Abstract
Progesterone receptor (PR) and ErbB-2 bidirectional cross talk participates in breast cancer development. Here, we identified a new mechanism of the PR and ErbB-2 interaction involving the PR induction of ErbB-2 nuclear translocation and the assembly of a transcriptional complex in which ErbB-2 acts as a coactivator of Stat3. We also highlighted that the function of ErbB-2 as a Stat3 coactivator drives progestin-induced cyclin D1 promoter activation. Notably, PR is also recruited together with Stat3 and ErbB-2 to the cyclin D1 promoter, unraveling a new and unexpected nonclassical PR genomic mechanism. The assembly of the nuclear Stat3/ErbB-2 transcriptional complex plays a key role in the proliferation of breast tumors with functional PR and ErbB-2. Our findings reveal a novel therapeutic intervention for PR- and ErbB-2-positive breast tumors via the specific blockage of ErbB-2 nuclear translocation.
Progesterone receptor (PR) and the ErbB family of receptor tyrosine kinases are major players in the breast cancer scenario. In its classical mechanism of action, PR acts as a ligand-induced transcription factor. Upon progestin binding, PR translocates to the nucleus and binds to specific progesterone response elements (PREs) in the promoter of target genes (31). In addition to its direct transcriptional effects, PR activates signal transduction pathways in breast cancer cells through a rapid or nongenomic mechanism (5, 22). On the other hand, the ErbB family of membrane receptor tyrosine kinases is composed of four members: epidermal growth factor (EGF) receptor (EGF-R) (ErbB-1), ErbB-2, ErbB-3, and ErbB-4. ErbB ligands include all isoforms of heregulins (HRGs), which bind to ErbB-3 and ErbB-4 and recognize EGF-R and ErbB-2 as coreceptors, and EGF, which binds to EGF-R (33). Upon ligand binding, ErbBs dimerize, and their intrinsic tyrosine kinase activity is stimulated, which leads to the activation of signal transduction pathways that mediate ErbB's proliferative effects. Although ErbB-2 is an orphan receptor, it participates in an extensive network of ligand-induced formation of ErbB dimers. Notably, this dogma of the ErbB-2 mechanism of action has been challenged by the most exciting findings of Wang and coworkers, demonstrating that ErbB-2 migrates to the nuclear compartment, where it binds DNA at specific sequences, which those authors named HER-2-associated sequences (HASs) (35). Through this function as a transcription factor, ErbB-2 modulates the expression of the cyclooxygenase-2 (COX-2) gene (35). The association of ErbB-2 with the COX-2 promoter was detected in breast cancer cell lines overexpressing ErbB-2 as well as in ErbB-2-positive human primary breast tumors (35).
Accumulating findings, including ours, have proven the presence of bidirectional interactions between PR and ErbB signaling pathways in breast cancer. On the one hand, we showed that PR activates the HRG/ErbB-2 pathway (2). On the other hand, we found that HRG induces PR transcriptional activation in breast tumors through a mechanism that requires functional ErbB-2 (16). Notwithstanding all these data, the identity of the common downstream targets of PR and HRG/ErbB-2 remains poorly known. Notably, our work revealed that signal transducer and activator of transcription 3 (Stat3) is indeed a downstream target of both PR and HRG/ErbB-2. First, we demonstrated that progestins induce the transcriptional activation of Stat3 in breast cancer (25). Most recently, we showed that Stat3 is activated by HRG via ErbB-2 and through the co-option of PR function as a signaling molecule (26). Particularly exciting is the fact that Stat3 itself has been found to play a key role in mammary cancer. Within the framework of the evidence revealing the function of ErbB-2 as a transcriptional regulator and of our previous data showing PR modulation of HRG/ErbB-2 signaling and considering on the other hand that Stat3, the nodal convergence point between PR and ErbB-2, acts as a transcription factor, we explored whether progestin induces ErbB-2 nuclear localization and its interaction with Stat3 in breast cancer. Our findings identified a new class of transcriptional complex in which ErbB-2 acts as a coactivator of Stat3 in progestin-induced breast tumor growth.
MATERIALS AND METHODS
Animals and tumors.
Experiments were carried out with female BALB/c mice raised at the Instituto de Biología y Medicina Experimental (IBYME). Animal studies were conducted as described previously (25), in accordance with the highest standards of animal care as outlined by the NIH Guide for the Care and Use of Laboratory Animals (22a), and were approved by the IBYME Animal Research Committee. The C4HD tumor line displays high levels of estrogen receptor (ER) and PR, overexpresses ErbB-2 and ErbB-3, exhibits low ErbB-4 levels, and lacks EGF-R expression (2). This tumor line does not express glucocorticoid receptor (GR) or androgen receptor (AR) (2).
Reagents.
Medroxyprogesterone acetate (MPA) and RU486 were purchased from Sigma-Aldrich (St. Louis, MO). 4-Amino-5-(4-chlorophenyl)-7-(t-butyl) pyrazolo[3,4-d]pyrimidine (PP2), tyrphostin AG825, and Jak inhibitor I were purchased from Calbiochem (San Diego, CA).
Antibodies.
The following antibodies were used for Western blots: phospho-Stat3 (pStat3) (Tyr 705) (B-7), total Stat3 (C-20), phospho-Jak1 (Tyr1022/1023), total Jak1 (HR-785), total Jak2 (C-20), phospho-p42/p44 mitogen-activated protein kinase (MAPK) (E-4), total p42 MAPK (C-14), ErbB-2 (C-18, raised against the C terminus), ErbB-2 (9G6, raised against the N terminus), and phosphotyrosine (PY99), all from Santa Cruz Biotechnology (Santa Cruz, CA); phospho-ErbB2 (Tyr 1221/1222), phospho-ErbB2 (Tyr 877), phospho-Jak2 (Tyr 1007/1008), c-Src, and phospho-Src (Tyr 416), all from Cell Signaling (Beverly, MA); cyclin D1, PR (clone hPRa7), and actin (clone ACTN05), all from Neomarkers (Freemont, CA); β-tubulin, from Sigma-Aldrich; histone H3, from Abcam (Cambridge, MA); phospho-PR (Ser294), from Affinity BioReagents (Rockford, IL); and horseradish peroxidase (HRP)-conjugated secondary antibody, from Vector Laboratories (Burlingame, CA). The antibodies used for immunoprecipitation experiments, chromatin immunoprecipitation (ChIP) assays, and sequential ChIP assays were rabbit polyclonal anti-ErbB-2, anti-Stat3, and anti-PR antibodies (C-18, C-20, and H-190, respectively; from Santa Cruz Biotechnology) and CBP/KAT3A (ab2832), p300 (clone RW128), acetyl histone H4 (06-866), and acetyl histone H3 (06-599) antibodies, from Millipore (Temecula, CA). Rabbit IgG (Sigma-Aldrich) was used as a negative control.
Cell cultures, treatments, and proliferation assays.
Primary cultures of epithelial cells from C4HD tumors were performed as described previously (2). T47D cells were obtained from the American Type Culture Collection, and T47D-Y cells were a generous gift from K. Horwitz (University of Colorado Health Sciences Center, Denver, CO). To evaluate the effects of the pharmacological inhibitors on MPA-induced protein phosphorylation or cyclin D1 expression, cells were starved in serum-free medium for 48 to 72 h and then also preincubated in serum-free medium for 90 min with RU486, PP2, tyrphostin AG825, or Jak inhibitor I before the addition of MPA. Cell proliferation was evaluated by a [3H]thymidine incorporation assay, and cell cycle distribution was analyzed by flow cytometry, as previously described (26), after a 48-h treatment in serum-free medium.
Western blots and immunoprecipitations.
Lysates were prepared from cells subjected to the different treatments, and proteins were subjected to SDS-PAGE as previously described (25). Membranes were immunoblotted with the antibodies detailed in each experiment. When phosphoprotein antibodies were used, filters were reprobed with total protein antibodies. Signal intensities of phospho-ErbB-2 (pErbB-2), pStat3, pSrc, pPR, pJak1, pJak2, and pp42/p44 MAPK bands were analyzed by densitometry and normalized to total protein bands. Similarly, signal intensities of PR, cyclin D1, Stat3, and ErbB-2 bands were normalized to actin or β-tubulin bands. Data analysis showed a significant increase in pErbB-2, pStat3, pp42/p44 MAPK, and pSrc levels by MPA treatment in comparison with untreated cells and a significant inhibition of MPA-induced protein phosphorylation when using the pharmacological inhibitors of ErbB-2 and Stat3 or PR and ErbB-2 small interfering RNAs (siRNAs) (P < 0.001). A similar data analysis showed that compared to control cells, the increase in cyclin D1 levels by MPA treatment from 12 to 72 h was significant, as was the inhibition of MPA effects by ErbB-2 and Stat3 inhibitors and siRNAs (P < 0.001). The NE-PER nuclear and cytoplasmic extraction reagent technique (Pierce Biotechnology) was performed according to the manufacturer's instructions. The use of this technique does not allow one to obtain the cytoplasmic membrane fraction. The nuclear association between ErbB-2 and Stat3 was studied by coimmunoprecipitation experiments using 200 μg of nuclear protein lysates as described previously (26).
Plasmids and transient transfections.
The luciferase reporter plasmid downstream of the cyclin D1 human promoter region (−1745 cyclin D1-luc) and constructs truncated at positions −963, −261, −141 were kindly provided by R. Pestell (Northwestern University Medical School, Chicago, IL). These constructs were generated by the truncation of the 1,745-bp-length promoter in order to sequentially exclude 5′ regions of the promoter. The −963 cyclin D1-luc construct excludes one gamma interferon-activated sequence (GAS) site (position −984), −261 cyclin D1-luc excludes three GAS sites (positions −984, −568, and −475), and −141 cyclin D1-luc excludes four GAS sites (positions −984, −568, −475, and −239). The empty vector pA3 Luc was also provided by R. Pestell. The luciferase reporter plasmid containing four copies of the m67 high-affinity binding site (p4xm67-tk-luc) and the pTATA-tk-Luc reporter lacking the m67 insertion were a gift from J. Darnell (Rockefeller University, New York, NY). The Renilla luciferase expression plasmid RL-CMV was obtained from Promega (Madison, WI). A dominant negative (DN) Stat3 expression vector, Stat3Y705-F, which carries a tyrosine-to-phenylalanine substitution at codon 705 that reduces the phosphorylation on tyrosine of the wild-type (WT) Stat3 protein, therefore inhibiting both the dimerization and DNA binding of Stat3 (6, 7, 18), was kindly provided by J. Darnell. The empty pcDNA3.1 vector was also a gift of J. Darnell. A human wild-type ErbB-2 expression vector (hErbB-2WT) as well as the empty pMe18SM vector were a gift from T. Yamamoto (University of Tokyo, Tokyo, Japan) (1). The green fluorescent protein (GFP)-tagged human ErbB-2 mutant, which lacks the putative nuclear localization signal sequence (676-KRRQQKIRKYTMRR-689), resulting in the sequence of KLM at the deletion junction (hErbB-2ΔNLS), was generously provided by M. C. Hung (University of Texas M. D. Anderson Cancer Center, Houston, TX) (13). The empty vector pEGFP-N1 was obtained from BD Biosciences-Clontech (Palo Alto, CA). The plasmid encoding human wild-type hPR-B was kindly provided by K. Horwitz. The plasmid encoding PR-B engineered to contain a point mutation in a conserved cysteine in the first zinc finger of the DNA binding domain (C587A-PR), which lacks the ability to bind to DNA, was also a gift of K. Horwitz (32). Mutant PR-B engineered to convert three key prolines (P422A, P423A, and P426A) to alanines (PR-BmPro), thus abolishing PR binding to all the SH3 domains and inhibiting the activation of c-Src family tyrosine kinases (5), was generously provided by D. Edwards (Baylor College of Medicine, Houston, TX). In experiments assessing the capacity of MPA to induce the transcriptional activation of Stat3, C4HD and T47D cells were transiently transfected for 48 h with 1 μg of −1745 cyclin D1-luc reporter plasmid or the truncated position −963, −261, and −141 constructs or with 1 μg p4xm67-tk-luc and 10 ng of RL-CMV, used to correct variations in transfection efficiency. As a control, cells were transfected with 1 μg of either the pA3 Luc or pTATA-tk-Luc reporter. Cells were cotransfected with 2 μg of Stat3Y705-F when indicated. The total amount of transfected DNA was standardized by the addition of the empty pcDNA3.1 vector. In experiments assessing the role of ErbB-2 in Stat3 transcriptional activation, cells were cotransfected with 2 μg of hErbB-2WT, hErbB-2ΔNLS, or the empty vectors pMe18SM and pEGFP-N1. Upon cotransfection with p4xm67-tk-luc, 400 ng was added instead of 2 μg. Cells were then starved in serum-free medium for 24 h and treated with MPA for 24 h or were left untreated. Fugene 6 transfection reagent (Roche Biochemicals) was used as described previously (26). Transfection efficiencies, evaluated by using the pEGFP-N1 vector and determined by the percentage of cells that exhibited GFP 4 days after transfection, varied between 60 and 70%. Transfected cells were lysed, and luciferase assays were carried out by using the Dual-Luciferase reporter assay system (Promega) in accordance with the manufacturer's instructions. Triplicate samples were analyzed for each datum point. Differences between experimental groups were analyzed by analysis of variance (ANOVA) followed by a Tukey test between groups.
siRNA transfections.
siRNAs targeting ErbB-2, Stat3, and PR were synthesized by Dharmacon, Inc. (Lafayette, CO) (ErbB-2 siRNA 5′-GAUGGUGCUUACUCAUUGA-3′, designed to specifically knock down mouse ErbB2 but not human ErbB-2; Stat3 siRNAs 5′-GGUCAAAUUUCCUGAGUUGUU-3′, which targets mouse Stat3, and 5′-GAGCAGAGAUGUGGGAAUGUU-3′, which targets human Stat3; and PR siRNA 5′-AUAGGCGAGACUACAGACGUU-3). A nonsilencing siRNA oligonucleotide from Dharmacon that does not target any known mammalian gene was used as a negative control. The transfection of siRNA duplexes was performed for 3 days by using DharmaFECT transfection reagent according to the manufacturer's directions. For reconstitution experiments, the cotransfection of 25 nM ErbB-2 siRNA with 2 μg of expression vectors was performed by using DharmaFECT Duo transfection reagent (Dharmacon).
Immunofluorescence and confocal microscopy.
Cells grown on glass coverslips were fixed and permeabilized in ice-cold methanol and were then blocked with phosphate-buffered saline (PBS)-1% bovine serum albumin (BSA). ErbB-2 was localized by using either a rabbit polyclonal (C-18) or a mouse monoclonal (F-11) ErbB-2 antibody (Santa Cruz Biotechnology), and Stat3 was detected by using a mouse monoclonal antibody (124H6; Cell Signaling), followed by incubation with a goat anti-rabbit IgG-Alexa 488 (Molecular Probes, Eugene, OR) secondary antibody for ErbB-2 (C-18) and with a rhodamine-conjugated goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for both ErbB2 (F-11) and Stat3. Negative controls were carried out by using PBS instead of primary antibodies or 5× competitive peptide (Santa Cruz Biotechnology) when ErbB-2 (C-18) was used. When cells were transfected with hErbB-2ΔNLS, the green fluorescent protein from this expression vector was visualized by direct fluorescence imaging. Approximately 100 to 200 cells were analyzed for each treatment, of which around 80% showed the same pattern of Stat3 and ErbB-2 cellular localization. Figures 2A and 3B and C, as well as Fig. S3B at http://www.ibyme.org.ar/laboratorios/elizalde.htm, illustrate a few cells representative of the ones examined. Cells were analyzed by using a Nikon Eclipse E800 confocal laser microscopy system (26).
ChIP and sequential ChIP assays.
ChIP was performed as described elsewhere previously (15), with minor modifications. Briefly, chromatin was sonicated to an average of about 500 bp. Sonicated chromatin was then immunoprecipitated by using 4 μg of the indicated antibodies and IgG as a control. The immunoprecipitate was collected by using either protein A or G beads (Millipore, Temecula, CA), which were washed repeatedly to remove nonspecific DNA binding. The chromatin was eluted from the beads, and cross-links were removed overnight at 65°C. DNA was then purified and quantified by using real-time PCR. For sequential ChIP experiments, chromatin immunoprecipitates were eluted with dithiothreitol (DTT) and then subjected to a second round of immunoprecipitation with the indicated antibodies or with IgG.
Real-time quantitative PCR.
ChIP DNA was amplified by real-time quantitative PCR (qPCR), performed with an ABI Prism 7500 sequence detector using SYBR green PCR master mix (Applied Biosystems, Foster City, CA). The primers used were as follows: 5′-TTCCGGTGGTCTGGTTCCT-3′ and 5′-GAGACACGATAGGCTCCTTCCTAA-3′, designed to amplify a region of the mouse cyclin D1 promoter containing two GAS sites (positions −971 and −874), and 5′-GGAACCTTCGGTGGTCTTGTC-3′ and 5′-GAATGGAAAGCTGAGAAACAGTGA-3′, designed to amplify a region of the human cyclin D1 promoter containing one GAS site (position −984). These primers were designed with Primer Express real-time PCR primer design software (Applied Biosystems). The primers used to amplify a transcribed region of the cyclin D1 gene (position +8000 in intron 4) were 5′-TGCCACACACCAGTGACTTT-3′ and 5′-ACAGCCAGAAGCTCCAAAAA-3′ (11). PCR was performed for 40 cycles with 15 s of denaturing at 95°C and annealing and extension at 60°C for 1 min.
RNA preparation and real-time RT-PCR.
Total RNA was isolated from C4HD cells treated or transfected as indicated by using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. One microgram of RNA was reverse transcribed by using SuperScript III reverse transcriptase (RT) (Invitrogen) according to the manufacturer's instructions. The following primers were used for mouse cyclin D1 cDNA: 5′-CGCCCTCCGTATCTTACT-3′ and 5′-CGCACTTCTGCTCCTCAC-3′. The primers used to amplify glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a normalization control were 5′-CCAGAACATCATCCCTGCAT-3′ and 5′-GTTCAGCTCTGGGATGACCTT-3′. These primers were designed with Primer Express software (Applied Biosystems), and PCR conditions were the same as those described above. The fold change of mRNA expression was calculated by normalizing the absolute cyclin D1 mRNA amounts to GAPDH mRNA levels, used as an internal control, and setting the value of untreated cells as 1.
In vivo inhibition of ErbB-2 nuclear localization.
C4HD cells were transiently transfected with the siRNAs and expression vectors detailed in Results. After transfection, 106 cells from each experimental group were inoculated subcutaneously (s.c.) into animals treated with a 40-mg MPA depot in the flank opposite of the cell inoculum. Tumor volume, growth rate, and growth delay were determined as previously described (25). Comparison of tumor volumes between the different groups at specific times was done by analysis of variance followed by Tukey's test among groups. Linear regression analysis was performed on tumor growth curves, and the slopes were compared by using analysis of variance followed by a parallelism test to evaluate the statistical significance of differences.
RESULTS
MPA induces rapid ErbB-2 activation and nuclear translocation.
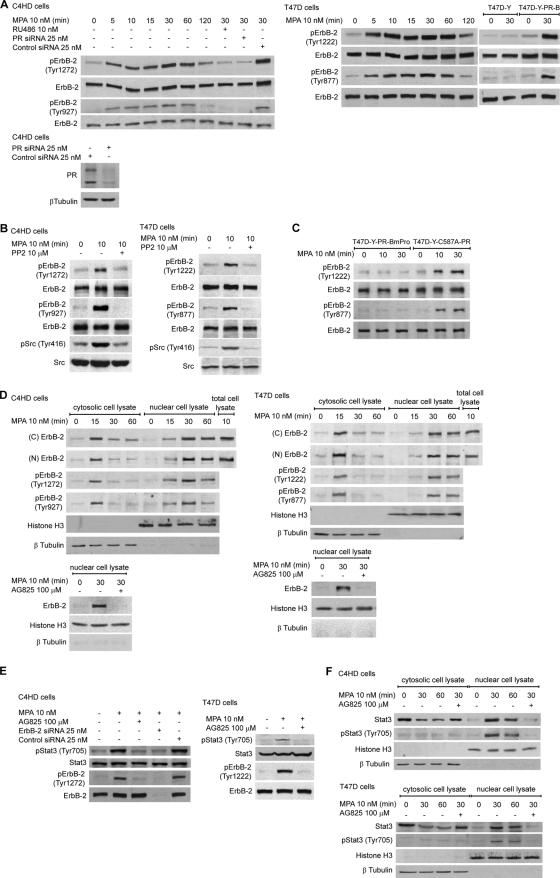
In this study we used primary cultures of C4HD epithelial cells from a model of mammary carcinogenesis induced by the synthetic progestin medroxyprogesterone acetate (MPA) in female BALB/c mice (2) and human breast cancer cell lines. C4HD cells display high levels of estrogen receptor (ER) and PR, overexpress ErbB-2 and ErbB-3, exhibit low ErbB-4 levels, and lack EGF-R expression (2). We have long demonstrated that prolonged MPA treatment of C4HD cells results in the upregulation of ErbB-2 expression as well as in the stimulation of ErbB-2 tyrosine phosphorylation (2). Here, we found that MPA treatment of C4HD cells induces a rapid phosphorylation of a major ErbB-2 autophosphorylation site, tyrosine (Tyr) 1272 (Tyr 1222 in the human protein) as well as of the residue Tyr 927 (Tyr 877 in human), a site different from the autophosphorylation ones (14, 36) (Fig. 1A). MPA effects were inhibited by preincubation with the antiprogestin RU486 (Fig. 1A). The same results were obtained by the knockdown of PR gene expression with PR small interfering RNAs (siRNAs) (Fig. 1A). Our findings with the human breast cancer cell line T47D also evidenced the rapid activation of ErbB-2 by PR (Fig. 1A). In order to further explore the role of PR, we used PR-null T47D cells (T47D-Y), in which we found that MPA had no effect on ErbB-2 phosphorylation at either Tyr 1222 or Tyr 877 (Fig. 1A). However, when we transfected T47D-Y cells with human PR-B (T47D-Y-PR-B), MPA treatment markedly enhanced the ErbB-2 phosphorylation of both residues (Fig. 1A). These results indicate that MPA regulates the rapid activation of ErbB-2 acting through the classical PR. Progestin induction of rapid c-Src activation in mammary tumor cells, including our C4HD tumor model, is well acknowledged (5, 22, 25). On the other hand, a series of recent findings, and ours as well, has shown that c-Src acts as an upstream effector of ErbB-2 (14, 26, 36). Therefore, we explored whether c-Src could be involved in MPA-induced ErbB-2 phosphorylation. We found that the inhibition of c-Src activity in C4HD and T47D cells with the c-Src kinase inhibitor PP2 abrogated MPA stimulation of ErbB-2 phosphorylation at Tyr 1272/1222 and Tyr 927/877 (Fig. 1B). In order to definitely demonstrate that the rapid effects of progestin mediate the activation of ErbB-2, we transfected T47D-Y cells with a mutant, PR-BmPro, in which three prolines (P422A, P423A, and P427A) were converted to alanines (T47D-Y-PR-BmPro cells). Previous works have defined the proline-rich domain of human PR as an absolute requirement for the progestin interaction with c-Src (5) and the consequent rapid activation of signaling cascades (5, 8). Consistent with our result showing that progestin-activated c-Src acts as an upstream activator of ErbB-2, we did not find ErbB-2 tyrosine phosphorylation in response to MPA in T47D-Y-PR-BmPro cells (Fig. 1C). In addition, in T47D-Y cells we restored the expression of a PR-B engineered to contain a point mutation in a conserved cysteine in the first zinc finger of the DNA binding domain (C587A), which is transcriptionally crippled (32). C587A-PR possesses a full ability to induce c-Src, p42/p44 MAPK, and Akt rapid activation in response to progestins, as reported previously by us and others (5, 8). Here, we found that MPA induces strong ErbB-2 phosphorylation in T47D-Y-C587A-PR cells (Fig. 1C). We then assessed whether MPA modulates ErbB-2 cellular localization. Subcellular fractionation and immunoblotting studies, using an antibody to the carboxy (C)-terminal region of ErbB-2, showed that MPA treatment of C4HD and T47D cells for 15 to 60 min induced strong ErbB-2 protein nuclear translocation (Fig. 1D). Similar results were found when we used an antibody against the amino (N) terminus of the receptor (Fig. 1D). Full-length ErbB-2 protein nuclear translocation was shown by the identical molecular mass of nuclear ErbB-2 compared to that of the ErbB-2 present in total cell extracts, corresponding to the entire 185-kDa protein (Fig. 1D), and was also shown by our findings with both the ErbB-2 carboxyl- and amino-terminal antibodies. Interestingly, this is the first report of steroid hormone receptor induction of endogenous ErbB-2 migration to the nucleus. Our findings also showed high levels of nuclear ErbB-2 phosphorylation at Tyr 1272/1222 and Tyr 927/877 in C4HD and T47D cells (Fig. 1D). The preincubation of cells with the specific ErbB-2 tyrosine kinase inhibitor AG825, which prevented MPA-induced ErbB-2 Tyr phosphorylation (see Fig. S1 at http://www.ibyme.org.ar/laboratorios/elizalde.htm), significantly inhibited ErbB-2 migration to the nucleus (Fig. 1D), indicating that ErbB-2 activation is an absolute requirement for this process. Our previous studies demonstrated that MPA induced rapid Stat3 Tyr 705 phosphorylation via a Jak- and c-Src-dependent pathway in breast cancer (25). Here, we found that the blockage of ErbB-2 activity in C4HD and T47D cells and the transfection of C4HD cells with ErbB-2 siRNAs designed to selectively knock down mouse ErbB-2 expression inhibited MPA-induced Stat3 phosphorylation (Fig. 1E), evidencing that ErbB-2 is also involved in MPA-induced Stat3 activation. To assess whether ErbB-2 and Stat3 are simultaneously present in the nucleus, we studied the kinetics of MPA-induced Stat3 nuclear translocation. We found that upon the stimulation of C4HD and T47D cells with MPA for 30 and 60 min, Stat3 is present at the nuclear compartment and is strongly phosphorylated at Tyr 705 (Fig. 1F). The inhibition of Stat3 tyrosine phosphorylation by blocking the activity of its upstream effector ErbB-2 with AG825 absolutely prevented Stat3 nuclear migration (Fig. 1F).
FIG. 1.
MPA effects on ErbB-2 and Stat3 activation and cellular localization. (A) MPA induces rapid ErbB-2 phosphorylation via the classical PR. Cells were treated with MPA or pretreated with RU486 and transfected with PR or control siRNAs before MPA stimulation. Western blots (WB) were performed with pErbB-2 antibodies, and filters were reprobed with a total ErbB-2 antibody. The Western blot of C4HD cells at the bottom shows the effects of siRNAs on PR expression. (B) c-Src mediates MPA-induced ErbB-2 activation. Cells were treated with MPA or preincubated with PP2 before MPA treatment. Western blots were performed with phosphoprotein antibodies, and membranes were reprobed with total protein antibodies. (C) Rapid progestin effects mediate the activation of ErbB-2. T47D-Y cells were transfected with either the PR-BmPro or C587A-PR mutant and were then treated with MPA. Western blots were performed with pErbB-2 antibodies, and filters were reprobed with total ErbB-2 antibody. (D) MPA induces ErbB-2 nuclear migration. (Top) Cells were treated with MPA for the time points shown, and nuclear and cytosolic protein extracts were analyzed by Western blotting. The pTyr 1272/1222 ErbB-2 blot was reprobed with the ErbB-2 carboxy-terminal region antibody, and the pTyr 927/877 blot was reprobed with the antibody to the ErbB-2 amino terminus. Total cell lysates were blotted in parallel. ErbB-2 in untreated cells remained in the cytoplasmic membrane, which was not analyzed in this Western blot (see Fig. 2A for an image of ErbB-2 membrane localization in cells not treated with MPA). (Bottom) Western blot showing that the inhibition of ErbB-2 phosphorylation with AG825 blocks ErbB-2 nuclear migration. Histone H3 and β-tubulin were used to control cellular fractionation efficiency. (E) MPA induces Stat3 activation via ErbB-2. Cells were treated with MPA or pretreated with AG825. C4HD cells were also transfected with ErbB-2 siRNAs targeting mouse ErbB-2 and with control siRNAs. Western blots were performed with phospho-antibodies, and filters were reprobed with the respective total protein antibody. (F) MPA stimulates Stat3 nuclear translocation. Nuclear and cytosolic protein extracts were analyzed by Western blotting with pStat3 antibody. Blots were reprobed with total Stat3 antibody. The experiments for which the results are shown were repeated five times, with similar results.
MPA induces ErbB-2 and Stat3 nuclear colocalization.
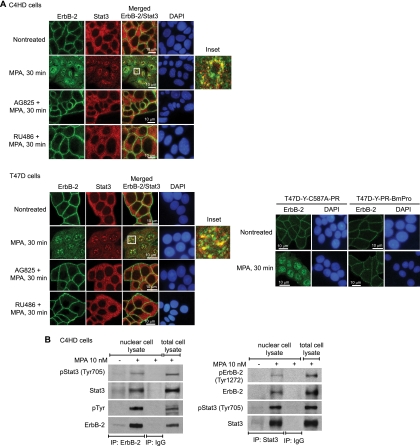
We then explored whether MPA treatment induces the nuclear colocalization of Stat3 and ErbB-2 by immunofluorescence staining and confocal microscopy. In the absence of MPA stimulation, the vast majority of ErbB-2 was localized in the cytoplasmic membrane of C4HD and T47D cells (Fig. 2A). MPA treatment of both cell types for 30 min resulted in ErbB-2 nuclear localization, detected as nuclear green foci (Fig. 2A). These results were obtained with an antibody against the ErbB-2 C terminus. The inhibition of ErbB-2 Tyr 1222/1272 and Tyr 877/927 phosphorylation by AG825 abrogated ErbB-2 nuclear translocation (Fig. 2A), which is consistent with results of our cellular fractionation studies. On the other hand, in the absence of MPA treatment, Stat3 was located diffusely throughout the cytoplasm (Fig. 2A). MPA stimulation induced the nuclear translocation of Stat3 in both cell lines (Fig. 2A). The inhibition of Stat3 tyrosine phosphorylation with AG825 absolutely prevented its nuclear migration (Fig. 2A). As expected, the abolishment of MPA-induced ErbB-2 and Stat3 activation with RU486 resulted in the abrogation of the migration of both proteins to the nucleus (Fig. 2A). Notably, our findings also demonstrated that MPA treatment of C4HD and T47D cells resulted in a strong nuclear colocalization of ErbB-2 and Stat3, as shown by the yellow foci in the merged images (Fig. 2A). Similar nuclear colocalization findings were obtained for T47D cells using an antibody raised against the NH2 terminus of ErbB-2 (data not shown). Significant ErbB-2 and Stat3 nuclear colocalization was also detected with up to 60 min of MPA stimulation (not shown). We did not observe Stat3 and ErbB-2 colocalization in the cytoplasm after MPA treatment for 30 min (Fig. 2A). Since we did not find significant levels of cytoplasmic phosphorylation in either protein at this time point (Fig. 1D and F), our results indicate that ErbB-2 and Stat3 colocalize only when both proteins are phosphorylated. To further demonstrate that PR's rapid, nongenomic activation of ErbB-2 induces its nuclear migration, we explored the ErbB-2 intracellular distribution in T47D-Y-PR-BmPro and T47D-Y-C587A-PR cells. While a clear MPA-stimulated ErbB-2 nuclear localization was detected in T47D-Y-C587A-PR cells, we did not observe ErbB-2 nuclear translocation upon MPA treatment of T47D-Y-PR-BmPro cells (Fig. 2A). The MPA-induced physical association between ErbB-2 and Stat3 in the nucleus was demonstrated through our coimmunoprecipitation studies with nuclear extracts from C4HD cells (Fig. 2B).
FIG. 2.
MPA induces Stat3 and ErbB-2 nuclear colocalization and physical association. (A) Cells were treated with MPA or pretreated with AG825 and RU486 before MPA stimulation. ErbB-2 (green) and Stat3 (red) were localized by immunofluorescence and confocal microscopy (see Materials and Methods for antibody specifications). Merged images in the third panels of the second rows show MPA-induced ErB-2 and Stat3 nuclear colocalization, evidenced by the yellow foci. The boxed areas are shown in detail in the right inset. (Bottom) T47D-Y cells were transfected with either PR-BmPro or C587A-PR mutants and were then treated with MPA. ErbB-2 (green) was localized by immunofluorescence as described above. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). (B) Nuclear extracts from C4HD cells treated and untreated with MPA for 30 min were immunoprecipitated (IP) with ErbB-2 or Stat3 antibodies and analyzed by Western blotting with the indicated phosphotyrosine antibodies. Membranes were reprobed with total protein antibodies. As a control for the specificity of these protein interactions, lysates were immunoprecipitated with rabbit immunoglobulin G (IgG). Total cell lysates were blotted in parallel. The experiments from which the results were obtained were repeated three times, with similar results.
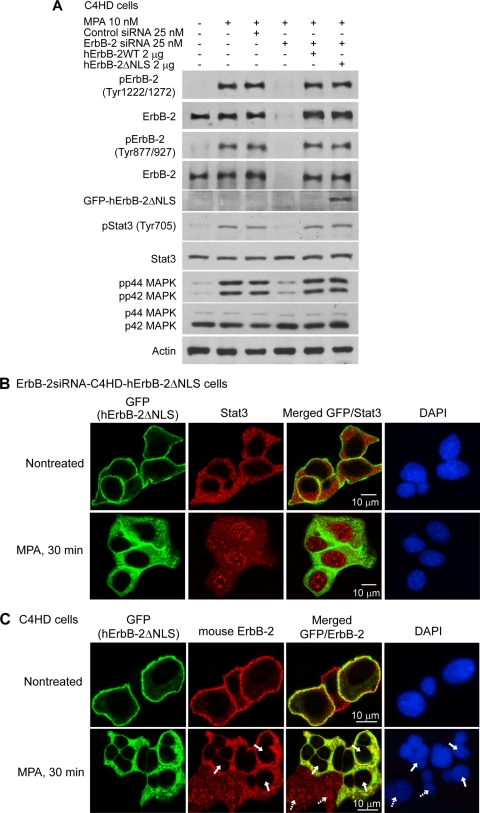
In order to study whether the inhibition of ErbB-2 nuclear localization affected Stat3 transport, we used an RNA interference (RNAi) reconstitution strategy. We transfected C4HD cells with ErbB-2 siRNAs specifically targeting mouse ErbB-2 (see Fig. S2A at http://www.ibyme.org.ar/laboratorios/elizalde.htm) in combination with either wild-type (WT) human ErbB-2 (ErbB-2siRNA-C4HD-hErbB-2WT cells) or a human ErbB-2 nuclear localization domain mutant (hErbB-2ΔNLS) (13), which is unable to translocate to the nucleus (ErbB-2siRNA-C4HD-hErbB-2ΔNLS cells) (see Fig. S2B at http://www.ibyme.org.ar/laboratorios/elizalde.htm). The characterization of the hErbB-2ΔNLS response to MPA showed levels of hErbB-2ΔNLS phosphorylation on Tyr 1222 and Tyr 877 comparable to those of hErbB-2WT and of endogenous ErbB-2 (Fig. 3A). Similarly, hErbB-2ΔNLS induced p42/p44 MAPK activation and Stat3 tyrosine phosphorylation upon MPA stimulation (Fig. 3A). On the one hand, these results indicate that ErbB-2ΔNLS retains its intrinsic tyrosine kinase activity, as described previously (13), as well as the capacity to activate classical ErbB-2 cascades, such as p42/p44 MAPKs, upon the treatment of mammary cancer cells with MPA. On the other hand, they also for the first time identify the role of ErbB-2ΔNLS as an upstream activator in the mechanism of MPA-induced Stat3 phosphorylation. In accordance with the pioneering work describing this mutant (13), our confocal microscopy studies revealed that hErbB-2ΔNLS did not translocate to the nucleus upon MPA treatment of ErbB-2siRNA-C4HD-hErbB-2ΔNLS cells, while a clear MPA-stimulated Stat3 migration to the nuclear compartment was detected in these cells (Fig. 3B). This finding indicates that the nuclear import of Stat3 mediated by MPA occurs independently of ErbB-2 nuclear localization. The merged image of MPA-treated cells, showing a lack of protein colocalization in the cytoplasm (Fig. 3B), further supports our finding that the phosphorylation of both ErbB-2 and Stat3 is mandatory for their colocalization. Thus, although both proteins are present in the cytoplasmic compartment, only hErbB-2ΔNLS is phosphorylated there, given that Stat3, which remains in the cytoplasm, is unphosphorylated, as shown in Fig. 1F. We then explored the effect of hErbB-2ΔNLS on the cellular localization of endogenous ErbB-2. For this purpose, we transfected the hErbB-2ΔNLS mutant into C4HD cells retaining endogenous ErbB-2 expression. Since hErbB-2ΔNLS is GFP tagged (13), this mutant was visualized through direct green fluorescence imaging. On the other hand, we visualized endogenous ErbB-2 by using an antibody that specifically recognizes mouse ErbB-2 and a rhodamine-labeled secondary antibody. Interestingly, our results showed that the expression of hErbB-2ΔNLS absolutely prevented the nuclear translocation of endogenous mouse ErbB-2 (Fig. 3C, bottom row, second panel; as an example, some cells are marked with solid arrows), for the first time revealing the function of hErbB-2ΔNLS as a dominant negative (DN) inhibitor of endogenous ErbB-2 nuclear migration. The merged image in Fig. 3C (bottom row, third panel) shows the cytoplasmic presence and the colocalization (yellow spots) of hErbB-2ΔNLS and mouse ErbB-2 in cells transfected with the hErbB-2ΔNLS (solid arrows), in contrast with the clear migration of mouse ErbB-2 to the nucleus in the cells that did not take up hErbB-2ΔNLS (dashed arrows). To explore whether Stat3 cellular localization regulates the nuclear import of ErbB-2 mediated by MPA, we inhibited Jak activity, which resulted in the abolishment of MPA-induced Stat3 phosphorylation without affecting ErbB-2 activation (see Fig. S3A at http://www.ibyme.org.ar/laboratorios/elizalde.htm). The inhibition of Stat3 tyrosine phosphorylation did not affect the migration of ErbB-2 to the nucleus (see Fig. S3B at http://www.ibyme.org.ar/laboratorios/elizalde.htm).
FIG. 3.
Nuclear import of Stat3 mediated by MPA occurs independently of ErbB-2 nuclear localization. (A) The ErbB-2ΔNLS mutant induces Stat3 phosphorylation in response to MPA. Cells were transfected with siRNAs targeting mouse ErbB-2 or with control siRNAs and cotransfected with hErbB-2WT or hErbB-2ΔNLS plasmids when indicated and then treated with MPA for 10 min. Cell lysates were analyzed by Western blotting with pTyr ErbB-2 and Stat3 antibodies and with pThr/Tyr p44/p42 MAPK antibody, and the membranes were then reprobed with the respective total protein antibody. Membranes were also probed with an anti-GFP antibody. (B) Cellular localization of Stat3 in ErbB-2-siRNA-C4HD-hErbB-2ΔNLS cells treated with MPA. Green fluorescent protein from the ErbB-2ΔNLS vector was visualized by direct fluorescence imaging (green). Nuclei were stained with DAPI (blue). (C) Effect of hErbB-2ΔNLS on endogenous ErbB-2 nuclear migration. C4HD cells retaining endogenous ErbB-2 expression were transfected with the hErbB-2ΔNLS mutant and treated with MPA. Green fluorescent protein from the hErbB-2ΔNLS expression vector was visualized as described above for B (green), and mouse ErbB-2 (red) was localized by using an antibody that specifically recognizes the mouse protein. Solid arrows indicate cells transfected with hErbB-2ΔNLS, and dashed arrows indicate wild-type C4HD cells that did not take up the hErbB-2ΔNLS mutant. See Materials and Methods for specifications of antibodies used in B and C. The experiments from which the data were obtained are representative of three independent experiments (data not shown).
ErbB-2 acts as a Stat3 coactivator.
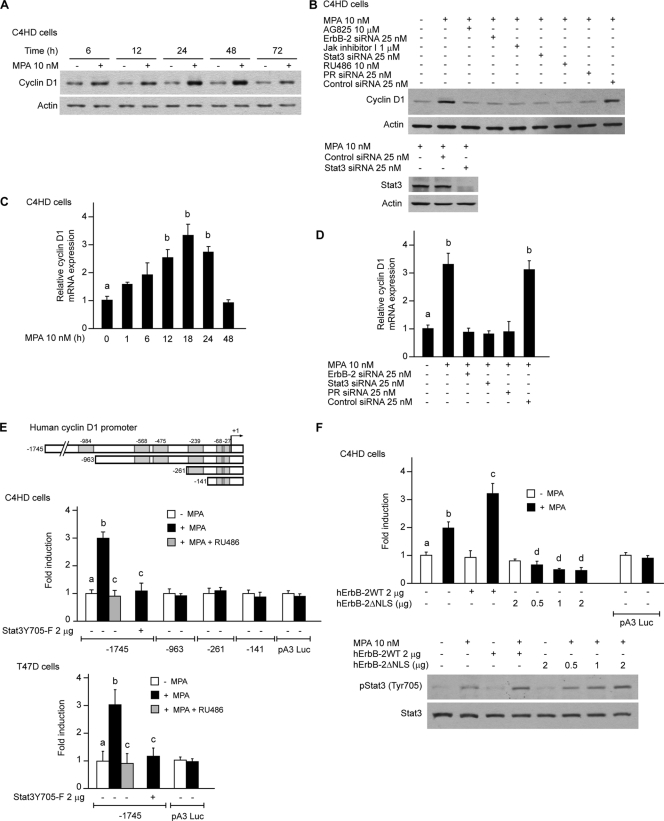
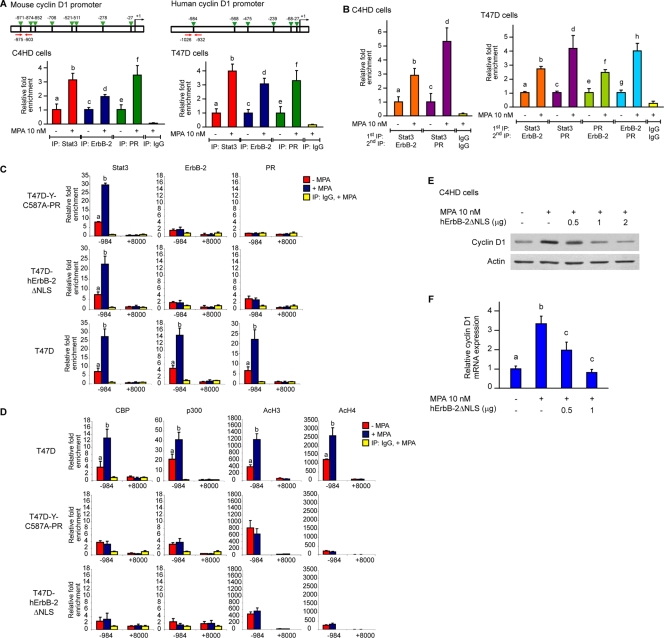
We then explored the nature of the nuclear interaction between ErbB-2 and Stat3. Although the Stat3 function as a transcription factor is well acknowledged, the coactivators that modulate Stat3 activity remain poorly studied. On the other hand, even though seminal findings unraveled the role of ErbB-2 as a transcription factor (35), the capacity of ErbB-2 to act as a transcriptional coactivator remains completely unknown. We consequently built up a novel hypothesis, namely, that ErbB-2 could modulate breast cancer growth acting as a coactivator of Stat3. Through database (MatInspector [http://www.genomatix.de]) and literature searches, we first identified cancer-related genes that contain Stat3 response elements but lack HAS sites. We found that cyclin D1 was a prospective gene to analyze, since it contains Stat3 binding sites in its proximal 1-kb promoter but lacks HASs. Cyclin D1 is a particularly attractive gene because its involvement in breast cancer growth as well as progestin induction of cyclin D1 gene expression have long been shown (4, 12, 27, 29). Importantly, the cyclin D1 promoter lacks a canonical PRE in its 1-kb promoter-proximal region. This turns cyclin D1 into an ideal model to investigate whether progestins may regulate gene expression through the assembly of a nonclassical transcriptional complex between Stat3 and ErbB-2, independently of PR binding to PREs. Here, we found that MPA treatment of C4HD cells induced a significant increase in cyclin D1 protein levels (Fig. 4A). Preincubation with RU486 and silencing of PR expression abrogated the effects of MPA (Fig. 4B). Constitutively activated Stat3 and ErbB-2 were recently found to stimulate cyclin D1 promoter activity in breast and prostate cancer cells, respectively (9, 17). Therefore, we sought to determine the participation of ErbB-2 and Stat3 in the upregulation of cyclin D1 expression by MPA. The inhibition of ErbB-2 activity or knockdown of ErbB-2 expression significantly inhibited the capacity of MPA to induce cyclin D1 expression (Fig. 4B). The abolishment of MPA-induced Stat3 activation or the silencing of Stat3 expression with Stat3 siRNAs also abrogated the upregulation of cyclin D1 protein levels by MPA (Fig. 4B). These findings demonstrate that both ErbB-2 and Stat3 are key players in the mechanism of MPA-induced cyclin D1 expression. We also found that MPA modulates cyclin D1 protein expression in T47D cells via ErbB-2 and Stat3 (see Fig. S4 at http://www.ibyme.org.ar/laboratorios/elizalde.htm). Next, we explored the regulation of cyclin D1 mRNA levels by MPA by quantitative real-time RT-PCR. MPA induced a 3- to 4-fold increase of cyclin D1 mRNA expression levels in C4HD cells (Fig. 4C), and this effect was abrogated by the silencing of the expression of ErbB-2, Stat3, and PR (Fig. 4D). We then assessed whether MPA regulates the transcriptional activity of the cyclin D1 promoter directly via the induction of Stat3 binding to its response elements. C4HD and T47D cells were transiently transfected with a 1,745-bp human cyclin D1 promoter luciferase construct containing Stat3 binding sites, named GAS sites, at positions −984, −568, −475, −239, −68, and −27 (Fig. 4E, top) (17). MPA treatment of both cell types resulted in a 3-fold increase of cyclin D1 promoter activity, which was completely abrogated by RU486 (Fig. 4E). Cotransfection with a DN Stat3 expression vector, Stat3Y705-F, absolutely inhibited the effects of MPA (Fig. 4E). In order to further demonstrate that MPA activates the cyclin D1 promoter via direct Stat3 binding to the GAS sequences, C4HD cells were transfected with cyclin D1 promoter constructs truncated at positions −963, −261, and −141, in which one, three, or four GAS sites, respectively, were excluded (Fig. 4E, top). Interestingly, the capacity of MPA to induce cyclin D1 promoter activation significantly decreased when the Stat3 binding site at position −984 was eliminated, and no further effects were found by the loss of the rest of the GAS sites (Fig. 4E).
FIG. 4.
ErbB-2 acts as a Stat3 coactivator in MPA-induced cyclin D1 promoter activation. MPA induces cyclin D1 expression at the protein and mRNA levels via ErbB-2 and Stat3. (A) Cyclin D1 protein expression was analyzed by Western blotting. (B) Cells were preincubated with the indicated pharmacological inhibitors or transfected with Stat3, ErbB-2, and PR siRNAs and were then treated with MPA for 48 h. Cyclin D1 levels were studied by Western blotting. (Bottom) Control of inhibition of Stat3 expression by siRNAs (data not shown). Experiments from which the results in A and B were obtained were repeated three times, with similar results. (C) Cyclin D1 mRNA expression levels were determined by RT-qPCR. The fold change of mRNA expression levels upon MPA treatment for the indicated times was calculated by normalizing the absolute levels of cyclin D1 mRNA to GAPDH levels, which were used as an internal control, and setting the value of untreated cells as 1. (D) C4HD cells were transfected with Stat3, ErbB-2, PR, and control siRNAs and were then treated with MPA for 18 h. Cyclin D1 mRNA levels were studied by RT-qPCR, and data analyses were performed as described above for C. Data shown in C and D represent the means of data from three independent experiments ± standard errors of the means (SEM) (P < 0.001 for b versus a). (E) MPA induces cyclin D1 promoter activation via Stat3. Cells were transfected with a 1,745-bp-length human cyclin D1 promoter luciferase construct containing the GAS sites indicated at the top. C4HD cells were also transfected with constructs truncated at positions −963, −262, and −141, as shown in the diagram. When indicated, cells were cotransfected with the Stat3Y705-F expression vector. After transfection, cells were treated with MPA for 24 h. Results are presented as the fold induction of luciferase activity with respect to control cells not treated with MPA. The data shown represent the means of data from six independent experiments for each cell type ± SEM (P < 0.001 for b versus a and for c versus b). (F) ErbB-2 acts as a Stat3 coactivator. (Top) C4HD cells were transfected with the 1,745-bp cyclin D1 promoter construct as described above for E and were also cotransfected with the hErbB-2WT or hErbB-2ΔNLS vector when indicated and treated with MPA as described above for E. The relative light units of luciferase obtained in the transient transfection assays were normalized by the arbitrary densitometric values of phosho-Tyr 705/total Stat3 obtained in the Western blot shown at the bottom, and data are presented as the fold induction of cyclin D1 promoter activity relative to cells not treated with MPA. Data shown represent the means of data from three independent experiments ± SEM (P < 0.001 for b versus a, c versus b, and d versus b). (Bottom) Cells were transfected with hErbB-2WT or hErbB-2ΔNLS and were then treated with MPA for 10 min. Stat3 phosphorylation was studied by Western blotting as described in the legend of Fig. 1E (data not shown).
We then specifically evaluated whether ErbB-2 acts as a transcriptional coactivator of Stat3 in the mechanism of MPA-induced cyclin D1 promoter activation. As shown in Fig. 4F, we found that the overexpression of hErbB-2WT significantly enhanced cyclin D1 promoter activation induced by MPA via Stat3. In the absence of MPA, ErbB-2WT did not modulate basal levels of Stat3 transcriptional activity under the assay conditions used. On the other hand, the transfection of C4HD cells with hErbB-2ΔNLS resulted in the abrogation of the MPA-stimulated Stat3 activation of the cyclin D1 promoter (Fig. 4F). This finding is consistent with the function of ErbB-2ΔNLS as a DN inhibitor of endogenous ErbB-2 nuclear migration, as we identified here (Fig. 3C), resulting in a scenario in which Stat3 is located in the nucleus and binds to the cyclin D1 promoter but in which ErbB-2 is not available to act as a coactivator. Notably, we are here defining a new class of transcriptional complex in which the transcription factor itself (Stat3) is a downstream target of its coactivator (ErbB-2). Therefore, simultaneously with the transient transfection assays, we also performed Western blots in which we studied Stat3 activation levels in cells transfected with hErbB-2WT or hErbB-2ΔNLS by assessing Stat3 Tyr 705 phosphorylation. As shown in Fig. 4F, the transfection of C4HD cells with hErbB-2WT or hErbB-2ΔNLS resulted in higher levels of Stat3 Tyr 705 phosphorylation upon MPA stimulation than those observed for wild-type C4HD cells also stimulated with MPA. To normalize for this modulation in Stat3 Tyr 705 phosphorylation levels, which is directly involved in Stat3 transcriptional activity (7), phospho-Stat3 bands in the immunoblots underwent densitometry analysis, and values were normalized to total Stat3 bands. The luciferase units obtained with the transfection assays were then divided by the densitometric values for phosho-Tyr 705/total Stat3. Figure 4F shows the data analysis thus performed, clearly evidencing that Stat3 activation of the cyclin D1 promoter was not due to an increase in Stat3 phosphorylation at Tyr 705 but to the ErbB-2 enhancement of MPA-induced Stat3 transcriptional activity (luciferase levels without normalization for differences in Stat3 phosphorylation are not shown). These findings identify a novel function of ErbB-2 as a Stat3 coactivator. In order to further explore the ErbB-2 action as a coactivator, we took advantage of our RNAi reconstitution model with C4HD cells. The expression of ErbB-2ΔNLS in C4HD cells in which endogenous ErbB-2 was abolished by ErbB-2 siRNAs failed to reconstitute the Stat3 activation of the cyclin D1 promoter (data not shown). To confirm that the role of ErbB-2 as a Stat3 coactivator is not restricted to the cyclin D1 promoter or to a specific cell line, we transfected C4HD and T47D cells with a luciferase reporter plasmid containing four copies of the m67 high-affinity Stat3 binding site (7). The MPA-induced Stat3 transcriptional activation measured using this reporter was significantly enhanced by cotransfection with hErbB-2WT (data not shown).
In vivo binding of a ternary transcriptional complex among Stat3, ErbB-2, and PR to the cyclin D1 promoter.
To assess the specific association of Stat3 and ErbB-2 in the context of living cells, we used a ChIP assay. Our findings with C4HD cells using primers spanning two GAS sites showed a significant and specific MPA-induced binding of both nuclear Stat3 and ErbB-2 to the mouse cyclin D1 promoter after 30 min of treatment (Fig. 5A). Importantly, both proteins associated with the cyclin D1 promoter at the same time, suggesting that they function together in the process of MPA-mediated cyclin D1 promoter activation. We also found that MPA caused a striking increase in the occupancy, by both Stat3 and ErbB-2, of the human cyclin D1 promoter in T47D cells using a pair of primers flanking the GAS site at position −984 (Fig. 5A). PR was found to induce the expression of genes that lack PREs in their promoters by a nonclassical transcriptional mechanism via PR tethering to other transcription factors in the promoter of said genes (23). Our present identification of a progestin-induced Stat3/ErbB-2 transcriptional complex raises the exciting question of whether PR is recruited along with Stat3 and ErbB-2 to the cyclin D1 promoter. ChIP analysis with C4HD and T47D cells demonstrated that, indeed, PR is recruited to the GAS sites in the cyclin D1 promoter along with Stat3 and ErbB-2 (Fig. 5A). We then assessed whether Stat3 and ErbB-2 bind simultaneously to the cyclin D1 gene promoter by using a sequential ChIP assay with a Stat3 antibody in the first immunoprecipitation and an ErbB-2 antibody in the sequential ChIp (re-ChIP). Quantitative real-time PCR analysis clearly showed that Stat3 and ErbB-2 co-occupy the cyclin D1 promoter after 30 min of stimulation of both cell types with MPA (Fig. 5B). Similarly, when Stat3-immunoprecipitated chromatin was reimmunoprecipitated with a PR antibody, we found a significant MPA-stimulated corecruitment of Stat3 and PR (Fig. 5B). The ErbB-2 and PR co-occupancy of the cyclin D1 promoter was shown by re-ChIPs using a PR antibody in the first chromatin immunoprecipitation and an ErbB-2 antibody in the sequential ChIP (re-ChIP), and vice versa (Fig. 5B illustrates the results with T47D cells). These findings clearly show that progestin induces the assembly of a ternary transcriptional complex among Stat3, ErbB-2, and PR at the GAS sites of the cyclin D1 promoter in breast cancer cells. We then evaluated whether PR tethering to Stat3 is an absolute requirement for the assembly of the Stat3/ErbB-2 transcriptional complex. For this purpose, we took advantage of the C587A-PR mutant. In the original description of this mutant (32), it was reported that PR tethering mechanisms require the two proteins to be involved, the one that binds DNA and its associated protein, to possess a DNA binding domain. Because the C587A-PR mutant lacks a functional DNA binding domain, we hypothesized that its capacity to be recruited to the GAS sites of the cyclin D1 promoter through tethering with Stat3 will be strongly impaired compared with that of wild-type PR-B. Figure 5C shows that while a clear Stat3 recruitment was observed upon the stimulation of T47D-Y-C587A-PR cells by MPA (left), C587A-PR was not loaded at this promoter (right). Interestingly, ErbB-2 was not recruited to the cyclin D1 promoter in T47D-Y-C587A-PR cells (Fig. 5C, middle). We then questioned whether ErbB-2 recruitment to the GAS sites of the cyclin D1 promoter is mandatory for PR tethering to Stat3 at this site. To address this issue, we transfected T47D cells with hErbB-2ΔNLS, which is unable to migrate to the nucleus and which functions as a DN inhibitor of endogenous ErbB-2 nuclear translocation (Fig. 3C). In the absence of ErbB-2 recruitment (Fig. 5C, middle), PR was not loaded at the GAS site at position −984 of the cyclin D1 promoter after MPA treatment of T47D-hErbB-2ΔNLS cells (Fig. 5C, right). MPA-induced Stat3 binding at this site remained unaffected (Fig. 5C, left). The recruitment of all three proteins to the site at bp +8000 was used as a negative control for transcription factor and coactivator binding, as described previously (11). Histone acetylation positively correlates with active gene transcription (21). Therefore, to gain insight into the mechanisms of the ErbB-2 coactivation of Stat3, we investigated whether coactivators with histone acetyltransferase (HAT) activity, such as p300 and CBP, are recruited along with Stat3, ErbB-2, and PR to the cyclin D1 promoter. We found that CBP and p300 were loaded at the GAS site at position −984 of the cyclin D1 promoter upon MPA treatment (Fig. 5D). Consistently, histone H3 and H4 acetylation at this site was significantly enhanced by MPA treatment (Fig. 5D). In T47D-hErbB-2ΔNLS and T47D-Y-C587A-PR cells, in which the Stat3/ErbB-2/PR transcriptional complex was not assembled, neither recruitment of CBP or p300 nor modification of histone acetylation levels was observed (Fig. 5D). To further confirm that the role of ErbB-2 as a Stat3 coactivator within the nuclear Stat3/ErbB-2/PR complex regulates cyclin D1 expression in breast cancer cells, we explored the levels of the cyclin D1 protein and mRNA in C4HD cells transfected with increasing amounts of hErbB-2ΔNLS. Our results showed that levels of MPA-induced cyclin D1 expression were significantly reduced by hErbB-2ΔNLS transfection compared to those found for wild-type C4HD cells (Fig. 5E and F).
FIG. 5.
MPA induces in vivo binding of Stat3, ErbB-2, and PR to the cyclin D1 promoter. (A) Recruitment of Stat3, ErbB-2, and PR to the cyclin D1 promoter was analyzed by ChIP of cells treated with MPA for 30 min. Immunoprecipitated DNA was amplified by qPCR using primers (red arrows) flanking the GAS sites (green arrows) indicated at the top. The arbitrary qPCR number obtained for each sample was normalized to the input, setting the value of the untreated sample as 1. Data are expressed as fold chromatin enrichment over untreated cells (P < 0.001 for b versus a, d versus c, and f versus e). (B) Sequential ChIP. Chromatins from cells treated as described above for panel A were first immunoprecipitated with Stat3, PR, or ErbB-2 antibodies, as indicated, and were then reimmunoprecipitated using either an ErbB-2 or a PR antibody. qPCR and data analysis were performed as detailed above for panel A (P < 0.001 for b versus a, d versus c, f versus e, and h versus g). Results in panels A and B are means ± SEM from three independent experiments. IgG was used as a negative control. (C, top) PR tethering to Stat3 at the GAS sites of the cyclin D1 promoter is required for ErbB-2 loading at these sites. T47D-Y cells were transfected with the C587A-PR mutant and were then treated with MPA for 30 min. (Middle) ErbB-2 recruitment to the GAS sites of the cyclin D1 promoter is mandatory for PR tethering to Stat3. T47D cells were transfected with the hErbB-2ΔNLS mutant and were treated with MPA for 30 min. (Bottom) T47D cells are shown as a control. The recruitment of Stat3, ErbB-2, and PR to the cyclin D1 promoter was analyzed by ChIP. Immunoprecipitated DNA was amplified by qPCR using primers flanking the GAS site at position −984 shown in panel A. A second pair of primers flanking the site at bp +8000 in intron 4 of the cyclin D1 gene was used as a negative control for transcription factor and coactivator binding. Amounts of immunoprecipitated DNA were normalized to inputs and are reported relative to the amount obtained by IgG immunoprecipitation, which was set to 1 (P < 0.001 for b versus a). (D) Recruitment of CBP and p300 and H3 and H4 acetylation levels (AcH3 and AcH4, respectively) at the sites described above for panel C for all three cell types was studied by ChIP, and data were also analyzed as described above for panel C (P < 0.001 for b versus a). Results in panels C and D are means ± SEM from two or three independent experiments. (E) C4HD cells were treated with MPA for 48 h or transfected with increasing amounts of the hErbB-2ΔNLS expression vector before MPA stimulation. Cyclin D1 protein levels were analyzed by Western blotting. (F) C4HD cells were treated with MPA for 18 h or transfected with increasing amounts of the hErbB-2ΔNLS expression vector before MPA stimulation, and cyclin D1 mRNA levels were studied by RT-qPCR. Data analysis was performed as described in the legend to Fig. 4C. Data represent the means of data from three independent experiments ± SEM (P < 0.001 for b versus a and c versus b).
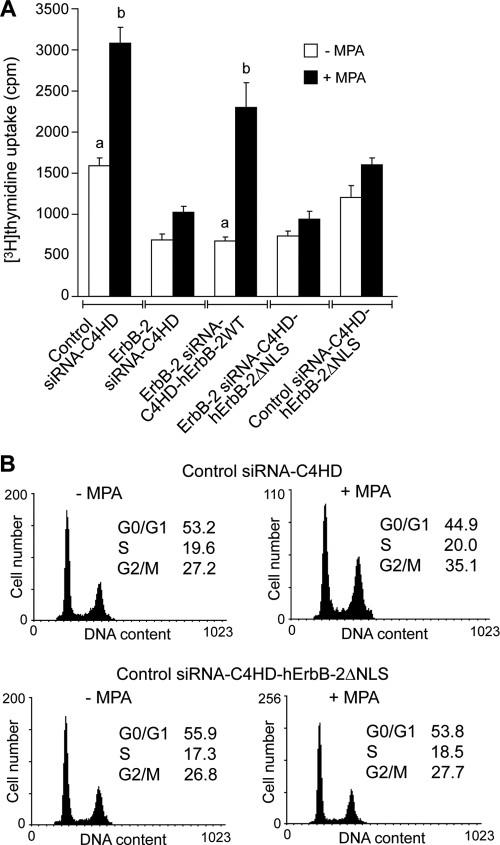
The nuclear Stat3/ErbB-2/PR complex regulates breast cancer cell proliferation.
To investigate the correlation between the MPA-induced assembly of the nuclear Stat3/ErbB-2/PR complex and cell growth, we examined the in vitro proliferative response of ErbB-2-siRNA-C4HD-hErbB-2ΔNLS cells to MPA. As shown in Fig. 6A, ErbB-2-siRNA-C4HD-ErbB-2ΔNLS cells were completely unresponsive to MPA stimulation. This finding reveals a direct correlation between ErbB-2 nuclear localization and progestin-induced breast cancer growth. Since we found that hErbB-2ΔNLS acts as a DN inhibitor of endogenous ErbB-2 nuclear translocation, we next addressed whether the transfection of hErbB-2ΔNLS into C4HD cells expressing ErbB-2 (control siRNA-C4HD-ErbB-2ΔNLS) affects MPA-induced growth. Our results showed that under these cell conditions, the response to MPA was abrogated (Fig. 6A), for the first time identifying the function of hErbB-2ΔNLS as a DN inhibitor of endogenous ErbB-2 proliferative effects in breast cancer cells. Proliferation was also evaluated by propidium iodide staining and flow cytometry analysis, with similar results. Figure 6B shows our results for control siRNA-C4HD-ErbB-2ΔNLS cells indicating their lack of a proliferative response to MPA.
FIG. 6.
The nuclear Stat3/ErbB-2 complex regulates in vitro breast cancer proliferation. (A) Endogenous ErbB-2 expression was silenced by transfection with ErbB-2 siRNAs, and expressions of either hErbB-2WT or hErbB-2ΔNLS were restored by cotransfection with the respective plasmids. Cells were treated with MPA for 48 h, and the incorporation of [3H]thymidine was used as a measure of DNA synthesis. Data are presented as means ± standard deviations (P < 0.001 for b versus a). (B) C4HD cells were transfected with control siRNA (top) and cotransfected with hErbB-2ΔNLS (bottom) before MPA stimulation for 48 h and were then stained with PI and analyzed for cell cycle distribution by flow cytometry. The data from the experiments shown are representative of data from a total of three experiments.
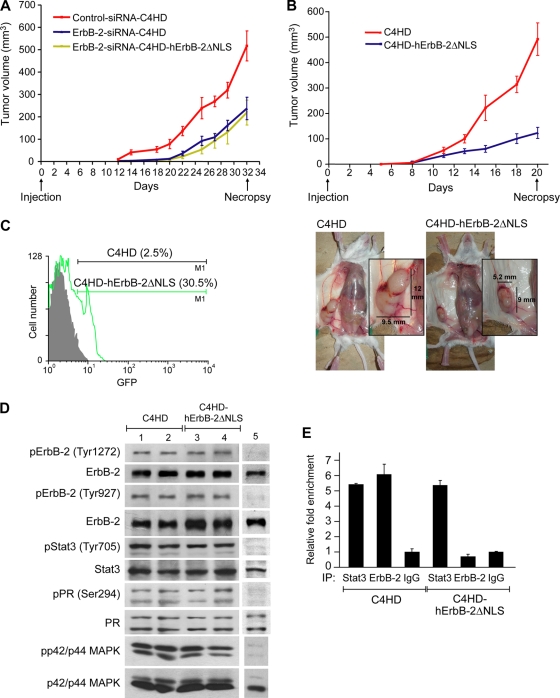
Abrogation of ErbB-2 nuclear localization inhibits in vivo growth of breast tumors expressing steroid hormone receptors and ErbB-2.
Our breast cancer model has unique features that make it particularly attractive for in vivo studies targeting ErbB-2. Since C4HD tumors overexpress ErbB-2 and also have high levels of ER and PR, they resemble a phenotype present in approximately 50% of human breast cancer cells that overexpress ErbB-2 and associated with resistance to hormonal treatment (24). In this study, control siRNA-C4HD, ErbB-2-siRNA-C4HD, and ErbB-2-siRNA-C4HD-hErbB-2ΔNLS cells were inoculated subcutaneously (s.c.) into mice treated with MPA. Here, we describe a representative experiment of a total of three. All mice (n = 6) injected with control siRNA-C4HD cells developed tumors, which became palpable after 12 days of inoculation. On the contrary, only four out of six mice injected with ErbB-2-siRNA-C4HD cells or with ErbB-2-siRNA-C4HD-hErbB-2ΔNLS cells developed tumors, with a delay of 4 days in tumor latency compared with tumors from the control group. The mean volumes (Fig. 7A) and growth rates (Table 1) of tumors that developed from either ErbB-2-siRNA-C4HD or ErbB-2-siRNA-C4HD-hErbB-2ΔNLS cells were significantly lower than those of tumors from the control group. We then used a second experimental protocol in which we addressed whether the transfection of hErbB-2ΔNLS into C4HD cells maintaining the expression of endogenous ErbB-2 could modulate the in vivo proliferative response to MPA. For this purpose, C4HD cells were transiently transfected with the hErbB-2ΔNLS vector (C4HD-hErbB-2ΔNLS) or with the empty pcDNA 3.1 vector (C4HD), and cells from each experimental group were inoculated s.c. into mice treated with MPA. Here, we show the results of a representative experiment of a total of four. All mice (n = 6) injected with C4HD-hErbB-2ΔNLS cells and with C4HD cells developed tumors that became palpable after 5 days of inoculation. As shown in Fig. 7B, the expression of hErbB-2ΔNLS in C4HD cells strongly inhibited MPA-induced proliferation. The mean volumes (Fig. 7B and Table 1) and growth rates (Table 1) of tumors that developed from C4HD-hErbB-2ΔNLS cells were significantly lower than those of tumors from the control group. Tumors were excised at day 32 in the first protocol and at day 20 in the second protocol, and the results are summarized in Table 1. Histopathological analysis revealed that tumors from mice receiving ErbB-2-siRNA-C4HD, ErbB-2-siRNA-C4HD-hErbB-2ΔNLS, or C4HD-hErbB-2ΔNLS cells showed a significantly lower histological grade (grade II), with 3 to 4 mitoses per 10 high-power fields (HPF), than tumors from animals receiving control siRNA-C4HD or C4HD cells, both of which showed histological grade III, with over 10 mitoses per 10 HPF (see Fig. S6 at http://www.ibyme.org.ar/laboratorios/elizalde.htm). The experimental strategies used here relied on transient transfections with the hErbB-2ΔNLS expression vector. Therefore, we explored its intratumoral expression at the end of the experiments. We chose to study samples of the second protocol because of the far-reaching implications of the use of hErbB-2ΔNLS as a single-agent therapy. Since hErbB-2ΔNLS is GFP tagged, we analyzed its content by flow cytometry. Figure 7C shows that at day 20, approximately 30% of the cells still expressed the hErbB-2ΔNLS mutant. Next, we examined the state of activation of ErbB-2, Stat3, p42/p44 MAPKs, and PR in tumor samples. Comparable ErbB-2, Stat3, and p42/p44 MAPK phosphorylation levels were found in tumors that developed in mice injected with C4HD-hErbB-2ΔNLS and C4HD cells (Fig. 7D). Similar levels of PR phosphorylation at Ser 294, which correlates directly with PR transcriptional activity (28), were present in tumors that developed from C4HD-hErbB-2ΔNLS and C4HD cells. ChIP analysis demonstrated comparable levels of Stat3 recruitment to the cyclin D1 promoter in tumors arising from C4HD-hErbB-2ΔNLS and C4HD cells (Fig. 7E). On the contrary, we found no ErbB-2 recruitment to the cyclin D1 promoter in C4HD-hErbB-2ΔNLS cells (Fig. 7E). These results further support the direct involvement of the nuclear Stat3/ErbB-2 transcriptional complex in the in vivo growth of breast tumors expressing both PR and ErbB-2.
FIG. 7.
In vivo blockage of ErbB-2 nuclear localization. (A and B) Cells (106) from each experimental group were inoculated subcutaneously (s.c.) into mice treated with MPA, and tumor volume was calculated as described in Materials and Methods. (Bottom) Decrease in tumor mass in mice injected with C4HD-hErbB-2ΔNLS cells compared to mice injected with C4HD cells. Each point represents the mean volume ± SEM for six independent tumors for all experimental groups except for the ErbB-2-siRNA-C4HD and ErbB-2-siRNA-C4HD-hErbB-2ΔNLS groups, which contained four tumors. (C) Content of hErbB-2ΔNLS. GFP expression levels were determined by flow cytometry. Shown is a representative sample of each tumor type. (D) Tumor lysates were analyzed by Western blotting with the indicated phosphoprotein antibodies, and membranes were reprobed with their respective total protein antibodies. Shown are two representative samples of mice injected with C4HD cells (lanes 1 and 2) and with C4HD-hErbB-2ΔNLS cells (lanes 3 and 4). Lane 5, C4HD cells not treated with MPA, used as a control for the protein phosphorylation state. (E) ChIP analysis of tumor samples. DNA-protein complexes were pulled down with Stat3 and ErbB-2 antibodies or with control IgG, and the resulting DNA was amplified by qPCR using primers indicated in Fig. 5. Results are expressed as n-fold over the IgG control and represent the averages of data from three replicates ± SEM. Shown are data for a representative sample of each tumor type.
TABLE 1.
Tumor growth ratesa
| Protocol and treatment | Mean tumor vol (mm3) ± SEM | Mean growth rate ± SEM (mm3/day) | % growth inhibition | Delay in tumor growth (days) |
|---|---|---|---|---|
| First protocol | ||||
| Control siRNA-C4HD | 516.7 ± 67.1* | 23.1 ± 1.5* | ||
| ErbB-2-siRNA-C4HD | 237.1 ± 50.1# | 11.2 ± 0.9# | 54.1b | 7b |
| ErbB-2-siRNA-C4HD-hErbB-2ΔNLS | 218.7 ± 55.5# | 10.2 ± 1.6# | 57.6b | 7b |
| Second protocol | ||||
| C4HD | 491.8 ± 64.0* | 32.1 ± 3.5* | ||
| C4HD-hErbB-2ΔNLS | 123.1 ± 21.8# | 8.5 ± 1.0# | 74.9c | 6.5c |
Growth rates were calculated as the slopes of growth curves. In the first protocol, volume, percentage of growth inhibition, and delay in tumor growth (days) in tumors from mice injected with ErbB-2-siRNA-C4HD or ErbB-2-siRNA-C4HD-hErbB-2ΔNLS cells with respect to mice injected with control siRNA-C4HD cells were calculated at day 32, as described in Materials and Methods. In the second protocol, comparisons between tumors that developed from C4HD-hErbB-2ΔNLS and C4HD cells were performed at day 20. # versus *, P < 0.001.
With respect to control siRNA cells, for growth inhibition, P < 0.001.
With respect to C4HD cells, for growth inhibition, P < 0.001.
DISCUSSION
Our present findings for breast cancer cells demonstrate that a steroid hormone receptor, PR, induces ErbB-2 nuclear translocation, its colocalization and physical association with Stat3 at the nuclear compartment, and the assembly of a transcriptional complex in which ErbB-2 acts as a coactivator of Stat3. In this newly discovered class of complex, the transcription factor (Stat3) is first phosphorylated at the cytoplasmic level via its coactivator (ErbB-2) function as an upstream effector. Notably, PR is also loaded onto the Stat3/ErbB-2 complex. Our results also highlight that in the frame of this Stat3/ErbB-2/PR transcriptional complex, the function of ErbB-2 as a Stat3 coactivator drives progestin-induced cyclin D1 promoter activation. Importantly, our findings also reveal a new and unexpected feature of the nonclassical PR genomic mechanisms. Thus, we showed that the corecruitment of ErbB-2 is an absolute requirement for PR tethering to Stat3. A model of PR action consistent with our findings is shown in Fig. S7 at http://www.ibyme.org.ar/laboratorios/elizalde.htm.
In addition to ErbB-2, all ErbB family members have been detected in the nucleus (34). Since ErbBs lack a putative DNA binding domain, it was proposed that other transcription factors with DNA binding capacities cooperate with ErbBs to regulate gene expression. Although pioneering findings demonstrated that ErbB-2 modulates COX-2 promoter activation functioning as a transcription factor (35), the capacity of ErbB-2 to act as a transcriptional coactivator had so far remained completely unknown. Our series of functional studies with mouse and human breast cancer cells have provided the first evidence that ErbB-2 indeed acts as a transcriptional coactivator of Stat3. As previously shown for constitutively activated ErbB-2 (35), our data now show that PR induces full-length ErbB-2 protein translocation to the nucleus. We also revealed a new feature of the ErbB-2 nuclear status, as we identified its specific phosphorylation at Tyr 1222/1272 and Tyr 877/927, induced by progestins via c-Src.
The nuclear interaction of EGF-R and Stat3 in the promoter of the inducible nitric oxide synthase (iNOS), containing both EGF-R binding sites (AT-rich sequences [ATRSs]) and Stat3 response elements, was identified in a seminal study (20). In that work, the nature of the EGF-R and Stat3 nuclear interplay was explored by a different strategy than that used here, since it relied on identifying genes containing both ATRS and Stat3 response elements in their promoters. The presence of two clusters of ATRS and Stat3 binding sites was essential for the EGF-R regulation of the iNOS promoter (20). This highlights a major difference with respect to the nuclear ErbB-2/Stat3 transcriptional complex function in the cyclin D1 promoter, which we found requires only Stat3 binding to the GAS sites and ErbB-2 recruitment to those sites in order to act as a Stat3 coactivator. A likely interpretation of this difference is that EGF-R/Stat3 and ErbB-2/Stat3 complexes regulate chromatin targets by distinct mechanisms as a general rule. It may also indicate that the nature of the interaction between ErbBs and Stat3 within intact cells depends on the set of Stat3/ErbB binding motifs available in the target gene promoter/enhancer regions as well as on the specific sequences and unique structural features of the DNA neighboring the Stat3/ErbB binding sites. Consistent with the latter, Stat3 and EGF-R do not associate at the cyclin D1 promoter, which was first found to be regulated by nuclear EGF-R (19) and which also contains a cluster of ATRS/Stat3 sites (20).
Our data showed that the nuclear import of Stat3 mediated by MPA occurs independently of ErbB-2 nuclear localization, as reported previously for Stat3 and EGF-R (20). The comigration of Stat3 and EGF from the cell surface to the perinuclear region via receptor-mediated endocytosis was previously described (3). Our results are consistent with those previous findings since we revealed here that hErbB-2ΔNLS moves from the cytoplasmic membrane to the perinuclear region in response to MPA and thus retains the potential capacity to cotransit with Stat3. Interestingly, our findings identified yet another level of interaction between Stat3 and ErbB-2 showing that the specific entrance of Stat3 to the nucleus, once located in the perinuclear cytoplasm, is not associated with ErbB-2 nuclear translocation.
It has long been acknowledged that progestins, acting through the classical PR, induce cyclin D1 gene expression in breast cancer cells (4, 12). However, the contribution of rapid PR signaling and of PR transcriptional mechanisms still remains to be elucidated. The cyclin D1 proximal promoter lacks a canonical PRE, for which this gene has become a model to investigate the mechanisms through which progestins/PR regulates the expression of genes independently of PR binding to PREs. Seminal works have demonstrated that the rapid progestin activation of p42/p44 mitogen-activated protein kinases (MAPKs) and of phosphatidylinositol (PI) 3-kinase (PI-3K)/Akt pathways mediates the PR regulation of cyclin D1 expression in breast cancer cells (4, 12, 27). Another study suggested that progestins induce cyclin D1 promoter activation via PR tethering to the AP-1 transcription factor at an AP-1 binding site encoded in the proximal promoter (10). Our data provide completely novel insights into the mechanism of PR induction of cyclin D1 expression in breast tumors, which integrates the rapid PR activation of ErbB-2 and Stat3 and a nonclassical PR transcriptional mechanism consisting of the assembly on the cyclin D1 promoter of a nuclear complex in which ErbB-2 acts as a coactivator of Stat3. Moreover, our finding that PR is recruited along with Stat3 and ErbB-2 to the cyclin D1 promoter reveals a new aspect of the nonclassical PR tethering mechanisms. Thus, we found here that ErbB-2 coloading is an absolute requirement for PR tethering to Stat3 at the GAS sites of the cyclin D1 promoter, for the first time revealing a functional cooperation between a steroid hormone receptor, PR, and a receptor tyrosine kinase, ErbB-2, to induce cyclin D1 promoter activation via Stat3 binding to its response elements in said promoter. We have also provided a mechanistic explanation for the mutual dependence of ErbB-2 and PR in Stat3 transcriptional activity at the cyclin D1 promoter. We showed that the corecruitment of coactivators with chromatin-remodeling activity, such as p300 and CBP, occurs only upon the assembly of the Stat3/ErbB-2/PR multiprotein complex.
The molecular mechanisms of the ErbB-2 and Stat3 interaction that lead to breast cancer growth remain almost completely unexplored. Most recently, we found that HRG-bound ErbB-2 activates Stat3 through the co-option of PR signaling (26). Activated Stat3 in turn acts as a downstream effector of both HRG/ErbB-2 and unliganded PR to induce the proliferation of mammary tumors (26). On the other hand, a startling study showed that the targeting of Stat3 inhibits the growth of ErbB-2-overexpressing mammary cancer cells (30). It has also been found that the overexpression of ErbB-2 correlates with Stat3 activation and binding to its response elements in the p21Cip1 promoter and that this is involved in chemotherapy resistance in breast tumors (15). An exciting and novel finding of our study is the demonstration of a direct correlation between nuclear ErbB-2 function as a Stat3 transcriptional coactivator and breast cancer growth. Indeed, we found that cells expressing the mutant hErbB-2ΔNLS showed a strongly reduced response to progestin-induced in vitro and in vivo proliferation. In support of a key role of nuclear ErbB-2 in mammary tumorigenesis, we found here that upon progestin stimulation, hErbB-2ΔNLS retains an intact, intrinsic tyrosine kinase activity and the capacity to activate p42/p44 MAPKs, a classical ErbB-2 signaling cascade, and induce Stat3 phosphorylation. This finding indicates that in spite of an intact function as a membrane tyrosine kinase and activator of mitogenic signaling cascades, the abolishment of ErbB-2 nuclear function significantly impairs its proliferative effects in breast cancer. Notably, the transfection of hErbB-2ΔNLS into C4HD cells expressing endogenous ErbB-2 (C4HD-hErbB-2ΔNLS cells) abrogated their proliferative response to progestins, consistent with our results identifying the role of hErbB-2ΔNLS as a DN inhibitor of wild-type ErbB-2 nuclear translocation. Our molecular studies of tumors from mice injected with C4HD-hErbB-2ΔNLS cells revealed high levels of ErbB-2, p42/p44 MAPK, and Stat3 tyrosine phosphorylation as well as a significant degree of PR phosphorylation at Ser294, which was found to correlate directly with PR transcriptional activity (28). We also detected strong Stat3 binding to the cyclin D1 promoter in tumors arising from C4HD-hErbB-2ΔNLS cells. Most challenging was our finding that ErbB-2 recruitment to the cyclin D1 promoter was completely abrogated in these tumors. These results have far-reaching therapeutic implications, since they indicate that the growth of breast tumors with intact ErbB-2 tyrosine kinase function and PR transcriptional activity can be abolished by the blockage of ErbB-2 nuclear translocation. At present, COX-2 is the only gene whose expression has been shown to be modulated through the role of ErbB-2 as a transcriptional activator (35). Interestingly, COX-2 inhibition in MCF-7 cells overexpressing ErbB-2 and in parental MCF-7 cells had no effect on the proliferation of the latter but suppressed the invasive activity of ErbB-2-overexpressing MCF-7 cells (35). Undoubtedly, other as-yet-unidentified genes regulated by ErbB-2 through its role as a transcription factor may be involved in ErbB-2 proliferative effects. On the other hand, our present results support the exciting notion that the function of ErbB-2 as a transcriptional coactivator may be the one directly involved in the ErbB-2 stimulation of breast cancer growth.
Approximately 50% of human breast cancer cells that overexpress ErbB-2 also display ER and PR, a phenotype associated with resistance to hormonal therapy whose clinical management still remains to be established (24). Although clinical data indicate that combined antihormonal and anti-ErbB-2 therapies, such as the blockage of ErbB-2 with the recombinant humanized anti-ErbB-2 monoclonal antibody trastuzumab (Herceptin), improve outcome compared to endocrine treatment alone, other studies suggested that this dual strategy might in fact render worse results than those obtained with the combination of trastuzumab with chemotherapy (24). This confronts us with a significant number of patients requiring new therapies for ErbB-2-overexpressing breast tumors. Our present findings provide a strong rationale for a potential novel gene therapy intervention in PR- and ErbB-2-positive breast tumors consisting of the transfer of hErbB-2ΔNLS to be used as a single-agent therapy.
Acknowledgments
We thank Mien-Chie Hung (M. D. Anderson Cancer Center, Houston, TX) for his generous gift of the hErbB-2ΔNLS, which indeed made this work possible, and A. A. Molinolo (NIH, Bethesda, MD) for his constant help and support.
This work was supported by IDB 1728 OC/AR PICT 0211 (2006) from the National Agency of Scientific Promotion of Argentina, PIP 737 from the Argentina National Council of Scientific Research, the Susan G. Komen for the Cure KG090250 investigator-initiated research grant, and Oncomed-Reno CONICET 1819/03, from the Henry Moore Institute of Argentina, all of them awarded to P.V.E.
Footnotes
Published ahead of print on 27 September 2010.
REFERENCES
- 1.Akiyama, T., S. Matsuda, Y. Namba, T. Saito, K. Toyoshima, and T. Yamamoto. 1991. The transforming potential of the c-erbB-2 protein is regulated by its autophosphorylation at the carboxyl-terminal domain. Mol. Cell. Biol. 11:833-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balana, M. E., R. Lupu, L. Labriola, E. H. Charreau, and P. V. Elizalde. 1999. Interactions between progestins and heregulin (HRG) signaling pathways: HRG acts as mediator of progestins proliferative effects in mouse mammary adenocarcinomas. Oncogene 18:6370-6379. [DOI] [PubMed] [Google Scholar]
- 3.Bild, A. H., J. Turkson, and R. Jove. 2002. Cytoplasmic transport of Stat3 by receptor-mediated endocytosis. EMBO J. 21:3255-3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boonyaratanakornkit, V., E. McGowan, L. Sherman, M. A. Mancini, B. J. Cheskis, and D. P. Edwards. 2007. The role of extranuclear signaling actions of progesterone receptor in mediating progesterone regulation of gene expression and the cell cycle. Mol. Endocrinol. 21:359-375. [DOI] [PubMed] [Google Scholar]
- 5.Boonyaratanakornkit, V., M. P. Scott, V. Ribon, L. Sherman, S. M. Anderson, J. L. Maller, W. T. Miller, and D. P. Edwards. 2001. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-Src family tyrosine kinases. Mol. Cell 8:269-280. [DOI] [PubMed] [Google Scholar]
- 6.Bromberg, J. F., C. M. Horvath, D. Besser, W. W. Lathem, and J. E. Darnell, Jr. 1998. Stat3 activation is required for cellular transformation by v-src. Mol. Cell. Biol. 18:2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bromberg, J. F., M. H. Wrzeszczynska, G. Devgan, Y. Zhao, R. G. Pestell, C. Albanese, and J. E. Darnell, Jr. 1999. Stat3 as an oncogene. Cell 98:295-303. [DOI] [PubMed] [Google Scholar]
- 8.Carnevale, R. P., C. J. Proietti, M. Salatino, A. Urtreger, G. Peluffo, D. P. Edwards, V. Boonyaratanakornkit, E. H. Charreau, J. E. B. de Kier, R. Schillaci, and P. V. Elizalde. 2007. Progestin effects on breast cancer cell proliferation, proteases activation, and in vivo development of metastatic phenotype all depend on progesterone receptor capacity to activate cytoplasmic signaling pathways. Mol. Endocrinol. 21:1335-1358. [DOI] [PubMed] [Google Scholar]
- 9.Casimiro, M., O. Rodriguez, L. Pootrakul, M. Aventian, N. Lushina, C. Cromelin, G. Ferzli, K. Johnson, S. Fricke, F. Diba, B. Kallakury, C. Ohanyerenwa, M. Chen, M. Ostrowski, M. C. Hung, S. A. Rabbani, R. Datar, R. Cote, R. Pestell, and C. Albanese. 2007. ErbB-2 induces the cyclin D1 gene in prostate epithelial cells in vitro and in vivo. Cancer Res. 67:4364-4372. [DOI] [PubMed] [Google Scholar]
- 10.Cicatiello, L., R. Addeo, A. Sasso, L. Altucci, V. B. Petrizzi, R. Borgo, M. Cancemi, S. Caporali, S. Caristi, C. Scafoglio, D. Teti, F. Bresciani, B. Perillo, and A. Weisz. 2004. Estrogens and progesterone promote persistent CCND1 gene activation during G1 by inducing transcriptional derepression via c-Jun/c-Fos/estrogen receptor (progesterone receptor) complex assembly to a distal regulatory element and recruitment of cyclin D1 to its own gene promoter. Mol. Cell. Biol. 24:7260-7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eeckhoute, J., J. S. Carroll, T. R. Geistlinger, M. I. Torres-Arzayus, and M. Brown. 2006. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 20:2513-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faivre, E., A. Skildum, L. Pierson-Mullany, and C. A. Lange. 2005. Integration of progesterone receptor mediated rapid signaling and nuclear actions in breast cancer cell models: role of mitogen-activated protein kinases and cell cycle regulators. Steroids 70:418-426. [DOI] [PubMed] [Google Scholar]
- 13.Giri, D. K., M. Ali-Seyed, L. Y. Li, D. F. Lee, P. Ling, G. Bartholomeusz, S. C. Wang, and M. C. Hung. 2005. Endosomal transport of ErbB-2: mechanism for nuclear entry of the cell surface receptor. Mol. Cell. Biol. 25:11005-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, W., Y. Pylayeva, A. Pepe, T. Yoshioka, W. J. Muller, G. Inghirami, and F. G. Giancotti. 2006. Beta 4 integrin amplifies ErbB2 signaling to promote mammary tumorigenesis. Cell 126:489-502. [DOI] [PubMed] [Google Scholar]
- 15.Hawthorne, V. S., W. C. Huang, C. L. Neal, L. M. Tseng, M. C. Hung, and D. Yu. 2009. ErbB2-mediated Src and signal transducer and activator of transcription 3 activation leads to transcriptional up-regulation of p21Cip1 and chemoresistance in breast cancer cells. Mol. Cancer Res. 7:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labriola, L., M. Salatino, C. J. Proietti, A. Pecci, O. A. Coso, A. R. Kornblihtt, E. H. Charreau, and P. V. Elizalde. 2003. Heregulin induces transcriptional activation of the progesterone receptor by a mechanism that requires functional ErbB-2 and mitogen-activated protein kinase activation in breast cancer cells. Mol. Cell. Biol. 23:1095-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leslie, K., C. Lang, G. Devgan, J. Azare, M. Berishaj, W. Gerald, Y. B. Kim, K. Paz, J. E. Darnell, C. Albanese, T. Sakamaki, R. Pestell, and J. Bromberg. 2006. Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. Cancer Res. 66:2544-2552. [DOI] [PubMed] [Google Scholar]
- 18.Li, L., and P. E. Shaw. 2002. Autocrine-mediated activation of STAT3 correlates with cell proliferation in breast carcinoma lines. J. Biol. Chem. 277:17397-17405. [DOI] [PubMed] [Google Scholar]
- 19.Lin, S. Y., K. Makino, W. Xia, A. Matin, Y. Wen, K. Y. Kwong, L. Bourguignon, and M. C. Hung. 2001. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3:802-808. [DOI] [PubMed] [Google Scholar]
- 20.Lo, H. W., S. C. Hsu, M. Ali-Seyed, M. Gunduz, W. Xia, Y. Wei, G. Bartholomeusz, J. Y. Shih, and M. C. Hung. 2005. Nuclear interaction of EGFR and STAT3 in the activation of the iNOS/NO pathway. Cancer Cell 7:575-589. [DOI] [PubMed] [Google Scholar]
- 21.MacDonald, V. E., and L. J. Howe. 2009. Histone acetylation: where to go and how to get there. Epigenetics 4:139-143. [DOI] [PubMed] [Google Scholar]
- 22.Migliaccio, A., D. Piccolo, G. Castoria, M. Di Domenico, A. Bilancio, M. Lombardi, W. Gong, M. Beato, and F. Auricchio. 1998. Activation of the Src/p21ras/Erk pathway by progesterone receptor via cross-talk with estrogen receptor. EMBO J. 17:2008-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.National Research Council. 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC.
- 23.Owen, G. I., J. K. Richer, L. Tung, G. Takimoto, and K. B. Horwitz. 1998. Progesterone regulates transcription of the p21(WAF1) cyclin-dependent kinase inhibitor gene through Sp1 and CBP/p300. J. Biol. Chem. 273:10696-10701. [DOI] [PubMed] [Google Scholar]
- 24.Prat, A., and J. Baselga. 2008. The role of hormonal therapy in the management of hormonal-receptor-positive breast cancer with co-expression of HER2. Nat. Clin. Pract. Oncol. 5:531-542. [DOI] [PubMed] [Google Scholar]
- 25.Proietti, C., M. Salatino, C. Rosemblit, R. Carnevale, A. Pecci, A. R. Kornblihtt, A. A. Molinolo, I. Frahm, E. H. Charreau, R. Schillaci, and P. V. Elizalde. 2005. Progestins induce transcriptional activation of signal transducer and activator of transcription 3 (Stat3) via a Jak- and Src-dependent mechanism in breast cancer cells. Mol. Cell. Biol. 25:4826-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proietti, C. J., C. Rosemblit, W. Beguelin, M. A. Rivas, M. C. Diaz Flaque, E. H. Charreau, R. Schillaci, and P. V. Elizalde. 2009. Activation of Stat3 by heregulin/ErbB-2 through the co-option of progesterone receptor signaling drives breast cancer growth. Mol. Cell. Biol. 29:1249-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitoh, M., M. Ohmichi, K. Takahashi, J. Kawagoe, T. Ohta, M. Doshida, T. Takahashi, H. Igarashi, A. Mori-Abe, B. Du, S. Tsutsumi, and H. Kurachi. 2005. Medroxyprogesterone acetate induces cell proliferation through up-regulation of cyclin D1 expression via phosphatidylinositol 3-kinase/Akt/nuclear factor-kappaB cascade in human breast cancer cells. Endocrinology 146:4917-4925. [DOI] [PubMed] [Google Scholar]
- 28.Shen, T., K. B. Horwitz, and C. A. Lange. 2001. Transcriptional hyperactivity of human progesterone receptors is coupled to their ligand-dependent down-regulation by mitogen-activated protein kinase-dependent phosphorylation of serine 294. Mol. Cell. Biol. 21:6122-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutherland, R. L., and E. A. Musgrove. 2004. Cyclins and breast cancer. J. Mammary Gland Biol. Neoplasia 9:95-104. [DOI] [PubMed] [Google Scholar]
- 30.Tan, M., K. H. Lan, J. Yao, C. H. Lu, M. Sun, C. L. Neal, J. Lu, and D. Yu. 2006. Selective inhibition of ErbB2-overexpressing breast cancer in vivo by a novel TAT-based ErbB2-targeting signal transducers and activators of transcription 3-blocking peptide. Cancer Res. 66:3764-3772. [DOI] [PubMed] [Google Scholar]
- 31.Tsai, M. J., and B. W. O'Malley. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 63:451-486. [DOI] [PubMed] [Google Scholar]
- 32.Tung, L., M. K. Mohamed, J. P. Hoeffler, G. S. Takimoto, and K. B. Horwitz. 1993. Antagonist-occupied human progesterone B-receptors activate transcription without binding to progesterone response elements and are dominantly inhibited by A-receptors. Mol. Endocrinol. 7:1256-1265. [DOI] [PubMed] [Google Scholar]
- 33.Tzahar, E., H. Waterman, X. Chen, G. Levkowitz, D. Karunagaran, S. Lavi, B. J. Ratzkin, and Y. Yarden. 1996. A hierarchical network of interreceptor interactions determines signal transduction by Neu differentiation factor/neuregulin and epidermal growth factor. Mol. Cell. Biol. 16:5276-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, S. C., and M. C. Hung. 2009. Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin. Cancer Res. 15:6484-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, S. C., H. C. Lien, W. Xia, I. F. Chen, H. W. Lo, Z. Wang, M. Ali-Seyed, D. F. Lee, G. Bartholomeusz, F. Ou-Yang, D. K. Giri, and M. C. Hung. 2004. Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell 6:251-261. [DOI] [PubMed] [Google Scholar]
- 36.Xu, W., X. Yuan, K. Beebe, Z. Xiang, and L. Neckers. 2007. Loss of Hsp90 association up-regulates Src-dependent ErbB2 activity. Mol. Cell. Biol. 27:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]