google Scholar]
- 14.Passmore, B. K., M. Sanger, L. S. Chin, B. W. Falk, and G. Bruening. 1993. Beet western yellows virus-associated RNA: an independently replicating RNA that stimulates virus accumulation. Proc. Natl. Acad. Sci. U. S. A. 90:10168-10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peremyslov, V. V., I. A. Andreev, A. I. Prokhnevsky, G. H. Duncan, M. E. Taliansky, and V. V. Dolja. 2004. Complex molecular architecture of beet yellows virus particles. Proc. Natl. Acad. Sci. U. S. A. 101:5030-5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peremyslov, V. V., Y. W. Pan, and V. V. Dolja. 2004. Movement protein of a closterovirus is a type III integral transmembrane protein localized to the endoplasmic reticulum. J. Virol. 78:3704-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peter, K. A., F. Gildow, P. Palukaitis, and S. M. Gray. 2009. The C terminus of the polerovirus P5 readthrough domain limits virus infection to the phloem. J. Virol. 83:5419-5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowhani, A., M. A. Maningas, L. S. Lile, S. D. Daubert, and D. A. Golino. 1995. Development of a detection system for viruses of woody-plants based on Pcr analysis of immobilized virions. Phytopathology 85:347-352. [Google Scholar]
- 19.Salem, N. M., A. Y. S. Chen, I. E. Tzanetakis, C. Mongkolsiriwattana, and J. C. K. Ng. 2009. Further complexity of the genus Crinivirus revealed by the complete genome sequence of Lettuce chlorosis virus (LCV) and the similar temporal accumulation of LCV genomic RNAs 1 and 2. Virology 390:45-55. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Satyanarayana, T., S. Gowda, M. A. Ayllon, and W. O. Dawson. 2004. Closterovirus bipolar virion: evidence for initiation of assembly by minor coat protein and its restriction to the genomic RNA 5′ region. Proc. Natl. Acad. Sci. U. S. A. 101:799-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian, T., L. Rubio, H. H. Yeh, B. Crawford, and B. W. Falk. 1999. Lettuce infectious yellows virus: in vitro acquisition analysis using partially purified virions and the whitefly Bemisia tabaci. J. Gen. Virol. 80:1111-1117. [DOI] [PubMed] [Google Scholar]
- 23.Voinnet, O., S. Rivas, P. Mestre, and D. Baulcombe. 2003. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33:949-956. [DOI] [PubMed] [Google Scholar]
- 24.Wang, J., M. Turina, L. R. Stewart, J. A. Lindbo, and B. W. Falk. 2009. Agroinoculation of the Crinivirus, Lettuce infectious yellows virus, for systemic plant infection. Virology 392:131-136. [DOI] [PubMed] [Google Scholar]
- 25.Wintermantel, W. M., G. C. Wisler, A. G. Anchieta, H. Y. Liu, A. V. Karasev, and I. E. Tzanetakis. 2005. The complete nucleotide sequence and genome organization of tomato chlorosis virus. Arch. Virol. 150:2287-2298. [DOI] [PubMed] [Google Scholar]
- 26.Yeh, H. H., T. Tian, V. Medina, and B. W. Falk. 2001. Green fluorescent protein expression from recombinant lettuce infectious yellows virus-defective RNAs originating from RNA 2. Virology 289:54-62. [DOI] [PubMed] [Google Scholar]
- 27.Yeh, H. H., T. Tian, L. Rubio, B. Crawford, and B. W. Falk. 2000. Asynchronous accumulation of lettuce infectious yellows virus RNAs 1 and 2 and identification of an RNA 1 trans enhancer of RNA 2 accumulation. J. Virol. 74:5762-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zinovkin, R. A., W. Jelkmann, and A. A. Agranovsky. 1999. The minor coat protein of beet yellows closterovirus encapsidates the 5′ terminus of RNA in virions. J. Gen. Virol. 80:269-272. [DOI] [PubMed] [Google Scholar]