Abstract
Autophagy can play an important part in protecting host cells during virus infection, and several viruses have developed strategies by which to evade or even exploit this homeostatic pathway. Tissue culture studies have shown that poliovirus, an enterovirus, modulates autophagy. Herein, we report on in vivo studies that evaluate the effects on autophagy of coxsackievirus B3 (CVB3). We show that in pancreatic acinar cells, CVB3 induces the formation of abundant small autophagy-like vesicles and permits amphisome formation. However, the virus markedly, albeit incompletely, limits the fusion of autophagosomes (and/or amphisomes) with lysosomes, and, perhaps as a result, very large autophagy-related structures are formed within infected cells; we term these structures megaphagosomes. Ultrastructural analyses confirmed that double-membraned autophagy-like vesicles were present in infected pancreatic tissue and that the megaphagosomes were related to the autophagy pathway; they also revealed a highly organized lattice, the individual components of which are of a size consistent with CVB RNA polymerase; we suggest that this may represent a coxsackievirus replication complex. Thus, these in vivo studies demonstrate that CVB3 infection dramatically modifies autophagy in infected pancreatic acinar cells.
Macroautophagy—henceforth referred to as autophagy—is an intracellular process that is important for cellular differentiation, homeostasis, and survival. Through autophagy, long-lived cytosolic proteins and organelles become encapsulated within double-membraned vesicles, called autophagosomes, which fuse with lysosomes to facilitate degradation of protein and cellular organelles and to promote nutrient recycling/regeneration. Autophagy plays a key role in the host immune response to infection by viruses, bacteria, fungi, and parasites (reviewed in references 10 and 62). Within virus-infected cells, whole virions and/or viral proteins and nucleic acids are captured inside autophagosomes and degraded (following lysosomal fusion) through the process of xenophagy. Moreover, autophagosome fusion with the endosomal/lysosomal pathway facilitates Toll-like receptor recognition of viral materials and delivers endogenous cytosolic viral proteins to the major histocompatibility complex (MHC) class II antigen presentation pathway, which in turn may help to trigger activation of innate immunity (and type I interferon production) and promote antigen presentation to virus-specific CD4+ T cells (reviewed in references 9, 41, 44, 47, 72, and 90). A recent study has shown that autophagy is also involved in the processing and presentation of MHC class I-restricted viral epitopes (13).
Given the importance of autophagy in antiviral immunity, it is perhaps not surprising that viruses have evolved mechanisms to evade and/or subvert this pathway (reviewed in references 9, 11, 14, 35, 37, 60, 61, and 77). Several members of the herpesvirus family, most notably herpes simplex virus type 1, inhibit autophagy within an infected cell and encode proteins that block and/or target intracellular signaling pathways that regulate autophagy (reviewed in references 60 and 61). However, some viruses not only evade autophagy but also appear to take advantage of the process; several RNA viruses induce autophagy and exploit the pathway during their replication (1, 12, 15, 31, 40, 43, 76, 93, 96). Viruses belonging to the Picornaviridae family and the Nidovirales order replicate their genomes on double-membraned vesicles that resemble autophagosomes; these vesicles are notably smaller in size than cellular autophagosomes and are decorated with proteins derived from the autophagic pathway (19, 21, 31, 37, 67, 68, 71, 92). Viral proteins encoded by poliovirus and equine arterivirus can trigger the formation of these autophagy-like vesicles (79, 80), and the expression of a single poliovirus protein, 2BC, is sufficient to induce lipidation of the host autophagy protein light chain 3 (LC3), encoded by the Atg8 gene (87). Taken together, these studies suggest that some viruses subvert the autophagy pathway to generate double-membraned vesicles that provide a surface for RNA replication (8, 37, 88). In addition, these vesicles may permit newly formed virions to escape from infected cells via a nonlytic route (36, 85).
Although studies have demonstrated that the autophagic pathway may play an important role in virus infection in vitro, either to promote or to restrict viral replication, we are just beginning to appreciate and understand the function and effects of autophagy for virus infections in vivo. Autophagy acts in an antiviral fashion to limit tobacco mosaic virus replication and programmed cell death in plants (46), to prevent a pathogenic infection with vesicular stomatitis virus in flies (73), and to protect against fatal encephalitis in Sindbis virus- or herpes simplex virus type 1-infected mice (45, 59, 63). Nonetheless, to date there is a dearth of in vivo studies; animal models of virus infection are needed in order to better define the antiviral role of autophagy in vivo (41, 62). In addition, studies that address the role of viral subversion of autophagy in vivo are warranted. Does this process occur within infected animals, and is it required for viral replication in particular cell types or for viral pathogenesis? Recent studies have shown that autophagy not only promotes the replication of hepatitis B virus and enterovirus 71 in vitro but also may be induced by infection in vivo, potentially to benefit the virus rather than the host (28, 78).
Type B coxsackieviruses (CVBs) are members of the Picornaviridae family and Enterovirus genus and, as such, are closely related to polioviruses. CVBs are important human pathogens that often induce severe acute and chronic diseases and cause morbidity and mortality (69, 91). CVBs are the most common cause of infectious myocarditis (38, 82) and frequently trigger pancreatitis and aseptic meningitis (7, 16, 29, 51). Tissue culture studies (93) have shown that CVB type 3 (CVB3) promotes LC3 conversion and autophagosome accumulation in virus-infected cells in vitro and that modulation of the autophagic pathway (using chemicals or small interfering RNA-mediated knockdown) to enhance or dampen autophagy results in an increase and a decrease, respectively, in viral protein expression and/or viral titers; however, the reported changes in viral titers were modest (2- to 4-fold). In the present study, we examine whether CVB3 activates the autophagic pathway in vivo, specifically in pancreatic acinar cells, which are a natural primary target for this virus. Using a mouse model of CVB3 infection, which faithfully recapitulates most aspects of CVB disease in humans, we demonstrate that this virus triggers LC3 conversion and also modulates other components of the autophagy machinery. In addition, using a recombinant CVB3 (rCVB3) that expresses Discosoma sp. red fluorescent protein (DsRed-CVB3), we identify virus-infected cells in situ and show that CVB3 infection increases autophagosome abundance in vivo. Lysosomal-associated membrane protein 1 (LAMP-1) immunostaining confirmed that amphisomes are generated in virus-infected cells but that autophagic flux was not substantially enhanced as the infection progressed; rather, there appears to be a substantial blockade in fusion with lysosomes. Finally, transmission electron microscopy (TEM) ultrastructural analysis of the infected pancreas confirmed that double-membraned autophagy-like vesicles as well as very large autophagic compartments (for which we have coined the term “megaphagosomes”) were generated in acinar cells following virus infection. Overall, these data provide compelling evidence that CVB3 induces autophagy in vivo and suggest that this picornavirus may subvert this process in a mammalian host.
MATERIALS AND METHODS
Mice.
GFP-LC3 mice contain a transgene encoding an enhanced green fluorescent protein (eGFP)-rat LC3 fusion protein (56). GFP-LC3 transgenic mice, backcrossed to the C57BL/6 background, were obtained from Riken BioResource Center (Koyadai, Japan) and maintained as homozygous transgenic (Tg/Tg) mice. Homozygous transgenic mice were bred with wild-type C57BL/6 mice to generate heterozygous transgenic (Tg/+) mice. C57BL/6J mice were purchased from The Scripps Research Institute (TSRI) breeding facility or from Jackson Laboratories. All experimental procedures involving mice were approved by the TSRI Animal Care and Use Committee and were carried out in accordance with NIH guidelines.
Viruses and infections.
The wild-type (wt) CVB3 used in these studies is a plaque-purified isolate (designated H3) of the myocarditic Woodruff variant of CVB3 (89). Plasmid pH3, encoding a full-length infectious clone of this virus (39), was provided by Kirk Knowlton (University of California, San Diego, CA). A recombinant CVB3 encoding DsRed was generated as described previously (81). wt CVB3 and DsRed-CVB3 were grown in HeLa cells, and virus stocks were generated as described previously (25). Naïve adult male GFP-LC3 heterozygous transgenic (Tg/+) mice (6 to 12 weeks old) were inoculated intraperitoneally with 104 PFU of wt CVB3 or 107 PFU of DsRed-CVB3.
Plaque assays.
Mice were perfused with Dulbecco's phosphate-buffered saline (DPBS; Invitrogen) to clear blood from the tissues and to prevent overestimation of organ virus titers due to CVB3 in the blood. Samples were weighed and homogenized in 1 ml Dulbecco modified Eagle medium (Invitrogen), and the titers of infectious virus in the lysates were determined. Plaque assays were performed on subconfluent HeLa cell monolayers, as described previously (30), and the virus titers (PFU/g) were calculated for each sample.
Analysis of protein expression by Western blotting.
Mice were perfused with DPBS, and the pancreas was collected and snap-frozen in liquid nitrogen. Samples were thawed, weighed, and homogenized in freshly prepared RIPA buffer (Millipore) containing the following inhibitors: 30 mM p-nitrophenylphosphate (MP Biomedicals), 1× protease inhibitor cocktail (Roche), 1 mM phenylmethylsulfonyl fluoride (EMD Calbiochem), 1 mM Na3VO4 (Sigma), and 10 mM NaF (Sigma). Homogenates were spun at maximum speed in a cold microcentrifuge for 30 min, and the clarified supernatant was collected. The total protein concentration of each lysate was determined by the Pierce bicinchoninic acid protein assay (Thermo Scientific), using bovine serum albumin (BSA) as a standard. Lysates were diluted in denaturing sample buffer and boiled for 5 min, and 20 to 40 μg of each sample was loaded on a bis-Tris NuPAGE gel (Invitrogen) and separated by SDS-PAGE. Proteins were then electroblotted onto a 0.45-μm-pore size Invitrolon polyvinylidene difluoride membrane (Invitrogen), following the manufacturer's instructions. Membranes were blocked with 5% BSA or nonfat milk in Tris-buffered saline (TBS) plus 0.1% Tween 20 (TBST) for 1 h at room temperature (RT). Next, the membranes were washed in TBST and incubated with primary antibody in 1% BSA (or milk) in TBST overnight at 4°C. The following primary antibodies were used: polyclonal rabbit anti-human LC3b (provided by Karla Kirkegaard, Stanford University), polyclonal rabbit Living Colors GFP antibody (Clontech), rabbit anti-human Atg5 (55), monoclonal mouse anti-p62 (clone 3/p62 lck ligand; BD Biosciences), or polyclonal guinea pig anti-p62 (C-terminal specific; Progen Biotechnik). Membranes were washed again and then incubated with horseradish peroxidase-conjugated secondary antibody (Amersham ECL donkey anti-rabbit IgG; sheep anti-mouse IgG [GE Healthcare Life Sciences], or rabbit anti-guinea pig IgG [Invitrogen]) in 1% BSA (or milk) in TBST for 1 h at RT. After additional washing of the membranes, the membranes were incubated with SuperSignal West Pico chemiluminescent substrate (Thermo Scientific) for 5 min and then exposed to BioMax MR film (Kodak). The membranes were then washed in TBST, stripped with Restore Western blot stripping buffer (Thermo Scientific) for 10 min at RT, and blocked with 5% milk in TBST for 1 h at RT. Finally, the membranes were washed in TBST, reprobed with mouse glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody (clone 6C5; Millipore) in 1% milk in TBST overnight at 4°C, and then processed as described above, which provided a control for protein loading of each sample.
Histology.
Mice were perfused with DPBS, and the pancreas was harvested and fixed in buffered zinc formalin (Z-FIX; Anatech, Ltd.) at RT overnight. Tissues were paraffin embedded, and 3-μm sections were cut and stained with hematoxylin-eosin or Masson's trichrome. Images were captured at ×32 magnification with an Axiovert 200 inverted microscope (Carl Zeiss, Inc.) using AxioVision (version 4.8.1) software (Carl Zeiss, Inc.).
Vibratome sectioning and immunostaining.
Mice were perfused with DPBS, and the pancreas was harvested and fixed in buffered zinc formalin (Z-FIX) at RT overnight. The fixative was replaced with PBS containing 0.1% sodium azide on the following day, and tissues were preserved at 4°C. Sections of pancreas (80 μm) were cut with a VT1000 S vibratome (Leica Microsystems Inc.). Some sections were stained with antibodies, as follows. Free-floating sections were washed 3 times in PBS and permeabilized in PBS with 0.5% Triton X-100 for 30 min at RT. After incubation, the sections were washed once in PBS with 0.3% Triton X-100 and then blocked in PBS with 5% normal goat serum (NGS) and 0.3% Triton X-100 for 2 h at RT. Next, the sections were washed 3 times in PBS with 0.3% Triton X-100 and then incubated with rat anti-mouse CD107a/LAMP-1 (clone 1D4B; BD Biosciences) in PBS with 1% NGS and 0.3% Triton X-100 for 1 h at RT and then for an additional 42 to 48 h at 4°C. The sections were then washed 3 times in PBS with 0.3% Triton X-100 and then incubated for 2 h at RT with goat anti-rat IgG Alexa Fluor 647 dye (Invitrogen). After incubation, the sections were washed once in PBS with 0.3% Triton X-100 and then 3 times in PBS. All sections were stained with Hoechst 33342 in PBS for 30 min at RT (to label nuclei) and then washed 3 times in PBS and mounted on glass slides with Vectashield (Vector Labs) or ProLong Gold (Invitrogen) mounting medium.
Confocal laser scanning microscopy.
Three or four color images were captured using an LSM 710 laser scanning confocal microscope equipped with Zen 2009 software (Carl Zeiss, Inc.). Representative regions within each vibratome section of pancreas were scanned at ×40 or ×63 magnification as serial 8-bit optical sections with 0.3- to 0.5-μm-interval step slices (either 512 by 512 or 1,024 by 1,024 image sizes). All images were then maximum projected for presentation using Image J software (NIH; http://rsbweb.nih.gov/ij/) or the Easy 3D function of Imaris software (BitPlane, Inc.). For quantitative analysis of autophagosome number and area, images were resampled to consist of one cell monolayer only and were then maximum projected/flattened and used for two-dimensional analysis in Image Pro Plus (Media Cybernetics Inc.) or Image J software. To determine the average feret of giant autophagosomes, each vesicle was analyzed independently by scrolling through each image z series of optical slices to find and determine the largest diameter of each autophagosome. Diameter values were measured in Image J software and corroborated in Image Pro Plus software.
Transmission electron microscopy.
Mice were perfused with 0.9% saline, followed by 4% paraformaldehyde-1.5% glutaraldehyde in 0.1 M cacodylate buffer with 1 mM CaCl2. The pancreas was removed and immersed in the above fixative on ice for 6 h and was transferred to 2.5% glutaraldehyde in 0.1 M cacodylate buffer with 1 mM CaCl2 for overnight fixation. After a buffer wash, the tissue was further fixed in 1% OsO4 with 1.5% potassium ferricyanide in 0.1 M sodium cacodylate and again washed in cacodylate buffer, dehydrated in graded ethanol series, and transitioned in propylene oxide. Pancreatic tissues were embedded in Embed 812/Araldite (Electron Microscopy Sciences, Hatfield, PA). Thick sections (1 to 2 μm) were cut, mounted on glass slides, and stained in toluidine blue for general assessment in the light microscope. Subsequently, 70-nm thin sections were cut, mounted on copper slot grids coated with Parlodion, and stained with uranyl acetate and lead citrate for examination on a Philips CM100 electron microscope (FEI, Hillsbrough, OR) at 80 kV, and images were collected using a Megaview III charge-couple-device (CCD) camera (Olympus Soft Imaging Solutions, Lakewood, CO).
Immunoelectron microscopy (immuno-EM).
Mice were perfused with 0.9% saline, followed by 4% paraformaldehyde-0.1% glutaraldehyde in PBS (pH 7.4). The pancreas was removed and immersed in the above buffer on ice for a further 6 h and then washed overnight in PBS. Pancreatic tissue was then treated with 50 mM glycine, washed in PBS, incubated in 1% sodium metaperiodate, and again washed in PBS, before being dehydrated in a freshly prepared graded ethanol series through to 90%. The tissue pieces were impregnated with L. R. White resin and embedded in predried gelatin capsules for polymerization at 40°C. Thin sections (70 nm) were cut and mounted on uncoated 300-mesh nickel grids. Immunostaining of the sections consisted first of blocking in 4% BSA with 2% whole goat serum in TBST, followed by an overnight incubation in the rabbit polyclonal anti-GFP antibody (catalog no. 6556; Abcam) diluted 1:250 in 2% BSA with 1% whole goat serum in TBST. The grids were then washed in 2% BSA in TBS, incubated in goat anti-rabbit secondary antibody tagged with 12-nm gold beads, (Jackson ImmunoResearch, West Grove, PA) diluted 1:20 in TBS, washed extensively in TBS, and then fixed in 2% glutaraldehyde, followed by a water wash, staining in 2% uranyl acetate, and finally, a brief water wash. The dried grids were examined on a Philips CM100 electron microscope (FEI) at 80 kV, and images were collected using a Megaview III CCD camera (Olympus Soft Imaging Solutions).
Statistical analyses.
An unpaired two-tailed t test (assuming equal variances) was used to determine statistical significance between 2 groups of data (GraphPad Prism software). Alternatively, one-way analysis of variance was used to determine statistical significance among 3 or more groups of data, and Tukey's post hoc test was used for multiple comparisons within each data set (GraphPad Prism software). A P value of <0.05 was considered statistically significant.
RESULTS
For in vivo studies of CVB infection, we took advantage of the GFP-LC3 transgenic mouse model, which facilitates the identification and analysis of autophagy in vivo. This transgenic mouse encodes a fusion protein of GFP and microtubule-associated protein 1 light chain 3 (LC3) (54, 56). The LC3 protein is encoded by the Atg8 gene, and its primary translation product, pro-LC3, undergoes posttranslational cleavage to remove residues from the C terminus, generating the stable cytosolic form, named LC3-I (33). During the process of autophagosome formation, LC3-I is lipidated and specifically associates with the membrane of autophagosomes (33); this membrane-bound form of LC3 (LC3-II) is, at present, considered the best marker of autophagosomes in mammalian cells. In GFP-LC3 transgenic mice, green fluorescent autophagosomes (tagged with GFP-LC3-II) are readily detected in many tissues and cell types, including the acinar cells of the pancreas (54, 56), as well as in Purkinje cells and cortical neurons in the brain (2, 3). These GFP-LC3 mice, which are on the H-2b background, support wt CVB3 replication; the virus reached a very high titer in the pancreas (∼1010 PFU/g) as early as day 1 postinfection (p.i.), while titers in the liver and heart were >3 log units lower at this time (see Fig. S1 in the supplemental material). Overall, the data demonstrated that wt CVB3 replicates rapidly to very high titers in GFP-LC3 mice, with the kinetics (and viral loads) being similar to those previously observed in nontransgenic C57BL/6 mice (18, 52). The identification of the pancreas as the dominant primary site of CVB3 replication in these mice is consistent with previous observations in wt mice, and for this reason, the analyses presented below focused mainly on this organ.
CVB3 infection in vivo is associated with dramatic modification of LC3 and Atg5.
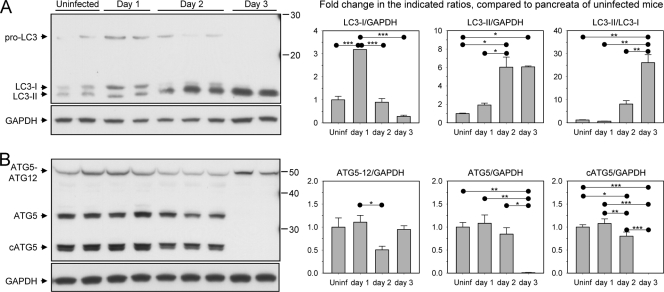
Given the rapid onset and extraordinarily high level of viral replication in the pancreas, we examined whether wt CVB3 infection of this organ was associated with changes to the normal homeostatic level of autophagy. In particular, we felt it to be important to determine if CVB3-induced conversion of LC3—previously documented in tissue culture (93)—occurs in vivo and/or whether other components of the autophagy machinery are also altered by virus infection. To this end, GFP-LC3 transgenic mice were infected with wt CVB3, and on days 1 to 3 p.i., the pancreata were harvested and analyzed by Western blotting (Fig. 1). First, we evaluated the effects of infection on LC3 (Fig. 1A). Pro-LC3 was detectable in the uninfected tissue and its level had increased at day 1 p.i., suggestive of increased Atg8 gene expression in response to CVB infection in vivo. The level of this protein declined at day 2 p.i., and it was undetectable at day 3. This is consistent with the well-established capacity of CVB3 to terminate the synthesis of most host cell proteins. Both the unlipidated (LC3-I) and lipidated (LC3-II) forms of endogenous LC3 were also present in uninfected pancreas. The level of LC3-I increased 3-fold on day 1 p.i. and then declined thereafter; by day 3 p.i., only a small amount of LC3-I was detectable. The drop in LC3-I coincided with a marked increase in LC3-II on day 2 p.i. On day 3 p.i., most of the LC3 present in the pancreas had been converted to LC3-II; by this time point, the ratio of LC3-II to LC3-I was ∼25:1 (Fig. 1A, right-hand graph), while pro-LC3 was no longer detected by Western blotting. Taken together, these data suggest that (i) CVB3 infection upregulates pro-LC3 expression in the pancreas and then (ii) terminates the production of pro-LC3, after which (iii) the existing LC3-I undergoes almost complete conversion to LC3-II. LC3 conversion was also evaluated in the livers of wt CVB-infected mice (in which virus titers reached ∼109 PFU/g; see Fig. S1 in the supplemental material), and similar results were obtained (data not shown).
FIG. 1.
Kinetic analysis of autophagic protein expression in CVB3-infected pancreas. Protein lysates were prepared from the pancreas of wt CVB3-infected mice (days 1, 2, and 3 p.i.) or from uninfected mice as a control, and the samples were subjected to SDS-PAGE and Western blotting to determine the expression kinetics of several proteins involved in the autophagic pathway. (A) A Western blot probed with an LC3-specific antibody is shown; 3 distinct bands are detected, including bands for pro-LC3 and endogenous LC3-I and -II. Each lane represents the pancreatic extract of an individual mouse. (B) Western blot analysis of Atg5 expression; 3 distinct bands are visible, including bands for the Atg5-Atg12 complex, monomeric Atg5, and a cleaved form of Atg5. All blots were stripped and reprobed with a GAPDH antibody as a protein loading control. For both panels, the fold change in protein expression over the course of infection was determined by densitometry analysis, and graphs are shown. Fold changes were normalized to the level of GAPDH expression, and data are represented as the means + standard errors of the means of 2 to 6 mice per group (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Another hallmark of autophagosome formation involves the covalent linkage, during the elongation step, of Atg5 to Atg12 by a ubiquitin E3-like protein conjugation system. The Atg5-Atg12 conjugate forms a complex with Atg16L, which recognizes the phagophore and localizes the lipidation of LC3 to this site (94). The presence of the Atg5-Atg12 complex in cells is one indicator of the activity of the elongation step of autophagy processing. Given that CVB3 infection promoted LC3 conversion, we questioned whether the virus might also affect these other components of the autophagy machinery (Fig. 1B). Three forms of Atg5 were identified in uninfected pancreas: free Atg5, the Atg5-Atg12 conjugate, and a cleaved form of Atg5 (cAtg5) that is generated by calpain proteases (95). Both the free and cleaved forms of Atg5 were detected in the pancreas on days 1 and 2 p.i. but were not detectable by Western blotting on day 3; by this time point, only the Atg5-Atg12 complex was present. These data suggest that, as we propose above for pro-LC3, de novo synthesis of Atg5 is no longer occurring at 3 days p.i. and that all existing Atg5 is complexed with Atg12, indicative of ongoing autophagic flux. As noted above for pro-LC3, we consider it likely that the reduced quantities of these other autophagy-related proteins result from the well-established phenomenon of coxsackievirus-induced shutoff of cap-dependent translation.
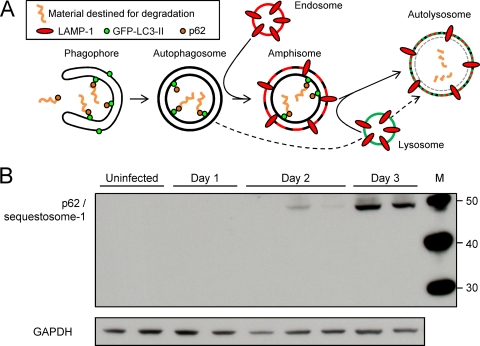
Kinetic analysis of autophagic flux in CVB3-infected pancreas.
The increase in LC3 conversion and the accumulation of Atg5-Atg12 in the pancreas (Fig. 1) suggest that there is enhanced autophagic activity in vivo following wt CVB3 infection. However, these are early events in the autophagy pathway, so we thought it important to carry out additional studies of autophagic flux. A diagrammatic outline of the autophagy pathway is shown in Fig. 2A. Polyubiquitinated protein aggregates are recognized by and attached to a protein named p62. This protein, also known as sequestosome-1, is a signaling adapter or scaffold protein that can bind to LC3. Complexes of aggregate-p62-LC3-II are recruited into the developing autophagosome (the phagophore), where they are attached, via the lipidated LC3-II, to the autophagosomal membrane (4, 65, 74, 75). The mature autophagosome then fuses with components of the endolysosomal pathway. As shown, an autophagosome may fuse with an endosome to form the amphisome, and this subsequently fuses with lysosomes, forming the autolysosome. Alternatively, the autophagosome may join directly with a lysosome (Fig. 2A, dashed line). Thus, the exact composition of an autolysosome varies according to the precursor vesicles from which it was formed. The fusion mechanism itself can add to this heterogeneity, because fusion often is incomplete, with each of the precursor vesicles contributing differently to the daughter autolysosome; this has been described as “kiss-and-run fusion” (32). p62 is a specific substrate that is efficiently degraded by autophagy; the amount of p62 within a cell inversely correlates with the cell's autophagic activity (57). Thus, the accumulation of p62 is a good indicator that autophagic flux is inhibited. Therefore, we used Western blotting to assess the quantity of p62 in pancreata at various times after CVB3 infection (Fig. 2B). Compared to uninfected pancreas, the amount of p62 increased markedly on day 2 p.i. and even more so on day 3. Note that, although it is not obvious in Fig. 2B, p62 was present in uninfected pancreatic acinar samples and could be visualized upon longer exposure of the Western blot (data not shown). Furthermore, similar data have been generated using a different antibody to p62. Thus, we conclude that autophagic flux is markedly reduced in the pancreas at day 3 p.i. These changes are most likely attributable to effects in acinar cells, which constitute the great majority of cells in the pancreas; the confocal microscopy studies reported below confirm that acinar cells are affected. Altered cleavage of GFP-LC3 is an alternative means by which to measure autophagy flux (57). Therefore, in addition to analyzing p62 in infected pancreata, we also assessed the stability of GFP-LC3. As shown in Fig. S2 in the supplemental material, this protein cleavage assay suggested that autophagy flux had not been entirely extinguished at day 3. However, in this case, interpretation is complicated by at least three factors: (i) the assay relies on a ratio of intact to fragmented protein, and the former may be reduced by CVB-induced host shutoff, as indicated in Fig. S2 in the supplemental material; (ii) CVB encodes two powerful proteases that may contribute to the degradation; and (iii) acinar cells are replete with proenzymes that may be activated during infection, further increasing nonspecific proteolysis.
FIG. 2.
Accumulation of p62 indicates blockade of autophagic flux in CVB3-infected acinar cells in vivo. (A) The autophagy pathway is shown in diagrammatic form. (B) Western blotting was carried out to determine the amount of p62/sequestosome-1 in the pancreas at the indicated times p.i. GAPDH is used as a gel loading control. Each lane represents an individual mouse. A p62-specific antibody (from BD Biosciences; see Materials and Methods) was used to generate the data shown. Data are representative of those from two independent experiments. Similar results (data not shown) were obtained using a different p62-specific antibody (from Progen Biotechnik; see Materials and Methods).
A recombinant CVB3 expressing DsRed triggers profound pancreatitis.
To allow us to identify infected cells in GFP-LC3 transgenic mice, in the remaining studies described in this report we used rCVB3 that expresses DsRed (DsRed-CVB3; see reference 81). This recombinant virus grows with nearly normal kinetics in vitro (see the one-step growth curve in Fig. S3A in the supplemental material), but before analyzing the effects of DsRed-CVB3 on autophagy in vivo, we considered it important to determine whether this recombinant virus caused pancreatic disease similar to that caused by the wt virus, which results in widespread infection of the exocrine pancreas and triggers severe pancreatitis (51). GFP-LC3 mice were inoculated with 107 PFU of this virus, and mice were killed at several times p.i. Paraffin sections of the pancreata were stained with hematoxylin-eosin or Masson's trichrome. As shown in Fig. S3B in the supplemental material, inflammatory infiltrates were abundant in the interstitial spaces by day 2 p.i., and acinar cells became hypochromic. One day later, infiltration was extensive, and acinar cell death was evident. As we have previously reported for wt CVB3, the islets of Langerhans showed no obvious histopathological changes. Thus, DsRed-CVB3 causes severe pancreatitis, validating the use of this agent in our subsequent studies.
Confocal microscopy of autophagosomes in DsRed-CVB3-infected pancreas.
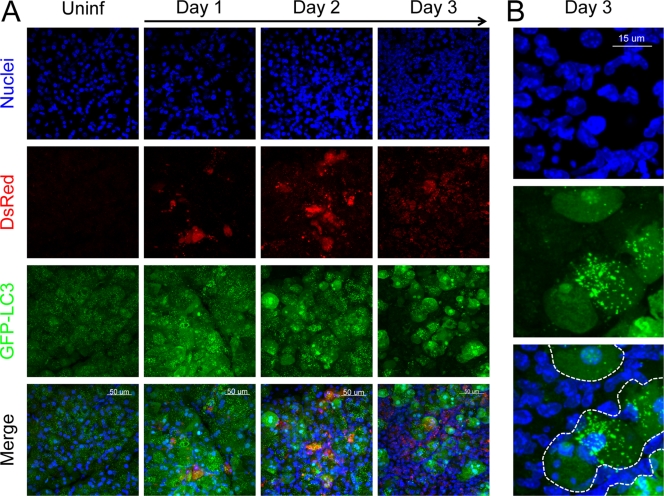
We have recently described a novel approach to visualize autophagosomes in vivo (2). This method combines two well-characterized techniques, vibratome sectioning and confocal microscopy, and identifies autophagosomes at a sufficiently high resolution to permit their enumeration and the evaluation of their physical characteristics (e.g., size, shape, and circularity). In light of the biochemical evidence for induction of autophagy in CVB3-infected pancreas (Fig. 1, Fig. 2), we next used this approach to investigate whether CVB3 infection led to a change in the number and/or characteristics of autophagosomes in acinar cells in vivo and to examine the interaction between viral protein and autophagic vesicles. We reasoned that using DsRed-CVB3 would be advantageous for these studies because both autophagosomes and virus-infected cells could be visualized directly. The use of fluorescence to identify both virus-infected cells and autophagosomes (i) obviated the need for antibody-based detection, thereby eliminating nonspecific/background signals from antibody staining, and (ii) avoided the need to permeabilize the tissue sections with detergent, helping to preserve the fluorescent signals of GFP and DsRed and ensuring that the native anatomical structure of the tissue was maintained. Pancreatic sections were first examined at a modest magnification (×40) to assess the overall pattern and distribution of fluorescence among a large number of acinar cells. Nuclear staining revealed a high density of cells within the exocrine tissue on day 3, with the large nuclei of acinar cells being intermingled with smaller, more numerous nuclei (Fig. 3A, top row); this reflects the inflammatory infiltrate shown in Fig. S3 in the supplemental material. Virus-infected acinar cells were visualized using DsRed (Fig. 3A, 2nd row); no signal was present in uninfected tissue, but foci of infected cells were visible on day 1 p.i., and by day 2 the extent of virus infection was widespread. In uninfected tissue, GFP-LC3 fluorescence in acinar cells consisted of a dim diffuse green signal in the cytoplasm (GFP-LC3-I) as well as more intense discrete punctae that represented GFP-LC3-II-positive autophagosomes (Fig. 3A, 1st panel, 3rd row) or, perhaps, autophagy-like vesicles in infected cells. The spreading virus infection was associated with an increase in the intensity of GFP-LC3 fluorescence from day 1 onward (Fig. 3A, 3rd row). In day 3 tissue, GFP-LC3 fluorescence was considerably stronger in acinar cells than in the infiltrating inflammatory cells (Fig. 3A and B).
FIG. 3.
CVB3 infection alters the intensity and distribution of GFP-LC3 in pancreatic acinar cells in vivo. Mice were infected with DsRed-CVB3; and on days 1, 2, and 3 p.i., the fluorescent GFP-LC3 and DsRed proteins were imaged in situ in vibratome sections of pancreas by confocal microscopy. (A) Representative images (×40 magnification) of nuclei, DsRed, GFP-LC3, and a merged view are shown for uninfected and infected tissues. (B) Higher-magnification images (nuclei, GFP-LC3, and merged) show that the increase in GFP-LC3 intensity occurs mainly in acinar cells (surrounded by dashed white lines in the merged image); the remaining nuclei belong to infiltrating cells, in which GFP-LC3 fluorescence is minimal.
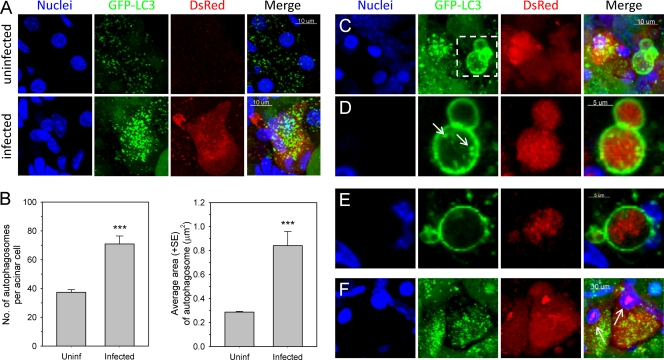
To better evaluate the effects of CVB3 on the abundance and physical characteristics of autophagosomes in vivo, we next performed high-magnification confocal imaging of infected pancreas. Both the number and size of GFP-LC3-positive (GFP-LC3+) punctae increased in strongly infected (DsRed bright) acinar cells compared to the number and size of the puntae in acinar cells in uninfected pancreas (Fig. 4A). The number of autophagosomes in these infected cells was ∼2-fold greater than that in acinar cells in uninfected pancreas, and the average area of an autophagosome was ∼3-fold greater in a virus-infected cell than in an uninfected acinar cell (Fig. 4B). In addition to these quantitative differences, several prominent phenotypic changes associated with autophagosomes were apparent in DsRed-CVB3-infected cells. First, we observed very large autophagosomes, often clustered together (Fig. 4C, 2nd panel, white dashed box), which have an average size (feret) of 6.85 ± 0.38 μm (mean ± standard error [SE]). Smaller autophagosomes were seen close to the membrane of the giant autophagosomes and/or were contained within the larger structures (Fig. 4D, 2nd panel, arrows). Intriguingly, some of these very large autophagosomes enclosed viral DsRed protein (Fig. 4D). Autophagosomes with these three features (large size, association with smaller autophagosomes, containing viral protein), although not ubiquitous, are not uncommon; a second example is shown (Fig. 4E). These virus-induced structures—some of which are as large as the cell nucleus—were also present, less frequently, in the pancreata of wt-infected mice; we propose that they be named megaphagosomes. Furthermore, in virus-infected acinar cells containing abundant GFP-LC3+ autophagosomes, DsRed was frequently observed not only in the cytoplasm but also in the nucleus; such nuclei were usually condensed and appeared pyknotic, suggesting that the virus infection had triggered apoptosis (Fig. 4F).
FIG. 4.
CVB3 infection of acinar cells in vivo increases the abundance and changes the physical characteristics of autophagosomes. Uninfected and DsRed-CVB3-infected pancreas sections were examined by confocal microscopy to detect the GFP-LC3 and DsRed proteins in situ at high magnification. (A) Representative z-stack images of nuclei, GFP-LC3, DsRed, and a merged view of uninfected and infected acinar cells. (B) Bar graphs displaying the average number of autophagosomes per acinar cell and the average total area of an autophagosome in uninfected and virus-infected acinar cells (days 1 and 2). An average of 70 cells/group was quantitated. Data are presented as the means + standard errors of the means; ***, P < 0.0001. (C) z-stack images showing the presence of large autophagosomes (megaphagosomes) in infected cells. (D) The region enclosed in a dashed white box in panel C is shown at higher magnification and as a narrow z-stack image. White arrows indicate the small autophagosomes that are often seen to be associated with the larger structures. The right-hand image shows DsRed within the megaphagosome. (E) Narrow z-stack images of a second megaphagosome; again, it contains DsRed, and smaller autophagosomes are found nearby. (F) z-stack image to show DsRed in two pyknotic cell nuclei (white arrows).
Amphisomes are formed in CVB3-infected acinar cells in vivo.
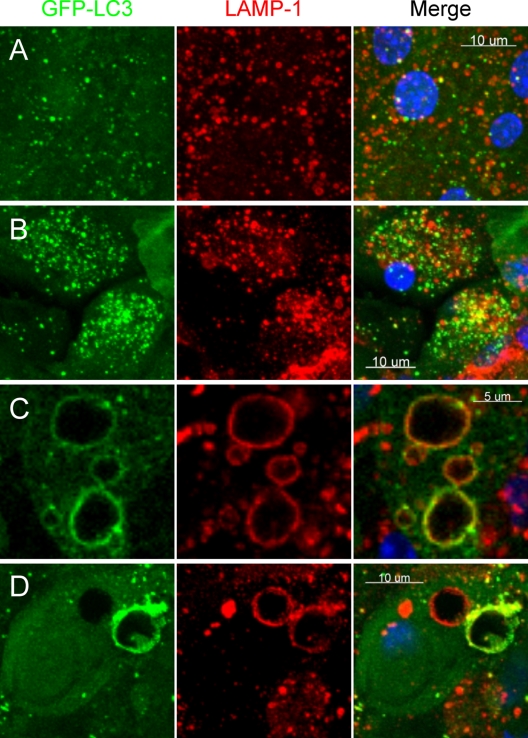
Several viruses are thought to inhibit autophagy by blocking the fusion of autophagosomes with endosomes and/or lysosomes, preventing the formation of amphisomes or autolysosomes (17, 40, 76, 88). To assess the effects of CVB3 on autophagosome fusion with endosomes/lysosomes in vivo, we evaluated the expression and distribution of LAMP-1, a protein that is found on endosomes and lysosomes. In GFP-LC3 mice, autophagosomes can be distinguished from amphisomes or autolysosomes as follows (see the diagrammatic outline of the autophagy pathway, Fig. 2A): green vesicles (GFP-LC3+ LAMP-1 negative [LAMP-1−]) will be autophagosomes; yellow vesicles (GFP-LC3+ LAMP-1 positive [LAMP-1+]) will be amphisomes; and red vesicles (GFP-LC3 negative [GFP-LC3−] LAMP-1+) cannot be categorized solely on the basis of LAMP-1 expression, as they may be endosomes, lysosomes, or autolysosomes (GFP-LC3-II is degraded in autolysosomes, and in addition, GFP fluorescence is quenched in an acidic environment [84]). Sections of uninfected and DsRed-CVB3-infected pancreas were stained with an antibody specific for LAMP-1, and the colocalization of GFP-LC3+ and LAMP-1+ vesicles was examined by confocal microscopy. In acinar cells from uninfected pancreas, endosomes (normal size range, 100 to 200 nm), lysosomes (normal size range, 0.1 to 1.2 μm), and autophagosomes were abundant and amphisomes were infrequent (yellow signal; Fig. 5A, right panel). Greater numbers of autophagosomes were seen in acinar cells of a DsRed-CVB3-infected pancreas, consistent with our previous findings, and a substantial number of amphisomes were present in the infected cells (Fig. 5B). Higher-magnification analyses of these double-positive vesicles showed them to vary in at least two ways (Fig. 5C and D). First, they varied in size; some were ∼2 μm in diameter, which is relatively normal for vesicles in the autophagy pathway, while others were extremely large (approaching 10 μm in diameter) and represent the aforementioned megaphagosomes. Second, they varied in the relative amounts of GFP-LC3 and LAMP-1; in some, GFP-LC3 appeared to be more abundant than LAMP-1, while in others, the opposite was true. This is consistent with kiss-and-run fusion (32). Most of the GFP-LC3+ megaphagosomes were also LAMP-1+ (Fig. 5C and D), but some large LAMP-1+ vesicles in infected cells were GFP-LC3 negative (Fig. 5D); we propose that these giant vesicles may represent megaphagosomes that have fused with lysosomes and in which, as a result, the GFP has been degraded. In summary, these data indicate that CVB3 permits amphisome formation but markedly limits subsequent steps in autophagy processing.
FIG. 5.
Fusion of autophagosomes with endosomes or lysosomes in CVB-infected acinar cells in vivo. Vibratome sections of uninfected and DsRed-CVB3-infected pancreas were stained with an antibody specific for the endosomal/lysosomal marker LAMP-1, and the colocalization of GFP-LC3 and LAMP-1 vesicles was examined by confocal microscopy. Representative z-stack images of uninfected (A) and virus-infected (B to D) tissues are shown. Single-color fluorescence images of GFP-LC3 and LAMP-1 are presented in the 1st and 2nd columns, and a merged view of these 2 proteins is shown in the 3rd column; a yellow signal signifies colocalization of GFP-LC3 and LAMP-1. In order to better visualize the fluorescent signals present in the borders of and within the large GFP-LC3+ vesicles, a narrow z stack of two cells containing several of these structures is shown (C and D).
Ultrastructural analysis of autophagosomes in CVB3-infected pancreas.
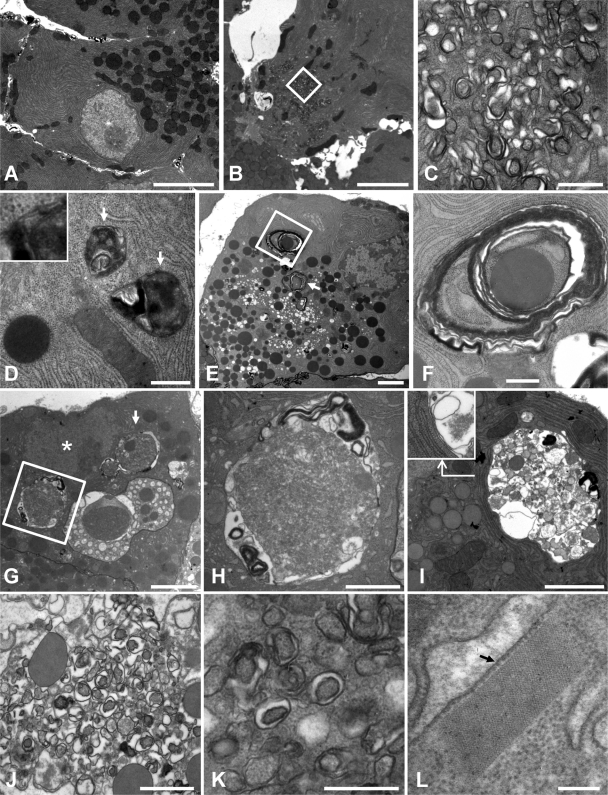
We next examined acinar cells by TEM (i) to confirm and extend the findings from our fluorescent microscopic analysis of GFP-LC3+ autophagosomes, (ii) to identify autophagosomes and/or autophagy-like vesicles, and (iii) to better characterize the giant GFP-LC3+ vesicles and assess their relationship to the autophagy pathway. Acinar cells in uninfected tissue contained abundant, tightly packed layers of rough endoplasmic reticulum (ER) along with numerous zymogen granules that were often concentrated in the apical region adjacent to the duct (Fig. 6A). Following infection with DsRed-CVB3, clusters of small double-membraned vesicles were seen in some acinar cells (Fig. 6B and C); these vesicles resembled conventional autophagosomes in that they contained cytoplasmic material, but they were smaller in size (∼250 nm). These structures, which resemble the autophagy-like vesicles that are induced by poliovirus (35, 88), have previously been identified in CVB-infected pancreas and were referred to as compound membrane-vesicle (CMV) complexes (5, 26). Autophagosomes of more normal size (500 to 1,500 nm) were also seen in acinar cells from infected pancreas (Fig. 6D); the double-membraned nature of these vesicles is apparent in the inset to Fig. 6D. Some acinar cells had larger (2- to 3-μm) multilamellar autophagosomes (Fig. 6E) that contained zymogen granules, rough ER, and/or ribosomes (Fig. 6F). Previous work has documented the presence of these three cellular components in autophagosomes, which have been termed reticulophagy, ribophagy, and zymophagy; in this study we have observed similar phenotypes in acinar cells from infected tissue. In addition, other cells contained very large autophagosomal compartments that were sometimes as large as the nucleus (Fig. 6G); these giant autophagosomes were located near the nucleus, and a double membrane was sometimes visualized (Fig. 6H and I). To ensure that the findings were representative of wt CVB3 infection, we carried out a limited set of TEM analyses of wt CVB-infected acinar cells. CMVs were abundant (Fig. 6J) and, as in DsRed-CVB-infected cells, these vesicles were smaller than normal autophagosomes but were often double membraned (Fig. 6K). This increased abundance of intracellular membranous structures in CVB-infected cells is consistent with the increase in LC3-II that is shown in Fig. 1A.
FIG. 6.
Ultrastructural analyses of the CVB3-infected pancreas. Pancreatic sections were examined by TEM. (A) Acinar cell in uninfected pancreas. (B) Acinar cell containing CMV complex at day 1 p.i. with DsRed-CVB3. The central area of the cell contains multiple autophagosome-like structures, some of which are enclosed in a white box and shown at higher magnification in (C). (D through I) From day 2 p.i., DsRed-CVB3. (D) Two autophagosomes are indicated with arrows, and a region from one is enlarged in the inset, demonstrating that the vesicles have two lipid bilayer membranes. (E) A series of images were reassembled to generate a montage of the whole acinar cell. Two large multilamellar autophagosomes are indicated, one with a white arrow and the other enclosed in a square. A higher-magnification image of the latter autophagosome is shown in panel F. (G) Low-magnification image of an acinar cell from infected pancreas. The nucleus is marked with an asterisk, and adjacent to it, two large autophagic compartments are indicated, one by a white arrow and the other enclosed in a square. A higher-magnification image of the latter is shown in panel H. An additional example of large autophagic compartments next to the nucleus is shown in panel I, and the region at the base of the angled arrow is displayed in the inset at a higher magnification to show that the compartment has a double membrane. (J) CMV complexes in cells at 2 days p.i. with wt CVB3. A separate field is shown at higher magnification (K), highlighting the fact that many of these small vesicles are double membraned. (L) A rectangular lattice structure present in wt CVB-infected cells adjacent to the ER. The approximate sizes of the individual components of this lattice can be estimated from comparison with a ribosome (arrowed, ∼20 nm in diameter). Scale bars, 5,000 nm (A and B), 500 nm (C, D, F, and K), 2,000 nm (E and I), 3,000 nm (G), 1,000 nm (H and J), and 200 nm (L).
Finally, during our analyses we observed a paracrystalline lattice in an infected acinar cell (rectangular structure in Fig. 6L). This structure, which was adjacent to the rough ER, appeared to be highly organized with tightly constrained boundaries. By comparison to a ribosome (diameter, ∼20 nm), the individual components of this lattice were estimated as having a diameter of ∼8 nm, which is similar to the reported size for enteroviral RNA polymerases (22, 24). Virus infection induced additional ultrastructural changes to acinar cells, including the formation of numerous single-membraned electron-lucent vacuoles, disorganized and highly dilated rough ER, and nuclei that appeared to be apoptotic and/or pyknotic (data not shown); these findings are consistent with those of previous ultrastructural analyses of CVB-infected pancreas (5, 20, 26). Overall, our EM analyses confirm that CVB3 infection triggered the formation of autophagy-like vesicles along with autophagosomes of various sizes. Furthermore, these data suggest that the giant GFP-LC3+ vesicles seen by confocal microscopy are autophagosomes of very large size.
Ultrastructural confirmation that the giant vesicles are related to the autophagy pathway.
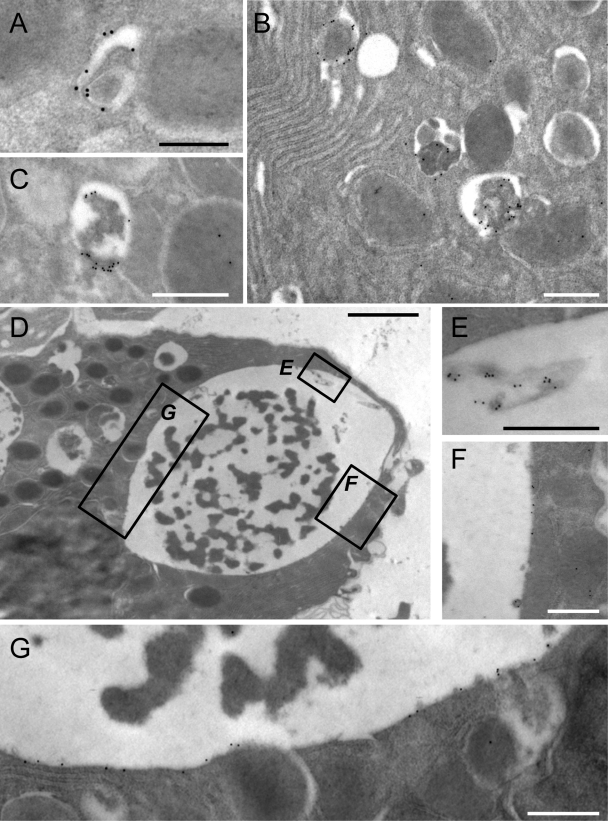
Themegaphagosomes appeared to be related to the autophagy pathway on the basis of three criteria: (i) as determined by confocal microscopy, many of them they were GFP+ (Fig. 4 and 5); and EM analyses (Fig. 6H and I) indicated that (ii) many were enclosed by at least two lipid bilayers and (iii) they contained cell debris. However, to further test this relationship, we used immunogold EM (immuno-EM) to evaluate the presence and distribution of GFP-LC3 in CVB-infected acinar cells. Occasional gold beads were observed scattered across the section, as would be expected from the background signal and/or the fact that GFP-LC3-I is distributed in the cytosol. In both uninfected (Fig. 7A) and infected (Fig. 7B and C) tissues, we observed structures whose morphology was consistent with autophagosomes. These vesicles labeled strongly with GFP-specific antibody, confirming their relationship to the autophagy pathway. Megaphagosomes were present only in infected pancreata and were also visualized by immuno-EM; an example of one such structure is shown (Fig. 7D). Consistent with the large GFP+ bodies shown in Fig. 5, the inner circumference of this structure is decorated by GFP (Fig. 7E to G), indicating that this vesicle, which measures approximately 8.2 by 5.7 μm, belongs to the autophagic pathway.
FIG. 7.
Immuno-EM analyses identify GFP associated with autophagosomes and megaphagosomes. Immuno-EM of the pancreata of GFP-LC3 mice was carried out as described in the text using an antibody specific for GFP. Acinar cells are shown. (A) Autophagosome in an uninfected mouse. (B to G) Two days p.i., DsRed-CVB3. (B and C) Autophagosomes in infected pancreata. (D) Megaphagosome in infected pancreas. This structure measures ∼8.2 by 5.7 μm. The boxed areas are enlarged in panels in E to G. Scale bars, 250 nm (A), 500 nm (B, C, and E to G), and 2,000 nm (D).
DISCUSSION
The autophagy pathway plays an important part in determining the nature and quantity of membranous structures within the cell, and autophagy-related vesicles are thought to serve as a scaffold for intracellular membrane-associated replication factories of RNA viruses that replicate and assemble in the cytoplasm (92). Members of the Picornaviridae, including poliovirus and coxsackieviruses, are known to induce spectacular rearrangements of intracellular membranes (35, 53); and although in vitro studies have demonstrated that these viruses can subvert components of the autophagic machinery for their own benefit, the extent to which this process occurs within infected tissues is uncertain, as is its role in viral replication or pathogenesis in vivo. In the present study, we have used a mouse model of CVB3 infection specifically to address whether this virus modifies the autophagic pathway in vivo.
In this study, we evaluated three distinct criteria in order to determine if CVB3 induces autophagy in pancreatic acinar cells. First, we sought a biochemical signature of autophagy induction by analyzing the conversion of LC3-I to LC3-II by Western blotting. The level of LC3-II increased significantly on day 2, and by day 3 nearly all of the endogenous LC3 present in pancreas was converted; this was accompanied by a loss of the monomeric and cleaved forms of Atg5; only the Atg5-Atg12 complex remained (Fig. 1A and B). Second, we assessed autophagic flux by monitoring, over the course of infection, the levels of p62 and fragmented GFP. The substantial increase in the autophagy substrate p62 by day 2 to 3 p.i. (Fig. 2B) suggests that autophagic flux is markedly reduced. Others have reported that p62 levels were unchanged in CVB-infected tissue culture cells (93); the reason for this apparent discrepancy is unclear but may reflect major differences between the experimental systems (tissue culture cells versus pancreatic tissue in vivo). We also found that the amount of cleaved GFP increased significantly by day 3 p.i. (see Fig. S2 in the supplemental material), but this is difficult to interpret for the reasons described above. Third, we performed high-resolution confocal imaging of vibratome-cut pancreas sections to quantitate the number of GFP-LC3+ punctae in uninfected and virus-infected tissue. DsRed-CVB3 triggered not only a 2-fold increase in the number of autophagosomes in strongly infected (DsRed bright) acinar cells but also an ∼3-fold increase in their average surface area (Fig. 4B). These data are consistent with the hypothesis that CVB3 recruits autophagosomes into virus factories to generate a scaffold for the replication complex.
Others have found that poliovirus RNA polymerase is associated with membranes isolated from infected tissue culture cells and that the purified protein can form a lattice in vitro (48); the authors proposed that poliovirus replication in vivo might take place on a “shell” of membrane-associated polymerases. The highly organized structure shown in Fig. 6L appears to be a paracrystalline lattice formed by individual components, each ∼8 nm in diameter. CVB RNA polymerase (22) is structurally very similar to poliovirus RNA polymerase, a single molecule of which is ∼8.8 by 8.8 by 15.8 nm in size (24). We suggest that the lattice shown in Fig. 6L represents an array of CVB RNA polymerases within an infected pancreatic acinar cell and speculate that CVB replication in vivo may take place on this sheet of polymerases, consistent with the aforementioned proposal.
In principle, the accumulation of autophagosomes that we observed in CVB-infected acinar cells could result from an increase in autophagosome formation, from a decrease in their degradation, or both. Previous studies in vitro have suggested that poliovirus and CVB3 can inhibit the maturation/degradation of virus-induced double-membraned vesicles (or autophagosomes) by blocking their fusion with lysosomes (86, 93). Indeed, coexpression of the poliovirus proteins 2BC and 3A is sufficient to induce the colocalization of LAMP-1 (a marker of the endolysosomal compartment) with GFP-LC3 (31). We therefore examined whether autophagosomes in CVB3-infected acinar cells were able to fuse with endosomes/lysosomes or if this process was inhibited by the virus in vivo. Many GFP+ vesicles in DsRed-CVB3-infected cells also contained LAMP-1 (Fig. 5), suggesting that amphisomes are generated; thus, CVB3 does not appear to prevent autophagosome-to-endosome fusion. It is more difficult to evaluate the effect (if any) of CVB3 on subsequent steps, i.e., the formation of autolysosomes. However, megaphagosomes have not been observed during the upregulation of autophagy that takes place in response to normal stimuli, such as food restriction, and their presence in CVB-infected cells therefore suggests that the virus may interrupt autophagic processing at some stage of the pathway. These very large vesicles, which were sometimes seen in clusters, appear to be related to autophagy because they were usually GFP+ by confocal microscopy (Fig. 4 and 5), and some also scored positive by immuno-EM (Fig. 7). Mice deficient in LAMP-2, a key constituent of the lysosomal membrane, develop large autophagic vacuoles in several cell types, including pancreatic acinar cells, and the authors suggested that these large vacuoles resulted from failure of fusion of autophagosomes with endosomes/lysosomes (83). Giant perinuclear autophagosomes have also been observed in cells from mice with nuclear envelopathies and in A549 human lung epithelial cells infected with influenza A virus (17, 49). For both of these models, it was suggested that clusters of smaller autophagosomes might fuse together to form a giant autophagosome; the influenza A virus M2 protein may play a role in this process by preventing autophagosome maturation (17, 66, 70). Likewise, CMVs were abundant in wt CVB3-infected acinar cells (Fig. 6), and small autophagosomes were often seen close to the membrane of the giant autophagosomes and/or were contained within the larger structures (Fig. 4D and E). Therefore, we suggest that CVB3 infection (i) increases the rate at which new autophagosomes are formed and (ii) inhibits their progress along the autophagy pathway. Consequently, (iii) these abundant smaller vesicles coalesce to form megaphagosomes. Where might CVB inhibit autophagy flux? The megaphagosomes usually express the endosomal/lysosomal marker LAMP-1 (Fig. 5), suggesting that they are very large amphisomes. These GFP+/LAMP-1+ vesicles are less likely to be autolysosomes because, as noted above (and see Fig. 2A), autolysosomes in GFP-LC3 mice should be LAMP-1+ but, because of their acidic environment and protease content, should show minimal GFP fluorescence. Thus, we suggest that CVB3 permits amphisome formation but limits the subsequent fusion of amphisomes with lysosomes. This is consistent with the observation that p62 accumulates in infected tissue by day 2 to 3 p.i. (Fig. 2B). Nevertheless, this inhibition, although profound, may be incomplete: we occasionally observed large vesicles that were predominantly LAMP-1+ with minimal GFP fluorescence (Fig. 5). Normal endosomes and lysosomes would be expected to show the same fluorescent qualities, but the unusually large size of these vesicles suggests that they may have been generated from megaphagosomes that have escaped the effects of CVB, becoming autolysosomes. In summary, CVB3 appears to block autophagosome fusion with lysosomes in vivo, as has been proposed for poliovirus in tissue culture cells (86).
What advantages might accrue to RNA viruses that increase autophagosome abundance and also prevent their maturation into autolysosomes? This strategy may benefit the virus in several ways: (i) by inducing the accumulation of intracellular vesicles on which the viral replication machinery is assembled; (ii) by preventing degradation of newly formed virions inside autolysosomes; and (iii) by limiting the interaction between viral RNA and the single-stranded sensor Toll-like receptor 7 (TLR-7), which is present in late endosomes and lysosomes and which is thought to play a role in innate responses to CVB. By terminating the autophagy pathway at the stage of amphisomes, CVB ensures that its RNA is separated from TLR-7 by the intact inner lipid bilayer that is derived solely from the TLR-deficient autophagosome; only upon degradation of this inner membrane (in an autolysosome) will the viral genome be introduced to TLR-7. Furthermore, (iv) others have proposed that the inhibition of autophagic flux and the generation of autophagy-like vesicles may permit the extracellular release of virus without cell lysis (6, 88). Finally, (v) autophagy can be important in antigen presentation via MHC class I (13) and MHC class II (42, 64); we speculate that by blocking the autophagy pathway, CVB3 infection may compromise the host's capacity to mount strong T-cell responses, possibly contributing to the weakness of the CVB-specific T-cell responses that this laboratory has recently reported (34).
Our findings may be relevant to other, nonviral forms of acute pancreatitis, in which autophagy also plays a key role. In the cerulein-induced model of experimental pancreatitis, acinar cells develop cytoplasmic vacuoles that are autophagic in origin; these vesicles are involved in the degradation of zymogen granules, leading to the conversion of trypsinogen to trypsin, which accumulates within the acinar cell, triggering cellular self-digestion (23, 27, 50, 58). Autophagy is required for this process, as trypsinogen activation is greatly reduced in mice that lack Atg5 specifically in acinar cells, and cerulein-induced acute pancreatitis does not develop in these mice (27). Our analysis of CVB3-infected acinar cells has shown that this virus induces autophagy and triggers the formation of numerous (and sometimes very large) autophagosomes that contain zymogen granules (e.g., Fig. 6F and I), as well as inducing substantial inflammation and profound acute pancreatitis. We have shown here that CVB appears to limit the subsequent fusion of these autophagic compartments with lysosomes, arguing that trypsinogen activation may not be increased within an infected cell. However, since CVB3 infection and chemically induced acute pancreatitis share several common features (in particular, LC3 conversion, accumulation of GFP-LC3 punctae, vacuole formation, and exocrine inflammation), we speculate that by inducing autophagy to promote its own replication, the virus may trigger a pathological process similar to that observed in cerulein-induced pancreatitis. We conclude that despite the apparent blockade in autolysosome formation, future studies are warranted to determine if CVB3 is able to modulate autolysosomal degradation and activation of intracellular proenzymes.
Supplementary Material
Acknowledgments
We thank Karla Kirkegaard (Stanford University) for providing LC3 antibody and Annette Lord for excellent secretarial support.
This work was supported by NIH grants R01 AI042314 and HL093177 (to J.L.W.) and grants T32 NS41219 and F32 AI078660 (to C.C.K.).
We have no conflicting financial interests.
Footnotes
Published ahead of print on 22 September 2010.
This is manuscript number 20701 from The Scripps Research Institute.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ait-Goughoulte, M., T. Kanda, K. Meyer, J. S. Ryerse, R. B. Ray, and R. Ray. 2008. Hepatitis C virus genotype 1a growth and induction of autophagy. J. Virol. 82:2241-2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alirezaei, M., C. C. Kemball, C. T. Flynn, M. R. Wood, J. L. Whitton, and W. B. Kiosses. 2010. Short-term fasting induces profound neuronal autophagy. Autophagy 6:702-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alirezaei, M., W. B. Kiosses, C. T. Flynn, N. R. Brady, and H. S. Fox. 2008. Disruption of neuronal autophagy by infected microglia results in neurodegeneration. PLoS One 3:e2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjorkoy, G., T. Lamark, A. Brech, H. Outzen, M. Perander, A. Overvatn, H. Stenmark, and T. Johansen. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171:603-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burch, G. E., and J. M. Harb. 1979. Electron microscopic studies of viral pancreatitis in coxsackie B4 virus infected mice. Exp. Mol. Pathol. 31:23-35. [DOI] [PubMed] [Google Scholar]
- 6.Choe, S. S., and K. Kirkegaard. 2004. Intracellular topology and epitope shielding of poliovirus 3A protein. J. Virol. 78:5973-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daley, A. J., D. Isaacs, D. E. Dwyer, and G. L. Gilbert. 1998. A cluster of cases of neonatal coxsackievirus B meningitis and myocarditis. J. Paediatr. Child Health 34:196-198. [DOI] [PubMed] [Google Scholar]
- 8.de Haan, C. A., and F. Reggiori. 2008. Are nidoviruses hijacking the autophagy machinery? Autophagy 4:276-279. [DOI] [PubMed] [Google Scholar]
- 9.Deretic, V. 2009. Multiple regulatory and effector roles of autophagy in immunity. Curr. Opin. Immunol. 21:53-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deretic, V., and B. Levine. 2009. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5:527-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreux, M., and F. V. Chisari. 2010. Viruses and the autophagy machinery. Cell Cycle 9:1295-1307. [DOI] [PubMed] [Google Scholar]
- 12.Dreux, M., P. Gastaminza, S. F. Wieland, and F. V. Chisari. 2009. The autophagy machinery is required to initiate hepatitis C virus replication. Proc. Natl. Acad. Sci. U. S. A. 106:14046-14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.English, L., M. Chemali, J. Duron, C. Rondeau, A. Laplante, D. Gingras, D. Alexander, D. Leib, C. Norbury, R. Lippe, and M. Desjardins. 2009. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat. Immunol. 10:480-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espert, L., P. Codogno, and M. Biard-Piechaczyk. 2007. Involvement of autophagy in viral infections: antiviral function and subversion by viruses. J. Mol. Med. 85:811-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Espert, L., M. Varbanov, V. Robert-Hebmann, S. Sagnier, I. Robbins, F. Sanchez, V. Lafont, and M. Biard-Piechaczyk. 2009. Differential role of autophagy in CD4 T cells and macrophages during X4 and R5 HIV-1 infection. PLoS One 4:e5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feuer, R., R. R. Pagarigan, S. Harkins, F. Liu, I. P. Hunziker, and J. L. Whitton. 2005. Coxsackievirus targets proliferating neuronal progenitor cells in the neonatal CNS. J. Neurosci. 25:2434-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gannage, M., D. Dormann, R. Albrecht, J. Dengjel, T. Torossi, P. C. Ramer, M. Lee, T. Strowig, F. Arrey, G. Conenello, M. Pypaert, J. Andersen, A. Garcia-Sastre, and C. Munz. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebhard, J. R., C. M. Perry, S. Harkins, T. Lane, I. Mena, V. C. Asensio, I. L. Campbell, and J. L. Whitton. 1998. Coxsackievirus B3-induced myocarditis: perforin exacerbates disease, but plays no detectable role in virus clearance. Am. J. Pathol. 153:417-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez, R. M., J. J. Lopez Costa, G. Pecci Saavedra, and M. I. Berria. 1993. Ultrastructural study of cell injury induced by coxsackievirus B3 in pancreatic and cardiac tissues. Medicina (Buenos Aires) 53:300-306. [PubMed] [Google Scholar]
- 21.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruez, A., B. Selisko, M. Roberts, G. Bricogne, C. Bussetta, I. Jabafi, B. Coutard, A. M. De Palma, J. Neyts, and B. Canard. 2008. The crystal structure of coxsackievirus B3 RNA-dependent RNA polymerase in complex with its protein primer VPg confirms the existence of a second VPg binding site on Picornaviridae polymerases. J. Virol. 82:9577-9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gukovsky, I., and A. S. Gukovskaya. 2010. Impaired autophagy underlies key pathological responses of acute pancreatitis. Autophagy 6:428-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen, J. L., A. M. Long, and S. C. Schultz. 1997. Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5:1109-1122. [DOI] [PubMed] [Google Scholar]
- 25.Harkins, S., C. T. Cornell, and J. L. Whitton. 2005. Analysis of translational initiation in coxsackievirus B3 suggests an alternative explanation for the high frequency of R+4 in the eukaryotic consensus motif. J. Virol. 79:987-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison, A. K., S. P. Bauer, and F. A. Murphy. 1972. Viral pancreatitis: ultrastructural pathological effects of coxsackievirus B3 infection in newborn mouse pancreas. Exp. Mol. Pathol. 17:206-219. [DOI] [PubMed] [Google Scholar]
- 27.Hashimoto, D., M. Ohmuraya, M. Hirota, A. Yamamoto, K. Suyama, S. Ida, Y. Okumura, E. Takahashi, H. Kido, K. Araki, H. Baba, N. Mizushima, and K. Yamamura. 2008. Involvement of autophagy in trypsinogen activation within the pancreatic acinar cells. J. Cell Biol. 181:1065-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, S. C., C. L. Chang, P. S. Wang, Y. Tsai, and H. S. Liu. 2009. Enterovirus 71-induced autophagy detected in vitro and in vivo promotes viral replication. J. Med. Virol. 81:1241-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huber, S., and A. I. Ramsingh. 2004. Coxsackievirus-induced pancreatitis. Viral Immunol. 17:358-369. [DOI] [PubMed] [Google Scholar]
- 30.Hunziker, I. P., C. T. Cornell, and J. L. Whitton. 2007. Deletions within the 5′UTR of coxsackievirus B3: consequences for virus translation and replication. Virology 360:120-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jackson, W. T., T. H. Giddings, Jr., M. P. Taylor, S. Mulinyawe, M. Rabinovitch, R. R. Kopito, and K. Kirkegaard. 2005. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 3:e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jahreiss, L., F. M. Menzies, and D. C. Rubinsztein. 2008. The itinerary of autophagosomes: from peripheral formation to kiss-and-run fusion with lysosomes. Traffic 9:574-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kemball, C. C., S. Harkins, J. K. Whitmire, C. T. Flynn, R. Feuer, and J. L. Whitton. 2009. Coxsackievirus B3 inhibits antigen presentation in vivo, exerting a profound and selective effect on the MHC class I pathway. PLoS Pathog. 5:e1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kirkegaard, K. 2009. Subversion of the cellular autophagy pathway by viruses. Curr. Top. Microbiol. Immunol. 335:323-333. [DOI] [PubMed] [Google Scholar]
- 36.Kirkegaard, K., and W. T. Jackson. 2005. Topology of double-membraned vesicles and the opportunity for non-lytic release of cytoplasm. Autophagy 1:182-184. [DOI] [PubMed] [Google Scholar]
- 37.Kirkegaard, K., M. P. Taylor, and W. T. Jackson. 2004. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2:301-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knowlton, K. U. 2008. CVB infection and mechanisms of viral cardiomyopathy. Curr. Top. Microbiol. Immunol. 323:315-335. [DOI] [PubMed] [Google Scholar]
- 39.Knowlton, K. U., E. S. Jeon, N. Berkley, R. Wessely, and S. A. Huber. 1996. A mutation in the puff region of VP2 attenuates the myocarditic phenotype of an infectious cDNA of the Woodruff variant of coxsackievirus B3. J. Virol. 70:7811-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kyei, G. B., C. Dinkins, A. S. Davis, E. Roberts, S. B. Singh, C. Dong, L. Wu, E. Kominami, T. Ueno, A. Yamamoto, M. Federico, A. Panganiban, I. Vergne, and V. Deretic. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J. Cell Biol. 186:255-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, H. K., and A. Iwasaki. 2008. Autophagy and antiviral immunity. Curr. Opin. Immunol. 20:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, H. K., L. M. Mattei, B. E. Steinberg, P. Alberts, Y. H. Lee, A. Chervonsky, N. Mizushima, S. Grinstein, and A. Iwasaki. 2010. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 32:227-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee, Y. R., H. Y. Lei, M. T. Liu, J. R. Wang, S. H. Chen, Y. F. Jiang-Shieh, Y. S. Lin, T. M. Yeh, C. C. Liu, and H. S. Liu. 2008. Autophagic machinery activated by dengue virus enhances virus replication. Virology 374:240-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levine, B., and V. Deretic. 2007. Unveiling the roles of autophagy in innate and adaptive immunity. Nat. Rev. Immunol. 7:767-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang, X. H., L. K. Kleeman, H. H. Jiang, G. Gordon, J. E. Goldman, G. Berry, B. Herman, and B. Levine. 1998. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 72:8586-8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu, Y., M. Schiff, K. Czymmek, Z. Talloczy, B. Levine, and S. P. Dinesh-Kumar. 2005. Autophagy regulates programmed cell death during the plant innate immune response. Cell 121:567-577. [DOI] [PubMed] [Google Scholar]
- 47.Lunemann, J. D., and C. Munz. 2009. Autophagy in CD4+ T-cell immunity and tolerance. Cell Death Differ. 16:79-86. [DOI] [PubMed] [Google Scholar]
- 48.Lyle, J. M., E. Bullitt, K. Bienz, and K. Kirkegaard. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218-2222. [DOI] [PubMed] [Google Scholar]
- 49.Marazziti, D., C. Di Pietro, E. Golini, S. Mandillo, R. Matteoni, and G. P. Tocchini-Valentini. 2009. Induction of macroautophagy by overexpression of the Parkinson's disease-associated GPR37 receptor. FASEB J. 23:1978-1987. [DOI] [PubMed] [Google Scholar]
- 50.Mareninova, O. A., K. Hermann, S. W. French, M. S. O'Konski, S. J. Pandol, P. Webster, A. H. Erickson, N. Katunuma, F. S. Gorelick, I. Gukovsky, and A. S. Gukovskaya. 2009. Impaired autophagic flux mediates acinar cell vacuole formation and trypsinogen activation in rodent models of acute pancreatitis. J. Clin. Invest. 119:3340-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mena, I., C. Fischer, J. R. Gebhard, C. M. Perry, S. Harkins, and J. L. Whitton. 2000. Coxsackievirus infection of the pancreas: evaluation of receptor expression, pathogenesis, and immunopathology. Virology 271:276-288. [DOI] [PubMed] [Google Scholar]
- 52.Mena, I., C. M. Perry, S. Harkins, F. Rodriguez, J. R. Gebhard, and J. L. Whitton. 1999. The role of B lymphocytes in coxsackievirus B3 infection. Am. J. Pathol. 155:1205-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller, S., and J. Krijnse-Locker. 2008. Modification of intracellular membrane structures for virus replication. Nat. Rev. Microbiol. 6:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mizushima, N., and A. Kuma. 2008. Autophagosomes in GFP-LC3 transgenic mice. Methods Mol. Biol. 445:119-124. [DOI] [PubMed] [Google Scholar]
- 55.Mizushima, N., A. Yamamoto, M. Hatano, Y. Kobayashi, Y. Kabeya, K. Suzuki, T. Tokuhisa, Y. Ohsumi, and T. Yoshimori. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizushima, N., A. Yamamoto, M. Matsui, T. Yoshimori, and Y. Ohsumi. 2004. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15:1101-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizushima, N., T. Yoshimori, and B. Levine. 2010. Methods in mammalian autophagy research. Cell 140:313-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohmuraya, M., and K. Yamamura. 2008. Autophagy and acute pancreatitis: a novel autophagy theory for trypsinogen activation. Autophagy 4:1060-1062. [DOI] [PubMed] [Google Scholar]
- 59.Orvedahl, A., D. Alexander, Z. Talloczy, Q. Sun, Y. Wei, W. Zhang, D. Burns, D. A. Leib, and B. Levine. 2007. HSV-1 ICP34.5 confers neurovirulence by targeting the beclin 1 autophagy protein. Cell Host Microbe 1:23-35. [DOI] [PubMed] [Google Scholar]
- 60.Orvedahl, A., and B. Levine. 2008. Viral evasion of autophagy. Autophagy 4:280-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orvedahl, A., and B. Levine. 2009. Autophagy in mammalian antiviral immunity. Curr. Top. Microbiol. Immunol. 335:267-285. [DOI] [PubMed] [Google Scholar]
- 62.Orvedahl, A., and B. Levine. 2009. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ. 16:57-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Orvedahl, A., S. Macpherson, R. Sumpter, Jr., Z. Talloczy, Z. Zou, and B. Levine. 2010. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe 7:115-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paludan, C., D. Schmid, M. Landthaler, M. Vockerodt, D. Kube, T. Tuschl, and C. Munz. 2005. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 307:593-596. [DOI] [PubMed] [Google Scholar]
- 65.Pankiv, S., T. H. Clausen, T. Lamark, A. Brech, J. A. Bruun, H. Outzen, A. Overvatn, G. Bjorkoy, and T. Johansen. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131-24145. [DOI] [PubMed] [Google Scholar]
- 66.Park, Y. E., Y. K. Hayashi, G. Bonne, T. Arimura, S. Noguchi, I. Nonaka, and I. Nishino. 2009. Autophagic degradation of nuclear components in mammalian cells. Autophagy 5:795-804. [DOI] [PubMed] [Google Scholar]
- 67.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romero, J. R. 2008. Pediatric group B coxsackievirus infections. Curr. Top. Microbiol. Immunol. 323:223-239. [DOI] [PubMed] [Google Scholar]
- 70.Rossman, J. S., and R. A. Lamb. 2009. Autophagy, apoptosis, and the influenza virus M2 protein. Cell Host Microbe 6:299-300. [DOI] [PubMed] [Google Scholar]
- 71.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmid, D., and C. Munz. 2007. Innate and adaptive immunity through autophagy. Immunity 27:11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shelly, S., N. Lukinova, S. Bambina, A. Berman, and S. Cherry. 2009. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity 30:588-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shvets, E., and Z. Elazar. 2008. Autophagy-independent incorporation of GFP-LC3 into protein aggregates is dependent on its interaction with p62/SQSTM1. Autophagy 4:1054-1056. [DOI] [PubMed] [Google Scholar]
- 75.Shvets, E., E. Fass, R. Scherz-Shouval, and Z. Elazar. 2008. The N-terminus and Phe52 residue of LC3 recruit p62/SQSTM1 into autophagosomes. J. Cell Sci. 121:2685-2695. [DOI] [PubMed] [Google Scholar]
- 76.Sir, D., W. L. Chen, J. Choi, T. Wakita, T. S. Yen, and J. H. Ou. 2008. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 48:1054-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sir, D., and J. H. Ou. 2010. Autophagy in viral replication and pathogenesis. Mol. Cells 29:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sir, D., Y. Tian, W. L. Chen, D. K. Ann, T. S. Yen, and J. H. Ou. 2010. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc. Natl. Acad. Sci. U. S. A. 107:4383-4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82:985-994. [DOI] [PubMed] [Google Scholar]
- 80.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tabor-Godwin, J. M., C. M. Ruller, N. Bagalso, N. An, R. R. Pagarigan, S. Harkins, P. E. Gilbert, W. B. Kiosses, N. A. Gude, C. T. Cornell, K. S. Doran, M. A. Sussman, J. L. Whitton, and R. Feuer. 2010. A novel population of myeloid cells responding to coxsackievirus infection assists in the dissemination of virus within the neonatal CNS. J. Neurosci. 30:8676-8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tam, P. E. 2006. Coxsackievirus myocarditis: interplay between virus and host in the pathogenesis of heart disease. Viral Immunol. 19:133-146. [DOI] [PubMed] [Google Scholar]
- 83.Tanaka, Y., G. Guhde, A. Suter, E. L. Eskelinen, D. Hartmann, R. Lullmann-Rauch, P. M. Janssen, J. Blanz, K. von Figura, and P. Saftig. 2000. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature 406:902-906. [DOI] [PubMed] [Google Scholar]
- 84.Tanida, I., N. Minematsu-Ikeguchi, T. Ueno, and E. Kominami. 2005. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy 1:84-91. [DOI] [PubMed] [Google Scholar]
- 85.Taylor, M. P., T. B. Burgon, K. Kirkegaard, and W. T. Jackson. 2009. Role of microtubules in extracellular release of poliovirus. J. Virol. 83:6599-6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taylor, M. P., and W. T. Jackson. 2009. Viruses and arrested autophagosome development. Autophagy 5:870-871. [DOI] [PubMed] [Google Scholar]
- 87.Taylor, M. P., and K. Kirkegaard. 2007. Modification of cellular autophagy protein LC3 by poliovirus. J. Virol. 81:12543-12553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Taylor, M. P., and K. Kirkegaard. 2008. Potential subversion of autophagosomal pathway by picornaviruses. Autophagy 4:286-289. [DOI] [PubMed] [Google Scholar]
- 89.van Houten, N., P. E. Bouchard, A. Moraska, and S. A. Huber. 1991. Selection of an attenuated coxsackievirus B3 variant, using a monoclonal antibody reactive to myocyte antigen. J. Virol. 65:1286-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Virgin, H. W., and B. Levine. 2009. Autophagy genes in immunity. Nat. Immunol. 10:461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Whitton, J. L. 2002. Immunopathology during coxsackievirus infection. Springer Semin. Immunopathol. 24:201-213. [DOI] [PubMed] [Google Scholar]
- 92.Wileman, T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312:875-878. [DOI] [PubMed] [Google Scholar]
- 93.Wong, J., J. Zhang, X. Si, G. Gao, I. Mao, B. M. McManus, and H. Luo. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J. Virol. 82:9143-9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoshimori, T., and T. Noda. 2008. Toward unraveling membrane biogenesis in mammalian autophagy. Curr. Opin. Cell Biol. 20:401-407. [DOI] [PubMed] [Google Scholar]
- 95.Yousefi, S., R. Perozzo, I. Schmid, A. Ziemiecki, T. Schaffner, L. Scapozza, T. Brunner, and H. U. Simon. 2006. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell Biol. 8:1124-1132. [DOI] [PubMed] [Google Scholar]
- 96.Zhou, Z., X. Jiang, D. Liu, Z. Fan, X. Hu, J. Yan, M. Wang, and G. F. Gao. 2009. Autophagy is involved in influenza A virus replication. Autophagy 5:321-328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.