Abstract
The Epstein-Barr virus immediate-early protein, BZLF1 (Z), initiates the switch between latent and lytic infection and plays an essential role in mediating viral replication. Z also inhibits expression of the major receptor for tumor necrosis factor (TNF), TNFR1, thus repressing TNF cytokine signaling, but the mechanism for this effect is unknown. Here, we demonstrate that Z prevents both C/EBPα- and C/EBPβ-mediated activation of the TNFR1 promoter (TNFR1p) by interacting directly with both C/EBP family members. We show that Z interacts directly with C/EBPα and C/EBPβ in vivo and that a Z mutant altered at alanine residue 204 in the bZIP domain is impaired for the ability to interact with both C/EBP proteins. Furthermore, we find that the Z(A204D) mutant is attenuated in the ability to inhibit the TNFR1p but mediates lytic viral reactivation and replication in vitro in 293 cells as well as wild-type Z. Although Z does not bind directly to the TNFR1p in EMSA studies, chromatin immunoprecipitation studies indicate that Z is complexed with this promoter in vivo. The Z(A204D) mutant has reduced interaction with the TNFR1p in vivo but is similar to wild-type Z in its ability to complex with the IL-8 promoter. Finally, we show that the effect of Z on C/EBPα- and C/EBPβ-mediated activation is promoter dependent. These results indicate that Z modulates the effects of C/EBPα and C/EBPβ in a promoter-specific manner and that in some cases (including that of the TNFR1p), Z inhibits C/EBPα- and C/EBPβ-mediated activation.
More than 90% of adults are infected with Epstein-Barr virus (EBV) (61). Although infection with this herpesvirus usually occurs during childhood and is asymptomatic, if acquired during adolescence or adulthood it can cause infectious mononucleosis. While the primary illness is usually short-lived, the virus persists in the host for life. EBV is also associated with several types of cancer, including Burkitt's lymphoma, the most common childhood cancer in equatorial Africa, and nasopharyngeal carcinoma (61, 80). EBV persists in the host by establishing a latent infection in memory B cells and reactivating periodically to undergo the lytic form of viral replication (38). The infectious viral particles released during lytic infection can be passed to new hosts via saliva (38, 61).
Expression of the two viral immediate-early proteins, BZLF1 (Z) and BRLF1 (R), leads to lytic reactivation in latently infected cells (11, 12, 62, 70, 71). Together, Z and R turn on the expression of the viral lytic proteins required for viral replication (12, 18, 24, 28, 36, 54, 59, 61, 62, 78). Z (also called Zta, ZEBRA, and EB1) is a member of the basic leucine zipper (bZIP) family of DNA binding proteins and is active only as a homodimer (70). Z transcriptionally activates lytic viral genes by binding to their promoters at AP-1-like sites called Z response elements (ZREs) (10, 17, 20, 41, 42). Furthermore, direct binding of Z to the EBV lytic origin of replication (oriLyt) is required for viral replication (19, 66, 79), and Z directly interacts with certain core viral replication proteins (40, 79).
In addition to activating other lytic viral genes, Z can also either activate or repress the expression of cellular genes. Some of the cellular genes inhibited by Z are important for the host immune response to virally infected cells (8). For example, Z downregulates expression of the genes encoding the receptors for the antiviral cytokines gamma interferon (IFN-γ) (50) and tumor necrosis factor (TNF; formerly TNF-α) (49) during lytic infection. However, the mechanisms by which Z inhibits transcription of certain cellular genes have not been well studied.
TNF is a multifunctional cytokine that is a key regulator of the host inflammatory and immune responses (44) and also mediates changes at the cellular level, including proliferation, differentiation, and apoptosis. The TNF protein, released by immune effector cells and epithelial cells, mediates its effects through binding to its major receptor, TNFR1 (also known as p55 TNFR, TR60, and CD120a), which is expressed on the surfaces of most cells. When TNF binds to its receptor, it starts a cascade of signals in the cell which ultimately leads to cell death or survival (5). Virally infected cells whose receptors are stimulated can be induced to start the apoptosis process, essentially preventing the virus from further replicating in these cells (4). The specific importance of TNF in the ability of the host to control the outcome of herpesvirus infections was shown in studies that used neutralizing antibodies against TNF in mouse models for herpes simplex virus type 1 (37) and murine cytomegalovirus (29) infection. Not surprisingly, many viruses have evolved ways to subvert steps in the TNFR1 signaling pathway (73). Cytomegalovirus, for example, disrupts TNFR1 signaling by relocalizing the receptor from the cell surface (3, 58).
We previously showed that the EBV Z protein decreases TNFR1 promoter (TNFR1p) activity in EBV-negative cells and demonstrated that induction of the lytic form of viral infection inhibits TNF-induced gene expression and cell killing in EBV-infected AGS cells (49). However, the mechanism for this effect has not been defined. To date, relatively little is known about how the TNFR1 promoter is normally regulated (35, 67, 72). However, our laboratory recently demonstrated that the promoter contains a C/EBP binding site and showed that both C/EBPα and C/EBPβ bind to and activate the TNFR1 promoter (7). Since the C/EBPα protein has been shown to directly interact with Z (76, 77), and the Z protein can bind to some consensus C/EBP sites (42, 75), we hypothesized that Z might inhibit transcription of the TNFR1 gene through its effects on C/EBPα and/or C/EBPβ.
C/EBPα and C/EBPβ, like Z, are bZIP proteins. These two transcription factors have a conserved leucine zipper (bZIP) domain that allows them to bind to similar DNA recognition sequences (53). Members of the C/EBP family have important roles in cell growth and differentiation (60). C/EBPα, which is highly expressed in liver, intestine, lung, and peripheral blood mononuclear cells, among others, plays a key role in monocyte and granulocyte differentiation (22, 74). C/EBPα also inhibits cell cycle progression in many cell types (51, 68). C/EBPβ, which is widely expressed, directs macrophage differentiation (56) and plays an important role in inducing immune responses in specific cell types (57). Furthermore, both C/EBPα and C/EBPβ contribute to keratinocyte differentiation (43).
Here, we report that Z decreases TNFR1 expression by preventing C/EBPα- and C/EBPβ-mediated activation of the TNFR1 promoter (TNFR1p). We show that Z interacts directly with both C/EBPα and C/EBPβ and that this interaction requires Z alanine residue 204 in the bZIP domain. Furthermore, we find that the mutant Z(A204D) protein, while transcriptionally competent and able to induce virus replication, is impaired in the ability to inhibit C/EBPα and C/EBPβ activation of TNFR1p in reporter gene assays. In addition, in comparison to wild-type (wt) Z, the mutant Z(A204D) protein has a reduced ability to decrease expression of the TNFR1 protein in transfected Hone cells. Although Z does not bind directly to the TNFR1 promoter in gel shift assays, chromatin immunoprecipitation (ChIP) assays reveal that the wild-type Z protein is complexed with the TNFR1 promoter in vivo and that the mutant Z(A204D) protein has reduced interaction with the TNFR1 promoter. Finally, we show that the effect of Z on C/EBPα- and C/EBPβ-mediated activation of promoters is dependent upon the specific promoter context. These results suggest that Z regulates transcription of certain genes indirectly through its effects on C/EBP family members.
MATERIALS AND METHODS
Cells.
HeLa cervical carcinoma cells, Hone nasopharyngeal carcinoma cells (EBV negative), AGS gastric carcinoma cells (EBV negative), and HEK-293 embryonic kidney cells were maintained in Dulbecco's modified Eagle medium (DMEM; Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gemini Bio-Products, West Sacramento, CA), 100 units/ml penicillin, and 100 μg/ml streptomycin. 293 cells infected with a BZLF1-deleted EBV mutant (293-ZKO cells), a gift from Henri-Jacques Delecluse, have previously been described (13, 18) and were maintained as described above and supplemented with 100 μg/ml hygromycin B. Raji cells (ATCC) were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, penicillin, and streptomycin. Cells were transiently transfected using FuGENE 6 (Roche, Indianapolis, IN) or Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Plasmids.
The TNFR1p-CAT plasmids contain the TNFR1 promoter region from position −338 through position +35 (relative to the transcriptional start site) inserted upstream of the chloramphenicol acetyltransferase (CAT) reporter gene (49). A TNFR1 promoter construct in which the C/EBP binding motif is specifically mutated (ΔC/EBP TNFR1p-CAT) was constructed as previously described (7). C/EBPα and C/EBPβ expression plasmids were gifts from Ormond MacDougald at the University of Michigan. Each plasmid has the full-length mouse C/EBPα or C/EBPβ gene sequence inserted into a pcDNA3.1+ expression vector. GST-C/EBPα and GST-C/EBPβ, containing the murine C/EBP gene sequences fused in frame to the glutathione S-transferase (GST) protein and inserted into pGEX, were gifts from Wen-Hwa Lee. The GST-Z vector was constructed as previously described (36) and contains the Z gene sequence inserted into pGEX. The Z and R expression vectors (pSG5-Z and pSG5-R) were gifts from S. Diane Hayward (63). These vectors contain genomic Z or R downstream of the simian virus 40 (SV40) promoter in the pSG5 vector (Stratagene). Site-directed mutagenesis was used to create the Z(A204D) mutation in the Z genomic pSG5 vector. Z cDNA (a gift from Paul Farrell) was cloned into the pSG5 vector to create pSG5-ZcDNA, which was used to in vitro translate the Z protein. The Z(A204D) mutation was also inserted into the Z cDNA in the pGEM vector (a gift from Erik Flemington), and this vector was used to in vitro translate the Z(A204D) protein. The Z(L214R/L218R) cDNA mutant (21) was also a gift from Erik Flemington. Z-FLAG (3xFLAG-Zta), a gift from Paul Lieberman, has Z cDNA inserted into a p3xFLAG-myc-CMV24 vector (Sigma, Saint Louis, MO) for mammalian cell expression of a FLAG-tagged Z protein. Site-directed mutagenesis was used to create the Z(A204D) mutation in the Z-FLAG vector. pRK-BALF4 encodes EBV glycoprotein 110 (gp110) and was a gift from Henri-Jacques Delecluse (52). Ob-LUC (a gift from Ormond MacDougald) contains the promoter from the obesity (Ob) gene inserted upstream of the luciferase gene in the pGL3-basic vector (Promega, Madison, WI). Zp-LUC contains the Z promoter (Zp) sequence (from position −495 to position +28 relative to the Z transcription start site) inserted upstream of the luciferase gene in pGL3-basic.
CAT assay.
Cells were transiently transfected with the TNFR1p-CAT plasmid (250 ng), along with combinations of C/EBPα (100 ng), C/EBPβ (100 ng), SG5-Z (250 ng), SG5-Z(A204D) (250 ng), and empty vector (control) plasmids. After 48 or 72 h, cells were harvested in reporter lysis buffer (Promega) plus protease inhibitors (Roche) and subjected to freeze-thawing and centrifugation per the manufacturer's instructions. The cell lysates were incubated at 37°C with acetyl coenzyme A and 14C-labeled chloramphenicol (Amersham Biosciences, Piscataway, NJ), as described previously (23). The activity of the TNFR1 promoter was measured by acetylation of chloramphenicol, and the percent acetylation was quantitated by thin-layer chromatography followed by phosphorimager screening using a Storm 840 phosphorimager (Molecular Dynamics, Piscataway, NJ). The results were quantified using ImageQuant software (Amersham Biosciences). Extracts in reporter lysis buffer were also subjected to immunoblotting to verify equivalent protein levels.
GST pulldown assays.
GST fusion proteins were expressed in bacteria and harvested as crude lysate as described previously (26). Crude lysate (40 μl) containing GST proteins was incubated with 40 μl 50% slurry glutathione (GSH)-agarose beads (Sigma) per reaction and rotated at 4°C for 1 h. The GST-coated beads were washed 3 times with 1× phosphate-buffered saline (PBS) and then resuspended in 500 μl binding buffer (140 mM NaCl, 0.2% NP-40, 100 mM NaF, 200 μM NaO3VO4, 50 mM Tris [pH 8], 5% bovine serum albumin [BSA]). Ten microliters of in vitro-translated protein (IVP) labeled with [35S]methionine (Amersham Biosciences) was added to the GST beads and rotated at 4°C for 2 h. The beads were then spun down and washed 5 times with 500 μl wash buffer (100 mM NaCl, 0.2% NP-40, 1 mM EDTA, 20 mM Tris [pH 8]). Beads were resuspended in 35 μl 2× sodium dodecyl sulfate (SDS) loading buffer and boiled for 5 min. The proteins (including 1 μl direct load of in vitro-translated proteins) were then separated on a 10% denaturing polyacrylamide gel. Gels were fixed, dried, and subjected to autoradiography. Quantity One software by Bio-Rad Laboratories (Hercules, CA) was used to quantify the results.
Immunoprecipitation.
HeLa cells were transiently transfected with Z and/or C/EBP expression vectors and then harvested 48 h later. Plates were washed with 1× PBS and then incubated on ice with occasional rocking for 30 min in NP-40 lysis buffer (150 mM NaCl, 1% NP-40, 50 mM Tris [pH 8], and protease inhibitors). Cells were scraped into microcentrifuge tubes, sonicated for 20 s, and then centrifuged at maximum speed for 10 min at 4°C. Normal rabbit serum was added to the supernatant and incubated on ice 1 h. Protein A/G PLUS agarose beads (sc-2003; Santa Cruz Biotechnology, Santa Cruz, CA) were added to preclear and rocked for an additional hour at 4°C. Beads were spun down, and the supernatant was divided for the appropriate conditions. NP-40 lysis buffer was added to each sample along with 1 μg antibody (or no antibody for the direct load) and rocked at 4°C for 1 h. The antibodies used were as follows: mouse anti-Flag (F1804; Sigma), mouse monoclonal anti-C/EBPβ (sc-7962; Santa Cruz), rabbit monoclonal anti-C/EBPα (1704-1; Epitomics, Burlingame, CA), control mouse IgG (sc-2025), and control rabbit IgG (sc-2027). A/G beads were added and rocked at 4°C for 2 h. Beads were spun down and washed three times in NP-40 lysis buffer. Sample buffer (6×) was added to each sample, and the samples were subjected to immunoblot analysis using the following antibodies: mouse monoclonal anti-C/EBPβ (sc-7962), mouse monoclonal anti-Z (sc-53904), and goat polyclonal anti-C/EBPα (sc-9314).
Immunoblot analysis.
Immunoblotting was performed as described previously (7). Briefly, cells were lysed in SUMO buffer plus protease inhibitors and then sonicated and centrifuged. Equivalent amounts of protein were separated in sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) gels and transferred to nitrocellulose membranes. Membranes were blocked in 5% milk and then incubated with the appropriate primary antibodies diluted in 5% milk in 1× PBS and 0.1% Tween 20 (PBS-T). The primary antibodies were as follows: mouse monoclonal anti-C/EBPβ (sc-7962; Santa Cruz), mouse monoclonal anti-Z (sc-53904), mouse monoclonal anti-TNFR1 (sc-8436), rabbit monoclonal anti-C/EBPα (1704-1; Epitomics), goat polyclonal anti-C/EBPα (sc-9314), and mouse monoclonal anti-β-actin (A5441; Sigma). After being washed, the membranes were incubated with the appropriate horseradish peroxidase-conjugated secondary antibodies (Pierce, Waltham, MA) in 5% milk-1× PBS-T for 1 h at room temperature and then washed again. Bound antibodies were visualized by use of enhanced chemiluminescent reagent (Pierce) according to the manufacturer's instructions.
Virus reactivation and lytic replication assays.
Virus reactivation and lytic replication titration assays were performed as previously described (30). 293-ZKO cells were transfected with vector control, SG5-Z (250 ng), or SG5-Z(A204D) (400 ng); for viral replication assays, expression vectors for the EBV gp110 protein (500 ng) and the EBV BRLF1 protein (500 ng) were also cotransfected. More Z(A204D) plasmid (versus wild-type Z) was transfected to correct for the fact that the mutant protein is somewhat less stable than the wild-type Z protein in 293-ZKO cells (data not shown). After 72 h, supernatant from the transfected cells was collected and filtered through a 0.8-μm-pore-size filter. The filtered virus was used to infect Raji cells (2 × 105 cells/infection). Phorbol-12-myristate-13-acetate (TPA; 20 ng/ml) and sodium butyrate (3 mM final concentration) were added at 24 h postinfection. Green fluorescent protein (GFP)-positive Raji cells were counted 72 h after infection to determine viral titer. The protein extracts of the transfected 293-ZO cells were analyzed in immunoblot assays to ensure equal levels of transfected wild-type and mutant Z proteins in each experiment.
Luciferase assay.
HeLa cells were transiently transfected with Ob-LUC or Zp-LUC plasmid (250 ng), along with combinations of C/EBPα (100 ng), C/EBPβ (100 ng), Zwt (250 ng), Z(A204D) (250 ng), and empty vector (control) plasmids. Cells were harvested 48 h later. Luciferase assays were performed by using extracts prepared by freeze-thawing the cell pellet in reporter lysis buffer plus protease inhibitors according to the instructions of the manufacturer (Promega). Luciferase activity was assayed using the luciferase reporter assay system (Promega), as suggested by the manufacturer, with a Monolight 3010 luminometer (Analytical Luminescence Laboratory, San Diego, CA).
EMSA.
Klenow fragment DNA polymerase I (Roche) and [γ-32P]dATP/dCTP (Amersham Biosciences) were used to label double-stranded, annealed DNA oligonucleotides for use in DNA-protein binding experiments. The oligonucleotides consisted of a 20-bp sequence containing a C/EBP site (underlined) in the TNFR1 promoter (TNFR1 positions −94 to −75 [5′-GAT CTC CCG CTG TTG CAA CAC TGC-3′ and 5′-GAT CGC AGT GTT GCA ACA GCG GGA-3′]). An oligonucleotide containing the consensus C/EBP binding sequence was also synthesized (5′-GAT CCT AGC TGC AGA TTG CGC AAT CTG CAG-3′ and 5′-GAT CCT GCA GAT TGC GCA ATC TGC AGC TAG-3′). The protein samples used in electrophoretic mobility shift assays (EMSAs) were either nuclear protein extracts harvested from transfected cells or in vitro-translated protein (IVP). In vitro-translated proteins were generated using TNT T7 or the SP6 quick coupled transcription/translation system (Promega) in accordance with the manufacturer's instructions. To make nuclear extracts, cells were transfected with plasmid DNA, harvested after 48 or 72 h, and prepared as described previously (7). Protein samples (2 μg nuclear extract or 2 μl IVP protein) were incubated with binding buffer [10 mM HEPES (pH 7.9), 50 mM KCl, 2.5 mM MgCl2, 10% glycerol, 1 μg BSA, 1 mM dithiothreitol, 2 μg poly(dI-dC)] for 5 min at room temperature before addition of radiolabeled probes (20,000 cpm/20-μl reaction mixture). After incubation for 20 min at room temperature, the samples were loaded onto a 4% polyacrylamide gel and run in 0.5× Tris-borate-EDTA buffer at room temperature. Supershift experiments adding 2 μg specific antibody to the protein-DNA complexes were performed to confirm the identity of the binding protein. The antibody used was goat polyclonal C/EBPα (sc-9314), mouse monoclonal C/EBPβ (sc-7962), or mouse monoclonal Z (11-007; Argene, Verniolle, France).
ChIP.
HeLa cells (3 × 107 cells per condition) or 293 cells were transfected with a control vector, Z, or Z(A204D) and harvested after 24 h for ChIP assays using a modified Upstate ChIP protocol (Millipore, Billerica, MA). Chromatin was sonicated to ∼500 bp. The sonicated DNA was precleared with a 50% slurry of protein A/G PLUS agarose beads in Triton lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 1% BSA, 10 μg/ml salmon sperm DNA) and then incubated overnight at 4°C with 2 μg of antibody. The antibodies used were control mouse IgG (sc-2025; Santa Cruz), mouse monoclonal anti-Z (sc-53904), and mouse monoclonal anti-C/EBPβ (sc-7962). Input (total) DNA was obtained from samples not incubated with antibody. ChIP DNA was analyzed using primers spanning TNFR1p positions −154 to −35 (5′-AGT TAA AGA ACG TTG GGC CTC CT-3′ and 5′-GCA GAG AGG AGG GGA GAG AAG G-3′), the β-globin gene (5′-AGG GCT GGG CAT AAA AGT CA-3′ and 5′-GCC TCA CCA CCA ACT TCA TC-3′), and the interleukin-8 promoter (IL-8p) (5′-ACC AAA TTG TGG AGC TTC AGT-3′ and 5′-AGC TTG TGT GCT CTG CTG TCT-3′). ChIP assay results were quantified using Quantity One software (Bio-Rad Laboratories) and results normalized relative to levels for input DNA for each condition.
RESULTS
Z inhibits C/EBPα- and C/EBPβ-mediated activation of TNFR1p.
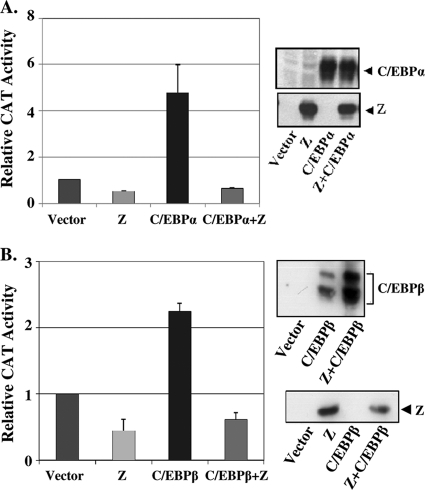
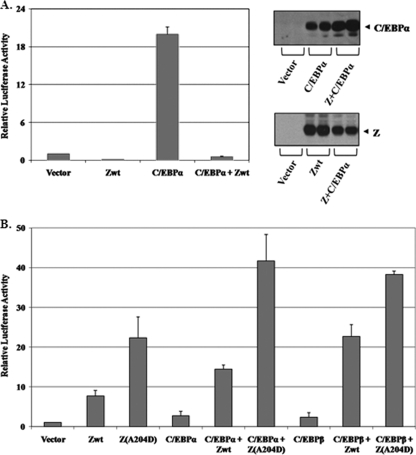
We recently showed that the TNFR1 promoter (TNFR1p) contains a C/EBP binding site located from position −88 to position −80 (relative to the transcriptional start site) and that both C/EBPα and C/EBPβ enhance the activity of TNFR1p through this binding site (7). Since Z has been reported to directly interact with the C/EBPα protein (76, 77), we examined whether Z affects the ability of cotransfected C/EBPα and C/EBPβ to activate the TNFR1 promoter. As shown in Fig. 1, both C/EBPα (Fig. 1A) and C/EBPβ (Fig. 1B) activated TNFR1p-CAT activity in the absence of Z, but this effect was greatly diminished in the presence of cotransfected Z. The protein levels of transfected C/EBPα and C/EBPβ were not reduced in the presence of cotransfected Z (Fig. 1A and B, right panels) but were instead increased in some experiments (consistent with a previous report indicating that Z stabilizes C/EBPα [76]). Although Z has been shown to enhance activation of its own promoter (Zp) by C/EBPα (77), the results here indicate that Z prevents activation of the TNFR1 promoter by both C/EBPα and C/EBPβ.
FIG. 1.
Z inhibits C/EBPα- and C/EBPβ-mediated activation of the TNFR1 promoter (TNFR1p). (A) (Left panel) HeLa cells were transfected with a TNFR1p-CAT construct in the presence or absence of cotransfected Z, C/EBPα, or control vectors. The relative CAT activity for each condition is shown; the value for the activity of the promoter construct plus the vector control is set at 1. Values are given as means ± standard deviations of results from three independent experiments. (Right panel) Immunoblot analyses were performed to determine the amounts of C/EBPα and Z in the extracts used for the CAT assays. (B) (Left panel) HeLa cells were transfected with TNFR1p-CAT in the presence or absence of cotransfected Z or C/EBPβ expression vectors as indicated. (Right panel) Immunoblot analyses were performed to determine the amounts of C/EBPβ and Z in the extracts used for the CAT assays.
Z alanine residue 204 is important for interaction of Z with C/EBPα and C/EBPβ.
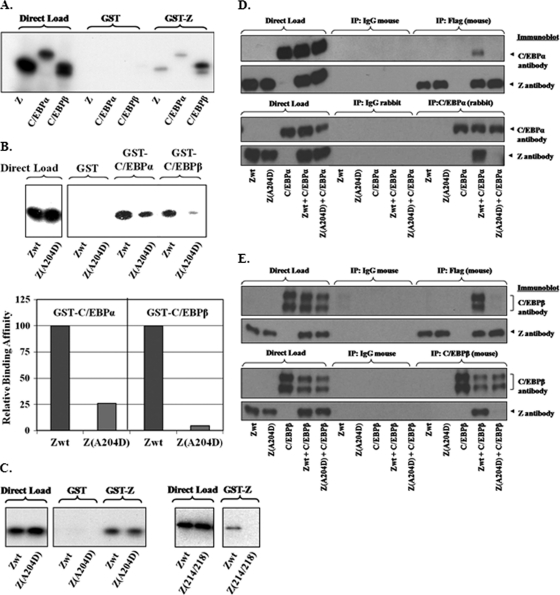
Z interacts directly with C/EBPα in vitro, and a Z mutant containing an aspartic acid (instead of alanine) at residue 204 within the bZIP domain is impaired for the ability to interact with C/EBPα in vitro (77). To determine if Z also directly interacts with C/EBPβ, we performed GST pulldown assays using in vitro-translated, 35S-labeled C/EBPα, C/EBPβ, or Z proteins and GST or GST-Z fusion proteins. As expected, in vitro-translated wild-type Z interacted much more strongly with the GST-Z protein than with the control GST protein (Fig. 2A). In vitro-translated C/EBPα and C/EBPβ also interacted with the GST-Z fusion protein much more strongly than with the GST protein (Fig. 2A). The results indicate that Z interacts with both C/EBPα and C/EBPβ in vitro.
FIG. 2.
Z alanine residue 204 is important for interaction of Z with C/EBPα and C/EBPβ. (A) GST pulldown assays were performed using GST or GST-Z fusion proteins incubated with 35S-labeled, in vitro-translated Z, C/EBPα, or C/EBPβ proteins. (B) GST pulldown assays were performed using GST, GST-C/EBPα, and GST-C/EBPβ proteins incubated with 35S-labeled, in vitro-translated wild-type or mutant Z protein (top panel); the results are quantified in the bottom panel, with the value for binding of wild-type Z to the GST-C/EBPα or GST-C/EBPβ protein set at 100. (C) GST and GST-Z proteins were incubated with 35S-labeled, in vitro-translated wild-type Z or mutant Z(A204D) protein (left panel) or with wild-type Z or mutant Z(L214R/L218R) protein (right panel). (D) HeLa cells were transfected with Flag-tagged wild-type or mutant Z protein in the presence or absence of cotransfected C/EBPα and then immunoprecipitated with a control mouse antibody or Flag antibody (top two rows) or a control rabbit antibody or C/EBPα rabbit antibody (bottom two rows). Immunoprecipitated proteins were then probed by immunoblot analysis using anti-Z or anti-C/EBPα antibodies as indicated. (E) HeLa cells were transfected with Flag-tagged wild-type or mutant Z proteins in the presence or absence of cotransfected C/EBPβ and then immunoprecipitated with a control mouse antibody or Flag antibody (top two rows) or a control mouse antibody or anti-C/EBPβ antibody (bottom two rows). Immunoprecipitated proteins were then probed by immunoblot analysis using anti-Z or anti-C/EBPβ antibodies as indicated.
To determine if Z alanine residue 204 is important for the ability of Z to interact with both C/EBPα and C/EBPβ, we next compared the abilities of in vitro-translated wild-type Z and a Z(A204D) mutant to interact with the GST-Z, GST-C/EBPα, and GST-C/EBPβ proteins in vitro. As shown in Fig. 2B, in comparison to wild-type Z, the Z(A204D) mutant had reduced interactions with both the GST-C/EBPα and GST-C/EBPβ proteins but was similar to wild-type Z with regard to its ability to interact with the GST-Z protein (Fig. 2C). As expected, a Z mutant previously shown to be unable to homodimerize, Z(L214R/L218R) (21), did not interact with GST-Z (Fig. 2C). These results suggest that the Z(A204D) mutant is deficient in its ability to interact with both C/EBPα and C/EBPβ in vitro but is not defective for homodimerization.
To determine if the Z(A204D) mutant is defective at interacting with C/EBP proteins in vivo, HeLa cells were transfected with expression vectors for wild-type Z-Flag, Z(A204D)-Flag, C/EBPα, and C/EBPβ (alone or in combination), and coimmunoprecipitation studies were performed using antibodies directed against Flag, C/EBPα, C/EBPβ, or control antibodies. As shown in Fig. 2D and E, wild-type Z directly interacts with cotransfected C/EBPα and C/EBPβ. In contrast, the Z(A204D) mutant is highly defective in the ability to interact with both C/EBPα and C/EBPβ. These results indicate that Z interacts with both the C/EBPα and the C/EBPβ proteins in vivo and that Z alanine residue 204 is required for efficient interaction with both C/EBP proteins.
The Z(A204D) mutant is attenuated in the ability to inhibit C/EBPα and C/EBPβ activation of the TNFR1 promoter.
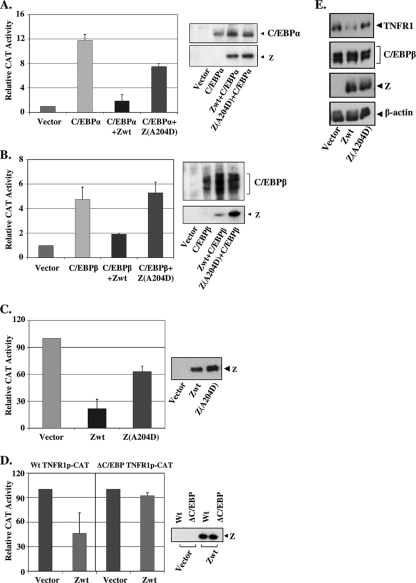
Given the poor ability of the Z(A204D) mutant to interact directly with the C/EBP proteins, we next determined if this mutant is also defective in the ability to inhibit C/EBPα and C/EBPβ activation of the TNFR1 promoter. In comparison to the wild-type protein, the Z(A204D) mutant had a reduced ability to block both C/EBPα and C/EBPβ transactivation of the TNFR1 promoter (Fig. 3A and B, left panels), although in some experiments the mutant Z(A204D) protein level was actually higher than that of wild-type Z (Fig. 3B, right panel). The Z(A204D) mutant was also impaired in comparison to wild-type Z in the ability to inhibit the constitutive activity of the TNFR1 promoter (Fig. 3C). To confirm that the C/EBP binding motif in the TNFR1 promoter is required for the ability of Z to inhibit its activity, we also compared the abilities of Z to inhibit the activity of the wild-type TNFR1 promoter and a promoter construct containing a mutated C/EBP binding motif (ΔC/EBP TNFR1p-CAT). As shown in Fig. 3D, the inhibitory effect of Z required the C/EBP binding site in the TNFR1 promoter.
FIG. 3.
The Z(A204D) mutant is attenuated in the ability to inhibit C/EBPα- and C/EBPβ-mediated activation of the TNFR1 promoter and decrease TNFR1 protein expression. (A) (Left panel) HeLa cells were transfected with the TNFR1p-CAT construct in the presence or absence of cotransfected expression vectors for C/EBPα, Z, or Z(A204D) in the various combinations indicated. The relative CAT activity for each condition is shown; the value for the activity of the promoter construct plus the vector control is set at 1. Values are given as means ± standard deviations of results from two independent experiments performed in duplicate. (Right panel) Immunoblot analyses were performed to determine the amounts of C/EBPα and Z in the extracts. (B) (Left panel) HeLa cells were transfected with the TNFR1p-CAT construct in the presence or absence of cotransfected expression vectors for C/EBPβ, Z, or Z(A204D) as indicated. The relative CAT activity for each condition is shown; the value for the activity of the promoter construct plus the vector control is set at 1. Values are given as means ± standard deviations of results from three independent experiments. (Right panel) Immunoblot analyses were performed to determine the amounts of C/EBPβ and Z in the extracts. (C) (Left panel) HeLa cells were transfected with the TNFR1p-CAT construct in the presence or absence of cotransfected expression vectors for Z or Z(A204D). The relative CAT activity for each condition is shown; the value for the activity of the promoter construct plus the vector control is set at 100. Values are given as means ± standard deviations of results from three independent experiments. (Right panel) Immunoblot analyses were performed to determine the amount of Z in the extracts. (D) (Left panel) HeLa cells were transfected with the wild-type TNFR1p-CAT construct (Wt TNFR1p-CAT) or a mutant promoter construct missing the C/EBP binding motif (ΔC/EBP TNFR1p-CAT) in the presence or absence of a cotransfected Z expression vector. The relative CAT activity for each condition is shown; the value for the activity of the promoter construct plus the vector control is set at 100. Values are given as means ± standard deviations of results from two independent experiments performed in duplicate. (Right panel) Immunoblot analyses were performed to determine the amounts of Z in the extracts. (E) Hone cells were transfected with 1 μg of vector control or wild-type Z versus Z(A204D) expression vectors, and immunoblot analysis was performed 2 days later to quantify the expression of TNFR1 protein, C/EBPβ, transfected Z, and β-actin.
Finally, to determine whether the ability of Z to downregulate endogenous TNFR1 protein expression in cells also requires its ability to interact with C/EBP proteins, we compared the effects of transfected wild-type Z and the Z(A204D) mutant on the level of TNFR1 protein expression in Hone cells. As shown in Fig. 3E, wild-type Z decreased the level of endogenous TNFR1 protein expression to a much greater extent than the Z(A204D) mutant, although the levels of Z expression and C/EBP expression were similar in cells transfected with the wild-type or mutant Z vectors. Together, these results suggest that a direct interaction between Z and the C/EBPα and C/EBPβ proteins contributes to its ability to inhibit the TNFR1 promoter as well as TNFR1 protein expression.
The Z(A204D) mutant is not defective in regard to its transcriptional or replicative functions in 293 cells.
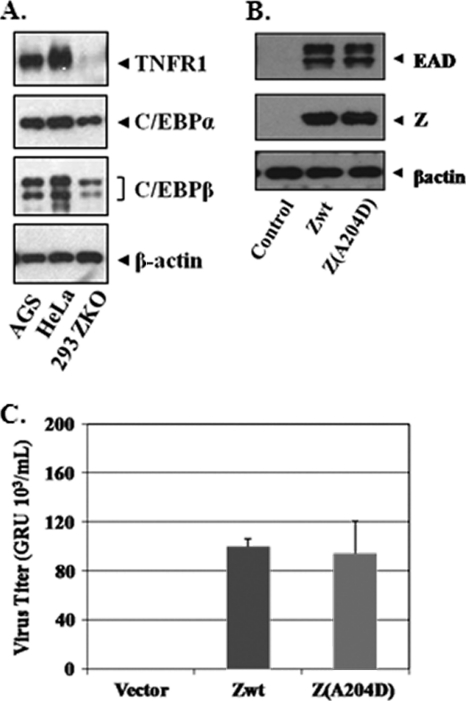
To determine if the direct interaction between Z and the C/EBPα and/or C/EBPβ protein is important for the ability of Z to induce lytic viral gene transcription or mediate lytic viral replication, we transfected the wild-type or mutant (A204D) Z protein into 293-ZKO cells, which are stably infected with a BZLF1-deleted EBV mutant (13, 18). Note that 293 cells, similar to many tumor cell lines, do not express the TNFR1 protein to any degree (presumably reflecting methylation of the TNFR1 promoter) but do express C/EBPα and C/EBPβ (Fig. 4A). Consistent with its ability to homodimerize (which is required for Z DNA binding activity and transcriptional function), we found that the Z(A204D) mutant reactivated expression of the EBV lytic viral protein BMRF1 (EAD) as efficiently as did the wild-type Z protein (Fig. 4B). This result indicates that the ability of Z to interact directly with C/EBP proteins is not required for its ability to activate lytic viral gene expression.
FIG. 4.
The Z(A204D) mutant is not defective in regard to its transcriptional activation or replicative functions in 293 cells. (A) Immunoblot analysis was performed to compare the expression levels of TNFR1, C/EBPα, C/EBPβ, and β-actin proteins in 293-ZKO cells with those in AGS and HeLa cells. (B) 293 cells stably infected with a BZLF1-deleted EBV mutant (293-Z-KO cells) were transfected with wild-type Z, the Z(A204D) mutant, or a vector control. Immunoblot analysis was performed 2 days after transfection to compare the levels of transfected Z protein, an early lytic EBV protein induced by Z (BMRF1 [EAD]), and β-actin (as loading control). (C) 293-Z-KO cells were transfected with wild-type Z, the Z(A204D) mutant, or a vector control (plus cotransfected BRLF1 and EBV gp110 expression vectors), and the amount of infectious virus produced by each condition was determined using the green Raji cell assay as previously described (30). The relative virus titer produced by each condition is shown; the value for the amount of virus produced in cells transfected with wild-type Z is set at 100.
Since certain Z mutants are transcriptionally competent but defective at inducing viral replication (16, 46), and the EBV lytic origin of replication contains C/EBP binding sites as well as Z binding sites (33), we next compared the abilities of wild-type Z and the Z(A204D) mutant to produce infectious viral particles following transfection of 293-ZO cells. As shown in Fig. 4C, 293-ZKO cells transfected with the wild-type and mutant Z expression vectors (along with BRLF1 and gp110) released similar amounts of infectious viral particles into the supernatant, as measured by the green Raji cell assay. These results indicate that the direct interaction between Z and C/EBP proteins is not required for the ability of Z to mediate lytic viral replication.
The effect of Z on C/EBPα- and C/EBPβ-mediated transcriptional activation is promoter dependent.
Although the results shown in Fig. 1 clearly suggest that Z inhibits the ability of C/EBPα and C/EBPβ to transactivate the TNFR1 promoter, a previous study reported that Z enhances the ability of the C/EBPα and β proteins to activate its own promoter (Zp) (33). To determine if the ability of Z to block C/EBP activation is unique to the TNFR1 promoter, we examined the effect of Z on C/EBPα-mediated activation of the well-studied, C/EBPα-responsive obesity (Ob) promoter (47). As shown in Fig. 5A, we found that similar to its effect on the TNFR1 promoter, Z also inhibited the ability of C/EBPα to activate the Ob promoter and decreased its constitutive activity. In contrast, as previously reported (33), we confirmed that Z interacts cooperatively with C/EBPα (as well as C/EBPβ) to activate its own promoter (Fig. 5B). These results indicate that while Z clearly inhibits the ability of C/EBPα and C/EBPβ to activate a subset of promoters, the effects of the interaction are promoter specific. Interestingly, although the Z(A204D) mutant was found to be impaired in the ability to inhibit C/EBPα- and C/EBPβ-mediated activation of the TNFR1 promoter (Fig. 3), it had no defect in the ability to mediate cooperative activation of the Zp promoter in conjunction with C/EBPα and C/EBPβ (Fig. 5B). This result suggests that the ability of the Z and C/EBPα/β proteins to cooperatively activate the Zp promoter does not involve direct protein-protein interactions between Z and C/EBP family members.
FIG. 5.
The effect of Z on C/EBPα- and C/EBPβ-mediated activation is promoter dependent. (A) (Left panel) HeLa cells were transfected with the Ob-luciferase construct in the presence or absence of Z or C/EBPα expression vectors as indicated. The relative luciferase activity for each condition is shown; the value for the activity of the Ob-luciferase construct plus the vector control DNA is set at 1. Values are given as means ± standard deviations of results from two independent experiments. (Right panel) Immunoblot analyses were performed to determine the amounts of C/EBPα and Z in the extracts. (B) HeLa cells were transfected with a Zp-luciferase construct in the presence or absence of various combinations of the wt Z, mutant Z(A204D), and C/EBPα expression vectors. The relative luciferase activity for each condition is shown; the value for the activity of the Zp-luciferase construct plus the vector control DNA is set at 1. Values are given as means ± standard deviations of results from two independent experiments performed in duplicate.
Z does not bind directly to the TNFR1 promoter C/EBP site.
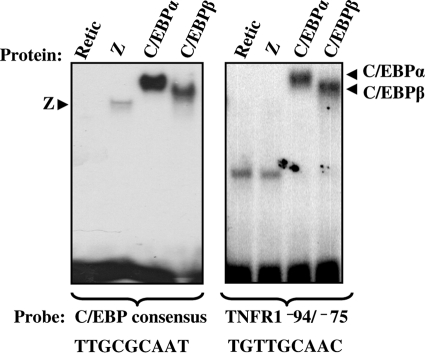
Since Z is known to bind to a variety of AP1-like DNA sequences, including some C/EBP binding motifs, we performed EMSAs (gel shifts) to determine if Z binds directly to the C/EBP site in the TNFR1 promoter (Fig. 6). While in vitro-translated Z bound to a labeled probe containing the consensus C/EBP sequence, it did not detectably bind to the specific C/EBP site in the TNFR1 promoter. As expected, C/EBPα and C/EBPβ both bound to the consensus C/EBP site and the previously identified C/EBP site in the TNFR1 promoter. Z also did not bind to a series of overlapping probes spanning the first 500 base pairs of the TNFR1 promoter in EMSA studies (data not shown). These results indicate that Z does not bind directly to the TNFR1 promoter.
FIG. 6.
Z does not bind directly to the TNFR1 promoter. An EMSA was performed by incubating in vitro-translated Z, C/EBPα, C/EBPβ, or reticulocyte lysate control with 32P-labeled DNA probes containing either a consensus C/EBP binding sequence (left panel) or the TNFR1 promoter sequence from position −94 to position −75 (right panel). Protein-DNA complexes are indicated by arrows.
Z does not inhibit binding of C/EBPα and C/EBPβ to the TNFR1 promoter in EMSAs.
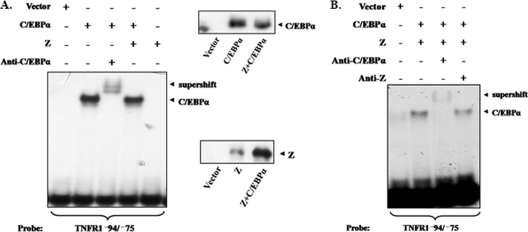
To determine if Z inhibits the ability of C/EBPα and/or C/EBPβ to bind to the TNFR1 promoter, we initially performed EMSAs using nuclear extracts obtained from HeLa cells transfected with C/EBPα or C/EBPβ expression vectors in the presence or absence of a cotransfected Z protein. As shown in Fig. 7A, C/EBPα bound to the TNFR1 C/EBP motif, as expected, and this binding was supershifted with an antibody against C/EBPα. In extracts obtained from cells cotransfected with both Z and C/EBPα (Fig. 7A), the amount of C/EBPα binding was similar to that observed in cells transfected with C/EBPα alone. To confirm that Z was not a part of the C/EBPα binding complex, we examined the ability of antibodies directed against C/EBPα or Z to supershift the C/EBPα binding complex in cells cotransfected with both C/EBPα and Z (Fig. 7B). Only the C/EBPα antibody was able to supershift the complex. Similar results were seen in cells transfected with C/EBPβ in place of C/EBPα (data not shown). These results suggest that Z does not inhibit the ability of C/EBPα or C/EBPβ to bind directly to the TNFR1 promoter.
FIG. 7.
Z does not inhibit binding of C/EBPα and C/EBPβ to the TNFR1 promoter in EMSAs. (A) (Left panel) An EMSA was performed by incubating nuclear extracts from 293 cells transfected with Z, C/EBPα, both proteins, or a vector control with the TNFR1p −94/−75 probe. The sample loaded in one lane had an additional incubation with an antibody against C/EBPα. Protein-DNA complexes, including those supershifted by antibody, are indicated by arrows. (Right panel) Immunoblot analyses were performed to compare the levels of C/EBPα and Z in the extracts. (B) An EMSA was performed by incubating nuclear extracts from 293 cells transfected with a vector control or Z plus C/EBPα with the TNFR1p −94/−75 probe. Samples loaded in some lanes had an additional incubation with an antibody against C/EBPα or Z. Protein-DNA complexes, including those supershifted by antibody, are indicated by arrows.
Wild-type Z is complexed more efficiently with the TNFR1 promoter in vivo than the Z(A204D) mutant is.
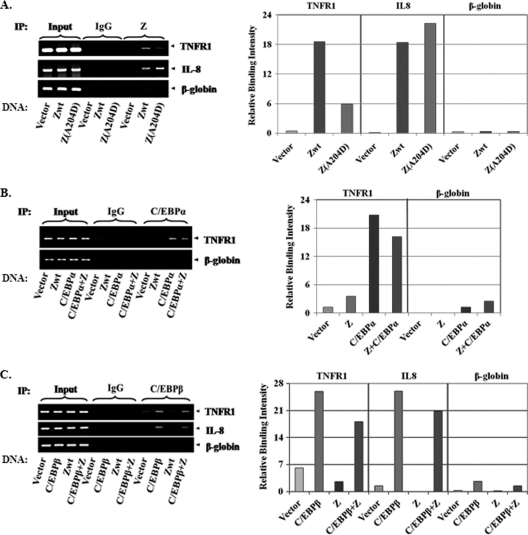
To determine if Z associates with the TNFR1 promoter in vivo, we performed ChIP assays on HeLa cells that were transfected with wild-type Z, the Z(A204D) mutant, or a vector control. Wild-type Z was found to be associated with the TNFR1p sequences (positions −154 to −35) and the cellular IL-8 promoter as previously described (32) but not the β-globin gene (Fig. 8A). In comparison to wild-type Z, the mutant Z(A204D) protein had a very weak association with the TNFR1 promoter but was associated with the IL-8 promoter at least as well as wild-type Z. These ChIP results, in contrast to the EMSA results, suggest that Z is complexed to the TNFR1 promoter in vivo through its direct interaction with C/EBP proteins bound to the TNFR1p DNA. Another group likewise found that an interaction between the Z and C/EBPα proteins which could be observed by ChIP assays over the C/EBPα promoter was not seen in gel shift assays, presumably because the complex does not survive the harsh EMSA conditions (77).
FIG. 8.
Wild-type Z is complexed more efficiently with the TNFR1 promoter in vivo than the Z(A204D) mutant is. (A) A ChIP assay was performed with HeLa cells transfected with vector control, wild-type Z, or the Z(A204D) mutant. Cross-linked DNA-protein complexes were immunoprecipitated using antibodies against Z or an IgG control (a nonimmunoprecipitated sample was saved as an input). Antibody-bound DNA sequences were then PCR amplified using primers spanning the TNFR1 promoter (−154/−35), the IL-8 promoter (which contains a Z binding site), or the β-globin gene. (B) A ChIP assay was performed with HeLa cells transfected with wild-type Z, C/EBPα, both proteins, or a vector control. Cross-linked DNA-protein complexes were immunoprecipitated using antibodies against C/EBPα or an IgG control, and PCR was performed using TNFR1p or β-globin primers. (C) A ChIP assay was performed with 293 cells transfected with wild-type Z, C/EBPβ, both proteins, or a vector control. Cross-linked DNA-protein complexes were immunoprecipitated using antibodies against C/EBPβ or an IgG control. PCR was performed using the TNFR1p, IL-8, or β-globin primer. The IL-8 promoter is known to have a C/EBP site. The results for each ChIP assay are quantitated on the right panels; y axis values were normalized relative to the level for input DNA for each condition.
To determine if Z inhibits the ability of C/EBPα to interact with the TNFR1 promoter in vivo, we performed ChIP assays using HeLa cells (which have very little endogenous C/EBPα) transfected with C/EBPα in the presence or absence of cotransfected Z. Z had a minimal effect on the ability of C/EBPα (Fig. 8B) and C/EBPβ (Fig. 8C) to bind to either the TNFR1 promoter or the IL-8 promoter. These results indicate that Z does not substantially inhibit binding of the C/EBP proteins to the TNFR1 promoter, consistent with the EMSA results.
In summary, our results indicate that wild-type Z, but not the Z(A204D) mutant, is tethered to the TNFR1p through its direct interactions with the C/EBPα and/or C/EBPβ protein. In the case of the TNFR1 promoter, this interaction presumably induces a transcriptionally incompetent complex.
DISCUSSION
The EBV Z immediate-early gene product is a multifunctional protein that not only plays an essential role in inducing lytic viral gene transcription and viral replication but also alters the host cell environment in multiple ways to promote efficient lytic viral replication. For example, Z disperses promyelocytic leukemia (PML) bodies (1), regulates cell cycle progression (9), and modulates p53 function (45, 64, 65). Z also inhibits a variety of different components of the innate immune response in cells, including the gamma interferon signaling pathway (50), interferon regulatory factor 7 (IRF-7) (27), NF-κB (14, 25, 31, 48), and TNF signaling (49). Although we previously reported that Z-mediated inhibition of TNF signaling is primarily mediated by loss of TNFR1 expression in Z-expressing cells and that Z decreases the constitutive activity of a TNFR1 promoter construct (49), the mechanism for this Z effect was unknown. In this report, we show that Z downregulates the TNFR1 promoter activity, as well as TNFR1 protein expression, by preventing C/EBPα- and C/EBPβ-mediated activation of the promoter. Furthermore, we demonstrate that this effect requires a direct interaction between Z and the C/EBP family members, since a Z mutant that cannot interact strongly with C/EBPα and -β is specifically impaired in the ability to inhibit the TNFR1 promoter as well as the ability to decrease TNFR1 protein expression. These results, combined with previous reports showing that the Z/C/EBP protein combination cooperatively activates the BZLF1 promoter (33, 77), suggest that Z modulation of C/EBP family members is yet another mechanism by which Z alters the host cell environment to promote successful lytic viral replication.
The transcriptional function of Z is essential for its ability to induce lytic viral gene transcription. Z initially activates the BRLF1 (Rp) promoter and then cooperates with the BRLF1 gene product (R) to activate all other lytic viral gene promoters. In addition, it has previously been suggested that C/EBPα cooperates with Z to help Z to autoactivate its own promoter (Zp) (33, 77). Interestingly, DNA methylation of lytic viral gene promoters in some cases enhances the ability of Z to bind to, and activate, these promoters (6). In this report, we have confirmed the finding by another group that Z directly interacts with C/EBPα (76, 77) and shown that Z also directly interacts with C/EBPβ. However, we found that a Z mutant that is highly impaired for interaction with both C/EBPα and C/EBPβ is similar to wild-type Z with regard to its induction of early lytic viral gene transcription in latently infected 293 cells (Fig. 4), which do not express the TNFR1 protein but do express C/EBPα and C/EBPβ. This result indicates that the direct interaction between Z and C/EBPα/β is not required for the ability of Z to induce early lytic viral gene expression, at least in the 293 cell environment.
In addition to its essential role in activating expression of the viral proteins that mediate lytic viral DNA replication, including the helicase (BBLF4), DNA polymerase (BALF5), DNA polymerase processivity factor (BMRF1), primase (BSLF1), primase-associated factor (BBLF2/3), and single-stranded DNA binding protein (BALF2) (19), Z plays another role in viral replication through its binding to the lytic origin of replication (oriLyt) and direct interactions with components of the viral replication machinery (19, 66, 79). The latter function is independent of its transcriptional activation potential, as certain mutations in Z prevent lytic viral replication but allow Z to activate expression from lytic viral gene promoters (16, 46). Interestingly, binding of C/EBPα and C/EBPβ to the EBV oriLyt was reported to promote viral replication (33). Therefore, we also compared the abilities of wild-type Z and the Z(A204D) mutant to mediate full viral replication in 293-ZKO cells. As shown in Fig. 4, we found that the wild-type and mutant Z proteins were similar in their ability to produce infectious viral particles. This result indicates that the direct interaction between Z and C/EBP family members is not required for the ability of Z to mediate viral replication and/or release of infectious viral particles.
Since we recently discovered that both C/EBPα and C/EBPβ activate the TNFR1 promoter, and we previously showed that Z inhibits the activity of this promoter, we explored the role of the interaction of Z with C/EBPα/β with regard to the ability of Z to turn off the TNFR1 promoter. We found that wild-type Z inhibits the constitutive activity of the TNFR1 promoter and greatly decreases the ability of both C/EBPα and C/EBPβ to activate the promoter. In contrast, the Z(A204D) mutant was impaired in the ability to inhibit the constitutive activity of the TNFR1 promoter as well as the ability to prevent C/EBPα- and/or β-mediated activation of the promoter. Likewise, the Z(A204D) mutant was defective in decreasing the endogenous level of TNFR1 protein in Hone cells. Together, these results indicate that the direct interaction between Z and C/EBPα and/or β may be primarily important for shutting down the expression of certain C/EBPα- and/or β-responsive cellular genes rather than modulating Z-mediated activation of viral genes.
Although we show here that Z prevents C/EBPα- and β-mediated induction of two different C/EBPα/β-responsive cellular promoters (the TNFR1 promoter and the Ob promoter) (Fig. 1 and 5A), another group previously reported that the combination of Z and C/EBPα actually increases the ability of Z to activate its own promoter (Zp) and also enhances C/EBPα-mediated activation of the cyclin-dependent kinase inhibitor p21 (76). We have confirmed here that the combination of Z and C/EBPα does indeed increase Zp activity compared to the effect of Z or C/EBPα alone (Fig. 5B). Thus, the effect of Z on C/EBPα/β-responsive promoters is clearly promoter dependent. Interestingly, in contrast to what was observed with the TNFR1 promoter [in which the Z(A204D) mutant is impaired for the ability to inhibit C/EBP-mediated activation], we found that Z(A204D) is not impaired for the ability to help Z activate its own promoter (Zp) (Fig. 5B). This result indicates that the cooperative activation of the Zp by Z and C/EBP family members does not require a direct interaction between these proteins.
Although the exact mechanism(s) by which Z inhibits C/EBPα- and C/EBPβ-mediated activation of the TNFR1 promoter is not yet completely unraveled, our results here suggest that this effect involves a direct protein-protein interaction between Z and the C/EBP proteins but is not mediated through decreased binding of C/EBPα and/or C/EBPβ to the promoter (Fig. 7 and 8). Instead, since we found in ChIP assays that wild-type Z is bound to the TNFR1 promoter to a greater extent than the Z(A204D) mutant (Fig. 8), we favor a model in which Z is tethered to the TNFR1 promoter not through a direct DNA binding mechanism but through its interactions with the DNA-bound C/EBPα and/or C/EBPβ protein. Similarly, Wu et al. found that the ability of Z to increase the C/EBPα-induced activation of certain promoters is likely mediated by Z “piggybacking” to DNA-bound C/EBPα (76). Interestingly, the same group showed that the direct interaction between Z and C/EBP proteins requires not only Z residue 204 but also residues within the basic DNA binding domain (77). Upon the basis of our results here, it appears the complex containing the Z/C/EBP proteins in the context of the TNFR1 promoter is not transcriptionally competent. As the effect of the Z/C/EBP combination on promoter activity is clearly context dependent, we speculate that the presence or absence of Z binding sites on a particular promoter (and perhaps their spacing relative to the C/EBP binding sites) may dictate the final outcome of the interaction and also determine the degree to which the proteins can directly interact when bound to that promoter.
The Z protein is increasingly recognized as playing a crucial role in protecting cells with the lytic form of EBV infection from being killed through the effects of the innate immune response. Z attenuates type I interferon (IFN-α and IFN-β) signaling by interacting directly with IRF-7 and preventing it from turning on IFN-α and IFN-β expression (27). Z also promotes a cellular environment conducive to lytic infection by dispersing PML bodies, which have antiviral activity (1). Z inhibits NF-κB-induced immune responses by directly interacting with the p65 component of NF-κB and inhibiting its transcriptional activity (48). In addition, Z decreases the expression of major histocompatibility complex (MHC) class I and II molecules, thereby inhibiting the ability of EBV-specific CD4 T cells and cytotoxic CD8 T cells to recognize and kill lytically infected cells (34, 39).
TNF is a master regulator of the immune response, and TNFR1 is its gatekeeper. In this paper, we have examined the mechanism(s) by which Z inhibits TNF-α signaling in lytically infected host cells and discovered that Z prevents TNF-α-mediated cell death through its direct protein-protein interactions with C/EBP family members. This ability of Z to interact with C/EBP family members may not only be important for inhibition of TNF signaling but could also promote viral success through regulation of additional cellular promoters. C/EBPβ in conjunction with NF-κB is a key activator of the immune system (2, 69), and thus, the ability of Z to block C/EBPβ activity may more globally help to hinder the immune response to infection. For example, the promoters driving the IL-1, TNF ligand, granulocyte colony-stimulating factor (G-CSF), and nitric oxide synthase genes are all also activated by C/EBPβ (15, 57). Inhibiting C/EBPβ activity could also provide a mechanism by which Z decreases MHC II expression (55) and antigen presentation. Determining if Z inhibits the ability of C/EBPβ to activate other critical components of the host immune response, in addition to TNFR1, will be an important area for future study.
Acknowledgments
This work was supported by grants R01-CA58853, R01-CA66519, and P01-CA022443 from the National Institutes of Health as well as Cancer Biology Training Grant T32 CA009135.
Footnotes
Published ahead of print on 22 September 2010.
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akira, S., H. Isshiki, T. Sugita, O. Tanabe, S. Kinoshita, Y. Nishio, T. Nakajima, T. Hirano, and T. Kishimoto. 1990. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 9:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baillie, J., D. A. Sahlender, and J. H. Sinclair. 2003. Human cytomegalovirus infection inhibits tumor necrosis factor alpha (TNF-alpha) signaling by targeting the 55-kilodalton TNF-alpha receptor. J. Virol. 77:7007-7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benedict, C. A., and C. F. Ware. 2001. Virus targeting of the tumor necrosis factor superfamily. Virology 289:1-5. [DOI] [PubMed] [Google Scholar]
- 5.Bertazza, L., and S. Mocellin. 2008. Tumor necrosis factor (TNF) biology and cell death. Front. Biosci. 13:2736-2743. [DOI] [PubMed] [Google Scholar]
- 6.Bhende, P. M., W. T. Seaman, H. J. Delecluse, and S. C. Kenney. 2004. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 36:1099-1104. [DOI] [PubMed] [Google Scholar]
- 7.Bristol, J. A., T. E. Morrison, and S. C. Kenney. 2009. CCAAT/enhancer binding proteins alpha and beta regulate the tumor necrosis factor receptor 1 gene promoter. Mol. Immunol. 46:2706-2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cayrol, C., and E. K. Flemington. 1995. Identification of cellular target genes of the Epstein-Barr virus transactivator Zta: activation of transforming growth factor beta igh3 (TGF-beta igh3) and TGF-beta 1. J. Virol. 69:4206-4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cayrol, C., and E. K. Flemington. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15:2748-2759. [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, Y. N., D. L. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, G., and D. V. Goeddel. 2002. TNF-R1 signaling: a beautiful pathway. Science 296:1634-1635. [DOI] [PubMed] [Google Scholar]
- 12.Countryman, J., H. Jenson, R. Seibl, H. Wolf, and G. Miller. 1987. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J. Virol. 61:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreyfus, D. H., M. Nagasawa, J. C. Pratt, C. A. Kelleher, and E. W. Gelfand. 1999. Inactivation of NF-kappaB by EBV BZLF-1-encoded ZEBRA protein in human T cells. J. Immunol. 163:6261-6268. [PubMed] [Google Scholar]
- 15.Dunn, S. M., L. S. Coles, R. K. Lang, S. Gerondakis, M. A. Vadas, and M. F. Shannon. 1994. Requirement for nuclear factor (NF)-kappa B p65 and NF-interleukin-6 binding elements in the tumor necrosis factor response region of the granulocyte colony-stimulating factor promoter. Blood 83:2469-2479. [PubMed] [Google Scholar]
- 16.El-Guindy, A., L. Heston, H. J. Delecluse, and G. Miller. 2007. Phosphoacceptor site S173 in the regulatory domain of Epstein-Barr Virus ZEBRA protein is required for lytic DNA replication but not for activation of viral early genes. J. Virol. 81:3303-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farrell, P. J., D. T. Rowe, C. M. Rooney, and T. Kouzarides. 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 8:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flemington, E., and S. H. Speck. 1990. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flemington, E., and S. H. Speck. 1990. Evidence for coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc. Natl. Acad. Sci. U. S. A. 87:9459-9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman, A. D. 2007. Transcriptional control of granulocyte and monocyte development. Oncogene 26:6816-6828. [DOI] [PubMed] [Google Scholar]
- 23.Gorman, C. M., L. F. Moffat, and B. H. Howard. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruffat, H., E. Manet, A. Rigolet, and A. Sergeant. 1990. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 18:6835-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutsch, D. E., E. A. Holley-Guthrie, Q. Zhang, B. Stein, M. A. Blanar, A. S. Baldwin, and S. C. Kenney. 1994. The bZIP transactivator of Epstein-Barr virus, BZLF1, functionally and physically interacts with the p65 subunit of NF-kappa B. Mol. Cell. Biol. 14:1939-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gutsch, D. E., K. B. Marcu, and S. C. Kenney. 1994. The Epstein-Barr virus BRLF1 gene product transactivates the murine and human c-myc promoters. Cell. Mol. Biol. (Noisy-le-Grand) 40:747-760. [PubMed] [Google Scholar]
- 27.Hahn, A. M., L. E. Huye, S. Ning, J. Webster-Cyriaque, and J. S. Pagano. 2005. Interferon regulatory factor 7 is negatively regulated by the Epstein-Barr virus immediate-early gene, BZLF-1. J. Virol. 79:10040-10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hardwick, J. M., P. M. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heise, M. T., and H. W. Virgin IV. 1995. The T-cell-independent role of gamma interferon and tumor necrosis factor alpha in macrophage activation during murine cytomegalovirus and herpes simplex virus infections. J. Virol. 69:904-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong, G. K., H. J. Delecluse, H. Gruffat, T. E. Morrison, W. H. Feng, A. Sergeant, and S. C. Kenney. 2004. The BRRF1 early gene of Epstein-Barr virus encodes a transcription factor that enhances induction of lytic infection by BRLF1. J. Virol. 78:4983-4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hong, Y., E. Holley-Guthrie, and S. Kenney. 1997. The bZip dimerization domain of the Epstein-Barr virus BZLF1 (Z) protein mediates lymphoid-specific negative regulation. Virology 229:36-48. [DOI] [PubMed] [Google Scholar]
- 32.Hsu, M., S. Y. Wu, S. S. Chang, I. J. Su, C. H. Tsai, S. J. Lai, A. L. Shiau, K. Takada, and Y. Chang. 2008. Epstein-Barr virus lytic transactivator Zta enhances chemotactic activity through induction of interleukin-8 in nasopharyngeal carcinoma cells. J. Virol. 82:3679-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang, J., G. Liao, H. Chen, F. Y. Wu, L. Hutt-Fletcher, G. S. Hayward, and S. D. Hayward. 2006. Contribution of C/EBP proteins to Epstein-Barr virus lytic gene expression and replication in epithelial cells. J. Virol. 80:1098-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keating, S., S. Prince, M. Jones, and M. Rowe. 2002. The lytic cycle of Epstein-Barr virus is associated with decreased expression of cell surface major histocompatibility complex class I and class II molecules. J. Virol. 76:8179-8188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kemper, O., and D. Wallach. 1993. Cloning and partial characterization of the promoter for the human p55 tumor necrosis factor (TNF) receptor. Gene 134:209-216. [DOI] [PubMed] [Google Scholar]
- 36.Kenney, S., J. Kamine, E. Holley-Guthrie, J. C. Lin, E. C. Mar, and J. Pagano. 1989. The Epstein-Barr virus (EBV) BZLF1 immediate-early gene product differentially affects latent versus productive EBV promoters. J. Virol. 63:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kodukula, P., T. Liu, N. V. Rooijen, M. J. Jager, and R. L. Hendricks. 1999. Macrophage control of herpes simplex virus type 1 replication in the peripheral nervous system. J. Immunol. 162:2895-2905. [PubMed] [Google Scholar]
- 38.Laichalk, L. L., and D. A. Thorley-Lawson. 2005. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J. Virol. 79:1296-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, D., L. Qian, C. Chen, M. Shi, M. Yu, M. Hu, L. Song, B. Shen, and N. Guo. 2009. Down-regulation of MHC class II expression through inhibition of CIITA transcription by lytic transactivator Zta during Epstein-Barr virus reactivation. J. Immunol. 182:1799-1809. [DOI] [PubMed] [Google Scholar]
- 40.Liao, G., F. Y. Wu, and S. D. Hayward. 2001. Interaction with the Epstein-Barr virus helicase targets Zta to DNA replication compartments. J. Virol. 75:8792-8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lieberman, P. M., and A. J. Berk. 1990. In vitro transcriptional activation, dimerization, and DNA-binding specificity of the Epstein-Barr virus Zta protein. J. Virol. 64:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lieberman, P. M., J. M. Hardwick, J. Sample, G. S. Hayward, and S. D. Hayward. 1990. The zta transactivator involved in induction of lytic cycle gene expression in Epstein-Barr virus-infected lymphocytes binds to both AP-1 and ZRE sites in target promoter and enhancer regions. J. Virol. 64:1143-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez, R. G., S. Garcia-Silva, S. J. Moore, O. Bereshchenko, A. B. Martinez-Cruz, O. Ermakova, E. Kurz, J. M. Paramio, and C. Nerlov. 2009. C/EBPalpha and beta couple interfollicular keratinocyte proliferation arrest to commitment and terminal differentiation. Nat. Cell Biol. 11:1181-1190. [DOI] [PubMed] [Google Scholar]
- 44.MacEwan, D. J. 2002. TNF receptor subtype signalling: differences and cellular consequences. Cell. Signal. 14:477-492. [DOI] [PubMed] [Google Scholar]
- 45.Mauser, A., S. Saito, E. Appella, C. W. Anderson, W. T. Seaman, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J. Virol. 76:12503-12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonald, C. M., C. Petosa, and P. J. Farrell. 2009. Interaction of Epstein-Barr virus BZLF1 C-terminal tail structure and core zipper is required for DNA replication but not for promoter transactivation. J. Virol. 83:3397-3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller, S. G., P. De Vos, M. Guerre-Millo, K. Wong, T. Hermann, B. Staels, M. R. Briggs, and J. Auwerx. 1996. The adipocyte specific transcription factor C/EBPalpha modulates human ob gene expression. Proc. Natl. Acad. Sci. U. S. A. 93:5507-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrison, T. E., and S. C. Kenney. 2004. BZLF1, an Epstein-Barr virus immediate-early protein, induces p65 nuclear translocation while inhibiting p65 transcriptional function. Virology 328:219-232. [DOI] [PubMed] [Google Scholar]
- 49.Morrison, T. E., A. Mauser, A. Klingelhutz, and S. C. Kenney. 2004. Epstein-Barr virus immediate-early protein BZLF1 inhibits tumor necrosis factor alpha-induced signaling and apoptosis by downregulating tumor necrosis factor receptor 1. J. Virol. 78:544-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morrison, T. E., A. Mauser, A. Wong, J. P. Ting, and S. C. Kenney. 2001. Inhibition of IFN-gamma signaling by an Epstein-Barr virus immediate-early protein. Immunity 15:787-799. [DOI] [PubMed] [Google Scholar]
- 51.Nerlov, C. 2007. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 17:318-324. [DOI] [PubMed] [Google Scholar]
- 52.Neuhierl, B., R. Feederle, W. Hammerschmidt, and H. J. Delecluse. 2002. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc. Natl. Acad. Sci. U. S. A. 99:15036-15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osada, S., H. Yamamoto, T. Nishihara, and M. Imagawa. 1996. DNA binding specificity of the CCAAT/enhancer-binding protein transcription factor family. J. Biol. Chem. 271:3891-3896. [DOI] [PubMed] [Google Scholar]
- 54.Packham, G., A. Economou, C. M. Rooney, D. T. Rowe, and P. J. Farrell. 1990. Structure and function of the Epstein-Barr virus BZLF1 protein. J. Virol. 64:2110-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pennini, M. E., Y. Liu, J. Yang, C. M. Croniger, W. H. Boom, and C. V. Harding. 2007. CCAAT/enhancer-binding protein beta and delta binding to CIITA promoters is associated with the inhibition of CIITA expression in response to Mycobacterium tuberculosis 19-kDa lipoprotein. J. Immunol. 179:6910-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pham, T. H., S. Langmann, L. Schwarzfischer, C. El Chartouni, M. Lichtinger, M. Klug, S. W. Krause, and M. Rehli. 2007. CCAAT enhancer-binding protein beta regulates constitutive gene expression during late stages of monocyte to macrophage differentiation. J. Biol. Chem. 282:21924-21933. [DOI] [PubMed] [Google Scholar]
- 57.Poli, V. 1998. The role of C/EBP isoforms in the control of inflammatory and native immunity functions. J. Biol. Chem. 273:29279-29282. [DOI] [PubMed] [Google Scholar]
- 58.Popkin, D. L., and H. W. Virgin IV. 2003. Murine cytomegalovirus infection inhibits tumor necrosis factor alpha responses in primary macrophages. J. Virol. 77:10125-10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ramji, D. P., and P. Foka. 2002. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365:561-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rickinson, A. B., and E. Kieff. 2007. Epstein-Barr virus, p. 2655-2700. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 62.Rooney, C. M., D. T. Rowe, T. Ragot, and P. J. Farrell. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 63:3109-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sato, Y., T. Kamura, N. Shirata, T. Murata, A. Kudoh, S. Iwahori, S. Nakayama, H. Isomura, Y. Nishiyama, and T. Tsurumi. 2009. Degradation of phosphorylated p53 by viral protein-ECS E3 ligase complex. PLoS Pathog. 5:e1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sato, Y., N. Shirata, T. Murata, S. Nakasu, A. Kudoh, S. Iwahori, S. Nakayama, S. Chiba, H. Isomura, T. Kanda, and T. Tsurumi. 2010. Transient increases in p53-responsible gene expression at early stages of Epstein-Barr virus productive replication. Cell Cycle 9:807-814. [DOI] [PubMed] [Google Scholar]
- 66.Schepers, A., D. Pich, and W. Hammerschmidt. 1993. A transcription factor with homology to the AP-1 family links RNA transcription and DNA replication in the lytic cycle of Epstein-Barr virus. EMBO J. 12:3921-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schling, P., C. Rudolph, S. Heimerl, S. Fruth, and G. Schmitz. 2006. Expression of tumor necrosis factor alpha and its receptors during cellular differentiation. Cytokine 33:239-245. [DOI] [PubMed] [Google Scholar]
- 68.Schuster, M. B., and B. T. Porse. 2006. C/EBPalpha: a tumour suppressor in multiple tissues? Biochim. Biophys. Acta 1766:88-103. [DOI] [PubMed] [Google Scholar]
- 69.Shirakawa, F., K. Saito, C. A. Bonagura, D. L. Galson, M. J. Fenton, A. C. Webb, and P. E. Auron. 1993. The human prointerleukin 1 beta gene requires DNA sequences both proximal and distal to the transcription start site for tissue-specific induction. Mol. Cell. Biol. 13:1332-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sinclair, A. J. 2003. bZIP proteins of human gammaherpesviruses. J. Gen. Virol. 84:1941-1949. [DOI] [PubMed] [Google Scholar]
- 71.Takada, K., N. Shimizu, S. Sakuma, and Y. Ono. 1986. trans activation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J. Virol. 57:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trefzer, U., M. Brockhaus, H. Lotscher, F. Parlow, A. Budnik, M. Grewe, H. Christoph, A. Kapp, E. Schopf, and T. A. Luger. 1993. The 55-kD tumor necrosis factor receptor on human keratinocytes is regulated by tumor necrosis factor-alpha and by ultraviolet B radiation. J. Clin. Invest. 92:462-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wallach, D., E. E. Varfolomeev, N. L. Malinin, Y. V. Goltsev, A. V. Kovalenko, and M. P. Boldin. 1999. Tumor necrosis factor receptor and Fas signaling mechanisms. Annu. Rev. Immunol. 17:331-367. [DOI] [PubMed] [Google Scholar]
- 74.Wang, D., J. D'Costa, C. I. Civin, and A. D. Friedman. 2006. C/EBPalpha directs monocytic commitment of primary myeloid progenitors. Blood 108:1223-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, P., L. Day, and P. M. Lieberman. 2006. Multivalent sequence recognition by Epstein-Barr virus Zta requires cysteine 171 and an extension of the canonical B-ZIP domain. J. Virol. 80:10942-10949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu, F. Y., H. Chen, S. E. Wang, C. M. ApRhys, G. Liao, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer binding protein alpha interacts with ZTA and mediates ZTA-induced p21(CIP-1) accumulation and G(1) cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu, F. Y., S. E. Wang, H. Chen, L. Wang, S. D. Hayward, and G. S. Hayward. 2004. CCAAT/enhancer binding protein alpha binds to the Epstein-Barr virus (EBV) ZTA protein through oligomeric interactions and contributes to cooperative transcriptional activation of the ZTA promoter through direct binding to the ZII and ZIIIB motifs during induction of the EBV lytic cycle. J. Virol. 78:4847-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. U. S. A. 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang, Q., Y. Hong, D. Dorsky, E. Holley-Guthrie, S. Zalani, N. A. Elshiekh, A. Kiehl, T. Le, and S. Kenney. 1996. Functional and physical interactions between the Epstein-Barr virus (EBV) proteins BZLF1 and BMRF1: effects on EBV transcription and lytic replication. J. Virol. 70:5131-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.zur Hausen, H., H. Schulte-Holthausen, G. Klein, W. Henle, G. Henle, P. Clifford, and L. Santesson. 1970. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature 228:1056-1058. [DOI] [PubMed] [Google Scholar]