Abstract
In comparison to other pseudomonads, Pseudomonas aeruginosa grows poorly in l-lysine as a sole source of nutrient. In this study, the ldcA gene (lysine decarboxylase A; PA1818), previously identified as a member of the ArgR regulon of l-arginine metabolism, was found essential for l-lysine catabolism in this organism. LdcA was purified to homogeneity from a recombinant strain of Escherichia coli, and the results of enzyme characterization revealed that this pyridoxal-5-phosphate-dependent decarboxylase takes l-lysine, but not l-arginine, as a substrate. At an optimal pH of 8.5, cooperative substrate activation by l-lysine was depicted from kinetics studies, with calculated Km and Vmax values of 0.73 mM and 2.2 μmole/mg/min, respectively. Contrarily, the ldcA promoter was induced by exogenous l-arginine but not by l-lysine in the wild-type strain PAO1, and the binding of ArgR to this promoter region was demonstrated by electromobility shift assays. This peculiar arginine control on lysine utilization was also noted from uptake experiments in which incorporation of radioactively labeled l-lysine was enhanced in cells grown in the presence of l-arginine but not l-lysine. Rapid growth on l-lysine was detected in a mutant devoid of the main arginine catabolic pathway and with a higher basal level of the intracellular l-arginine pool and hence elevated ArgR-responsive regulons, including ldcA. Growth on l-lysine as a nitrogen source can also be enhanced when the aruH gene encoding an arginine/lysine:pyruvate transaminase was expressed constitutively from plasmids; however, no growth of the ldcA mutant on l-lysine suggests a minor role of this transaminase in l-lysine catabolism. In summary, this study reveals a tight connection of lysine catabolism to the arginine regulatory network, and the lack of lysine-responsive control on lysine uptake and decarboxylation provides an explanation of l-lysine as a poor nutrient for P. aeruginosa.
Decarboxylation of amino acids, including lysine, arginine, and glutamate, is important for bacterial survival under low pH (2, 7, 19). Lysine is abundant in the rhizosphere where fluorescent Pseudomonas preferentially resides, and serves as a nitrogen and carbon source to these organisms (28). In microbes, lysine catabolism can be initiated either through monooxygenase, decarboxylase, or transaminase activities. The monooxygenase pathway has been considered the major route for l-lysine utilization in Pseudomonas putida, and davBATD encoding enzymes for the first four steps of the pathway have been characterized (25, 26). In contrast, Pseudomonas aeruginosa cannot use exogenous l-lysine efficiently for growth (5, 24). It has been reported that enzymatic activities for the first two steps of the monooxygenase pathway are not detectable in P. aeruginosa, and no davBA orthologs can be identified from this organism (24, 25).
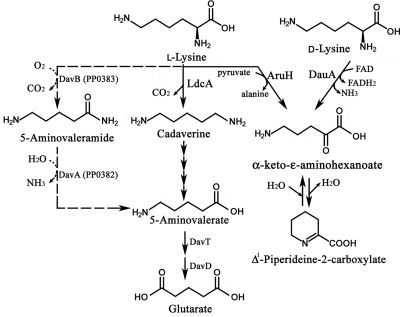
Mutants of P. aeruginosa with improved growth on l-lysine and a high level of lysine decarboxylase activity can be isolated by repeated subcultures in l-lysine (5). This suggests that in P. aeruginosa, l-lysine utilization might be mediated by the lysine decarboxylase pathway with cadaverine and 5-aminovalerate as intermediates (Fig. 1). Alternatively, conversion of l-lysine into 5-aminovalerate may also be accomplished by a coupled reaction catalyzed by AruH and AruI. The AruH and AruI enzymes were reported as arginine:pyruvate transaminase and 2-ketoarginine decarboxylase, respectively (36). Interestingly, transamination by AruH using l-lysine as an amino group donor can also be detected in vitro (35). The reaction product α-keto-ɛ-aminohexanonate can potentially be decarboxylated into 5-aminovalerate by AruI, providing an alternative route for lysine degradation.
FIG. 1.
Lysine catabolic pathways. l-lysine decarboxylase pathway is shown at center. Broken arrows represent lysine monooxygenase pathway from P. putida which is not present in P. aeruginosa.
In this study, we showed that the lysine decarboxylase pathway is the main route for lysine utilization under arginine control. Expression of the ldcAB operon encoding l-lysine decarboxylase and a putative lysine/cadaverine antiporter was analyzed regarding its response to l-lysine, l-arginine, and the arginine-responsive regulator ArgR. Enzyme characterization was performed to verify the function of LdcA as l-lysine decarboxylase. Arginine control on lysine incorporation was also investigated by genetic studies and uptake experiments. The peculiar role of ArgR controlling arginine and lysine uptake and catabolism provides the explanation for poor growth in lysine, and it implies a higher level of complexity in metabolic networks of pseudomonads.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1 . Escherichia coli and P. aeruginosa strains were grown in Luria-Bertani (LB) medium supplemented with antibiotics when indicated at conventional concentrations (36). The minimal medium P (MMP) containing the indicated carbon and nitrogen sources at 20 mM and 5 mM, respectively, was used for the growth of P. aeruginosa (8).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− φ80dlac ΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17 (rK− mK−) supE44 λ−thi-1 gyrA96 relA | Bethesda Research Laboratories |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Smr) endA1 nupG | Invitrogen |
| SM10 | thi-1 thr leu tonA lacY supE recA::RP4-2-Tc::Mu (Kmr) | 31 |
| P. aeruginosa strains | ||
| PAO1 | Wild type | 8 |
| PAO1-Smr | Spontaneous Smr mutant strain of PAO1 | 11 |
| PAO501 | argR::Gmr | 23 |
| PAO1214 | aotJ::Tn5-751 insertion mutant of PAO1 | 23 |
| PAO5715 | ΔaruCFGDBE::Gmr | This study |
| PAO5716 | ldcA::Tcr | This study |
| PAO5717 | ldcB::Tcr | This study |
| PAO5602 | aruH::Tcr ΔaruF | 36 |
| PAO5718 | ΔaruCFGDBE::GmrldcA::Tcr | This study |
| PAO5720 | aruH::Tcr | This study |
| PAO5721 | aruI::Tcr | This study |
| PAO5802 | dauA::Tcr | 8, 14 |
| Plasmids | ||
| pRTP1 | Ampr Sms conjugation vector | 32 |
| pRTP2 | pRTP1 derivative; EcoRI site deleted | 13 |
| pGMΩ1 | bla gen; gentamicin resistance gene cassette with omega loop on both ends | 29 |
| pUCP18 | Escherichia-Pseudomonas shuttle vector | 30 |
| pHC5306 | pUCP18 derivative carrying ldcA gene | This study |
| pHC5307 | pUCP18 derivative carrying aruH gene | This study |
| pBAD-His6 | Modified protein expression vector inducible by arabinose | 13 |
| pBAD-ldcA | 6His-tagged LdcA protein expression vector | This study |
| pQF50 | bla lacZ transcriptional fusion vector | 4 |
| pQF52 | bla lacZ translational fusion vector derived from pQF50 | 23 |
| pST500 | aotJ::lacZ translational fusion of pQF52 | 22 |
| pZY5 | PA4980::lacZ translational fusion of pQF52 | 36 |
| pHT1818 | ldcA::lacZ transcriptional fusion of pQF50 | This study |
Kmr, kanamycin resistance; Smr, streptomycin resistance; Sms, streptomycin sensitive; Tcr, tetracyclin resistance; Gmr, gentamicin resistance; Ampr, ampicillin resistance.
Construction of ldcA::lacZ fusion.
The upstream regulatory region of the ldcAB operon covering 635 bp upstream of the start codon and the first 24 bp of the structural gene was PCR amplified from the genomic DNA of P. aeruginosa PAO1 using the following primers: 5′-TCACTCTGGGCGCAAGCTTAGGCGCCGGTCGGC-3′ and 5′-GGGAAATTTGAGGTCTTT-3′. PCR product was cloned into pQF50 and confirmed by DNA sequencing.
Construction of mutant strains.
For tetracycline-resistant mutants, DNA fragments covering the genes of interest were PCR amplified from PAO1 genomic DNA using the following primers: for ldcAB mutants, 5′-ATGTATAAAGACCTCAAATTTCCCGTCCTC-3′ and 5′-TCAGTCATTGGCTTTGAGCGTCGGCACTCC-3′; and for aruHI mutants, 5′-GTCTAAGCTTGACTGGCCTGGCGCGCGTCG-3′ and 5′-CGCAAGCTTCGGGCAGTCCGGCGTGACCCT-3′. PCR products were cloned into a conjugation vector, pRTP1 (32), and the tetracycline resistance cassette was introduced by using the EZ-Tn5 <TET-1> insertion system (Epicentre). For gentamicin deletion mutants, two flanking regions of a targeted gene were amplified by PCR with the following primers: 5′-GAGGGATCACTCGGGTGCATACTTCTTCTACG-3′, 5′-GAGGAATTCAGCGCT TCCAGGTCGTTGTAG-3′, 5′-GAGGAATTCGAGCGCTACGTCGAACAGGACATGA-3′ and 5′-GAGAAGCTTCAATCCGAGCAGGTTGCTCATGGTC-3′. The PCR products were cloned into pRTP1. A cassette carrying the gentamicin resistance gene from pGMΩ1 was inserted into the conjunction of the two DNA fragments (29). For gene replacement, E. coli SM10 was served as the donors in biparental mating with PAO1-Smr (6). The desired knockout mutants were selected on LB plates containing streptomycin and either gentamicin or tetracycline, and the mutation was confirmed by PCR.
Constitutive expression from pUCP18.
Full-length ldcA (PA1818) and aruIH (PA4976 and PA4977) genes were successfully PCR amplified and subcloned into pUCP18 to construct pHC5306 and pHC5307 using the following primers: PA1818F, 5′-TGAAGATCTGAGGAGTCAACAATGTATAAAGACCTC-3′; PA1818R, 5′-CCCAAGCTTTCATTCCTTTATGCATTCAACGGT-3′; PA4976F 5′-CACGGATCCGCTTGTGGGAATGGGAGCAAGAGC-3′; and PA4976R, 5′-CACAAGCTTCATCGTGGTTTCCGAATCG TGGTGAC-3′. These plasmids can reach 10 to 25 copy numbers per cell with constitutive expression in P. aeruginosa (30).
Measurements of β-galactosidase activity.
The cells were grown in the minimal medium P containing the carbon and nitrogen sources as indicated. Cells in the mid-log phase were harvested by centrifugation and then passed through a French press cell at 8,500 lb/in2. The cell debris were removed by centrifugation at 20,000 × g for 15 min, at 4°C, and protein concentrations in the crude extracts were determined by the Bradford method (1). The levels of β-galactosidase activity were measured using Miller's method (18).
Gel retardation assay.
A DNA fragment covering 635 bp upstream of the ATG start codon and the first 24-bp coding sequence of ldcA was PCR amplified from P. aeruginosa PAO1 chromosome using the designed primers. The DNA probe was labeled with [γ-32P]dATP by polynucleotide kinase. The radioactively labeled DNA probe (0.1 nM) was allowed to interact with different concentrations of the purified ArgR in a 20-μl reaction mixture containing 20 mM Tris-HCl (pH 7.6), 50 mM KCl, 1 mM EDTA, 5% (vol/vol) glycerol, and 50 μg/ml bovine serum albumin. The reaction mixtures were incubated for 15 min at room temperature before applying them to a 5% polyacrylamide gel in Tris-acetate-EDTA running buffer. After being dried, the gel was autoradiographed by exposure to a phosphorimager plate (Fuji).
Overexpression and purification of LdcA.
Full-length ldcA was amplified from the genomic DNA of P. aeruginosa using the following designed primers: 1818F, 5′-TATAAAGACCTCAAATTTCCCGTC-3′; and 1818R, 5′-TCATTCCTTTATGCATTCAACGGT-3′. The PCR product was subcloned into pBAD-HisD, a modified pBAD expression vector (13, 35). Recombinant 6×His-LdcA was expressed in Escherichia coli TOP10 (Invitrogen Life Technologies) by arabinose induction. Cell extract was obtained by passage through a French pressure cell at 8,500 lb/in2 followed by centrifugation at 25,000 × g for 30 min at 4°C. Soluble His-LdcA protein was purified from a HisTrap HP column (GE Healthcare) at the concentration of 100 mM imidazole. Eluted fractions detected by UV were analyzed by SDS-PAGE, pooled together, and concentrated by an Amicon Ultra-15 centrifugal filter unit (Millipore). Active fractions were determined by a lysine decarboxylase assay as described below. Protein concentration was determined by the method of Bradford (1).
Measurements of lysine decarboxylase activity.
Purified His-LdcA was used to test l-lysine decarboxylation in vitro. Enzyme-catalyzed decarboxylation was assayed by measuring the liberated 14CO2 at 37°C as previously described, with slight modification (34). The assay was carried out in a standard scintillation vial, and the liberated 14CO2 was trapped by a filter paper preimpregnated with 0.1 ml barium hydroxide in the cap. One milliliter of standard reaction mixture contains 125 μM pyridoxal 5′-phosphate, 100 μg/ml acetylated bovine serum albumin, 0.2 μCi [14C]-l-lysine, and 3 mM cold l-lysine in 100 mM Tris buffer at pH 8.5. The mixture was incubated at 37°C in a water bath, and the reaction was started by adding 1.5 μg of purified LdcA. Reaction was stopped by adding seven drops of 1N H2SO4, and the incubation was continued for 30 min more for complete absorption of CO2. Labeled CO2 was measured in a liquid scintillation counter. Apparent kinetics parameters were determined using nonlinear regression equations of the kinetics module of SigmaPlot 9.0 software.
l-lysine uptake experiments.
Radiolabeled l-lysine was used for uptake assays as previously described, with slight modification (15). Cultures were grown in glutamate-MMP in the absence or presence of 20 mM l-arginine or l-lysine. Cells were harvested during logarithmic growth (optical density at 600 nm [OD600] of 0.5 to 0.8), washed twice, and resuspended at a concentration of ca. 108 cells/ml (OD600 = 0.1) using MMP containing chloramphenicol (250 μg/ml). After incubation of the cell suspension for 5 min in a 37°C water bath, 14C-labeled l-lysine was added to a final concentration of 20 μM (10 mCi/mmole), and samples (0.5 ml) were withdrawn at various time intervals. Cells were collected on a cellulose membrane filter (0.22-μm-pore size, type GS; Millipore) and washed with 10 ml of MMP. Membranes were air dried in clean scintillation vials overnight. Incorporated radioactivity was measured using adequate scintillation liquid and a spectrometer (Beckman).
RESULTS
ArgR-dependent induction of the ldcA promoter by arginine but not by lysine.
The putative ldcAB operon was initially identified as a member of the ArgR regulon from transcriptome analysis (16). ArgR is autoinducible from the aot-argR operon and is initially identified as the major regulator of aerobic arginine catabolism and biosynthesis in response to arginine (23). The ldcA promoter in response to exogenous l-arginine and l-lysine was tested by measurements of β-galactosidase activities in PAO1 harboring pHT1818, a PldcA::lacZ fusion. As shown in Table 2, exogenous arginine but not lysine exerts a strong induction effect (12-fold) on the ldcA promoter.
TABLE 2.
Promoter activities of aotJ and ldcA in response to arginine and lysine
| Strain | Description or genotype | Nutrientsa | Sp act (mmole/mg/min) for:b |
|
|---|---|---|---|---|
| PldcA::lacZ | PaotJ::lacZ | |||
| PAO1 | Wild type | G | 0.8 (1) | 14 (1) |
| A + G | 9.5 (12) | 61 (4) | ||
| L + G | 1.2 (1.5) | 18 (1.2) | ||
| PAO5715 | ΔaruCFGDBE::Gmr | G | 4.1 (1) | 58 (1) |
| A + G | 16.0 (4) | 105 (2) | ||
| L + G | 4.0 (1) | 26 (0.5) | ||
Cells were grown in minimal medium P supplemented with 20 mM lysine (L), arginine (A), or glutamate (G).
Specific activities represent the averages from three measurements with standard errors below 5%. Fold changes are in parentheses.
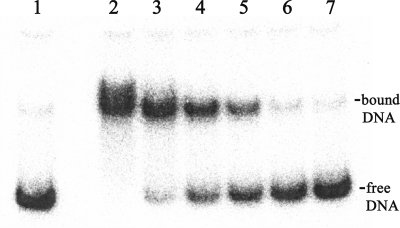
A possible ArgR binding site (from 120 bp to 81 bp upstream of the ATG start codon) was identified previously in the ldcA regulatory region (9) based on its sequence similarity to the consensus ArgR binding site (16). Electro-mobility shift assays were conducted to demonstrate the binding of ArgR to the ldcA regulatory region. As shown in Fig. 2, ArgR binding to the ldcAB regulatory region can be detected using this approach, with an estimated dissociation constant of 1.0 nM.
FIG. 2.
Gel retardation assay for the binding of ArgR to the ldcA promoter. Binding reactions of radioactively labeled regulatory region with different concentrations of purified ArgR were performed in vitro and subjected to nondenaturing polyacrylamide gel electrophoresis analysis. DNA probe concentration was 0.1 nM. Protein concentrations from lane 2 to lane 7 were 45 nM, 18 nM, 9 nM, 4.5 nM, 2.2 nM, and 1 nM. Lane 1 served as negative control by removing ArgR from the reaction.
The ldcA gene is essential for lysine utilization.
The results of ldcA expression in response to arginine from the above-described experiments led us to propose that ldcA encodes an arginine-inducible arginine decarboxylase (ADC), the first enzyme of the ADC pathway (21). Arginine succinyltransferase (AST) and arginine transaminase (ATA) pathways were reported to sustain for aerobic growth in P. aeruginosa (36). In order to test for the ADC pathway as an alternative route for arginine utilization, we used a double mutant strain, PAO5602, devoid of the AST (by aruF deletion) and ATA (by aruH::Tcr) pathways, while the putative ADC pathway remained intact (Table 3). However, we observed that growth on l-arginine as the sole source of carbon and nitrogen was completely abolished in PAO5602 and growth on l-arginine could be restored when the mutant strain was complemented by pHC5307, which expressed aruH from the lac promoter on the pUCP18 vector (Table 4). Contrarily, PAO5602 harboring pHC5306 (ldcA in pUCP18) still showed no growth on l-arginine. From these results we concluded that ldcA is not involved in arginine utilization in P. aeruginosa PAO1.
TABLE 3.
Lysine decarboxylase pathway is the main route for lysine catabolism
| Strain | Description of genotypeb | Affected pathway | Growth with indicated supplementa |
||||
|---|---|---|---|---|---|---|---|
| Lc | Ln | Ac | An | GN | |||
| PAO1 | WT | None | + | ++ | +++ | +++ | +++ |
| PAO5716 | ldcA::Tcr | LDC | − | − | +++ | +++ | +++ |
| PAO5717 | ldcB::Tcr | LDC | +/− | ++ | +++ | +++ | +++ |
| PAO5720 | aruH::Tcr | ATA | + | ++ | +++ | +++ | +++ |
| PAO 501 | argR::Gmr | AST/LDC | − | + | + | ++ | +++ |
| PAO5602 | aruH::Tcr ΔaruF | AST/ATA | ++ | +++ | − | − | +++ |
| PAO5715 | ΔaruCFGDBE::Gmr | AST | ++ | +++ | + | ++ | +++ |
| PAO5718 | ΔaruCFGDBE::GmrldcA::Tcr | LDC/AST | M | M | + | ++ | +++ |
Logarithmically growing cells were plated on MMP solid agar media supplemented with the following: Lc, 20 mM lysine + 5 mM ammonia; Ln, 5 mM lysine + 20 mM glucose; Ac, 20 mM arginine + 5 mM ammonia; An, 5 mM arginine + 20 mM glucose; GN, 20 mM glucose + 5 mM ammonia. Growth at 37°C was recorded daily during 3 days of incubation period as follows: +++, growth in 1 day; ++, growth in 2 days; +, growth in 3 days; +/−, faint growth in 3 days; −, no growth in 3 days; M, growth under stress with high mutation rate.
WT, wild type.
TABLE 4.
Effects of constitutive expression of ldcA or aruH
| Strain | Affected pathway | Plasmid | Description or genotype | Growth with indicated supplementa |
|||
|---|---|---|---|---|---|---|---|
| Lc | Ln | Arg | Agm | ||||
| PAO1 | None | pUCP18 | WT, pUCP18 | + | ++ | +++ | +++ |
| pHC5307 | WT, pUCP18 aruH | + | +++ | +++ | +++ | ||
| pHC5306 | WT, pUCP18 ldcA | +++ | +++ | +++ | +++ | ||
| PAO5716 | LDC | pUCP18 | ldcA::Tcr, pUCP18 | − | − | +++ | +++ |
| pHC5307 | ldcA::Tcr, pUCP18 aruH | − | +++ | +++ | +++ | ||
| pHC5306 | ldcA::Tcr, pUCP18 ldcA | +++ | +++ | +++ | +++ | ||
| PAO5602 | ATA/AST | pUCP18 | aruH::Tcr ΔaruF, pUCP18 | +++ | +++ | − | +++ |
| pHC5307 | aruH::Tcr ΔaruF, pUCP18 aruH | +++ | +++ | +++ | +++ | ||
| pHC5306 | aruH::Tcr ΔaruF, pUCP18 ldcA | +++ | +++ | − | +++ | ||
Logarithmically growing cells were subjected to growth phenotype test in MMP media supplemented with the following: Lc, 20 mM lysine + 5 mM ammonia; Ln, 5 mM lysine + 20 mM glucose; Arg, 20 mM arginine; Agm, 20 mM agmatine; and GN, 20 mM glucose + 5 mM ammonia. Aerobic growth at 37°C was monitored during 3 days as follows: +++, prominent growth in 1 day; ++, growth in 2 days; +, growth in 3 days; −, no growth in 3 days.
Surprisingly, the ldcA mutant strain, PAO5716, lost the ability to grow on lysine, while arginine utilization remained unaffected (Table 3). Introducing pHC5306 to PAO5716 complemented the growth defect on lysine. Even growth of the wild-type strain PAO1 on lysine can be enhanced by the presence of pHC5306. These data support that ldcA may encode a lysine decarboxylase that is essential for l-lysine catabolism via the decarboxylase pathway (5).
LdcA as lysine-specific PLP-dependent decarboxylase.
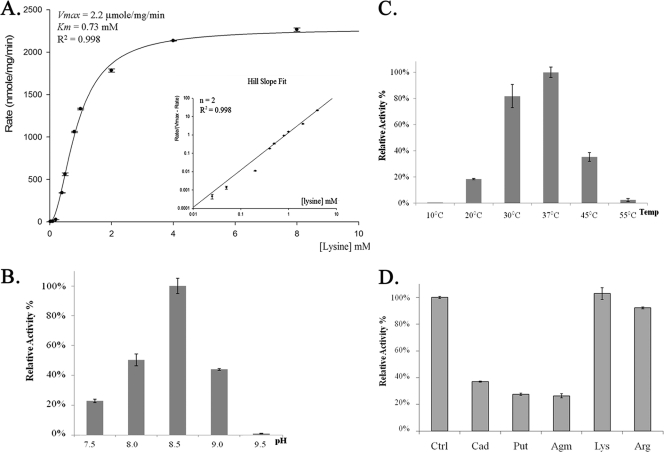
To further confirm the enzymatic function of LdcA in lysine degradation, we performed an initial characterization of purified His-tagged LdcA with radioactive-labeled l-lysine to determine the reaction rate by measuring the liberated 14CO2. As shown in Fig. 3A, LdcA possesses calculated Km and Vmax values of 0.73 mM and 2.2μmole/mg/min, respectively. An estimated Hill slope of 2.0 indicated cooperative substrate activation by l-lysine. The decarboxylase activity of LdcA was also checked with 14C-labeled l-arginine as a substrate, and no activity on l-arginine could be detected.
FIG. 3.
Lysine decarboxylase enzyme characterization. His-tagged LdcA protein was purified from E. coli. Enzyme-catalyzed lysine decarboxylation was assayed by measuring the liberated 14CO2 as previously described in Materials and Methods. (A) Kinetics studies exhibiting cooperative substrate activation pattern with the corresponding Hill slope plot. Assays with increasing lysine concentrations were performed at pH 8.5, 37°C. (B and C) Optimal conditions for lysine decarboxylation were tested as described in Materials and Methods. (D) Cold polyamines or amino acids (5 mM) were added into the standard reaction mixture to test for allosteric effects.
A pyridoxal 5′-phosphate (PLP) binding site of LdcA was predicted for A387THSTHKMLAAF398, which shows 90% and 91% similarities to the PLP binding sites of E. coli lysine decarboxylase (CadA; ETESTHKLLAAF) and arginine decarboxylase (AdiA; ATHSTHKLLNAF), respectively (20). Indeed, when PLP is removed from the reaction mixture, enzyme activity is abolished for LdcA (data not shown).
The optimal pH and temperature for LdcA were also determined. As shown in Fig. 3C and D, LdcA exhibits a maximum activity around 37°C, pH 8.5, with l-lysine as a substrate. At pH 6, almost no activity was detectable for LdcA in this acidic environment (data not shown). These results strongly suggest that LdcA may not be involved in acid stress as E. coli CadA and AdiA are (2, 17, 33) but rather is suitable for lysine catabolism under alkaline conditions.
The potential effects of l-arginine and compounds derived from l-arginine/ornithine/lysine decarboxylation (agmatine, putrescine, and cadaverine) on LdcA were tested. An inhibition effect was detected by the last three compounds but not by l-arginine (Fig. 3B). These results imply that arginine intervention in lysine utilization is exerted only at the transcriptional level and that lysine decarboxylation is subjected to product inhibition by cadaverine and other polyamines.
Lysine uptake is enhanced by exogenous arginine but not lysine.
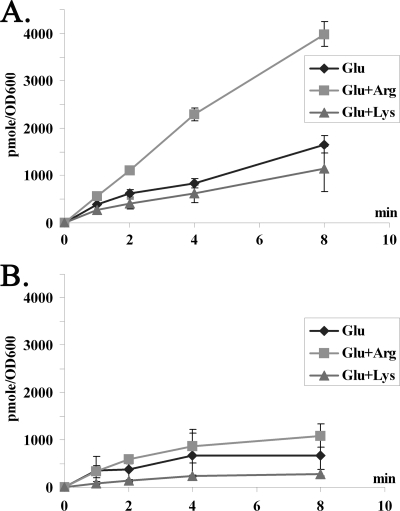
The arginine effect on lysine utilization may range from uptake to degradation. In order to investigate this hypothesis, lysine uptake was examined in a pair of isogenic strains of P. aeruginosa, PAO1 (wild type) and PAO501 (the argR::Gmr mutant). Cells were grown in minimal medium P with l-glutamate as the background nutrient with or without addition of exogenous l-arginine or l-lysine. The uptake of lysine in cell suspensions was examined. As shown in Fig. 4, uptake of lysine is significantly induced by the presence of exogenous arginine but not by lysine. Interestingly, this arginine-inducible lysine uptake is also ArgR dependent (Fig. 4B).
FIG. 4.
Enhanced lysine uptake by arginine. Induction of l-lysine uptake by exogenous arginine in P. aeruginosa PAO1 (A) and its argR mutant (B). Cultures grown in glutamate-MMP in the absence (diamond) or in the presence of l-arginine (square) or l-lysine (triangle) were harvested during exponential growth and used for l-lysine transport assays as described in Materials and Methods.
Exogenous arginine improves the growth on lysine in PAO1.
All our data indicate that expression of ldcA, the essential gene for lysine catabolism, is controlled by ArgR in response to l-arginine but not to l-lysine. In the wild-type strain PAO1, the estimated generation time was about 122 min when grown on l-lysine; however, it experienced a long lag phase of 44 ± 2 h. We reasoned that this growth curve was due to a lack of ldcA induction by exogenous lysine and hypothesized that growth of PAO1 on lysine can be improved by a trace amount of l-arginine to kick off ldcA expression. Indeed, addition of 0.15 mM l-arginine removed completely the otherwise long lag phase, while the generation time remained unchanged (data not shown).
The basal level of ldcAB expression is elevated in the mutant devoid of the major arginine catabolic pathway.
In analysis of the growth phenotype of PAO5602 (an arginine nongrower), we observed significant improvement of this strain on lysine in comparison to PAO1 (Table 3). A similar growth behavior on lysine was also detected in PAO5715, an aruCFGDBE deletion mutant devoid of the major pathway (AST) for arginine catabolism (11). One hypothesis was that the intracellular level of arginine may be elevated when the major catabolic pathway is blocked, which in turn activates ArgR to induce the ArgR regulon (16), including the aotJQMOP-argR operon for uptake and regulation, ldcAB, and many others. To test this hypothesis, we compared the activities of the aotJ and ldcA promoters from lacZ fusions in PAO1 and PAO5715. As shown in Table 2, the basal level of these two promoters was increased 4- or 5-fold in PAO5715, and they were still responsive to the presence of arginine but not lysine in both strains. These results support the hypothesis of an activated ArgR regulon and hence high lysine uptake and catabolism in AST mutant strains.
Potential lysine catabolism by transamination.
While the LdcA-catalyzed decarboxylation appeared to be the major route for lysine catabolism, l-lysine can potentially be degraded by other approaches, e.g., transamination. One possible candidate was AruH, which has been studied and characterized in vitro as a transaminase able to remove the α-amino group from arginine and lysine (35). Although the ldcA mutant lost completely the capability to grow on l-lysine as a sole source of carbon and/or nitrogen (Table 3), introducing aruH carried on pHC5307 to this mutant indeed restored growth on lysine as a sole source of nitrogen but not carbon (Table 4).
DISCUSSION
The bottleneck of lysine catabolism.
The most intriguing finding in this study was that both lysine uptake and degradation are inducible by l-arginine and show no response to l-lysine. Arginine-dependent ldcA expression was revealed from its promoter activities, and electro-mobility shift assays also revealed interactions of the arginine-responsive regulator ArgR to its putative binding site in the promoter region. The lack of lysine-responsive induction on ldcA is likely the major limiting factor of growth on l-lysine. Growth of P. aeruginosa on l-lysine exhibited a long lag phase of 44 ± 2 h. In comparison, growth on cadaverine, the decarboxylation product of l-lysine by LdcA, had a normal lag phase. Indeed, putting ldcA under the control of the constitutive lac promoter in a plasmid improved growth significantly.
Conditions that increase the level of intracellular arginine and ArgR could also induce ldcA expression and hence better growth on l-lysine. While ldcA was induced under these conditions, at least two ABC transporter systems for l-arginine were also increased (aotJQMP and PA5152 to PA5155). Interestingly, counterparts of these two systems in P. putida KT2440 have been reported as candidate l-lysine transport systems in recent studies (26). PP0283 to PP0280 and PP4486 to PP4482 show high similarities in gene organization as well as in amino acid sequences (82% and 76%, respectively) with P. aeruginosa PA5152 to PA5155 and aotJQMP-argR loci. Together with LdcB, a candidate lysine/cadaverine antiporter, all three candidate transport systems in P. aeruginosa are regulated by ArgR in response to l-arginine but not to l-lysine. This may explain the lack of lysine-inducible uptake as another limiting factor for growth on l-lysine in P. aeruginosa PAO1.
The lysine decarboxylase LdcA is conserved among pseudomonads.
In the Pseudomonas Genome Database (www.pseudomonas.com), orthologs of LdcA can be found in strains of P. entomophila, P. fluorescens, P. mendocina, P. putida, and P. stutzeri, but not in P. syringae. For P. putida KT2440, the corresponding gene (PP4140) contains an authentic mutation that causes a frameshift of the translated product. While gene organization of ldcA and upstream dnaQ encoding the epsilon subunit of DNA polymerase III is conserved among these strains, downstream ldcB can be found only in strains of P. aeruginosa. In conclusion, it is reasonable to predict that most species of pseudomonads possess the l-lysine decarboxylase.
Other pathways of lysine catabolism.
The aruH gene encodes for an l-arginine and l-lysine:pyruvate transaminase (36). Contribution of this enzyme in lysine utilization as the sole nitrogen source can be detected only when constitutively expressed from a plasmid. However, AruH may not have any physiological implication on lysine catabolism in PAO1, as the ldcA mutant showed no growth on l-lysine as the sole source of carbon or nitrogen. Perhaps it was because of low l-lysine uptake and the low affinity of AruH on l-lysine.
The aruH and aruI genes encode the first two enzymes of the arginine transaminase pathway in P. aeruginosa (36). While the aruHI genes are conserved in P. putida, an extra gene encoding an amino acid racemase is located between aruH and aruI of this organism. The amino acid sequence of this racemase contains a predicted signal peptide at the N terminus, suggesting its secretion to the periplasm. This implies the role of this additional gene as the racemase to convert l-lysine into d-lysine before being channeled into the d-lysine catabolic pathway in P. putida (3, 10, 25).
P. aeruginosa can utilize d-lysine only as a nitrogen source and not as a carbon source by the flavin adenine dinucleotide (FAD)-dependent dehydrogenase DauA (13). Transamination of l-lysine by AruH or d-lysine by DauA leads to α-keto-ɛ-aminohexanoate, which is spontaneously converted into its cyclic form Δ1-piperideine-2-carboxylate at a physiological pH (12). Recent studies of P. putida suggest further degradation of this molecule into l-pipecolate by the reductase DpkA (PP3591) (27). However, viability of this pathway is still undefined in P. aeruginosa since the dpkA orthologue is not present on the chromosome, the corresponding biochemical reaction cannot be detected, and no growth is detectable using d-lysine as a carbon source (5).
Lysine decarboxylation is the main route of l-lysine catabolism in P. aeruginosa.
With the purified enzyme, we demonstrated that LdcA is an l-lysine decarboxylase with no activity toward l-arginine. Genetic studies also indicated that ldcA is essential for l-lysine utilization in P. aeruginosa. Therefore, we concluded that the lysine decarboxylase LdcA catalyzes the pivotal step of l-lysine catabolism in this organism.
Acknowledgments
This work was supported by National Science Foundation MCB-0950217 and by the Molecular Basis of Diseases Program at Georgia State University.
Footnotes
Published ahead of print on 10 September 2010.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, Y. F., and E. Adams. 1974. d-lysine catabolic pathway in Pseudomonas putida: interrelations with l-lysine catabolism. J. Bacteriol. 117:753-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farinha, M. A., and A. M. Kropinski. 1990. Construction of broad-host-range plasmid vectors for easy visible selection and analysis of promoters. J. Bacteriol. 172:3496-3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fothergill, J. C., and J. R. Guest. 1977. Catabolism of l-lysine by Pseudomonas aeruginosa. J. Gen. Microbiol. 99:139-155. [DOI] [PubMed] [Google Scholar]
- 6.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong, S., H. Richard, and J. W. Foster. 2003. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J. Bacteriol. 185:4402-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas, D., B. W. Holloway, A. Schambock, and T. Leisinger. 1977. The genetic organization of arginine biosynthesis in Pseudomonas aeruginosa. Mol. Gen. Genet. 154:7-22. [DOI] [PubMed] [Google Scholar]
- 9.Hegazy, M. 2004. Characterization of the arginine decarboxylase pathway in Pseudomonas aeruginosa. Ph.D. dissertation. Georgia State University, Atlanta, GA.
- 10.Ichihara, A., S. Furiya, and M. Suda. 1960. Metabolism of l-lysine by bacterial enzymes. III. Lysine racemase. J. Biochem. 48:277-283. [Google Scholar]
- 11.Itoh, Y. 1997. Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J. Bacteriol. 179:7280-7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ko, K. C., B. Wang, P. C. Tai, and C. D. Derby. 2008. Identification of potent bactericidal compounds produced by escapin, an l-amino acid oxidase in the ink of the sea hare Aplysia californica. Antimicrob. Agents Chemother. 52:4455-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, C., and C. D. Lu. 2009. Arginine racemization by coupled catabolic and anabolic dehydrogenases. Proc. Natl. Acad. Sci. U. S. A. 106:906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, C., X. Yao, and C. D. Lu. 2009. Regulation of the dauBAR operon and characterization of d-amino acid dehydrogenase DauA in arginine and lysine catabolism of Pseudomonas aeruginosa PAO1. Microbiology 156:60-71. [DOI] [PubMed] [Google Scholar]
- 15.Lu, C. D., Y. Itoh, Y. Nakada, and Y. Jiang. 2002. Functional analysis and regulation of the divergent spuABCDEFGH-spuI operons for polyamine uptake and utilization in Pseudomonas aeruginosa PAO1. J. Bacteriol. 184:3765-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu, C. D., Z. Yang, and W. Li. 2004. Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J. Bacteriol. 186:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meng, S. Y., and G. N. Bennett. 1992. Regulation of the Escherichia coli cad operon: location of a site required for acid induction. J. Bacteriol. 174:2670-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 19.Moreau, P. L. 2007. The lysine decarboxylase CadA protects Escherichia coli starved of phosphate against fermentation acids. J. Bacteriol. 189:2249-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris, D. R., and E. A. Boeker. 1983. Biosynthetic and biodegradative ornithine and arginine decarboxylases from Escherichia coli. Methods Enzymol. 94:125-134. [DOI] [PubMed] [Google Scholar]
- 21.Morris, D. R., and A. B. Pardee. 1966. Multiple pathways of putrescine biosynthesis in Escherichia coli. J. Biol. Chem. 241:3129-3135. [PubMed] [Google Scholar]
- 22.Nishijyo, T., S. M. Park, C. D. Lu, Y. Itoh, and A. T. Abdelal. 1998. Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J. Bacteriol. 180:5559-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, S. M., C. D. Lu, and A. T. Abdelal. 1997. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J. Bacteriol. 179:5309-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman, M., and P. H. Clarke. 1980. Genes and enzymes of lysine catabolism in Pseudomonas aeruginosa. J. Gen. Microbiol. 116:357-369. [DOI] [PubMed] [Google Scholar]
- 25.Revelles, O., M. Espinosa-Urgel, T. Fuhrer, U. Sauer, and J. L. Ramos. 2005. Multiple and interconnected pathways for l-lysine catabolism in Pseudomonas putida KT2440. J. Bacteriol. 187:7500-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revelles, O., M. Espinosa-Urgel, S. Molin, and J. L. Ramos. 2004. The davDT operon of Pseudomonas putida, involved in lysine catabolism, is induced in response to the pathway intermediate delta-aminovaleric acid. J. Bacteriol. 186:3439-3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Revelles, O., R. M. Wittich, and J. L. Ramos. 2007. Identification of the initial steps in d-lysine catabolism in Pseudomonas putida. J. Bacteriol. 189:2787-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revelles, O., and M. Espinosa-Urgel. 2004. Proline and lysine metabolism, p. 273-292. In J. L. Ramos (ed.), Pseudomonas, vol. 3. Biosynthesis of macromolecules and molecular metabolism. Kluwer Academic/Plenum Publishers, New York, NY. [Google Scholar]
- 29.Schweizer, H. D. 1993. Small broad-host-range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. Biotechniques 15:831-834. [PubMed] [Google Scholar]
- 30.Schweizer, H. P. 1991. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene 97:109-121. [DOI] [PubMed] [Google Scholar]
- 31.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotech. 1:784-791. [Google Scholar]
- 32.Stibitz, S., W. Black, and S. Falkow. 1986. The construction of a cloning vector designed for gene replacement in Bordetella pertussis. Gene 50:133-140. [DOI] [PubMed] [Google Scholar]
- 33.Watson, N., D. S. Dunyak, E. L. Rosey, J. L. Slonczewski, and E. R. Olson. 1992. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J. Bacteriol. 174:530-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wertheimer, S. J., and Z. Leifer. 1983. Putrescine and spermidine sensitivity of lysine decarboxylase in Escherichia coli: evidence for a constitutive enzyme and its mode of regulation. Biochem. Biophys. Res. Commun. 114:882-888. [DOI] [PubMed] [Google Scholar]
- 35.Yang, Z., and C. D. Lu. 2007. Characterization of an arginine:pyruvate transaminase in arginine catabolism of Pseudomonas aeruginosa PAO1. J. Bacteriol. 189:3954-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang, Z., and C. D. Lu. 2007. Functional genomics enables identification of genes of the arginine transaminase pathway in Pseudomonas aeruginosa. J. Bacteriol. 189:3945-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]