Abstract
Herpesviruses are members of a diverse family of viruses that colonize all vertebrates from fish to mammals. Although more than one hundred herpesviruses exist, all are nearly identical architecturally, with a genome consisting of a linear double-stranded DNA molecule (100 to 225 kbp) protected by an icosahedral capsid made up of 162 hollow-centered capsomeres, a tegument surrounding the nucleocapsid, and a viral envelope derived from host membranes. Upon infection, the linear viral DNA is delivered to the nucleus, where it circularizes to form the viral episome. Depending on several factors, the viral cycle can proceed either to a productive infection or to a state of latency. In either case, the viral genetic information is maintained as extrachromosomal circular DNA. Interestingly, however, certain oncogenic herpesviruses such as Marek's disease virus and Epstein-Barr virus can be found integrated at low frequencies in the host's chromosomes. These findings have mostly been viewed as anecdotal and considered exceptions rather than properties of herpesviruses. In recent years, the consistent and rather frequent detection (in approximately 1% of the human population) of human herpesvirus 6 (HHV-6) viral DNA integrated into human chromosomes has spurred renewed interest in our understanding of how these viruses infect, replicate, and propagate themselves. In this review, we provide a historical perspective on chromosomal integration by herpesviruses and present the current state of knowledge on integration by HHV-6 with the possible clinical implications associated with viral integration.
Integration of viral genomes into the host's chromosomes is mandatory for the successful completion of the life cycles of several viruses, including retroviruses and adeno-associated viruses (AAV). In contrast, herpesviruses maintain their genomes as extrachromosomal circular episomes in the nuclei of infected cells without the need for integration. However, there have been several reports of chromosomally integrated herpesvirus (CIHHV) DNA over the years, suggesting that herpesviruses can indeed integrate into the host's chromosomes under certain conditions. In addition, for a virus such as human herpesvirus 6 (HHV-6), found integrated into the germ lines of approximately 1% of the world's population, integration may represent more than a sporadic or anecdotal event.
Considering that replication of nonintegrated herpesvirus DNA occurs through the well-accepted rolling-circle mechanism, yielding long DNA concatemers that are subsequently cleaved into single-genome equivalents during nucleic acid encapsidation, how replication of linear CIHHV DNA can occur (if it does) remains unknown. In this document, we review cases and reports of integrated nonhuman and human herpesviruses and discuss the outcomes of such events on the life cycles of the viruses and the potential medical consequences of integration.
Chromosomal insertions of alphaherpesvirus DNA segments, including those from herpes simplex viruses and equine herpesvirus types 1 and 3, have been reported on numerous occasions in the past (10, 11, 71, 77, 81, 87, 106). In most instances, these events were detected following infection with defective interfering particles or UV-irradiated viral preparations or transfection of sheared or subgenomic viral DNA fragments. The integrated viral genome therefore consists mostly of subgenomic fragments, and there is no possibility for the production of infectious viral particles to occur. Many of the cells carrying integrated viral DNA displayed a transformed phenotype, fueling hypotheses on the oncogenic nature of these viruses. Although the integration of foreign (viral) DNA into chromosomes can cause several anomalies, the intent of this review is to focus on viruses for which integration of full-length viral DNA is documented and to raise, at least theoretically, the possibility that viral replication can occur following integration. Viruses that meet these criteria include Marek's disease virus (MDV), Epstein-Barr virus (EBV), and HHV-6.
MAREK'S DISEASE VIRUS
Marek's disease virus is an oncogenic alphaherpesvirus responsible for the development of T cell lymphomas in chickens. MDV shares the closest homology with the varicella-zoster virus. The genomic structure of MDV is analogous to that of herpes simplex virus, consisting of a long unique sequence (UL) and a short unique sequence (US), each flanked by a terminal repeat (TR) sequence and an internal repeat (IR) sequence, TRL to IRL and TRS to IRS, respectively (15). Within the TRL and IRL are short, repeated sequences containing telomere-like sequences (TAACCC) typically found at the ends of chromosomes (52). It has been demonstrated that infection by MDV commonly leads to the integration of full-length MDV DNA into chicken chromosomes, with no preference for a particular chromosome (27). The integration sites are preferentially located near the ends of the chromosomes within the telomeric region. The precise mechanisms by which the MDV genome integrates into the chicken chromosomes are unknown. Interestingly, oncogenic MDV strains, unlike nononcogenic MDV strains, encode an RNA telomerase subunit (viral TR [vTR]) that shares 88% sequence identity with the chicken TR (cTR) gene (33). The secondary structure of the vTR RNA is very similar to that of cTR RNA and can complement telomerase reverse transcriptases (TERTs) in functional assays (33). TERTs are enzymes that create single-stranded DNA using single-stranded RNA (TR) as a template. In doing so, TERTs add a six-nucleotide repeating sequence, 5′-TTAGGG (in all vertebrates), to the 3′ strands of chromosomes (reviewed in reference 76). It is speculated that vTR might play a role in MDV genome integration by aiding in the generation of telomeric elongations at the ends of the viral genome as a prerequisite for integration.
MDV can be found integrated at multiple sites in the chromosomes of cells derived from MDV-related T cell lymphomas, which is suggestive of a correlation between integration and oncogenicity (27). Considering that herpesvirus replication typically occurs through a rolling-circle mechanism, using the circularized viral DNA as a template, whether a linear and integrated herpesvirus can replicate and generate infectious virions is of great interest. In the case of MDV, the work by Delecluse et al. (28) teaches us that once integrated, MDV can replicate its DNA and generate new linear viral DNA molecules, but the genomic structure replicating the viral genome remains unknown. Based on the limited number of cell lines and tumors studied and results generated from a single laboratory, the significance of the studies is not clear at present, due to the paucity of reports in this area.
EPSTEIN-BARR VIRUS
Epstein-Barr virus is a ubiquitous oncogenic human gammaherpesvirus whose primary infection is often asymptomatic in young children but which can lead to infectious mononucleosis if contracted by young adults. Cancers associated with EBV infection include Burkitt's lymphoma (BL), nasopharyngeal carcinomas (NPCs), Hodgkin's lymphoma, and posttransplant lymphoproliferative disorders (reviewed in reference 58). EBV infects two main cell types, B lymphocytes and epithelial cells. The first reports on the possible integration of the EBV genome into human chromosomes date back to the mid- to late 1970s (3, 4, 50, 54, 89, 104). At that time, the conclusion that EBV was integrated was derived mostly from isopycnic centrifugation experiments. Subsequent DNA hybridization experiments using multiple probes and sequencing confirmed the integration of the full-length EBV genome (40, 70). Interestingly, Namalwa cells that contain 2 integrated EBV DNAs in the absence of episomal viral DNA cannot replicate the virus (53), although expression of latent genes (EBNA1, EBNA2, and latent membrane proteins [LMPs]) can be documented (9, 64). Similar results were reported with the IB4 cell line, which contains 4 to 5 copies of full-length integrated EBV (51), and the NAB-2 cell line, which carries a single integrated EBV copy (64). These results suggest that even though some genes are expressed, reactivation from integrated EBV does not occur. An interesting article by Hurley et al. (45) provides evidence that extrachromosomal circularization of the EBV genome is detected mostly after infection of resting primary cells, while in infection of activated primary B cells, the EBV genome persists solely as a single integrated copy. Furthermore, in every example studied, the viral DNA appeared to have integrated as a linear genome (45). These results point to cellular factors present in activated cells that could negatively influence the formation or stable retention of the viral episome (such as proteases and nucleases) or, alternatively, that integration occurs more efficiently in activated cells, as replicating DNA is more permissive to integration. This is further supported by infection of established EBV-negative (EBV−) Burkitt's lymphoma (BL) cell lines in which EBV has to integrate in order to persist (45). It thus appears that during in vitro infection of EBV− BL cell lines by EBV, integration and episomal maintenance are mutually exclusive (4, 45, 100). This is not to say that the viral episome and integrated viral DNA cannot coexist, as several endogenously EBV-infected BL-derived cell lines carrying both episomal and integrated viral DNA have been described (5, 26). Integration of EBV DNA is by no mean restricted to B lymphocytes, as epithelial cells derived from NPCs are also found to carry CIEBV (16, 57). EBNA1 was expressed in NPCs carrying CIEBV, indicating that unlike viral replication, which appears to necessitate a circularized viral genome, viral gene transcription can occur despite the absence of episomal viral DNA (16). A central question is whether EBV integration occurs subsequently to in vitro cultivation of EBV-infected cells or whether it occurs in vivo. Using NPC biopsy specimen samples, Kripalani-Joshi and Law reported the detection of integrated EBV in 4 of 17 samples, which argues that integration can occur in vivo (57). This is somewhat analogous to chicken T cell lymphoma cells in which MDV is found integrated in primary tumors (27).
As chromosomal integration may lead to genetic abnormalities, whether EBV integration into human chromosomes occurs randomly or not is of importance. It is clear from the literature that EBV does not integrate at a single site. Whether the integration sites are randomly distributed is still debatable, due in part to the relatively small number of integration sites determined. By analyzing 12 in vitro-immortalized B cell lines, 2 in vivo-infected cell lines, and one BL cell line (EB2), Lestou et al. concluded that EBV integration is nonrandom, with involvement of bands 1p31, 1q43, 2p22, 3q28, 4q13, 5p14, 5q12, and 11p15 in the majority of cell lines examined (60). Interestingly, EBV integration was located mainly in G-band-positive material. G-band-positive material refers to regions of chromosomes that stain with Giemsa reagent and are generally associated with heterochromatin, regions that contain many repeats and no functional genes. Note that integration does not occur exclusively in regions devoid of genes, as integration sites occasionally overlap with bona fide cellular genes such as MACF1 (cell motility factor) in Namalwa cells (70), BACH2 (cell differentiation and/or tumor suppressor) in Raji cells (88), and REL and BCL-11A (myeloid and B cell proto-oncogene) in NAB-2 cells (64), raising the possibility that oncogenesis can arise through expression of EBV oncogenes as well as through alterations in cellular gene expression.
HUMAN HERPESVIRUS 6
HHV-6 is one of the most widespread herpesviruses, infecting the human population with a prevalence approaching 100%. HHV-6 primary infection, which generally occurs during the first 2 years of life, develops into roseola infantum, a disease characterized by intense fever for 2 to 3 days followed for a minority of infants (25 to 30%) by a nonvesicular cutaneous rash on the trunk and back (105). Following the primary infection, HHV-6 establishes latency, most likely in monocytes and/or macrophages (55, 56). HHV-6 reactivation from latency can cause serious secondary infections such as encephalitis, primarily in immunosuppressed patients such as bone marrow transplant recipients (96, 97, 107, 108). There are two variants of HHV-6 present within the population. The B variant is the most prevalent and is associated with virtually all cases of roseola. The epidemiology and disease association of the rarer A variant are much less well defined.
CHROMOSOMAL INTEGRATION OF HHV-6
Among herpesvirus family members, HHV-6 is certainly the one for which integration into the host's genome is most consistently observed. In fact, over the last 15 years several independent research groups have published more than 30 articles on the subject. Most of these are case reports of individuals carrying integrated HHV-6 in their genomes (Table 1). The first reports date to the early to mid-1990s, when Luppi et al. made the first demonstration of the presence of a partial and possibly full-length integrated HHV-6 genome in the DNA of freshly isolated peripheral blood mononuclear cells (PBMCs) (66, 68, 94). Subsequent work by researchers around the world give estimates of the prevalence of CIHHV-6 in the world population of approximately 1%, with no apparent disease association, although the number of cases reported remains limited: 1.5% of Czech Republic childhood leukemia patients (43), 0.9% of the Italian transplant population (78), 0.8% of United Kingdom blood donors (up to 2.9% of hospital patients) (59), 1.6% of patients with suspected encephalitis in the United Kingdom (found in the cerebrospinal fluid) (102), and 0.21% of the general Japanese population (90). Both HHV-6 variants A and B have the ability to integrate the human genome. By extrapolation, considering that HHV-6 infects close to 100% of the human population, this suggests that nearly 70 million individuals carry CIHHV-6. Not only is the prevalence of integration much higher than for any other human herpesvirus, but a major difference lies in the observation that CIHHV-6 is present in every nucleated cell of the body, from hair follicles to PBMCs (37, 78, 101). Because the medical consequences of CIHHV-6 are not known, there is a lot of concern as to whether cells, tissues, and organs from individuals with CIHHV-6 are to be considered acceptable and “medically safe” for transplant purposes.
TABLE 1.
Census of CIHHV-6 cases published in the literature
| Variant | No. of cases/chromosomal locationa | References |
|---|---|---|
| A | 1/10q26.3 | 75 |
| 2/17p13.3 | 6, 17, 75, 101 | |
| 1/18q23 | 6 | |
| 29/N/A | 37, 39, 42, 43, 44, 72, 78, 90, 95, 102 | |
| B | 1/1q44 | 20, 24 |
| 2/9q34.3 | 75, 101 | |
| 1/11p15.5 | 17, 75, 101 | |
| 5/17p13.3 | 68, 74, 75, 94, 101 | |
| 1/18p11.3 | 44 | |
| 2/19q13.4 | 75, 101 | |
| 3/22q13 | 6, 90 | |
| 1/22q13 and 1q44 | 23, 25 | |
| 51/N/A | 6, 37, 39, 43, 48, 59, 78, 90, 102 | |
| N/A | 1/1q44 | 103 |
| 1/9q | 26 | |
| 1/22q | 63 | |
| 2/N/A | 47, 49 |
Cases from the same family are reported as a unique event. N/A, not available.
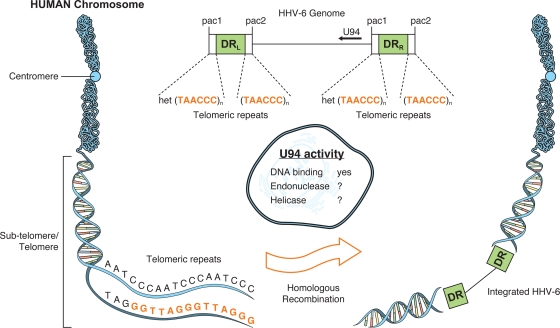
The mechanisms involved in HHV-6 integration are still largely unknown. The HHV-6 genome consists of a single unique component ([U] ∼145 kbp) flanked by identical direct repeats ([DR] ∼9 kbp) (32, 36, 46, 69) that are in turn bracketed by pac1 and pac2 sequences involved in the cleavage and packaging of the HHV-6 genome (29, 91) (Fig. 1). Adjacent to the pac2 sequences are serial TAACCC motifs that are identical to the human telomeric repeat sequence (TRS), while those adjacent to pac1 are imperfect and are referred to as het(TAACCC)n sequences (35, 91) (Fig. 1). It is presumed that through homologous recombination between the TRS present within the HHV-6 genome and the telomeres, the HHV-6 genome, or part of it, gets integrated within human chromosomes (Fig. 1). In fact, all integration sites identified to date (1q44, 9q34.3, 10q26, 11p15.5, 17p13.3, 18p11.3, 18q23, 19q13.4, and 22q13.3) have been localized in the telomeric regions (Table 1). Although there is no clear-cut evidence suggesting that viral integration occurs preferentially in certain chromosomes, some, such as 17p13.3, appear to be targeted preferentially (Table 1). More cases need to be studied to validate this information, but it is possible that HHV-6 integration at sites other than the telomeres is not tolerated or that the virus is not viable, explaining why integration at other sites has not been observed. Having telomeric sequences within its extremities does not appear to be sufficient to promote integration, as HHV-7, the closest relative of HHV-6, possesses such sequences but integration of HHV-7 has never been reported (38). HHV-6 has a distinctive feature not possessed by other human herpesviruses that might favor viral integration: an open reading frame (U94) coding for a protein that shares homologies (24% amino acid identity) with the REP68/78 protein of the parvovirus adeno-associated virus type 2 (AAV-2) (92). The AAV-2 REP68/78 is a nonstructural protein essential for the virus to integrate within chromosome 19 (61, 62). The REP68/78 protein has at least three basic activities that play a role in AAV-2 integration: DNA-binding, endonuclease, and helicase-ATPase activities. Partial characterization of U94 demonstrated that the protein possesses DNA binding activities (31, 73). Although the endonuclease and helicase activities of U94 have not been determined experimentally, mutational analysis of the AAV-2 REP68 has identified several key amino acids that are essential for such functions and that are conserved within U94 (98, 99), suggesting that U94 might possess these activities, especially considering the fact that HHV-6 U94 can functionally complement an AAV-2 REP68/78 deletion mutant (93). These facts argue that U94 retains at least some basic functional activities similar to those of REP68/78. Interestingly, when expressed, U94 is reported to inhibit viral replication, suggesting that it may play a role in the establishment of latency (13, 82) or, alternatively, that U94 might favor viral integration.
FIG. 1.
Hypothetical model of the integration of the HHV-6 genome into the subtelomeric and/or telomeric region of human chromosomes, along with the possible contributions of the U94 gene product in homologous recombination events. Drawing is not to scale.
The HHV-6 genome is present in at least three different forms during a productive infection. In the virion and upon entry, the viral genome is linear. Once in the nucleus, it circularizes into an episome, and during replication, head-to-tail concatemers are formed. Can all three viral genomic forms (linear, episomal, and concatemeric) integrate, and if so, what is the structure of the integrated genome and the fate of the integrated provirus? We present some of the structures of the integrated HHV-6 genome that depend upon whether integration occurs when the viral genome is linear or episomal or in concatemeric form (Fig. 2). As shown in Fig. 2A, integration of the linear viral genome through a single homologous recombination event between the TRS present in DRR and chromosomal TRS results in an integrated provirus that has lost the pac2 sequence from DRR. Without additional recombination events, the genome can no longer fuse its termini to generate the pac2/x/pac1 site (where x denotes a putative cleavage site) required for packaging and cleavage (29, 91). Under such circumstances, integration constitutes a viral dead end, yielding a nonexcisable, incomplete viral genome. The structure of an integrated HHV-6 whose recombination occurred within DRL would be slightly different (not shown), but the result would be the same. We do not imply that integration of a linear genome is without consequences for the host, since following a single recombination event, the end of the chromosome would be lost, possibly affecting the chromosome's stability or causing premature senescence. Furthermore, viral gene expression remains possible (see below for possible consequences). The next possible scenario is integration of the HHV-6 episome. As presented in Fig. 2B, the circularized HHV-6 genome contains a head-to-tail junction that results in the generation of one pac2/x/pac1 site. Recombination through the TRS of DRL or DRR and the TRS of the chromosome results in two distinct viral genomic organizations (Fig. 2B). Assuming that the HHV-6 genome could be replicated by the cellular DNA polymerases, excision would remain an issue, and cleavage at the pac2/x/pac1 junction would result in incomplete viral genomes. If replication were to be assumed by the HHV-6 polymerase complex, with initiation starting at the origin of replication located within the unique region, incomplete genomes would also be generated. Integration of the viral episome is also predictive of a virus that is incapable of excision and replication. In the last scenario, integration of viral concatemers occurs (Fig. 2C). Provided that a concatemer containing at least two pac2/x/pac1 junctions gets integrated, the possibility exists that a full-length HHV-6 genome could be excised from the chromosome and initiate an infectious cycle. As shown, the cleavage of two consecutive pac2/x/pac1 junctions within the concatemer would result in the release of a full-length genome that could circularize, regenerating the viral episome that is essential for viral replication. In our opinion, only integration of viral concatemers is compatible with the excision and generation of a full-length genome. Based on the number of integrated HHV-6 copies/genome, it might therefore be possible to predict which individuals are theoretically susceptible to the production of infectious virions. Integration of linear or episomal viral DNA would yield one copy of CIHHV-6/cell, while integration of viral concatemers would yield >1 copy of CIHHV-6/cell. The work of Leong et al. (59) indicates that some subjects have 2 to 5 copies of integrated HHV-6/cell, which suggests that these individuals may have integrated concatemeric HHV-6 DNA.
FIG. 2.
Structure, integrity, and orientation of the HHV-6 genome following integration of linear (A), episomal (B), or concatemeric (C) viral DNA. For simplicity, only one recombination event with the chromosome's telomeric region, yielding a chromosome ending with the viral genome, is presented. (A) Recombination of linear viral DNA through the (TAACCC)n repeats in DRR results in the loss of the pac2 at the right end of the genome. Recombination within (TAACCC)n of DRL (not shown) would result in the loss of the majority of the genome. Without additional recombination events, these structures would not be compatible with a replication/packaging-competent HHV-6. (B) Recombination of episomal viral DNA would result in two different viral genomic architectures, depending on whether recombination occurred within DRL or DRR. Once again, in the absence of additional recombination events, these structures would not be compatible with a replication/packaging-competent HHV-6. (C) Recombination of concatemeric viral DNA through the (TAACCC)n of one DRL is presented. Considering that the pac1-pac2 junction forms a viral genomic cleavage site provided that two such junctions are present, the possibility exists that a full-length genome could be excised once integrated. Similar conclusions could be drawn if the recombination event were to occur within the DRR (not shown). Please refer to main text for further details.
It is important to distinguish the concept of genetic HHV-6 transmission (inherited) from chromosomal integration that occurs following infection. From the literature, it is clear that CIHHV-6 can be transmitted between generations, with CIHHV-6 found integrated at the same chromosomal sites in the progeny and the parent (6, 72). This germ line transmission may come from either the mother or the father according to Mendel's laws, since not all offspring carry CIHHV-6 (24). At first, the hypothesis that CIHHV-6 could be inherited did not reach consensus. Daibata et al. first reported the presence of CIHHV-6 at similar chromosomal locations in 3 subjects from the same family and suggested that CIHHV-6 was transmitted through inheritance (24). Others have proposed alternative models for apparent transmission through inheritance, such as a possible tendency for HHV-6 to integrate the same chromosomal locus (67). In 1999, Daibata et al. reported the study of a child carrying two HHV-6 chromosomal integrations, with one site (22q13) matching her mother's and the other (1q44) matching her father's (23). Once again, this important observation has been criticized, since all experiments were made using only peripheral blood cells (41). This argument was frequently used, as peripheral blood cells could be infected and/or integrated during the course of an individual's lifetime or during embryogenesis, yielding cells that harbor, at least for some subjects, less than one copy of HHV-6 DNA per cell, which is not compatible with the inherited transmission of CIHHV-6 (65, 67). However, considering the current body of evidence, the demonstration of positive fluorescent in situ hybridization of HHV-6 probes in the chromosomes of fibroblasts (22), the detection of viral DNA in the hair follicles of CIHHV-6 patients (≥1 copy/hair follicle) (101), and the fact that high loads of viral DNA can be found in several tissues throughout the bodies of subjects with CIHHV-6 (44), little doubt persists that CIHHV-6 can be inherited. The child conceived from CIHHV-6-positive gametes shows at least one HHV-6 genome copy in each nucleated cell of his or her body. We know little about the long-term transmission (over several generations) of CIHHV-6 within the population but can affirm that it occurs over at least 3 generations (24). That HHV-6 can be transmitted from one parent to a child suggests that at some point HHV-6 can infect and integrate into germ cells, yielding gametes with CIHHV-6. Alternatively, CIHHV-6 could arise by direct infection and integration of HHV-6 into sperm or ova once gametogenesis has occurred. Further work is needed in this area to better understand this phenomenon. Following fertilization with a CIHHV-6-containing gamete, the zygote develops into an embryo and then a fetus, carrying one copy of CIHHV-6 into every somatic cell. For the vast majority of children with reported congenital HHV-6 infections, as diagnosed by the presence of HHV-6 DNA in cord blood cells, CIHHV-6 was transmitted through inheritance (37). Interestingly, Hall et al. reported that HHV-6A represents 32% of congenitally transmitted CIHHV-6, an incidence much greater than that for postnatally acquired infections, which are caused almost exclusively by HHV-6B (37) (there is an exception in Africa [8]). Computation of all cases reported in the literature supports the 1:3 ratio of HHV-6A to HHV-6B in patients with CIHHV-6 (Table 1).
In addition to inherited CIHHV-6, HHV-6 infection of somatic cells may lead to the chromosomal integration of HHV-6. In such cases, the number of cells carrying CIHHV-6 would be relatively small and thus would not be detected using conventional assays. In this context, Arbuckle et al. successfully demonstrated HHV-6 integration following infection of JJHan and 293 cells with viral DNA maintained in the absence of viral episomes (6). Whether similar results can be obtained following in vitro infection of primary cells is yet to be demonstrated. Arbuckle et al. suggested that chromosomal integration by HHV-6 would not constitute a viral dead end but rather represent a means to achieve latency in a fraction of cells during active infection (6). Another group also supported the idea of an unconventional latent form of HHV-6 (23, 24). By definition, viral latency is a phase in the virus's life cycle in which, after initial infection, virus production ceases. In latency, the viral genome is not eradicated, and the virus can reactivate and begin producing new virions without the host being de novo infected. As presented in Fig. 2, integration is not without consequences for the virus, since sequences at the extremities are lost upon integration. This has been verified experimentally by sequencing the cellular-viral junctions, which revealed a 79-nucleotide deletion at the far right extremity of the HHV-6 genome (6). Whether CIHHV-6 can reactivate and cause secondary infections does not meet consensus in the literature. To demonstrate reactivation from an integrated virus, one first needs to exclude the presence of nonintegrated viral episomes. The absence or presence of viral episomes is generally determined using Gardella gel analysis (34) or accomplished by PCR, using primers positioned within the DR regions such that successful amplification occurs only when the viral DNA circularizes and joins the genomic extremities together. Event though PCR is more sensitive that Gardella gels, proving that viral episomes are absent remains a difficult and delicate task. Second, one needs to be able to transmit infection from CIHHV-6-carrying cells to uninfected cells, and third, to demonstrate that the newly infecting virus matches the one integrated. Attempts by several groups have failed at demonstrating HHV-6 reactivation from CIHHV-6-infected cells (7, 14, 21, 24). However, in a recent paper, Arbuckle et al. suggest that reactivation does occur, since treating T cells from individuals carrying CIHHV-6 with the histone deacetylase inhibitor trichostatin A results in a 1.5- to 2-fold increase in the copy numbers of HHV-6A (6). Furthermore, HHV-6 can be transmitted from activated primary T cells of CIHHV-6-positive donors to Molt3 cells. We feel that without additional experimental data, the conclusion that CIHHV-6 can reactivate should be interpreted with caution for the following reasons. First, although U94-sequencing results were provided to support that the reactivated virus infecting Molt3 cells was the same as the one integrated into the primary T cells, there were several divergent nucleotides between the reactivated HHV-6 and the one integrated, making it difficult to draw firm conclusions; second, several nucleotide polymorphisms between CIHHV-6 from the same family members were also observed; third, the reactivated virus is presumed to be an A variant, but HHV-6A is well known for being unable to infect Molt3 cells (1), raising the possibility that nonintegrated HHV-6B was rescued. The choice of U94 to confirm the source of the reactivated HHV-6 is also somewhat questionable, as this gene is among the most conserved (>95%) between the two variants. There are several HHV-6 variant discriminatory genes, such as the gH, gB, or U90 genes, that would have unquestionably identified the source of the reactivated HHV-6 transmitted to Molt3 cells (2, 32, 46). More studies will be necessary to confirm CIHHV-6 reactivation and to elucidate the associated mechanisms. If chromosomal integration represents an unusual latent mechanism for HHV-6, the virus has little interest in integrating in actively transcribed genomic regions. In that regard, telomeres would represent adequate regions. Regardless of whether HHV-6 can reactivate or not, the virus can certainly express some of its genes once integrated. In fact, there have been reports that expression of several HHV-6 genes covering all kinetic classes from immediate-early to late genes could be measured, while protein expression was either not detected or not determined (18, 25).
CONSEQUENCES OF CIHHV-6 FOR DIAGNOSIS
Clark and Ward have written an excellent review on the consequences of CIHHV-6 for diagnosis (19). Perhaps the most frequent consequence of CIHHV-6 is the wrongful diagnosis of active HHV-6 infection. Typically, HHV-6 DNA is not present in the serum and/or plasma of healthy non-CIHHV-6 individuals (83). Thus, when HHV-6 DNA is detected in plasma or serum, one might assume that there is active viral replication. However, for CIHHV-6 individuals whose cells contain at least one copy of the HHV-6 genome/cell, loads of HHV-6 DNA of 104 to 105copies/ml of plasma are detected. The simple act of drawing blood causes a certain amount of cellular lysis. The inability of practitioners to distinguish bona fide viral loads originating from active viral reactivation from high DNA loads due to CIHHV-6 is probably the first established consequence of viral integration. CIHHV-6 patients have high DNA loads (and not high viral loads) that are unaffected by antiviral medications (14, 42). This misdiagnosis results in patients receiving unnecessary treatments, and antiviral therapies can have potentially serious consequences for some patients (e.g., bone marrow transplant recipients). When high DNA loads in plasma and leukocytes of cerebrospinal fluids are detected, hair follicle DNA should be tested to determine whether patients carry CIHHV-6 (19).
TRANSMISSION OF CIHHV-6 THROUGH TRANSPLANTATION
Several case reports have shown the possibility of CIHHV-6 transmission during transplants of hematopoietic and cord blood cells from healthy CIHHV-6-positive donors to transplant recipients (17, 30, 48, 49). Except for the increase in HHV-6 DNA load in the blood and the possibility of misdiagnosis confusion during monitoring of the recipient (for CIHHV-6 and HHV-6 reactivation related to the immunosuppression therapy), the impacts of this kind of acquisition, if there are any, are still unknown. On the other hand, transfer of hematopoietic stem cells from a healthy donor to a CIHHV-6-positive recipient may lead to the complete elimination of HHV-6 DNA from the leukocyte pool (44). At present, cord blood and banked tissues are not screened for CIHHV-6. Considering that the possibility that CIHHV-6 might reactivate or be associated with chromosomal or genetic anomalies might exist, should blood/tissue/hematopoietic stem cell banks be screened for CIHHV-6? In our opinion and considering the current body of knowledge (or lack thereof), we feel that until it is proven that CIHHV-6 does not pose a threat or is not associated with genetic abnormalities, samples positive for CIHHV-6 should be eliminated from biobanks. Considering the prevalence of HHV-6 in the population (nearly 100%), with an estimated 1% of individuals carrying an integrated form, the number of tainted samples is not trivial, and they could easily be identified by a simple quantitative PCR.
CIHHV-6-ASSOCIATED PATHOLOGIES
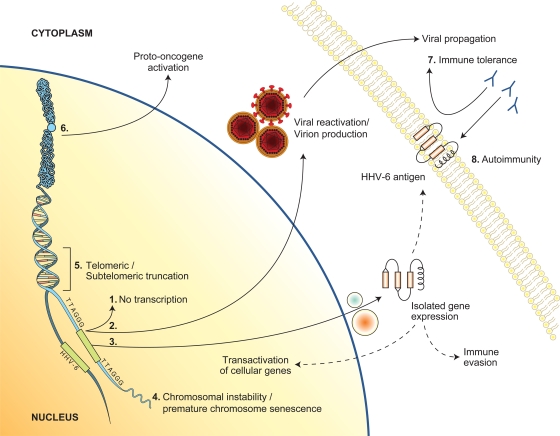
As mentioned previously, no definite pathology is associated with a CIHHV-6 genotype. Interestingly, when cases from the literature are analyzed carefully, some pathologies, like hematologic neoplasia, appear overrepresented. However, it would be premature to conclude that there is a direct association, since many of these studies were done with unhealthy cohorts and detection of CIHHV-6 was not the first objective. In this context, we take the liberty to propose several hypothetical scenarios or pathologies associated with CIHHV-6 (schematically represented in Fig. 3). The telomeric regions, sites of HHV-6 insertion, are not devoid of activities. Not only do they contribute to chromosomal integrity and to cellular homeostasis, but they also encode several transcripts (12, 80). Interestingly, the telomeric regions where the majority of HHV-6 insertions occur are also associated with rare syndromes (e.g., Miller-Dieker syndrome, which is associated with microdeletion and/or LIS-1 gene alteration in chromosome 17p13.3; chromosome 9q subtelomere deletion syndrome; and 18q syndrome, which is associated with an 18q23 deletion [79, 84-86]). CIHHV-6 is likely not responsible for these diseases, but the possibility of similar chromosomal alterations caused by HHV-6 integration in these areas is of interest. Furthermore, a 170-kb insertion (HHV-6 genome size) within the telomeric areas can affect the integrity of telomeres and possibly induce premature chromosome senescence (Fig. 3, pathway 4). Alternatively, could HHV-6 integration cause a truncation in telomeric regions (Fig. 3, pathway 5)? Can CIHHV-6 influence activation of proto-oncogenes (Fig. 3, pathway 6)? We and others (6) think that many important questions related to the immune system should also be considered. For instance, since CIHHV-6 is transmitted from mother to child, can immune tolerance toward certain HHV-6 antigens be induced when and if viral proteins get expressed during embryogenesis (Fig. 3, pathway 7)? Lastly, if genes from CIHHV-6 get expressed in a cell-specific manner, could an autoimmune reaction be induced following the primary infection (Fig. 3, pathway 8)?
FIG. 3.
Schematic representation of the hypothetical cellular consequences associated with CIHHV-6 (see text for details), as follows: (1) no viral gene transcription; (2) viral gene expression, replication, and virion production; (3) expression of a subset of HHV-6 genes; (4 and 5) impact of HHV-6 integration on telomere function, architecture, and chromosome stability; (6) trans and/or cis activation of cellular gene expression following integration; (7) immune tolerance due to expression of HHV-6 genes during embryogenesis; (8) destruction of tissues or cells expressing HHV-6 antigens (from integrated HHV-6) by immune defense mechanisms developed in response to natural HHV-6 infection.
CONCLUSIONS
Chromosomal integration by herpesviruses has been known for many years but has mostly been considered an exception rather than a true mechanism of viral persistence. Considering that chromosomal integration may occur in a fraction of infected cells, if one is not specifically looking for it one might miss it. Detection of in vivo chromosomally integrated HHV is even more challenging except in cases such as that of HHV-6, where the integrated virus can be transmitted through the gametes, generating offspring with a viral copy in every cell. Furthermore, scientific reports addressing the consequences of herpesvirus integration on cellular functions are very scarce, and this review certainly raises more questions than it answers. However, that chromosomal integration by certain herpesviruses does occur is a certainty. Whether it represents a bona fide viral persistence mode or a viral dead end is yet to be resolved, but it constitutes one of the challenges that need to be addressed by virologists for a more complete understanding of the biology of these complex viruses.
Footnotes
Published ahead of print on 15 September 2010.
REFERENCES
- 1.Ablashi, D. V., N. Balachandran, S. F. Josephs, C. L. Hung, G. R. Krueger, B. Kramarsky, S. Z. Salahuddin, and R. C. Gallo. 1991. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology 184:545-552. [DOI] [PubMed] [Google Scholar]
- 2.Achour, A., I. Malet, F. Le Gal, A. Dehee, A. Gautheret-Dejean, P. Bonnafous, and H. Agut. 2008. Variability of gB and gH genes of human herpesvirus-6 among clinical specimens. J. Med. Virol. 80:1211-1221. [DOI] [PubMed] [Google Scholar]
- 3.Adams, A., T. Lindahl, and G. Klein. 1973. Linear association between cellular DNA and Epstein-Barr virus DNA in a human lymphoblastoid cell line. Proc. Natl. Acad. Sci. U. S. A. 70:2888-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersson-Anvret, M., and T. Lindahl. 1978. Integrated viral DNA sequences in Epstein-Barr virus-converted human lymphoma lines. J. Virol. 25:710-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anvret, M., A. Karlsson, and G. Bjursell. 1984. Evidence for integrated EBV genomes in Raji cellular DNA. Nucleic Acids Res. 12:1149-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arbuckle, J. H., M. M. Medveczky, J. Luka, S. H. Hadley, A. Luegmayr, D. Ablashi, T. Lund, J. Tolar, K. DeMeirleir, J. G. Montoya, A. L. Komaroff, P. F. Ambros, and P. G. Medveczky. 2010. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 107:5563-5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bandobashi, K., M. Daibata, M. Kamioka, Y. Tanaka, I. Kubonishi, H. Taguchi, Y. Ohtsuki, and I. Miyoshi. 1997. Human herpesvirus 6 (HHV-6)-positive Burkitt's lymphoma: establishment of a novel cell line infected with HHV-6. Blood 90:1200-1207. [PubMed] [Google Scholar]
- 8.Bates, M., M. Monze, H. Bima, M. Kapambwe, D. Clark, F. C. Kasolo, and U. A. Gompels. 2009. Predominant human herpesvirus 6 variant A infant infections in an HIV-1 endemic region of sub-Saharan Africa. J. Med. Virol. 81:779-789. [DOI] [PubMed] [Google Scholar]
- 9.Bernasconi, M., C. Berger, J. A. Sigrist, A. Bonanomi, J. Sobek, F. K. Niggli, and D. Nadal. 2006. Quantitative profiling of housekeeping and Epstein-Barr virus gene transcription in Burkitt lymphoma cell lines using an oligonucleotide microarray. Virol. J. 3:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biegeleisen, K., and M. G. Rush. 1976. Association of herpes simplex virus type 1 DNA with host chromosomal DNA during productive infection. Virology 69:246-257. [DOI] [PubMed] [Google Scholar]
- 11.Biegeleisen, K., K. Yanagi, and M. G. Rush. 1977. Further studies on the association of herpes simplex virus type 1 DNA with host DNA during productive infection. Virology 83:221-225. [DOI] [PubMed] [Google Scholar]
- 12.Calado, R. T., and N. S. Young. 2009. Telomere diseases. N. Engl. J. Med. 361:2353-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caselli, E., A. Bracci, M. Galvan, M. Boni, A. Rotola, C. Bergamini, C. Cermelli, P. Dal Monte, U. A. Gompels, E. Cassai, and D. Di Luca. 2006. Human herpesvirus 6 (HHV-6) U94/REP protein inhibits betaherpesvirus replication. Virology 346:402-414. [DOI] [PubMed] [Google Scholar]
- 14.Caserta, M. T., C. B. Hall, K. Schnabel, G. Lofthus, A. Marino, L. Shelley, C. Yoo, J. Carnahan, L. Anderson, and H. Wang. 2010. Diagnostic assays for active infection with human herpesvirus 6 (HHV-6). J. Clin. Virol. 48:55-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cebrian, J., C. Kaschka-Dierich, N. Berthelot, and P. Sheldrick. 1982. Inverted repeat nucleotide sequences in the genomes of Marek disease virus and the herpesvirus of the turkey. Proc. Natl. Acad. Sci. U. S. A. 79:555-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang, Y., S. D. Cheng, and C. H. Tsai. 2002. Chromosomal integration of Epstein-Barr virus genomes in nasopharyngeal carcinoma cells. Head Neck 24:143-150. [DOI] [PubMed] [Google Scholar]
- 17.Clark, D. A., E. P. Nacheva, H. N. Leong, D. Brazma, Y. T. Li, E. H. Tsao, H. C. Buyck, C. E. Atkinson, H. M. Lawson, M. N. Potter, and P. D. Griffiths. 2006. Transmission of integrated human herpesvirus 6 through stem cell transplantation: implications for laboratory diagnosis. J. Infect. Dis. 193:912-916. [DOI] [PubMed] [Google Scholar]
- 18.Clark, D. A., E. H. Tsao, H. N. Leong, K. N. Ward, E. P. Nacheva, and P. D. Griffiths. 2006. Reply to Boutolleau et al. and Luppi et al. J. Infect. Dis. 194:1021-1023. [Google Scholar]
- 19.Clark, D. A., and K. N. Ward. 2008. Importance of chromosomally integrated HHV-6A and -6B in the diagnosis of active HHV-6 infection. Herpes 15:28-32. [PubMed] [Google Scholar]
- 20.Daibata, M., T. Taguchi, M. Kamioka, I. Kubonishi, H. Taguchi, and I. Miyoshi. 1998. Identification of integrated human herpesvirus 6 DNA in early pre-B cell acute lymphoblastic leukemia. Leukemia 12:1002-1004. [DOI] [PubMed] [Google Scholar]
- 21.Daibata, M., T. Taguchi, I. Kubonishi, H. Taguchi, and I. Miyoshi. 1998. Lymphoblastoid cell lines with integrated human herpesvirus type 6. J. Hum. Virol. 1:475-481. [PubMed] [Google Scholar]
- 22.Daibata, M., T. Taguchi, K. Miyoshi, H. Taguchi, and I. Miyoshi. 2000. Presence of human herpesvirus 6 DNA in somatic cells. Blood 95:1108-1109.10691335 [Google Scholar]
- 23.Daibata, M., T. Taguchi, Y. Nemoto, H. Taguchi, and I. Miyoshi. 1999. Inheritance of chromosomally integrated human herpesvirus 6 DNA. Blood 94:1545-1549. [PubMed] [Google Scholar]
- 24.Daibata, M., T. Taguchi, T. Sawada, H. Taguchi, and I. Miyoshi. 1998. Chromosomal transmission of human herpesvirus 6 DNA in acute lymphoblastic leukaemia. Lancet 352:543-544. [DOI] [PubMed] [Google Scholar]
- 25.Daibata, M., T. Taguchi, H. Taguchi, and I. Miyoshi. 1998. Integration of human herpesvirus 6 in a Burkitt's lymphoma cell line. Br. J. Haematol. 102:1307-1313. [DOI] [PubMed] [Google Scholar]
- 26.Delecluse, H. J., S. Bartnizke, W. Hammerschmidt, J. Bullerdiek, and G. W. Bornkamm. 1993. Episomal and integrated copies of Epstein-Barr virus coexist in Burkitt lymphoma cell lines. J. Virol. 67:1292-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delecluse, H. J., and W. Hammerschmidt. 1993. Status of Marek's disease virus in established lymphoma cell lines: herpesvirus integration is common. J. Virol. 67:82-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delecluse, H. J., S. Schuller, and W. Hammerschmidt. 1993. Latent Marek's disease virus can be activated from its chromosomally integrated state in herpesvirus-transformed lymphoma cells. EMBO J. 12:3277-3286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng, H., and S. Dewhurst. 1998. Functional identification and analysis of cis-acting sequences which mediate genome cleavage and packaging in human herpesvirus 6. J. Virol. 72:320-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Pagter, P. J., A. Virgili, E. Nacheva, D. van Baarle, R. Schuurman, and J. J. Boelens. 2010. Chromosomally integrated human herpesvirus 6: transmission via cord blood-derived unrelated hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 16:130-132. [DOI] [PubMed] [Google Scholar]
- 31.Dhepakson, P., Y. Mori, Y. B. Jiang, H. L. Huang, P. Akkapaiboon, T. Okuno, and K. Yamanishi. 2002. Human herpesvirus-6 rep/U94 gene product has single-stranded DNA-binding activity. J. Gen. Virol. 83:847-854. [DOI] [PubMed] [Google Scholar]
- 32.Dominguez, G., T. R. Dambaugh, F. R. Stamey, S. Dewhurst, N. Inoue, and P. E. Pellett. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J. Virol. 73:8040-8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fragnet, L., M. A. Blasco, W. Klapper, and D. Rasschaert. 2003. The RNA subunit of telomerase is encoded by Marek's disease virus. J. Virol. 77:5985-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardella, T., P. Medveczky, T. Sairenji, and C. Mulder. 1984. Detection of circular and linear herpesvirus DNA molecules in mammalian cells by gel electrophoresis. J. Virol. 50:248-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gompels, U. A., and H. A. Macaulay. 1995. Characterization of human telomeric repeat sequences from human herpesvirus 6 and relationship to replication. J. Gen. Virol. 76:451-458. [DOI] [PubMed] [Google Scholar]
- 36.Gompels, U. A., J. Nicholas, G. Lawrence, M. Jones, B. J. Thomson, M. E. Martin, S. Efstathiou, M. Craxton, and H. A. Macaulay. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209:29-51. [DOI] [PubMed] [Google Scholar]
- 37.Hall, C. B., M. T. Caserta, K. Schnabel, L. M. Shelley, A. S. Marino, J. A. Carnahan, C. Yoo, G. K. Lofthus, and M. P. McDermott. 2008. Chromosomal integration of human herpesvirus 6 is the major mode of congenital human herpesvirus 6 infection. Pediatrics 122:513-520. [DOI] [PubMed] [Google Scholar]
- 38.Hall, C. B., M. T. Caserta, K. C. Schnabel, C. Boettrich, M. P. McDermott, G. K. Lofthus, J. A. Carnahan, and S. Dewhurst. 2004. Congenital infections with human herpesvirus 6 (HHV6) and human herpesvirus 7 (HHV7). J. Pediatr. 145:472-477. [DOI] [PubMed] [Google Scholar]
- 39.Hall, C. B., M. T. Caserta, K. C. Schnabel, L. M. Shelley, J. A. Carnahan, A. S. Marino, C. Yoo, and G. K. Lofthus. 2010. Transplacental congenital human herpesvirus 6 infection caused by maternal chromosomally integrated virus. J. Infect. Dis. 201:505-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henderson, A., S. Ripley, M. Heller, and E. Kieff. 1983. Chromosome site for Epstein-Barr virus DNA in a Burkitt tumor cell line and in lymphocytes growth-transformed in vitro. Proc. Natl. Acad. Sci. U. S. A. 80:1987-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hermouet, S., and S. Minvielle. 2000. Inheritance of chromosomally integrated viral DNA? Blood 95:1108-1109. [PubMed] [Google Scholar]
- 42.Hubacek, P., J. Maalouf, M. Zajickova, M. Kouba, O. Cinek, K. Hyncicova, I. Fales, and P. Cetkovsky. 2007. Failure of multiple antivirals to affect high HHV-6 DNAaemia resulting from viral chromosomal integration in case of severe aplastic anaemia. Haematologica 92:e98-e100. [DOI] [PubMed] [Google Scholar]
- 43.Hubacek, P., K. Muzikova, A. Hrdlickova, O. Cinek, K. Hyncicova, H. Hrstkova, P. Sedlacek, and J. Stary. 2009. Prevalence of HHV-6 integrated chromosomally among children treated for acute lymphoblastic or myeloid leukemia in the Czech Republic. J. Med. Virol. 81:258-263. [DOI] [PubMed] [Google Scholar]
- 44.Hubacek, P., A. Virgili, K. N. Ward, D. Pohlreich, P. Keslova, B. Goldova, M. Markova, M. Zajac, O. Cinek, E. P. Nacheva, P. Sedlacek, and P. Cetkovsky. 2009. HHV-6 DNA throughout the tissues of two stem cell transplant patients with chromosomally integrated HHV-6 and fatal CMV pneumonitis. Br. J. Haematol. 145:394-398. [DOI] [PubMed] [Google Scholar]
- 45.Hurley, E. A., S. Agger, J. A. McNeil, J. B. Lawrence, A. Calendar, G. Lenoir, and D. A. Thorley-Lawson. 1991. When Epstein-Barr virus persistently infects B-cell lines, it frequently integrates. J. Virol. 65:1245-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Isegawa, Y., T. Mukai, K. Nakano, M. Kagawa, J. Chen, Y. Mori, T. Sunagawa, K. Kawanishi, J. Sashihara, A. Hata, P. Zou, H. Kosuge, and K. Yamanishi. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J. Virol. 73:8053-8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeulin, H., M. Guery, L. Clement, A. Salmon, M. Beri, P. Bordigoni, and V. Venard. 2009. Chromosomally integrated HHV-6: slow decrease of HHV-6 viral load after hematopoietic stem-cell transplantation. Transplantation 88:1142-1143. [DOI] [PubMed] [Google Scholar]
- 48.Jeulin, H., A. Salmon, A. Gautheret-Dejean, H. Agut, P. Bordigoni, B. Fortier, and V. Venard. 2009. Contribution of human herpesvirus 6 (HHV-6) viral load in whole blood and serum to investigate integrated HHV-6 transmission after haematopoietic stem cell transplantation. J. Clin. Virol. 45:43-46. [DOI] [PubMed] [Google Scholar]
- 49.Kamble, R. T., D. A. Clark, H. N. Leong, H. E. Heslop, M. K. Brenner, and G. Carrum. 2007. Transmission of integrated human herpesvirus-6 in allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 40:563-566. [DOI] [PubMed] [Google Scholar]
- 50.Kaschka-Dierich, C., L. Falk, G. Bjursell, A. Adams, and T. Lindahl. 1977. Human lymphoblastoid cell lines derived from individuals without lymphoproliferative disease contain the same latent forms of Epstein-Barr virus DNA as those found in tumor cells. Int. J. Cancer 20:173-180. [DOI] [PubMed] [Google Scholar]
- 51.King, W., A. L. Thomas-Powell, N. Raab-Traub, M. Hawke, and E. Kieff. 1980. Epstein-Barr virus RNA. V. Viral RNA in a restringently infected, growth-transformed cell line. J. Virol. 36:506-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kishi, M., G. Bradley, J. Jessip, A. Tanaka, and M. Nonoyama. 1991. Inverted repeat regions of Marek's disease virus DNA possess a structure similar to that of the a sequence of herpes simplex virus DNA and contain host cell telomere sequences. J. Virol. 65:2791-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klein, G., and L. Dombos. 1973. Relationship between the sensitivity of EBV-carrying lymphoblastoid lines to superinfection and the inducibility of the resident viral genome. Int. J. Cancer 11:327-337. [DOI] [PubMed] [Google Scholar]
- 54.Koliais, S. I. 1979. Mode of integration of Epstein-Barr virus genome into host DNA in Burkitt lymphoma cells. J. Gen. Virol. 44:573-576. [DOI] [PubMed] [Google Scholar]
- 55.Kondo, K., T. Kondo, T. Okuno, M. Takahashi, and K. Yamanishi. 1991. Latent human herpesvirus 6 infection of human monocytes/macrophages. J. Gen. Virol. 72:1401-1408. [DOI] [PubMed] [Google Scholar]
- 56.Kondo, K., T. Kondo, K. Shimada, K. Amo, H. Miyagawa, and K. Yamanishi. 2002. Strong interaction between human herpesvirus 6 and peripheral blood monocytes/macrophages during acute infection. J. Med. Virol. 67:364-369. [DOI] [PubMed] [Google Scholar]
- 57.Kripalani-Joshi, S., and H. Y. Law. 1994. Identification of integrated Epstein-Barr virus in nasopharyngeal carcinoma using pulse field gel electrophoresis. Int. J. Cancer 56:187-192. [DOI] [PubMed] [Google Scholar]
- 58.Kutok, J. L., and F. Wang. 2006. Spectrum of Epstein-Barr virus-associated diseases. Annu. Rev. Pathol. 1:375-404. [DOI] [PubMed] [Google Scholar]
- 59.Leong, H. N., P. W. Tuke, R. S. Tedder, A. B. Khanom, R. P. Eglin, C. E. Atkinson, K. N. Ward, P. D. Griffiths, and D. A. Clark. 2007. The prevalence of chromosomally integrated human herpesvirus 6 genomes in the blood of UK blood donors. J. Med. Virol. 79:45-51. [DOI] [PubMed] [Google Scholar]
- 60.Lestou, V. S., M. De Braekeleer, S. Strehl, G. Ott, H. Gadner, and P. F. Ambros. 1993. Non-random integration of Epstein-Barr virus in lymphoblastoid cell lines. Genes Chromosomes Cancer 8:38-48. [DOI] [PubMed] [Google Scholar]
- 61.Linden, R. M., P. Ward, C. Giraud, E. Winocour, and K. I. Berns. 1996. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. U. S. A. 93:11288-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Linden, R. M., E. Winocour, and K. I. Berns. 1996. The recombination signals for adeno-associated virus site-specific integration. Proc. Natl. Acad. Sci. U. S. A. 93:7966-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lohi, O., M. Arola, I. Lautenschlager, E. P. Nacheva, and K. Vettenranta. 29 June 2010. A high circulating copy number of HHV-6 due to chromosomal integration in a child with acute lymphoblastic leukemia. Pediatr. Blood Cancer [Epub ahead of print.] doi: 10.1002/pbc.22671. [DOI] [PubMed]
- 64.Luo, W. J., T. Takakuwa, M. F. Ham, N. Wada, A. Liu, S. Fujita, E. Sakane-Ishikawa, and K. Aozasa. 2004. Epstein-Barr virus is integrated between REL and BCL-11A in American Burkitt lymphoma cell line (NAB-2). Lab. Invest. 84:1193-1199. [DOI] [PubMed] [Google Scholar]
- 65.Luppi, M., P. Barozzi, R. Bosco, D. Vallerini, L. Potenza, F. Forghieri, and G. Torelli. 2006. Human herpesvirus 6 latency characterized by high viral load: chromosomal integration in many, but not all, cells. J. Infect. Dis. 194:1020-1021. [DOI] [PubMed] [Google Scholar]
- 66.Luppi, M., P. Barozzi, R. Marasca, and G. Torelli. 1994. Integration of human herpesvirus-6 (HHV-6) genome in chromosome 17 in two lymphoma patients. Leukemia 8(Suppl 1):S41-S45. [PubMed] [Google Scholar]
- 67.Luppi, M., P. Barozzi, C. M. Morris, E. Merelli, and G. Torelli. 1998. Integration of human herpesvirus 6 genome in human chromosomes. Lancet 352:1707-1708. [DOI] [PubMed] [Google Scholar]
- 68.Luppi, M., R. Marasca, P. Barozzi, S. Ferrari, L. Ceccherini-Nelli, G. Batoni, E. Merelli, and G. Torelli. 1993. Three cases of human herpesvirus-6 latent infection: integration of viral genome in peripheral blood mononuclear cell DNA. J. Med. Virol. 40:44-52. [DOI] [PubMed] [Google Scholar]
- 69.Martin, M. E., B. J. Thomson, R. W. Honess, M. A. Craxton, U. A. Gompels, M. Y. Liu, E. Littler, J. R. Arrand, I. Teo, and M. D. Jones. 1991. The genome of human herpesvirus 6: maps of unit-length and concatemeric genomes for nine restriction endonucleases. J. Gen. Virol. 72:157-168. [DOI] [PubMed] [Google Scholar]
- 70.Matsuo, T., M. Heller, L. Petti, E. O'Shiro, and E. Kieff. 1984. Persistence of the entire Epstein-Barr virus genome integrated into human lymphocyte DNA. Science 226:1322-1325. [DOI] [PubMed] [Google Scholar]
- 71.Moore, D. F., and D. T. Kingsbury. 1980. Integration and transcription of virus DNA in herpes simplex virus transformed cell lines. J. Gen. Virol. 48:123-133. [DOI] [PubMed] [Google Scholar]
- 72.Mori, T., K. Tanaka-Taya, H. Satoh, Y. Aisa, R. Yamazaki, J. Kato, Y. Ikeda, and S. Okamoto. 2009. Transmission of chromosomally integrated human herpesvirus 6 (HHV-6) variant A from a parent to children leading to misdiagnosis of active HHV-6 infection. Transpl. Infect. Dis. 11:503-506. [DOI] [PubMed] [Google Scholar]
- 73.Mori, Y., P. Dhepakson, T. Shimamoto, K. Ueda, Y. Gomi, H. Tani, Y. Matsuura, and K. Yamanishi. 2000. Expression of human herpesvirus 6B rep within infected cells and binding of its gene product to the TATA-binding protein in vitro and in vivo. J. Virol. 74:6096-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morris, C., M. Luppi, M. McDonald, P. Barozzi, and G. Torelli. 1999. Fine mapping of an apparently targeted latent human herpesvirus type 6 integration site in chromosome band 17p13.3. J. Med. Virol. 58:69-75. [DOI] [PubMed] [Google Scholar]
- 75.Nacheva, E. P., K. N. Ward, D. Brazma, A. Virgili, J. Howard, H. N. Leong, and D. A. Clark. 2008. Human herpesvirus 6 integrates within telomeric regions as evidenced by five different chromosomal sites. J. Med. Virol. 80:1952-1958. [DOI] [PubMed] [Google Scholar]
- 76.Osterhage, J. L., and K. L. Friedman. 2009. Chromosome end maintenance by telomerase. J. Biol. Chem. 284:16061-16065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Padgett, R. A., D. F. Moore, and D. T. Kingsbury. 1978. Herpes simplex virus nucleic acid synthesis following infection of non-permissive XC cells. J. Gen. Virol. 40:605-614. [DOI] [PubMed] [Google Scholar]
- 78.Potenza, L., P. Barozzi, M. Masetti, M. Pecorari, P. Bresciani, A. Gautheret-Dejean, G. Riva, D. Vallerini, S. Tagliazucchi, M. Codeluppi, F. Di Benedetto, G. E. Gerunda, F. Narni, G. Torelli, and M. Luppi. 2009. Prevalence of human herpesvirus-6 chromosomal integration (CIHHV-6) in Italian solid organ and allogeneic stem cell transplant patients. Am. J. Transplant. 9:1690-1697. [DOI] [PubMed] [Google Scholar]
- 79.Reiner, O., R. Carrozzo, Y. Shen, M. Wehnert, F. Faustinella, W. B. Dobyns, C. T. Caskey, and D. H. Ledbetter. 1993. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature 364:717-721. [DOI] [PubMed] [Google Scholar]
- 80.Riethman, H. 2008. Human telomere structure and biology. Annu. Rev. Genomics Hum. Genet. 9:1-19. [DOI] [PubMed] [Google Scholar]
- 81.Robinson, R. A., and D. J. O'Callaghan. 1983. A specific viral DNA sequence is stably integrated in herpesvirus oncogenically transformed cells. Cell 32:569-578. [DOI] [PubMed] [Google Scholar]
- 82.Rotola, A., T. Ravaioli, A. Gonelli, S. Dewhurst, E. Cassai, and D. Di Luca. 1998. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc. Natl. Acad. Sci. U. S. A. 95:13911-13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Secchiero, P., D. R. Carrigan, Y. Asano, L. Benedetti, R. W. Crowley, A. L. Komaroff, R. C. Gallo, and P. Lusso. 1995. Detection of human herpesvirus 6 in plasma of children with primary infection and immunosuppressed patients by polymerase chain reaction. J. Infect. Dis. 171:273-280. [DOI] [PubMed] [Google Scholar]
- 84.Singh, R., R. J. Gardner, K. M. Crossland, I. E. Scheffer, and S. F. Berkovic. 2002. Chromosomal abnormalities and epilepsy: a review for clinicians and gene hunters. Epilepsia 43:127-140. [DOI] [PubMed] [Google Scholar]
- 85.Stewart, D. R., and T. Kleefstra. 2007. The chromosome 9q subtelomere deletion syndrome. Am. J. Med. Genet. C Semin. Med. Genet. 145C:383-392. [DOI] [PubMed] [Google Scholar]
- 86.Strathdee, G., R. Sutherland, J. J. Jonsson, R. Sataloff, M. Kohonen-Corish, D. Grady, and J. Overhauser. 1997. Molecular characterization of patients with 18q23 deletions. Am. J. Hum. Genet. 60:860-868. [PMC free article] [PubMed] [Google Scholar]
- 87.Sullivan, D. C., S. S. Atherton, G. B. Caughman, J. Staczek, and D. J. O'Callaghan. 1986. Oncogenic transformation of primary hamster embryo cells by equine herpesvirus type 3. Virus Res. 5:201-212. [DOI] [PubMed] [Google Scholar]
- 88.Takakuwa, T., W. J. Luo, M. F. Ham, F. Sakane-Ishikawa, N. Wada, and K. Aozasa. 2004. Integration of Epstein-Barr virus into chromosome 6q15 of Burkitt lymphoma cell line (Raji) induces loss of BACH2 expression. Am. J. Pathol. 164:967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tanaka, A., and M. Nonoyama. 1974. Latent DNA of Epstein-Barr virus: separation from high-molecular-weight cell DNA in a neutral glycerol gradient. Proc. Natl. Acad. Sci. U. S. A. 71:4658-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tanaka-Taya, K., J. Sashihara, H. Kurahashi, K. Amo, H. Miyagawa, K. Kondo, S. Okada, and K. Yamanishi. 2004. Human herpesvirus 6 (HHV-6) is transmitted from parent to child in an integrated form and characterization of cases with chromosomally integrated HHV-6 DNA. J. Med. Virol. 73:465-473. [DOI] [PubMed] [Google Scholar]
- 91.Thomson, B. J., S. Dewhurst, and D. Gray. 1994. Structure and heterogeneity of the a sequences of human herpesvirus 6 strain variants U1102 and Z29 and identification of human telomeric repeat sequences at the genomic termini. J. Virol. 68:3007-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thomson, B. J., S. Efstathiou, and R. W. Honess. 1991. Acquisition of the human adeno-associated virus type-2 rep gene by human herpesvirus type-6. Nature 351:78-80. [DOI] [PubMed] [Google Scholar]
- 93.Thomson, B. J., F. W. Weindler, D. Gray, V. Schwaab, and R. Heilbronn. 1994. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology 204:304-311. [DOI] [PubMed] [Google Scholar]
- 94.Torelli, G., P. Barozzi, R. Marasca, P. Cocconcelli, E. Merelli, L. Ceccherini-Nelli, S. Ferrari, and M. Luppi. 1995. Targeted integration of human herpesvirus 6 in the p arm of chromosome 17 of human peripheral blood mononuclear cells in vivo. J. Med. Virol. 46:178-188. [DOI] [PubMed] [Google Scholar]
- 95.Troy, S. B., B. G. Blackburn, K. Yeom, A. K. Caulfield, M. S. Bhangoo, and J. G. Montoya. 2008. Severe encephalomyelitis in an immunocompetent adult with chromosomally integrated human herpesvirus 6 and clinical response to treatment with foscarnet plus ganciclovir. Clin. Infect. Dis. 47:e93-e96. [DOI] [PubMed] [Google Scholar]
- 96.Vu, T., G. Carrum, G. Hutton, H. E. Heslop, M. K. Brenner, and R. Kamble. 2007. Human herpesvirus-6 encephalitis following allogeneic hematopoietic stem cell transplantation. Bone Marrow Transplant. 39:705-709. [DOI] [PubMed] [Google Scholar]
- 97.Wainwright, M. S., P. L. Martin, R. P. Morse, M. Lacaze, J. M. Provenzale, R. E. Coleman, M. A. Morgan, C. Hulette, J. Kurtzberg, C. Bushnell, L. Epstein, and D. V. Lewis. 2001. Human herpesvirus 6 limbic encephalitis after stem cell transplantation. Ann. Neurol. 50:612-619. [DOI] [PubMed] [Google Scholar]
- 98.Walker, S. L., R. S. Wonderling, and R. A. Owens. 1997. Mutational analysis of the adeno-associated virus Rep68 protein: identification of critical residues necessary for site-specific endonuclease activity. J. Virol. 71:2722-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Walker, S. L., R. S. Wonderling, and R. A. Owens. 1997. Mutational analysis of the adeno-associated virus type 2 Rep68 protein helicase motifs. J. Virol. 71:6996-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang, F., A. Marchini, and E. Kieff. 1991. Epstein-Barr virus (EBV) recombinants: use of positive selection markers to rescue mutants in EBV-negative B-lymphoma cells. J. Virol. 65:1701-1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ward, K. N., H. N. Leong, E. P. Nacheva, J. Howard, C. E. Atkinson, N. W. Davies, P. D. Griffiths, and D. A. Clark. 2006. Human herpesvirus 6 chromosomal integration in immunocompetent patients results in high levels of viral DNA in blood, sera, and hair follicles. J. Clin. Microbiol. 44:1571-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ward, K. N., H. N. Leong, A. D. Thiruchelvam, C. E. Atkinson, and D. A. Clark. 2007. Human herpesvirus 6 DNA levels in cerebrospinal fluid due to primary infection differ from those due to chromosomal viral integration and have implications for diagnosis of encephalitis. J. Clin. Microbiol. 45:1298-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Watanabe, H., M. Daibata, M. Tohyama, J. Batchelor, K. Hashimoto, and M. Iijima. 2008. Chromosomal integration of human herpesvirus 6 DNA in anticonvulsant hypersensitivity syndrome. Br. J. Dermatol. 158:640-642. [DOI] [PubMed] [Google Scholar]
- 104.Yamamoto, K., F. Mizuno, T. Matsuo, A. Tanaka, M. Nonoyama, and T. Osato. 1978. Epstein-Barr virus and human chromosomes: close association of the resident viral genome and the expression of the virus-determined nuclear antigen (EBNA) with the presence of chromosome 14 in human-mouse hybrid cells. Proc. Natl. Acad. Sci. U. S. A. 75:5155-5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yamanishi, K., T. Okuno, K. Shiraki, M. Takahashi, T. Kondo, Y. Asano, and T. Kurata. 1988. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet i:1065-1067. [DOI] [PubMed] [Google Scholar]
- 106.Yanagi, K., M. G. Rush, and K. Biegeleisen. 1979. Integration of herpes simplex virus type 1 DNA into the DNA of growth-arrested BHK-21 cells. J. Gen. Virol. 44:657-667. [DOI] [PubMed] [Google Scholar]
- 107.Yoshikawa, T., and Y. Asano. 2000. Central nervous system complications in human herpesvirus-6 infection. Brain Dev. 22:307-314. [DOI] [PubMed] [Google Scholar]
- 108.Zerr, D. M. 2006. Human herpesvirus 6 and central nervous system disease in hematopoietic cell transplantation. J. Clin. Virol. 37(Suppl. 1):S52-S56. [DOI] [PubMed] [Google Scholar]