Abstract
A set of genome-wide screens for proteins whose absence exacerbates growth defects due to pseudo-haploinsufficiency of ribosomal proteins in Saccharomyces cerevisiae identified Dom34 as being particularly important for cell growth when there is a deficit of 40S ribosomal subunits. In contrast, strains with a deficit of 60S ribosomal proteins were largely insensitive to the loss of Dom34. The slow growth of cells lacking Dom34 and haploinsufficient for a protein of the 40S subunit is caused by a severe shortage of 40S subunits available for translation initiation due to a combination of three effects: (i) the natural deficiency of 40S subunits due to defective synthesis, (ii) the sequestration of 40S subunits due to the large accumulation of free 60S subunits, and (iii) the accumulation of ribosomes “stuck” in a distinct 80S form, insensitive to the Mg2+ concentration, and at least temporarily unavailable for further translation. Our data suggest that these stuck ribosomes have neither mRNA nor tRNA. We postulate, based on our results and on previously published work, that the stuck ribosomes arise because of the lack of Dom34, which normally resolves a ribosome stalled due to insufficient tRNAs, to structural problems with its mRNA, or to a defect in the ribosome itself.
Efficient growth requires efficient ribosome biosynthesis, since ribosome synthesis consumes a large proportion of the cell's resources (38). Efficient ribosome biosynthesis requires the equimolar production of the many ribosomal proteins (RPs) matched to the transcription of rRNA. While the cell has evolved mechanisms to do this effectively, experimental manipulations that bring about an imbalance in the components reveal systems for the rapid degradation of excess RPs or rRNAs (19, 35, 37, 39). The development of resources and methods for the systematic genome-wide screening of Saccharomyces cerevisiae provides an opportunity to examine the surveillance and degradation of ribosomal subunits in greater detail. Such surveillance mechanisms should come into play under conditions where a single ribosomal protein is limiting. Our approach therefore was to identify gene products that would exaggerate the growth defect in such a situation. We anticipated identifying not only proteins potentially involved in ribosome surveillance but also those involved in processes downstream of ribosome synthesis, namely, translation.
To set up a screen in which the synthesis of one ribosomal subunit was limiting, we made use of the fact that most RPs in yeast are encoded by two genes with nearly identical gene products. Strains with a deletion of one of those, while viable, will generate substantial amounts of preribosomes that are missing the protein in question. Predicting that a cell's failure to properly deal with such defective preribosomes, or their consequences, could be deleterious for growth, we carried out synthetic genetic array (SGA) analysis against an array of nonessential gene deletion strains.
SGA screens using strains with a deletion of one of the genes encoding Rpl1, Rpl4, or Rps6 identified a number of genes whose products might be involved in the turnover of aberrant ribosomes. The investigation of these genetic interactions is ongoing. In this communication we focus on DOM34, a gene whose deletion resulted in reduced fitness in combination with rps6aΔ but not rpl1bΔ or rpl4aΔ. Dom34 is an interesting protein, having been implicated circumstantially in a number of aspects of translation. It is important for the growth of cells deficient in Rps30 (4) and in instances where translation initiation is compromised by constitutively active Gcn2 (1). Having been implicated in the “no-go” decay of mRNA whose translation has been halted, e.g., by a strong secondary structure (5), it has now been shown to release a ribosome, stuck at a “hungry” codon, from both its peptidyl-tRNA and its mRNA (32). Dom34 is involved in the elimination of 40S subunits that carry nonfunctional mutations in the decoding site of 18S rRNA, although in this case, 40S subunit turnover is exceedingly slow (2). The lack of Dom34 suppresses the phenotype of cells depleted of late-acting 40S assembly factors such as Nob1, apparently by allowing more time for the final steps of 40S maturation to occur and even for immature 40S particles to become associated with functional polyribosomes (33). Our results suggest that the lack of Dom34 leads to an unusual class of 80S ribosomes that are resistant to dissociation even though they appear to lack both mRNA and tRNAs. We suggest that these “stuck” ribosomes represent incompletely resolved products of defective translation, likely related to no-go situations. We conclude that the lack of Dom34 inhibits the growth of cells pseudo-haploinsufficient for a RPS gene by exacerbating the already short supply of 40S subunits.
MATERIALS AND METHODS
Strains.
Strains used in this study are described in Table 1. The query strains were constructed by the integrative transformation of a PCR product in which the nourseothricin (clonNAT) resistance gene under the Ashbya gossypii TEF1 promoter replaces the open reading frame (ORF) of the gene in question (34).
TABLE 1.
Strain list
| Strain | Genotype | Reference or Source |
|---|---|---|
| Y7092 (wt) | matα can1Δ::STE2pr-Sp_his5 lyp1Δ his3Δ1 leu2Δ0Δ ura3Δ0 met15Δ0 LYS2+ | 34 |
| ABY4 (rpl1bΔ) | Y7092+rpl1bΔ::natR | This study |
| ABY2 (ura3Δ) | Y7092+ura3Δ::natR | This study |
| ABY7 (rps6aΔ) | Y7092+rps6aΔ::natR | This study |
| ABY5 (rpl4aΔ) | Y7092+rpl4aΔ::natR | This study |
| ABY8 (dom34Δ) | Y7092+dom34Δ::natR | This study |
| ABY10 (rps6aΔ dom34Δ) | Y7092+rps6aΔ::natR dom34Δ::G418R | This study |
| ABY9 (hbs1Δ) | Y7092+hbs1Δ::natR | This study |
| ABY12 (dom34Δ rpl4aΔ) | Y7092+rpl4aΔ::natR dom34Δ::G418R | This study |
| ABY11 (rps6aΔ hbs1Δ) | Y7092+rps6aΔ::natR hbs1Δ::G418R | This study |
| ABY13 (hcr1Δ) | Y7092+hcr1Δ::natR | This study |
| ABY14 (hcr1Δ dom34Δ) | Y7092+hcr1Δ::natR dom34Δ::G418R | This study |
| KBM26 (fun12Δ) | Y7092+fun12Δ::natR | This study |
| ABY15 (fun12Δ dom34Δ) | Y7092+fun12Δ::natR | This study |
| ABY16 (rps6aΔ dom34Δ rpl4aΔ) | Y7092+rpl4aΔ::natR dom34Δ::G418R rps6aΔ::ura3 | This study |
| BY4741 | matahis3Δ1 leu2Δ0 lys2Δ0 met15Δ0 ura3Δ0 | 34 |
| ABY17 (bud21Δ) | BY4741+bud21Δ::G418R | This study |
| ABY20 (bud21Δ dom34Δ) | Y7092+dom34Δ::natR bud21Δ::G418R | This study |
| ABY18 (tsr2Δ) | BY4741+tsr2Δ::G418R | This study |
| ABY21 (tsr2Δ dom34Δ) | Y7092+dom34Δ::natR tsr2Δ::G418R | This study |
| ABY19 (yar1Δ) | BY4741+yar1Δ::G418R | This study |
| ABY22 (yar1Δ dom34Δ) | Y7092+dom34Δ::natR yar1Δ::G418R | This study |
Buffers.
LHB buffer contains 0.1 M NaCl, 0.01 M Tris-Cl (pH 7.4), and 30 mM MgCl2 (10). TMN buffer contains 0.05 M Tris-acetate (Ac) (pH 7.0), 0.05 M NH4Cl, and 12 mM MgCl2. AE buffer contains 50 mM NaAc (pH 5.3) and 1 mM EDTA.
SGA methods.
Duplicate SGA screens of strains ABY2 (ura3Δ), ABY4 (rpl1bΔ), ABY5 (rpl4aΔ), and ABY7 (rps6aΔ) (Table 1) were performed against the nonessential gene deletion array (∼4,300 strains). Each screen was conducted with duplicate copies of the array in a 1,536-colony-per-plate format according to standard SGA protocols (34). Subsequently, other query strains used to perform SGA screens were derived from the parental Y7092 strain. Visual inspection and computer-based analysis of digital images were used to identify double mutant strains exhibiting fitness (growth) defects. Interactions were confirmed by random spore analysis and by growth rate determinations.
Growth curve analysis.
Individual strains were streaked onto appropriate selection plates at 30°C, and single colonies were inoculated after appropriate dilution into 150 μl yeast extract-peptone-dextrose (YPD) in a 100-well plate for the Bioscreen C instrument and grown at 30°C. Light scattering was determined at 30-min intervals over 48 to 60 h, and the doubling time during log phase was determined algebraically. The analyses were performed at least in triplicate with a standard error of generally <5%. The fitness phenotypes of double mutant strains were calculated according to methods described previously (20), using the multiplicative model for genetic interactions.
Sucrose gradient analysis.
Cycloheximide was added to a final concentration of 100 μg/ml to a 50-ml culture in YPD at an optical density at 600 nm (OD600) of 0.5 to 1.0. The culture was swirled rapidly and immediately poured over crushed ice. The cells were collected by centrifugation and washed with LHB buffer (except where indicated) containing 100 μg/ml cycloheximide and 200 μg/ml heparin. The washed cells were resuspended in 0.5 ml of the same solution and disrupted by shaking with an equal volume of glass beads in a bead beater. Next, 0.5 ml of buffer was added, and the sample was centrifuged at 10,000 rpm for 5 min to remove debris. The supernatant was layered over a 10% to 50% (wt/vol) sucrose gradient in TMN buffer (except where indicated) and centrifuged at 4°C for 150 min at 39,000 rpm in an SW41 rotor. The gradient was analyzed by using a density gradient fractionator (ISCO model 640), recording UV absorbance at 260 nm. Where indicated, the extracts were prepared in LHB buffer containing 1.5 mM MgCl2 and centrifuged in TMN buffer containing 1.5 mM MgCl2.
As an alternative approach to freezing polyribosomes on mRNA, we analyzed sucrose gradients of extracts prepared with 1% formaldehyde (HCHO) instead of cycloheximide (36), with the following modifications. HCHO was added to a final concentration of 1% to a 50-ml culture in YPD, which was immediately poured over crushed ice. After 1 h, HCHO cross-linking was terminated by the addition of glycine to a final concentration of 0.1 M. Subsequent steps were identical to those described above using LHB and TMN buffers containing 1.5 mM MgCl2 but no cycloheximide.
RNA extraction, RNA slot blots, and Northern and qPCR analysis.
RNA was extracted from different populations of ribosomes using AE buffer-saturated phenol and AE buffer-saturated phenol-chloroform-isoamyl alcohol (25:24:1). For RNA slot blots, an equal volume of RNA from individual fractions was loaded. 32P-labeled antisense oligodeoxynucleotides were used to detect mRNAs. For Northern analysis, the preparation of yeast total RNA and blotting was performed as described previously (17a). The distribution of no-go PGK1 mRNAs in polysome gradients was determined as follows: wild-type (wt) and dom34Δ strains were transformed with galactose-inducible plasmids (a kind gift from Roy Parker) carrying the PGK1(sl) gene with a poly(G) tract in the 3′ untranslated region (UTR) (used in our case to distinguish these transcripts from the endogenous PGK1 transcripts) with a stem-loop structure inserted into its ORF (5). These constructs were confirmed by sequencing. Polysome profiling and RNA slot blotting for individual gradient fractions were monitored with hybridization using specific oligonucleotide probes. (Oligonucleotide sequences are available upon request.)
Quantitative PCR (qPCR) was carried out on RNA prepared from pooled fractions of sucrose gradients run under low-Mg2+ conditions (see Table 4). Two micrograms of RNA isolated from polysome fractions and equivalent volumes RNA from other gradient fractions were treated with RQ1 DNase (Promega), and one-half of the resultant reaction mixture (5.5 μl) was used for reverse transcription with SuperScript III (Invitrogen) using both oligo(dT) and random hexamer primers. cDNA levels were analyzed by using an ABI Prism 7900HT Fast real-time PCR system (Applied Biosystems). Reactions were prepared by using Thermo Scientific ABsolute Blue QPCR SYBR green 6-carboxyl-X-chatamine (ROX) mix and run in triplicate on a 384-well plate. Primers were designed by using NCBI Primer BLAST.
RESULTS
SGA screens using rpl1bΔ, rpl4aΔ, and rps6aΔ strains.
Synthetic genetic array screens were set up by using “query” strains missing one of a pair of genes encoding a particular RP. The rationale was that cells missing a gene whose product plays a role either in identifying and disposing of incomplete ribosomal subunits or in effecting efficient translation would have substantially compromised growth. We constructed three query strains, missing genes encoding the ribosomal protein L1, L4, or S6. These proteins occupy distinct regions of the complete ribosomal particle. L1 is a peripheral protein, added late in the assembly of the 60S subunit, and is involved in E-site tRNA movement and exit (7). L4 associates with rRNA early in the assembly of 60S subunits and is important for many subsequent assembly steps (17). S6 has been localized by chemical and UV cross-linking studies to a region of the 40S subunit termed the “beak” (23) and lies at the interface between the large subunits and small subunits (SSUs) (22), being one of the few proteins of the 40S subunit to contact 25S rRNA.
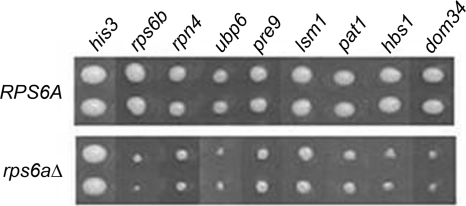
In brief, the rpl1bΔ, rpl4aΔ, or rps6aΔ strain was screened against an ordered array of ∼4,300 viable yeast gene deletion strains, and the relative growth of the double mutants was scored both visually and by computer-based image analysis (34). A number of potential synthetic sick/synthetic lethal interactions scored highly upon both visual inspection and computer-based image analysis in the three individual screens. A representative set from the rps6aΔ strain screen is shown in Fig. 1. All interactions from the three screens subsequently confirmed by random spore analysis are listed in Table 2. Satisfyingly, since both S6 and L1 are essential proteins (16, 26), lethal interactions were observed for the rps6bΔ strain (in the rps6aΔ strain screen) and rpl1aΔ (in the rpl1bΔ strain screen). (The rpl4bΔ strain was not present on the array.) The LSM1 and PAT1 genes, implicated in mRNA degradation (11), were recovered in all three screens. Both S6 and L1 also shared interactions with several genes involved in proteasome function, RPN4, PRE9, and UBP6, suggesting an interesting relationship between the ribosome, the proteasome, and mRNA degradation. L1 interacts with a large number of genes, especially in the proteasomal pathway. Studies of these relationships are ongoing (K B. McIntosh, A. Bhattacharya, and J. R. Warner, manuscript in preparation). A gene that was synthetic sick uniquely with the rps6aΔ strain, DOM34, is the subject of the remainder of this communication.
FIG. 1.
Genetic interactions between rps6aΔ and different nonessential gene deletions. Shown is a composite image taken from the final plates of the SGA screen, comparing nine deletion strains (in duplicate). (Top) The query strain is the ura3Δ strain. (Bottom) The query strain is the rps6aΔ strain. The deletion strains are listed at the top, with the His3Δ strain as a control.
TABLE 2.
List of SS/SL interactions obtained from SGA
| Function | Interaction(s) for strain used as a query for SGA |
||
|---|---|---|---|
| rps6aΔ | rpl1bΔ | rpl4aΔ | |
| mRNA degradation | LSM1 | LSM1 | LSM1 |
| PAT1 | PAT1 | PAT1 | |
| KEM1 | KEM1 | ||
| Translation | HBS1 | RPL1A | |
| DOM34 | ASC1 | ||
| RPS6B | |||
| Proteasome, ubiquitin, | RPN4 | RPN4, UBC4 | |
| SUMO, and URM | UBP6 | UBP6, CUE3 | |
| PRE9 | PRE9, DOA1 | ||
| RPN10, RUP1 | |||
| SEM1, UBI4 | |||
| UBP2, PRE9 | |||
| UBP8, UBA4 | |||
| ULS1, URM1 | |||
| Other categories | RVS161, RVS167 | ||
| EAF7, SRO1 | |||
| SGF11, STB5 | |||
Note that a recent genome-wide comprehensive SGA analysis was published, identifying tens of thousands of positive and negative genetic interactions (3). These were based strictly on the size of robotically displayed colonies. While our data are in rough agreement with the data of that study, we identified fewer interactions from our screens because we counted only strains that passed a series of additional tests, including random spore analysis and growth rate determination in liquid medium.
Deletion of DOM34 affects cells deficient in 40S but not 60S synthesis.
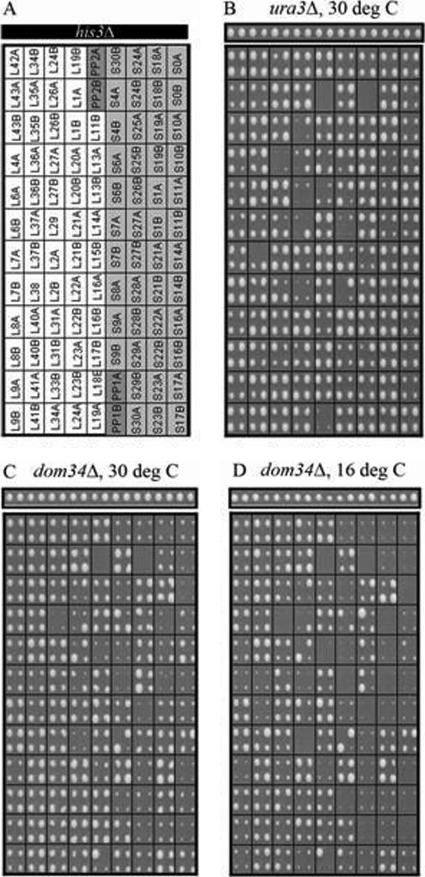
To ask whether the defective growth of strains that combine dom34Δ with rps6aΔ is a general property of all ribosomal proteins, we constructed a miniarray of strains comprising all of the RP gene deletions in our initial screens along with additional RP gene deletions from the haploid yeast knockout collection and his3Δ as a control (Fig. 2A) (8). A dom34Δ strain and a ura3Δ control strain were then used as queries to score synthetic sick interactions with the miniarray (Fig. 2). While a few strains failed to grow in the control strain (ura3Δ) (Fig. 2B), usually because of poor sporulation, the lack of Dom34 along with a deficiency in a majority of the 40S subunit RPs led to a clear slow-growth phenotype (Fig. 2C). Remarkably, this was rarely the case for deficiencies of 60S subunit RPs. The synthetic phenotype between dom34Δ and 40S RP genes was enhanced at 37°C (not shown) and was even more severe at 16°C (Fig. 2D), beyond the cold-sensitive phenotype of dom34Δ itself (top row) (4). Qualitative examination showed that most genes encoding 40S RPs (the A and/or B copy) were synthetic sick with DOM34.
FIG. 2.
SGA screens using a miniarray of RP gene deletion strains. A miniarray in quadruplicate of strains carrying G418-resistant deletions of individual RP genes (A) was crossed to clon-NAT-resistant ura3Δ (B) and dom34Δ (C and D) query strains. Genetic interactions of haploid double-drug-resistant strains were scored according to standard SGA protocols (34). The figures show the final selection plates incubated at either 30°C or 16°C. The narrow panels above show the his3Δ controls on the same plates. The ura3Δ screen served as a control. Strains lacking one of the two copies of an RP gene have a relatively high probability of pseudoreversion through the duplication of the remaining gene. This is evident for the rps6aΔ strain (B to D) as well as from the occasional substantial variations in colony size among the quadruplicates.
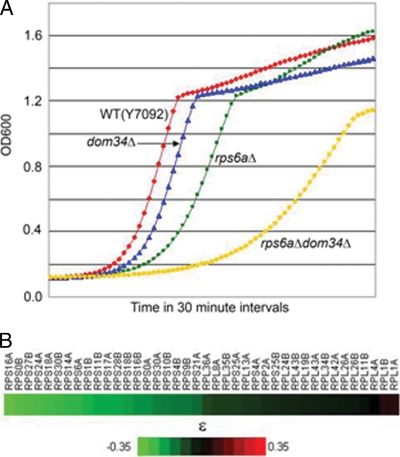
The fitness defects in such double mutants were quantitated by subjecting a number of individual RP-dom34 double-deletion strains from the SGA screen to growth analysis in a Bioscreen C instrument. While the dom34Δ strain grew at a wild-type rate, the rps6aΔ strain grew more slowly, and the dom34Δ rps6aΔ strain was substantially slower (Fig. 3A and Table 3). A quantitative analysis of the strength of genetic interactions (20) of dom34Δ with a number of RP genes is presented as a “heat map” (Fig. 3B), which reflects the greater-than-expected loss of fitness when dom34Δ is combined with the deletion of any of several RP genes. The quite striking result is that dom34Δ interacted genetically with essentially all the tested genes encoding proteins of the 40S subunit but barely at all with those of the 60S subunit. A similar identification of many RPS genes but few RPL genes as synthetic sick with dom34Δ was made in the comprehensive SGA analysis (3). We conclude that DOM34 shares the strongest synthetic sick interactions with RPS genes. In general, the degree of synthetic sickness observed was correlated with the relative contribution of that RPS gene to the mRNA transcriptome (12); i.e., the more highly expressed the gene copy, the more severe the synthetic sickness with DOM34.
FIG. 3.
(A) Growth curves from the Bioscreen C analysis. The growths of the wild-type (Y7092), dom34Δ, rps6aΔ, and dom34Δ rps6aΔ strains were determined from light-scattering measurements at 30-min intervals (see Materials and Methods). (B) Quantitation of genetic interactions between the DOM34 and RP genes. Doubling times of dom34Δ, rpxΔ, and dom34Δ rpxΔ mutants measured by Bioscreen C analysis at 30°C were used to calculate strain fitness. The fitness (W) of a strain deleted for a given gene, “x,” is defined as the ratio of the doubling time (D) of the wild-type strain to the deletion strain (Wx = Dwt/Dx). The value of ɛ is the deviation in the fitness phenotype of the double mutant (Wxy) from Wx × Wy, as predicted for noninteracting gene pairs by the multiplicative model (20). The values of ɛ for strains representing double mutants of dom34Δ and 40 individual RP deletion mutants are presented as a “heat map.” ɛ values of <0 indicate synthetic sick genetic interactions.
TABLE 3.
Doubling time values of key strains
| Strains | Mean t1/2 (min) ± SDa | ɛb |
|---|---|---|
| ABY1 (wt) | 89.7 ± 3.8 | NA |
| ABY2 (ura3Δ) | 90.5 ± 0.1 | NA |
| ABY7 (rps6aΔ) | 128.7 ± 5.3 | NA |
| ABY5 (rpl4aΔ) | 105.4 ± 1.5 | NA |
| ABY8 (dom34Δ) | 92.5 ± 5.5 | NA |
| ABY10 (rps6aΔ dom34Δ) | 202.3 ± 4.0 | −0.24 |
| ABY12 (rpl4aΔ dom34Δ) | 105.4 ± 1.1 | −0.03 |
| ABY16 (rps6aΔ dom34Δ rpl4aΔ) | 161.8 ± 4.5 | −0.04 |
| ABY17 (bud21Δ) | 140.5 ± 1.2 | NA |
| ABY20 (bud21Δ dom34Δ) | 217.0 ± 2.8 | −0.21 |
| ABY18 (tsr2Δ) | 112.4 ± 4.9 | NA |
| ABY21 (tsr2Δ dom34Δ) | 162.7 ± 3.1 | −0.23 |
| ABY19 (yar1Δ) | 135.0 ± 1.4 | NA |
| ABY22 (yar1Δ dom34Δ) | 190.5 ± 0.7 | −0.17 |
| ABY13 (hcr1Δ) | 134.3 ± 0.7 | NA |
| ABY14 (hcr1Δ dom34Δ) | 145.2 ± 1.2 | −0.03 |
t1/2, doubling time.
ɛ is a measure of the phenotypic interaction of two genes, as defined in the legend to Fig. 3. NA, not applicable.
Genetic interactions of DOM34 with three genes implicated in small ribosomal subunit biogenesis.
To identify interactions between DOM34 and additional genes, we screened the dom34Δ strain against the full array of nonessential gene deletion strains. In addition to the genes encoding 40S ribosomal proteins that we had previously identified, three additional genes displayed synthetic sick interactions with DOM34, namely, BUD21, TSR2, and YAR1. These interactions were confirmed by random spore (not shown) and growth curve (Table 3) analyses. All are involved in 40S assembly: Bud21 is a component of the U3 snoRNA complex (SSU processome) required for 18S rRNA biogenesis (6, 21), Tsr2 has a potential role in 20S pre-rRNA processing (25), and Yar1 is a cytoplasmic ankyrin repeat protein proposed previously to couple 40S biogenesis to environmental stresses (18). These observations provide independent confirmation that cells deficient for 40S ribosomal subunits are particularly sensitive to the loss of Dom34. Since most of the proteins involved in 40S subunit assembly and maturation are essential, few are represented in the SGA analysis.
Lack of Dom34 reduces polysome/monosome ratios.
We next asked whether the severity of growth phenotypes among double mutants of DOM34 and genes encoding proteins of the 40S subunit was reflected in the translational apparatus of the cells. Sucrose gradient analysis of the ribosome complement of the dom34Δ strain shows that the loss of Dom34 has little effect on the level of free 40S and 60S subunits but results in decreased levels of polysomes and a concomitant increase in levels of the 80S monosomes, suggesting an impaired initiation of translation (compare Fig. 4A and B), as was shown previously (4). Thus, a smaller proportion of the ribosome complement is engaged in translation. Nevertheless, the growth rate of the dom34Δ strain is the same as that of the wild type (Table 3). Thus, an interesting conclusion is that the degree of impairment in translation initiation caused by the absence of Dom34 must not be limiting for growth in such cells.
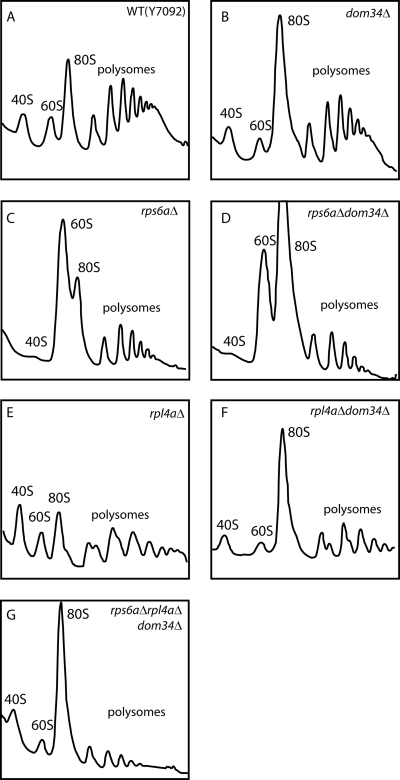
FIG. 4.
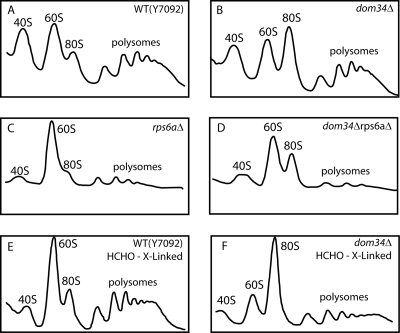
Polysome profiles of extracts of various strains. (A) Y7092; (B) dom34Δ; (C) rps6aΔ; (D) rps6aΔ dom34Δ; (E) rpl4aΔ; (F) rpl4aΔ dom34Δ; (G) rps6aΔ rpl4aΔ dom34Δ (in the presence of 12 mM MgCl2). Cultures of the six strains were grown at 30°C to mid-log phase. Cell extracts were prepared and polysome profiles were analyzed as described in Materials and Methods. Sedimentation is from left to right.
The deletion of RPS6A leads to a substantial deficiency of 40S subunits, as shown by the lack of free 40S subunits in the gradient as well as by the accumulation of a large peak of free 60S subunits (Fig. 4C). Indeed, by Northern analysis, the overall ratio of 18S to 25S rRNAs in these cells is only 73% of that of the wild type. The polysome profile of the dom34Δ rps6aΔ double mutant combines characteristics of each of the single mutants (Fig. 4D). In this case, the 18S/25S ratio is 84% of that of wild-type cells. The paucity of free 40S subunits and the very large peak of 60S subunits are combined with an exaggerated 80S monosome peak and substantially reduced polysomes.
Lack of Dom34 results in accumulation of stuck ribosomes.
What is the nature of the 80S ribosomes observed by gradient analysis of wild-type cells, of dom34Δ cells, or of cells in which the inhibition of initiation leads to a near-total conversion of polysomes to 80S monosomes (9)? Sucrose gradients of yeast extracts have conventionally been analyzed with solutions with a high Mg2+ concentration (9). However, it seems likely that this Mg2+ concentration drives the equilibrium of 40S + 60S ⇋ 80S far to the right. Therefore, we reanalyzed the extracts of our strains under conditions of 1.5 mM Mg2+, which is close to the apparent physiological level (29). Even this analysis may not replicate the in vivo situation, since in a sucrose gradient, the >100-fold dilution of the ribosomes relative to the cytoplasm (31) will influence the equilibrium between the 80S and separated subunits.
As expected, conditions of physiological Mg2+ concentrations shift the equilibrium toward the free subunits. For wild-type and rps6aΔ cells, most of the 80S peak dissociates into subunits (Fig. 5A and C). We suggest that the 80S peak seen at high Mg2+ concentrations is a mixture of ribosomes translating short mRNAs, such as those encoding the 24-amino-acid Rpl41 (40), together with a majority of inactive couples. In contrast, in the dom34Δ and dom34Δ rps6aΔ strains, a substantial amount of the 80S peak is resistant to dissociation (Fig. 5B and D). Such stuck 80S monosomes are a characteristic feature of cells missing Dom34. To exclude the possibility that stuck ribosomes are caused by the cycloheximide used to freeze polysomes, we employed 1% formaldehyde as an alternative approach (36). Polysome gradients showed that stuck ribosomes are still evident in dom34Δ strains under such conditions (Fig. 5E and F). Thus, the 80S peak from a dom34Δ strain analyzed at high Mg2+ concentrations contains three classes of ribosomes: ribosomes translating short mRNAs, 80S couples in equilibrium with free 60S and 40S subunits, and stuck 80S ribosomes.
FIG. 5.
Polysome profiles of extracts of various strains. (A and E) Y7092; (B and F) dom34Δ; (C) rps6aΔ; (D) rps6aΔ dom34Δ (in the presence of 1.5 mM MgCl2). A to D are as described in the legend of Fig. 4 except that the extraction and the sucrose gradient buffers contained 1.5 mM MgCl2. E and F were prepared using HCHO but without cycloheximide (see Materials and Methods).
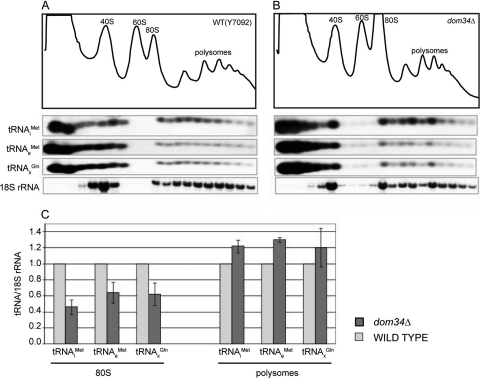
We conclude that in cells lacking Dom34, a substantial amount of ribosomes is tied up in stuck 80S monosomes. Analysis by Northern blotting of the increased fraction of total 18S rRNA that is in monosomes of the dom34Δ strain showed that stuck ribosomes represent about 10% of the total ribosomes (Fig. 6). What is the origin of these stuck ribosomes?
FIG. 6.
Distribution of several tRNAs in 80S and polysomes. Extracts of Y7092 (A) and dom34Δ (B) were analyzed on sucrose gradients in the presence of 1.5 mM MgCl2. RNA was prepared from individual fractions and separated on a 1.5% agarose gel. Northern analysis was done with 32P-end-labeled oligonucleotides complementary to sequences specific for 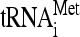 ,
, 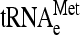 , and tRNAGln. (C) The ratio of tRNA/18S rRNA present in a particular fraction was calculated as a percentage of the total in the entire gradient. The values shown represent the ratio of tRNA/18SrRNA in 80S and polysome fractions of the dom34Δ strain normalized to that in the wild-type strain.
, and tRNAGln. (C) The ratio of tRNA/18S rRNA present in a particular fraction was calculated as a percentage of the total in the entire gradient. The values shown represent the ratio of tRNA/18SrRNA in 80S and polysome fractions of the dom34Δ strain normalized to that in the wild-type strain.
Stuck ribosomes are not stalled at the start site of translation initiation.
One possible origin of stuck ribosomes is that they are stalled on mRNA at or near the translation initiation site, due perhaps to a defect in the initiation process that mimics no-go translation. If so, they should be enriched specifically for initiator tRNA-methionine (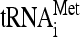 ) compared to elongator tRNA-methionine (
) compared to elongator tRNA-methionine (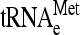 ) and tRNA-glutamine (tRNAGln). RNA extracted from gradient fractions of wild-type and dom34Δ strains was measured by using probes that anneal specifically to
) and tRNA-glutamine (tRNAGln). RNA extracted from gradient fractions of wild-type and dom34Δ strains was measured by using probes that anneal specifically to 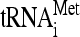 ,
, 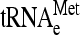 , tRNAGln, as well as 18S rRNA (Fig. 6A and B). The results show that
, tRNAGln, as well as 18S rRNA (Fig. 6A and B). The results show that 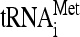 /18S rRNA levels were in fact lower in stuck ribosome fractions in strains lacking Dom34 (Fig. 6C). Indeed, levels of
/18S rRNA levels were in fact lower in stuck ribosome fractions in strains lacking Dom34 (Fig. 6C). Indeed, levels of 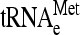 and tRNAGln were also lower, suggesting that stuck ribosomes in dom34Δ strains lack tRNAs, although we cannot exclude the possibility that they are occupied by a special class of tRNAs for which we did not probe. The lack of tRNAs on a ribosome could occur after the release of the peptidyl tRNA following translation termination or after translational stalling. On the other hand, in the polysomes of the dom34Δ strain, tRNA/ribosome levels are actually 20% higher than those in wild-type cells. This observation suggests that tRNA on these ribosomes may be associated with the mRNA for longer periods of time due to a nonresolution of translational stalling events. Hence, stuck ribosomes may be the consequence of an incomplete resolution of translational stalling events.
and tRNAGln were also lower, suggesting that stuck ribosomes in dom34Δ strains lack tRNAs, although we cannot exclude the possibility that they are occupied by a special class of tRNAs for which we did not probe. The lack of tRNAs on a ribosome could occur after the release of the peptidyl tRNA following translation termination or after translational stalling. On the other hand, in the polysomes of the dom34Δ strain, tRNA/ribosome levels are actually 20% higher than those in wild-type cells. This observation suggests that tRNA on these ribosomes may be associated with the mRNA for longer periods of time due to a nonresolution of translational stalling events. Hence, stuck ribosomes may be the consequence of an incomplete resolution of translational stalling events.
The 80S monosomes that accumulate in Dom34-deficient strains are not associated with no-go mRNA.
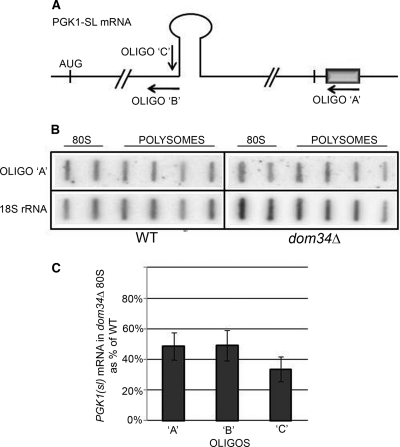
Since Dom34 has been implicated in the resolution of no-go translation complexes (5), we asked whether stuck ribosomes were attached to no-go mRNA. The distribution of a stem-loop no-go PGK1(sl) mRNA across a polysome gradient was determined by using slot blotting so that we could identify fragmented, as well as intact, no-go mRNAs, using the three oligonucleotides indicated in Fig. 7A. Representative data are shown in Fig. 7B. The analysis of three independent experiments, using the three probes, is summarized in Fig. 7C. From the distribution of stem-loop mRNAs between 80S monosomes and polysomes, it is clear that there is no specific enrichment of the stem-loop mRNA in the monosomes that accumulate in the absence of Dom34 (Fig. 7B and C). Indeed, it is the wt strain that seems to accumulate stem-loop RNA (or its 3′ fragment) in monosomes, perhaps during the Dom34-catalyzed resolution of the stem-loop obstruction. Subsequently, the slot blot was probed with oligonucleotides complementary to the sequences within (oligonucleotide C) or just upstream of (oligonucleotide B) the stem-loop, regions that might be sequestered within the ribosome as a result of no-go decay. The monosomes of the dom34Δ strain had substantially less signal from either probe (Fig. 7C). Thus, neither intact PGK1(sl) mRNA nor fragments in the vicinity of the no-go impediment accumulate in the 80S monosomes when Dom34 is missing, leading us to conclude that stuck ribosomes are not held together by a fragment of mRNA.
FIG. 7.
Distribution of “no-go” mRNA in 80S and polysomes. (A) The wild-type or dom34Δ strain was transformed with plasmids containing the PGK1(sl) gene under the control of the GAL1 promoter and tagged at the 3′ end with a string of G residues (gray box) and containing a strong stem-loop within the ORF (5). The sites of oligonucleotides A, B, and C are indicated. (B) RNA was prepared from fractions of 1.5 mM Mg2+ sucrose gradients of each strain and was applied onto slot blots, which were probed with oligonucleotide A, complementary to the tagged PGK1(sl) transcripts, and subsequently with an oligonucleotide complementary to 18S rRNA as a measure of ribosome content. The intensity of the individual bands was quantitated with a phosphorimager, and the values for the 80S and polysomal regions were individually totaled. (C) The proportion of PGK1(sl) mRNA in 80S monosomes, normalized to 18S rRNA, is shown compared to that present in the combined 80S and polysome fractions, with values for the wild-type strain set to 1.0. The slot blot was also probed with oligonucleotides complementary to the sequences within (oligonucleotide C) and just upstream of (oligonucleotide B) the stem-loop. Note that the latter will identify not only the no-go transcripts but also those from the endogenous PGK1.
To ask more generally whether stuck ribosomes are associated with mRNA, we asked whether there is an increased level of mRNA in the 80S peak of extracts of the dom34Δ strain analyzed in low-Mg2+ gradients (Table 4). For the 10 mRNAs analyzed, the amount of mRNA in the 80S peak rose only from about 5% of the total in the wt to 7% in the dom34Δ strain. This is far too little to furnish each stuck ribosome with an mRNA (see Discussion).
TABLE 4.
Accumulation of mRNA in 80S ribosomesa
| Gene | Avg % mRNA ± SD |
|||
|---|---|---|---|---|
| 80S |
Polysome |
|||
| wt | dom34Δ | wt | dom34Δ | |
| ACT1 | 3.3 ± 0.5 | 4.5 ± 1.8 | 90.2 ± 2.0 | 93.1 ± 4.0 |
| TUB1 | 4.8 ± 0.0 | 7.0 ± 0.6 | 81.8 ± 3.4 | 85.9 ± 5.0 |
| TDH3 | 2.0 ± 0.3 | 2.6 ± 0.8 | 93.5 ± 2.4 | 96.0 ± 0.1 |
| CPR1 | 4.2 ± 0.9 | 6.1 ± 1.0 | 90.3 ± 3.4 | 91.7 ± 3.3 |
| ZEO1 | 6.9 ± 0.8 | 9.1 ± 1.9 | 80.8 ± 6.4 | 86.8 ± 5.2 |
| RPL1B | 8.3 ± 0.2 | 10.5 ± 0.4 | 75.0 ± 2.2 | 80.8 ± 7.3 |
| RPL3 | 4.3 ± 0.7 | 5.7 ± 0.8 | 86.2 ± 0.1 | 91.8 ± 3.5 |
| RPL4B | 4.6 ± 0.1 | 6.1 ± 0.4 | 83.1 ± 0.5 | 89.7 ± 2.4 |
| RPL28 | 8.1 ± 1.6 | 11.2 ± 0.5 | 80.3 ± 3.9 | 84.4 ± 4.1 |
| RPS6B | 6.5 ± 0.4 | 7.8 ± 0.6 | 83.9 ± 3.0 | 88.6 ± 3.4 |
| Avg | 5.3 | 7.1 | 84.5 | 88.9 |
Extracts of wild-type and Δdom34 strains were analyzed in low-Mg2+ sucrose gradients, which were divided into four fractions, representing the top, the 40 and 60S subunits, the 80S monosomes, and the polysomes. RNA was isolated from each fraction and subjected to qPCR analysis using oligonucleotides specific for the indicated genes. The percentages of total mRNA found in the 80S peak and in the polysome fraction are shown. The data represent the averages of data from two independent experiments.
A triple-null rpl4aΔ rps6aΔ dom34Δ mutant rescues the slow-growth phenotype of the rps6aΔ dom34Δ strain.
In contrast to the dom34Δ rps6aΔ strain, the polysome profile of the dom34Δ rpl4aΔ strain (Fig. 4F) shows an availability of 40S subunits, no accumulation of 60S subunits, and the presence of half-mer polysomes, with an initiated 40S subunit awaiting a scarce 60S subunit (30). The lack of Dom34 does not influence the level of half-mers in an rpl4aΔ strain (Fig. 4E and F). The additional 60S subunits sequestered in the stuck ribosomes appear to be balanced by the sequestered 40S subunits in the stuck ribosomes of the double mutant. It is interesting that the near-normal growth rate of the dom34Δ rpl4aΔ strain (Table 3) suggests again that there is a substantial excess capacity in the translation system during favorable growth conditions.
Is the slow growth of the dom34Δ rps6aΔ strain due to the accumulation of 60S subunits that could sequester the scarce 40S subunits to form translationally inactive 80S couples? Indeed, an rpl4aΔ rps6aΔ dom34Δ triple mutant (ɛ = −0.04) (Table 3) substantially rescues the synthetic growth defect of the dom34Δ rps6aΔ strain (ɛ = −0.24) (Table 3). This notion was confirmed by comparing the polysome profiles of the double and triple mutants (Fig. 4D and G). The triple mutant showed an increase in the level of free 40S subunits at the expense of 80S monosomes, thereby promoting higher rates of translation initiation. Overall, it appears that the balance of large and small subunits is important for maximal translational efficiency, particularly under circumstances where the concentration of free 40S subunits available for translation initiation is limiting.
These results explain the apparent anomaly that Dom34 is far more important for cells depleted of 40S subunits than for cells depleted of 60S subunits. The accumulation of stuck 80S subunits exacerbates a shortage of 40S subunits already depleted by insufficient synthesis and by the sequestering of 40S subunits in transient 80S monosomes due to the high concentration of free 60S subunits.
DISCUSSION
Large-scale genetic screens such as SGA analysis have generally been used to reveal unexpected relationships between genes, between pathways, or between biological processes. Here we found a surprising genetic interaction between DOM34 and the members of a single large complex, the 40S small ribosomal subunit.
Dom34 and 40S subunits.
The role of the ribosome in translation can be subdivided into four events: initiation, elongation, termination, and recycling (14, 15). Synthetic sickness between DOM34 and RPS genes appears to be due primarily to a deficiency in the rate of translation initiation, as evident from the very low polysome-to-monosome ratio of double mutants (Fig. 4). This conclusion is strengthened by the observation that the synthetic sick phenotype is exacerbated by low temperatures (Fig. 2D) at which the initiation/elongation balance disfavors initiation.
The deficiency of translation initiation in the dom34Δ rps6aΔ strain is caused by the combination of three factors. First, the deletion of RPS6A reduces small-subunit biosynthesis, leading to a substantial deficit of 40S subunits. This limits the availability of small subunits to initiate on mRNA, resulting in a 30% reduction in the growth rate (Table 3). Second, the accumulated free 60S subunits sequester the rare free 40S subunits in nonproductive 80S couples due to the natural equilibrium between free and paired subunits (see Fig. 4 versus Fig. 5). Thus, growth is partially restored by the reduction of 60S subunits when an RPS deletion is combined with an RPL deletion (Table 3). Finally, the lack of Dom34 results in an accumulation of monosomes (Fig. 4 and 5) (4). We have shown above that these stuck ribosomes are resistant to dissociation under conditions of low Mg2+ concentrations and are basically inactive, apparently with little tRNA (Fig. 6).
Stuck ribosomes.
An important unresolved issue is whether stuck 80S ribosomes have mRNA. On the one hand, the presence of mRNA, or a fragment of mRNA, would explain their resistance to dissociation at low Mg2+ concentrations. Furthermore, in vitro analysis of recycling after normal (eRF1/eRF3) termination suggests that both initiation factors and the ATPase ABCE1 (Rli1 in yeast) are necessary for separating the subunits before mRNA is released (28). However, the occurrence of 80S runoff ribosomes in the presence of a genetic or thermal inhibition of translation initiation suggests that there may be exceptions to the rule of stepwise dissociation (13).
On the other hand, the mRNA in the 80S peak at low Mg2+ concentrations, determined by qPCR analysis (Table 4), is increased only from ∼5% of the total in wt cells to 7% in dom34Δ cells for the dozen or so mRNAs that we have measured. The cell contains about 200,000 ribosomes (38) and 50,000 mRNAs (41). If 10% of the ribosomes are stuck on an mRNA, then 20,000 mRNAs, nearly 40% of the total, should be in the 80S peak. It is clearly unlikely for each stuck ribosome to be associated with an mRNA. Another possibility, since Dom34 has been implicated in resolving no-go translation, is that stuck ribosomes could retain fragments of mRNA. While we cannot exclude that general possibility, we found no evidence for no-go mRNA or for fragments near the no-go site in the stuck ribosomes (Fig. 7C). Thus, the question remains open.
The origin of stuck ribosomes.
What is the basis for the accumulation of stuck ribosomes in cells lacking Dom34? We considered the possibility that stuck ribosomes represent aberrant initiation complexes. If so, one would expect them to be enriched in the initiator tRNA. On the contrary, they seem to lack not only initiator tRNA but elongator tRNAs as well (Fig. 6), suggesting that aberrant initiation is not the source of stuck ribosomes. Since we have tested only three tRNA species, it is possible that stuck ribosomes could contain rare or specialized tRNAs.
Stuck ribosomes could have defects in subunit separation after the termination of translation during ribosome recycling. Little is known about this process in vivo. In vitro, recycling after the translation of a very short ORF appears to be catalyzed by initiation factors, particularly eIF3, which is known to bind to 40S subunits to prevent them from joining 60S subunits prematurely (27). While in vitro analysis implicates eIF3j in releasing 40S subunits from the mRNA (27), the yeast homologue of eIF3j, Hcr1, is not essential for growth, nor did we find any genetic interaction between HCR1 and DOM34 in a double-deletion strain (Table 3). Thus, although the mechanism for normal recycling remains somewhat unclear, there has been no evidence that Dom34 is involved in this process.
Dom34 has been implicated in resolving no-go translation (5, 24) as well as in the identification or the degradation of defective 40S ribosomal subunits (2). Cryo-electron microscopy (EM) analysis finds that a portion of Dom34 itself fills the A site of a ribosome whose progress has stalled because of a strong stem-loop in the mRNA and that its presence seems to loosen the mRNA-ribosome interaction (T. Becker and R. Beckmann, personal communication). In vitro, for a translating ribosome with a “hungry” codon, i.e., when there is no available amino-acyl tRNA, Dom34 can cause the release of its peptidyl tRNA and the separation of the 40S and 60S subunits (32).
Based on such considerations, we postulate that stuck ribosomes result from the failure of Dom34 to resolve a ribosome that has become stalled during translation due to insufficient tRNAs, to structural problems with its mRNA, or to a defect in the ribosome itself. In the absence of Dom34, a ribosome which has stalled for any of a number of reasons may have no physiological release and may remain as a stuck monosome. What is unclear is whether the 10% of the ribosomes that are stuck as free 80S ribosomes represent only the tip of the iceberg; i.e., does the absence of Dom34 cause many stuck ribosomes to be present in polyribosomes, awaiting release?
Effects on cell growth.
The absence of Dom34 in otherwise wild-type cells has a relatively minor effect on overall growth. Since in these cells, about 10% of the ribosomes are stuck, this observation suggests that a log-phase cell has an excess of ribosomes. Indeed, there is little understanding of the factors that determine the level of ribosomes in a cell and the mRNA/ribosome ratio.
Why does DOM34 display synthetic sick interactions with RPS but not RPL genes? The situations are not parallel. With a deficit of 60S subunits, most of the excess 40S subunits are associated with mRNA in the form of “half-mers” (30) awaiting the rare free 60S subunit. Thus, excess 40S subunits do not sequester the scarce free 60S subunits. In contrast, the excess free 60S subunits accumulate in a form that can sequester the rare free 40S subunits. We propose a role for Dom34 in the resolution/separation of stalled ribosomes and provide physiological evidence that the nonresolution of stalled translation events, when accompanied by an insufficient production of 40S ribosomal subunits and the formation of inactive couples, leads to a severe limitation in translation initiation and, necessarily, to reduced growth.
What are 80S ribosomes?
While much attention has been showered on polyribosomes hard at work translating mRNAs and on 40S and 60S subunits, either as newly formed additions to the cytoplasm or as recycled actors ready for the next stage, the ever-present 80S monomers have usually been ignored. While some are clearly translating very short mRNAs, most are not (data not shown). Do such 80S couples actually exist in the cell, with its apparently low concentration of Mg ions? Does the >100-fold-higher concentration of ribosomes in the cytoplasm (versus a sucrose gradient) force the subunits into union? Do these ribosomes indeed serve as a pool, ready to release subunits on the demand of the initiation factors, particularly eIF3, one of whose jobs is to keep them apart? Do these excess ribosomes permit normal growth when ribosome numbers are reduced? Do the stuck ribosomes eventually recycle to the free subunit pool? Attention should be paid!
Acknowledgments
We are grateful to Roy Parker for the no-go plasmids, to Uma Maitra for fruitful discussions, to Robyn Moir for advice on SGA, and to Thomas Becker, Roland Beckmann, Chris Shoemaker, and Rachel Green for personal communications.
This research was supported in part by the following grants from the NIH: grants GM25532 and ARRAGM 25532-S1 to J.R.W., GM085177 to I.M.W., and CAI-3330 to the Albert Einstein Cancer Center.
Footnotes
Published ahead of print on 27 September 2010.
REFERENCES
- 1.Carr-Schmid, A., C. Pfund, E. A. Craig, and T. G. Kinzy. 2002. Novel G-protein complex whose requirement is linked to the translational status of the cell. Mol. Cell. Biol. 22:2564-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole, S. E., F. J. LaRiviere, C. N. Merrikh, and M. J. Moore. 2009. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol. Cell 34:440-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costanzo, M., A. Baryshnikova, J. Bellay, Y. Kim, E. D. Spear, C. S. Sevier, H. Ding, J. L. Koh, K. Toufighi, S. Mostafavi, J. Prinz, R. P. St. Onge, B. VanderSluis, T. Makhnevych, F. J. Vizeacoumar, S. Alizadeh, S. Bahr, R. L. Brost, Y. Chen, M. Cokol, R. Deshpande, Z. Li, Z. Y. Lin, W. Liang, M. Marback, J. Paw, B. J. San Luis, E. Shuteriqi, A. H. Tong, N. van Dyke, I. M. Wallace, J. A. Whitney, M. T. Weirauch, G. Zhong, H. Zhu, W. A. Houry, M. Brudno, S. Ragibizadeh, B. Papp, C. Pal, F. P. Roth, G. Giaever, C. Nislow, O. G. Troyanskaya, H. Bussey, G. D. Bader, A. C. Gingras, Q. D. Morris, P. M. Kim, C. A. Kaiser, C. L. Myers, B. J. Andrews, and C. Boone. 2010. The genetic landscape of a cell. Science 327:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis, L., and J. Engebrecht. 1998. Yeast dom34 mutants are defective in multiple developmental pathways and exhibit decreased levels of polyribosomes. Genetics 149:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doma, M. K., and R. Parker. 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 440:561-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fei, J., P. Kosuri, D. D. MacDougall, and R. L. Gonzalez, Jr. 2008. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol. Cell 30:348-359. [DOI] [PubMed] [Google Scholar]
- 8.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 9.Hartwell, L. H., and C. S. McLaughlin. 1968. Temperature-sensitive mutants of yeast exhibiting a rapid inhibition of protein synthesis. J. Bacteriol. 96:1664-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hartwell, L. H., and C. S. McLaughlin. 1969. A mutant of yeast apparently defective in the initiation of protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 62:468-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, W., and R. Parker. 2000. Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 12:346-350. [DOI] [PubMed] [Google Scholar]
- 12.Holstege, F. C. P., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 13.Jackson, R. J. 2007. The missing link in the eukaryotic ribosome cycle. Mol. Cell 28:356-358. [DOI] [PubMed] [Google Scholar]
- 14.Jackson, R. J., C. U. Hellen, and T. V. Pestova. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kapp, L. D., and J. R. Lorsch. 2004. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 73:657-704. [DOI] [PubMed] [Google Scholar]
- 16.Kruse, C., S. P. Johnson, and J. R. Warner. 1985. Phosphorylation of the yeast equivalent of ribosomal protein S6 is not essential for growth. Proc. Natl. Acad. Sci. U. S. A. 82:7515-7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lecompte, O., R. Ripp, J. C. Thierry, D. Moras, and O. Poch. 2002. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. 30:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Li, B., C. R. Nierras, and J. R. Warner. 1999. Transcriptional elements involved in the repression of ribosomal protein synthesis. Mol. Cell. Biol. 19:5393-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loar, J. W., R. M. Seiser, A. E. Sundberg, H. J. Sagerson, N. Ilias, P. Zobel-Thropp, E. A. Craig, and D. E. Lycan. 2004. Genetic and biochemical interactions among Yar1, Ltv1 and Rps3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics 168:1877-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maicas, E., F. G. Pluthero, and J. D. Friesen. 1988. The accumulation of three yeast ribosomal proteins under conditions of excess mRNA is determined primarily by fast protein decay. Mol. Cell. Biol. 8:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mani, R., R. P. St. Onge, J. L. Hartman, G. Giaever, and F. P. Roth. 2008. Defining genetic interaction. Proc. Natl. Acad. Sci. U. S. A. 105:3461-3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ni, L., and M. Snyder. 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12:2147-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nygard, O., and H. Nika. 1982. Identification by RNA-protein cross-linking of ribosomal proteins located at the interface between the small and the large subunits of mammalian ribosomes. EMBO J. 1:357-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nygard, O., and L. Nilsson. 1990. Translational dynamics. Interactions between the translational factors, tRNA and ribosomes during eukaryotic protein synthesis. Eur. J. Biochem. 191:1-17. [DOI] [PubMed] [Google Scholar]
- 24.Passos, D. O., M. K. Doma, C. J. Shoemaker, D. Muhlrad, R. Green, J. Weissman, J. Hollien, and R. Parker. 2009. Analysis of Dom34 and its function in no-go decay. Mol. Biol. Cell 20:3025-3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, V. Jojic, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113:919-933. [DOI] [PubMed] [Google Scholar]
- 26.Petitjean, A., N. Bonneaud, and F. Lacroute. 1995. The duplicated Saccharomyces cerevisiae gene SSM1 encodes a eucaryotic homolog of the eubacterial and archaebacterial L1 ribosomal proteins. Mol. Cell. Biol. 15:5071-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pisarev, A. V., C. U. Hellen, and T. V. Pestova. 2007. Recycling of eukaryotic posttermination ribosomal complexes. Cell 131:286-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pisarev, A. V., M. A. Skabkin, V. P. Pisareva, O. V. Skabkina, A. M. Rakotondrafara, M. W. Hentze, C. U. Hellen, and T. V. Pestova. 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol. Cell 37:196-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romani, A. M., and A. Scarpa. 2000. Regulation of cellular magnesium. Front. Biosci. 5:D720-D734. [DOI] [PubMed] [Google Scholar]
- 30.Rotenberg, M. O., M. Moritz, and J. L. Woolford, Jr. 1988. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 2:160-170. [DOI] [PubMed] [Google Scholar]
- 31.Sethy-Coraci, I., R. D. Moir, A. Lopez-de-Leon, and I. Willis. 1998. A differential response of wild type and mutant promoters to TFIIIB70 overexpression in vivo and in vitro. Nucleic Acids Res. 26:2344-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shoemaker, C. J., D. E. Eyler, and R. Green. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop off to initiate no-go decay. Science, in press. [DOI] [PMC free article] [PubMed]
- 33.Soudet, J., J. P. Gelugne, K. Belhabich-Baumas, M. Caizergues-Ferrer, and A. Mougin. 2010. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 29:80-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tong, A. H., and C. Boone. 2006. Synthetic genetic array analysis in Saccharomyces cerevisiae. Methods Mol. Biol. 313:171-192. [DOI] [PubMed] [Google Scholar]
- 35.Tsay, Y. F., J. R. Thompson, M. O. Rotenberg, J. C. Larkin, and J. L. Woolford, Jr. 1988. Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes Dev. 2:664-676. [DOI] [PubMed] [Google Scholar]
- 36.Valasek, L., B. Szamecz, A. G. Hinnebusch, and K. H. Nielsen. 2007. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol. 429:163-183. [DOI] [PubMed] [Google Scholar]
- 37.Warner, J. R. 1977. In the absence of ribosomal RNA synthesis, the ribosomal proteins of HeLa cells are synthesized normally and degraded rapidly. J. Mol. Biol. 115:315-333. [DOI] [PubMed] [Google Scholar]
- 38.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 39.Warner, J. R., G. Mitra, W. F. Schwindinger, M. Studeny, and H. M. Fried. 1985. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol. Cell. Biol. 5:1512-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, X., and J. R. Warner. 2001. Expression of a micro-protein. J. Biol. Chem. 276:33821-33825. [DOI] [PubMed] [Google Scholar]
- 41.Zenklusen, D., D. R. Larson, and R. H. Singer. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15:1263-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]