Abstract
Human antibody recognition of Chlamydia trachomatis plasmid-encoded Pgp3 protein is dependent on the native conformation of Pgp3. The structural basis for the conformation dependence and the function of Pgp3 remain unknown. Here, we report that Pgp3 trimerization is required for the recognition of Pgp3 by human antibodies. In a native polyacrylamide gel, Pgp3 purified from a bacterial expression system migrated as stable trimers that were dissociated into monomers only by treatment with urea or sodium dodecyl sulfate (SDS) but not nonionic detergents. Human antibodies recognized trimeric but not monomeric Pgp3, suggesting that Pgp3 is presented to the human immune system as trimers during C. trachomatis infection. The endogenous Pgp3 secreted into the chlamydial outer membrane complex or host cell cytosol is always trimerized. Intact Pgp3 trimers were eluted from the outer membrane complex by a combination of nonionic detergents with reducing agents but not by the presence of either alone. These observations have provided important information for further understanding the role of Pgp3 in chlamydial pathogenesis and potentially optimizing Pgp3 as a subunit vaccine candidate antigen.
Chlamydia trachomatis consists of multiple serovars and causes various human diseases. Serovars A to C primarily infect ocular epithelial cells, potentially leading to blinding trachoma (23, 42). Serovars D to K mainly target urogenital epithelial cells (39) whereas serovars L1 to L3 can invade lymphatic tissue, potentially resulting in systematic infection (40). Despite the differences in tissue tropism, all C. trachomatis organisms share a conserved biphasic growth cycle that has to be completed in a cytoplasmic vacuole called an inclusion (21, 46). Chlamydial infection starts with the entry of an infectious elementary body (EB) into an epithelial cell via pathogen-induced endocytosis (8, 19). The endocytosed EB differentiates to a noninfectious but metabolically active reticulate body (RB). After replication, the progeny RBs differentiate to EBs that exit the infected cells to invade adjacent cells (25). The C. trachomatis organisms also share a highly conserved cryptic plasmid that encodes 8 open reading frames (ORFs) designated pORF 1 to 8 (28, 35, 43).
Urogenital tract infection with C. trachomatis is a leading cause of sexually transmitted diseases worldwide (11) and, if left untreated, can lead to severe complications such as pelvic inflammatory diseases, ectopic pregnancy, and infertility (15). Due to the lack of symptoms exhibited by individuals infected with C. trachomatis, it is not possible to effectively control C. trachomatis infection with antibiotics. Prophylactic vaccines may be among the most effective approaches for preventing C. trachomatis-induced pathologies (34). However, the pathogenic mechanisms of C. trachomatis remain unclear and there is no licensed C. trachomatis vaccine, probably due to limited knowledge of the roles of individual C. trachomatis antigens in pathogenesis and protective immunity. The cryptic plasmid has been considered a virulence factor of C. trachomatis, because plasmid-free variants have been found to be less invasive and to cause pathologies of lesser severity in mouse upper genital tract tissues (7, 32). However, the roles of the plasmid-encoded or regulated proteins in either chlamydial pathogenesis or protective immunity remain largely unknown. Pgp3, one of the plasmid-encoded proteins, was found to be recognized by human antibodies in enzyme-linked immunosorbent assays (ELISAs) but not in Western blot assays (9). We further confirmed that Pgp3 was an immunodominant antigen in woman urogenitally infected with C. trachomatis and that the human antibody recognition of Pgp3 was dependent on the native conformation of Pgp3 (30). We also observed that among the 8 ORFs encoded by the cryptic plasmid, only Pgp3 was secreted into the cytosol of the infected cells (28). Furthermore, various groups demonstrated that immunization with pgp3-encoding plasmid DNA induced protective immunity in mice (16, 29). However, the molecular basis for the conformation dependence of human antibody recognition of Pgp3 remains uncharacterized and the function of Pgp3 is unknown. Here, we present evidence that Pgp3 forms stable trimers that are responsible for the native conformation-dependent recognition of Pgp3 by human antibodies. The current study has provided important information for further understanding the roles of Pgp3 in C. trachomatis pathogenesis and protective immunity.
MATERIALS AND METHODS
(i) Chlamydial organisms and infection.
C. trachomatis serovar D (strain UW-3/Cx) and L2 (strain 434/Bu) organisms were propagated in HeLa-229 cells (human cervical carcinoma epithelial cells; ATCC catalog no. CCL2), and the stocks were prepared as described previously (20). Briefly, HeLa cells grown in either 24-well plates with coverslips or tissue flasks containing Dulbecco's modified essential medium (DMEM) (Gibco BRL, Rockville, MD) with 10% fetal calf serum (FCS; Gibco BRL) at 37°C in an incubator supplied with 5% CO2 were inoculated with C. trachomatis stock organisms. The infected cultures were harvested at different time points postinfection for either organism purification and outer membrane complex (OMC) preparation or Western blot analyses as described below.
(ii) Preparation of C. trachomatis outer membrane complex.
The EBs were purified from 48-h cultures as described previously (27), and the chlamydial outer membrane complex (COMC) was extracted from the purified EB organisms as described elsewhere (6, 31, 41) with minor modifications. Approximately 2 to 3 mg of purified EBs was resuspended in 5 ml of phosphate-buffered saline (PBS; pH 7.4) containing 2% (wt/vol) N-lauroylsarcosine sodium salt (Sarkosyl [catalog no. L5777]; Sigma, St. Louis, MO). The suspension was sonicated, incubated at 37°C for 1 h, and centrifuged at 100,000 × g for 30 min at 4°C. The insoluble pellet (defined as 1× extraction insoluble fraction/pellet) was resuspended in the Sarkosyl buffer, incubated for another 15 min, and centrifuged as described above. The insoluble fraction (defined as 2× pellet) was similarly extracted with the Sarkosyl buffer four more times. A portion of each insoluble fraction was saved for monitoring the quality of COMC in a sodium dodecyl sulfate (SDS) polyacrylamide gel. After 6 extractions, the final pellet was resuspended in PBS (pH 7.4) and labeled as COMC.
(iii) Purification of Pgp3 from an Escherichia coli expression system.
The pgp3 gene (also referred to as pORF5) encoded by the pCHL1 plasmid from C. trachomatis serovar D organisms was cloned into a pGEX-6P2 vector (Amersham Pharmacia Biotech, Inc., Piscataway, NJ) and expressed as a fusion protein with glutathione S-transferase (GST) fused to the N terminus as previously described (28). The GST-Pgp3 fusion protein was bound to glutathione-conjugated agarose beads (Pharmacia), and a precision protease in the form of a GST fusion protein (Pharmacia) was used to cleave the Pgp3 protein from the beads. The cleaved Pgp3 protein was thus released into the solution while the GST-precision enzyme fusion protein was absorbed onto the glutathione beads. The eluent containing cleaved Pgp3 was collected and further purified using a HiTrap Q high performance ion exchange column (catalog no. 17-1154-01; GE Healthcare, Piscataway, NJ).
(iv) Analytical ultracentrifugation.
All sedimentation velocity experiments were performed at 20°C using a Beckman Optima XL-I centrifuge equipped with an eight-hole AN-50 rotor (Beckman Coulter Inc., Brea, CA). Pgp3 samples with initial readings of the optical density at 280 nm (OD280) of 0.3, 0.5, and 0.8, corresponding to protein concentrations of ∼0.6, 1.0, and 1.5 mg/ml, respectively, were centrifuged at 40,000 rpm. Sedimentation velocity data were acquired using absorbance optics at 280 nm. Hydrodynamic corrections for buffer density, viscosity, and partial specific volume were applied according to methods outlined by Laue (26). Sedimentation velocity data were analyzed using the method of van Holde and Weischet (44), followed by two-dimensional spectrum analyses (3) (with simultaneous removal of time-invariant noise) and then by genetic algorithm refinement (4), followed by Monte Carlo analyses (14) as implemented in the ULTRASCAN software package (12, 13). Analysis of sedimentation velocity data by the van Holde and Weischet method effectively removes the contribution of diffusion to boundary spreading to yield G(s), the integral distribution of S20,w of all species in the sample. Consequently, a G(s) plot of the boundary fraction versus S20,w is vertical when the sample is homogenous and has a positive slope when the sample is heterogeneous (44). The S value is directly proportional to molecular mass and inversely proportional to the frictional ratio (f/f0). The frictional ratio is 1 for a sphere and increases for elongated molecules (with values usually in the range of 1.25 to 2.5) (5).
(v) Polyacrylamide gel electrophoresis and Western blot.
Polyacrylamide gels used in this study were prepared following the procedure provided by the manufacturer (Bio-Rad, Hercules, CA). The running buffer used for the native gels also contained 0.01% (wt/vol) SDS, which did not affect the stability of Pgp3 trimers but allowed clear separation of protein bands. Protein samples run on denaturing gels were subjected to heat denaturing by boiling for 5 min in an SDS sample buffer (63 mM Tris-HCl [pH 6.8], 10% glycerol, 0.0025% bromophenol blue, 2% [wt/vol] SDS, 1% [vol/vol] 2-mercaptoethanol [2-ME]). Protein samples run on native gels were treated with or without various reducing and denaturing agents in the individual experiments as indicated. The native gel sample buffer was the same as the denaturing gel sample buffer except without SDS.
In addition to the purified Pgp3 protein sample, the following samples were also subjected to polyacrylamide gel electrophoresis analyses: whole-cell lysates from either normal HeLa cells or C. trachomatis-infected HeLa cells, cytosolic fractions of the infected-HeLa cells, purified EBs, or outer membrane complexes. The whole-cell lysates were prepared by resuspending the cell samples at 1 × 107 cells per ml of 1× MLB buffer (1% [wt/vol] Igepal, 10 mM MgCl, 1 mM EDTA, 150 mM NaCl, 10% [vol/vol] glycerol, 1 mM Na3VO4, 25 mM HEPES, pH 7.4) with a protease inhibitor cocktail (1 mM phenylmethylsulfonyl fluoride [PMSF; catalog no. P7626], 20 μM leupeptin [catalog no. L2884], 1.6 μM pepstatin A [catalog no. P5318], 1.7 μg of aprotinin/ml [catalog no. A6279], all from Sigma) and incubated on ice for 30 min. The mixtures were centrifuged at 21,000 × g for 15 min at 4°C, and the remaining supernatant was collected for loading into gels. The cytosolic fractions of the chlamydia-infected HeLa cells (defined as S100) were prepared as previously described (20). Briefly, the infected cells were harvested via low-speed centrifugation and the cell pellets were resuspended in a Dounce homogenization buffer (10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol [DTT], 250 mM sucrose, 20 mM HEPES-KOH [pH 7.5], all from Sigma) containing a protease inhibitor cocktail as described above. Limited Dounce homogenization was applied to ensure that >70% cells are broken without damage to either the nuclei or inclusions. The supernatants were harvested after a series of centrifugations, including a final centrifugation at 100,000 × g using an Airfuge Ultracentrifuge (Beckman Coulter). For some COMC experiments, various Sarkosyl-insoluble fractions (after serial Sarkosyl extractions) were loaded to denaturing gels, while for other COMC experiments, the COMC samples obtained after 6 consecutive Sarkosyl extractions were treated with either 2% (wt/vol) Sarkosyl or 2% (wt/vol) SDS in the presence or the absence of 20 mM DTT and the treatments were carried out at 37°C for 30 min. The soluble fractions collected after high-speed centrifugation (21,000 × g) at 4°C for 30 min were used for gel loading.
After electrophoresis, proteins resolved in the polyacrylamide gels were either visualized by staining with a Coomassie blue dye (Sigma) or transferred onto nitrocellulose membranes for antibody detection of antigens in Western blot assays. The following primary antibodies were used in the Western blot assays: human antisera were collected from women diagnosed with C. trachomatis urogenital tract infection by ligase chain reaction detection of C. trachomatis DNA in vaginal swabs (36-38). An IRB (Institutional Review Board)-exempt protocol is in place for the current study. A pooled serum sample collected from 8 healthy individuals was used as a negative control. A mouse polyclonal antibody (pAb) raised with a GST-Pgp3 fusion protein was used to detect both native and denatured Pgp3 (30), and 2H4, a mouse monoclonal antibody (MAb), was used to detect native Pgp3 (30). Mouse 100a mAb against the C terminus of CPAF secreted serine protease (47), BC7.1 against chlamydial HSP60 (47), MC22, and a rabbit antiserum against the chlamydial major outer membrane protein (MOMP) (47) were also used. Since CPAF is secreted into the host cell cytosol and is not associated with chlamydial organisms, the anti-CPAF antibody was used to indicate cytosolic fractions of the chlamydia-infected cells. Since MOMP is a structural protein of C. trachomatis organisms, the anti-MOMP antibody was used to detect fractions containing chlamydial organisms.
The primary antibody binding was probed with a horseradish peroxidase (HRP)-conjugated goat anti-human, anti-mouse, or anti-rabbit IgG secondary antibody (Jackson ImmunoResearch Laboratories, Inc., Westgrove, PA) and visualized using an enhanced chemiluminescence (ECL) kit (Santa Cruz Biotech).
(vi) Immunofluorescence assay.
HeLa cells grown on coverslips with or without chlamydial infection were processed for chemical and antibody staining. The Hoechst dye (blue; Sigma) was used to visualize DNA. A rabbit anti-chlamydial organism antibody plus a goat anti-rabbit IgG secondary antibody conjugated with Cy2 (green; Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) was used to visualize chlamydial organisms. Mouse anti-CPAF MAb 100a (47) or anti-Pgp3 MAb 2H4 (28) plus a goat anti-mouse IgG conjugated with Cy3 (red; Jackson ImmunoResearch) were used to visualize the corresponding antigens. The immunofluorescence images were acquired using an Olympus AX-70 fluorescence microscope equipped with multiple filter sets and Simple PCI imaging software (Olympus, Melville, NY) as described previously (20). The images were processed using Adobe Photoshop software (Adobe Systems, San Jose, CA).
RESULTS
(i) Pgp3 purified from a bacterial expression system is a stable trimer.
It was previously reported that human antibody recognition of GST-Pgp3 fusion proteins was abolished by SDS plus heat treatment in denaturing gels (9, 30). In the current study, we compared the mobility of purified Pgp3 with or without SDS plus heat treatment in a native gel (Fig. 1A). The purified Pgp3 was free of the GST fusion tag. We found that the SDS-denatured Pgp3 protein migrated with an apparent mass of ∼28 kDa, corresponding to the expected molecular mass of a Pgp3 monomer. In contrast, Pgp3 without the denaturation treatment migrated at ∼84 kDa, suggesting that the purified Pgp3 was a trimer. The purified Pgp3 was further investigated using analytical ultracentrifugation sedimentation velocity analysis (Fig. 1B), which provides information about macromolecule purity, mass, and oligomeric status (44). The vertical G(s) plots reveal that the purified Pgp3 was homogenous, with an S20,w value of ∼4.2, for all the Pgp3 concentrations tested. The theoretical S20,w values for monomeric, dimeric, and trimeric Pgp3 proteins with molecular masses of 28, 56, and 84 kDa are predicted to be 2.3, 3.7, and 4.2, respectively. The Monte Carlo fit analysis of the Pgp3 sedimentation velocity data revealed a molecular mass of 84 kDa and a frictional ratio of 1.6 (Fig. 1C), strongly suggesting that Pgp3 is a trimer with an elongated shape. The sedimentation velocity data confirmed the native gel results. Thus, the native gel electrophoresis can be used as a convenient method for monitoring the oligomeric status of Pgp3.
FIG. 1.
Pgp3 purified from an E. coli expression system is trimeric. (A) Pgp3 was purified from E. coli expressing a GST-Pgp3 fusion protein via affinity binding to glutathione agarose beads and cleaving of the fusion tag GST. The purified Pgp3 was either left untreated (lane 1) or treated with 2% SDS plus 5 min of boiling (lane 2), and the samples were subjected to electrophoresis in a 12% polyacrylamide native gel. The resolved protein bands were visualized by staining with Coomassie blue dye. Note that the untreated Pgp3 migrated at the ∼84 kDa position (corresponding to the molecular mass of trimeric Pgp3) whereas the SDS-denatured Pgp3 migrated at the ∼28 kDa position (corresponding to the monomeric form of Pgp3), as indicated on the right of the gel image. (B) Sedimentation velocity analyses of Pgp3 at concentrations of 0.6 (panel a), 1 (panel b), and 1.5 (panel c) mg/ml. The G(s) plot was obtained using the van Holde and Weischet method (44). Note that, regardless of the protein concentration, most readings (open circles) formed a vertical line with an S20,w value of ∼4.2 (x axis). Assuming that Pgp3 maintains a similar overall shape, the theoretic S20,w values for the trimeric, dimeric, and monomeric forms of Pgp3 are expected to be 4.2 (triple arrows on the top of the figure), 3.7 (double arrows), and 2.3 (single arrow), respectively. (C) Monte Carlo analysis of the sedimentation velocity of Pgp3 revealed a molecular mass of 84 kDa, corresponding to the molecular mass of a trimeric Pgp3, and a frictional ratio of 1.6, suggesting that the Pgp3 trimers were in an elongated form. (D) The stability of Pgp3 trimers was analyzed by treating Pgp3 with various combinations of denaturing agents under various conditions as indicated on top of the figure. The mobility of the treated Pgp3 molecules was monitored using a 12% native gel plus a Western blot assay with an anti-Pgp3 mouse polyclonal antibody known to recognize both native and denatured Pgp3. Note that neither of the reducing agents (2% 2-ME or 20 mM DTT, for 30 min of exposure) altered the ratio of the trimeric versus monomeric Pgp3 molecules (lanes 2 and 3). Treatment with 8 M urea and 2% SDS for 30 min converted ∼50% of the Pgp3 into monomers (lanes 4 and 5). Inclusion of DTT in the SDS treatment did not further increase the monomer/trimer ratio (lane 6). Boiling for 1 min caused aggregation of Pgp3, and most Pgp3 molecules stayed in the stacking gel (lane 7). Heating for 5 min in the presence of SDS completely converted all Pgp3 trimers into monomers (lane 9).
We evaluated the stability of trimeric Pgp3 under various reducing and denaturing conditions using the native gel plus Western blot assay (Fig. 1D). A mouse anti-Pgp3 antibody known to detect both native and denatured Pgp3 (30) was used to monitor the oligomeric status of Pgp3. The untreated control Pgp3 sample was detected as a trimer. Treatment with the reducing agents (2% [vol/vol] 2-ME or 20 mM DTT for 30 min) alone did not affect the Pgp3 trimers, while treatment with 8 M urea or 2% (wt/vol) SDS for 30 min converted more than 50% of the trimers into monomers. Interestingly, the combination of SDS with the reducing agent DTT did not further increase the monomer/trimer ratio, suggesting that subunits in the Pgp3 trimer are not cross-linked via disulfide bonds. Boiling the samples in the absence of detergents caused protein aggregation. However, boiling in the presence of SDS significantly increased the monomer/trimer ratio. The observations above, considered together, suggest that ionic and/or hydrophobic interactions might be the major forces responsible for stabilizing the trimers.
(ii) The Pgp3 trimer is preferentially recognized by human antibodies.
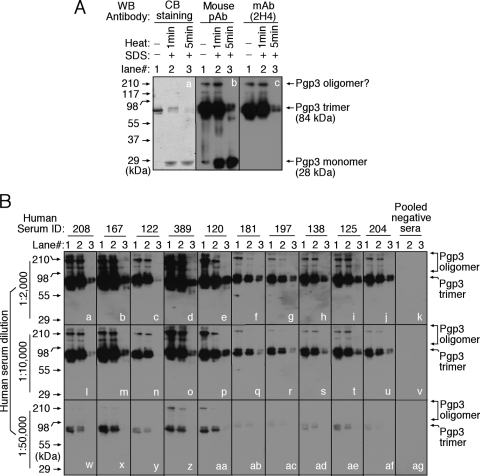
To test whether Pgp3 trimerization contributes to the conformation dependence of human antibody recognition of Pgp3, we compared the characteristics of human antibody binding to Pgp3 monomers and trimers by the use of the native gel-Western blot assay (Fig. 2). The mouse anti-Pgp3 polyclonal antibody detected all forms of Pgp3, while the conformation-dependent MAb recognized only the oligomerized Pgp3, which is consistent with what was previously reported (30). Importantly, all human antibodies at various dilutions recognized the oligomerized but not monomeric Pgp3 (Fig. 2B). This was also true even when the antiserum dilution was reduced to 1:400 (data not shown). These observations suggest that Pgp3 trimerization is likely responsible for the previously described conformation dependence of human antibody recognition of Pgp3 (30). The preferential recognition of trimeric Pgp3 by human antibodies suggests that Pgp3 is presented to the human immune system in the form of trimers during chlamydial infection in humans.
FIG. 2.
Human antibody recognition of trimeric Pgp3. (A) Purified Pgp3, left untreated (lane 1) or treated with 2% SDS plus boiling for 1 min (lane 2) or 5 min (lane 3), was loaded onto a 12% native gel. After electrophoresis, the gel was either stained with Coomassie blue (CB) dye (panel a) or transferred onto nitrocellulose membrane for Western blot (WB) detection with a mouse anti-Pgp3 polyclonal antibody (pAb) (panel b) that is known to recognize both native and denatured Pgp3 or with a monoclonal antibody (mAb), clone 2H4 (panel c), that is known to recognize only native Pgp3. (B) The antigen sets described in Fig. 2A were also reacted with 10 human antibodies (as indicated on top of the figure) on a Western blot. Each serum was tested at three different dilutions as indicated on the left of the figure. A serum sample pooled from 8 different healthy individuals was used as a negative control (panels k, v, and ag). Note that all serum samples from C. trachomatis-infected individuals recognized trimeric or oligomeric but not monomeric Pgp3 even at the lowest dilution.
(iii) Endogenous Pgp3 produced during chlamydial infection is trimeric.
To determine whether the chlamydial organism-produced Pgp3 is also a trimer, we monitored the oligomeric state of Pgp3 during C. trachomatis infection in cell cultures (Fig. 3). The conformation-dependent MAb detected strong signals in both the chlamydial inclusion and the cytoplasm of the C. trachomatis-infected cells in an immunofluorescence assay (Fig. 3A), suggesting that chlamydia-produced Pgp3 is trimeric. We further used the native gel-Western blot assay to visualize the oligomeric status of Pgp3 produced in C. trachomatis-infected cells (Fig. 3B). We found that the Pgp3 that was both secreted into the host cell cytosol and associated with chlamydial EB organisms was in the form of trimers, suggesting that C. trachomatis-produced Pgp3 is an obligate trimer regardless of its intracellular location. Pgp3 trimerization was also detected in cultures infected with other serovars of C. trachomatis (data not shown), indicating that trimer formation is a common feature of Pgp3 produced by all C. trachomatis organisms.
FIG. 3.
Detection of Pgp3 trimers in chlamydia-infected cell samples. (A) HeLa cells infected with C. trachomatis were processed for antibody labeling in an immunofluorescence assay. CPAF and Pgp3 were labeled with MAbs 100a and 2H4, respectively (red), and the chlamydial organisms were labeled with a rabbit antibody (green) and DNA with Hoechst dye (blue). Note that MAb 2H4, known to recognize only native trimeric Pgp3, detected signals in both inclusions (white star) and cytoplasms (white arrows) of chlamydia-infected cells, suggesting that the Pgp3 in the chlamydia-infected cells also represented trimers. (B) Lysates of HeLa cells alone (lane 1) or HeLa cells with C. trachomatis serovar D infection (D-HeLa; lane 2), cytosolic fractions of D-HeLa cells (S100; lane 3), and purified EB extracts (lane 4) were loaded to native gradient (panel a) or denaturing (panels b to d) gels for Western blot analyses with a mouse anti-Pgp3 polyclonal antibody (pAb; panels a and b), an anti-CPAF MAb (100a; panel c), or a rabbit anti-chlamydial major outer membrane protein (MOMP) polyclonal antibody (pane d) as indicated on the left of the figure. Note that the Pgp3 that both secreted into the host cytosol and associated with the purified EBs was in the form of trimers. CPAFc, C-terminal half of CPAF.
(iv) The association of Pgp3 trimers with the chlamydial outer membrane complexes requires disulfide bond interactions.
Previous studies showed an association of Pgp3 with the Sarkosyl-insoluble fraction that mainly contains the chlamydial outer membrane complex (COMC) (9), indicating that the EB organism-associated Pgp3 is a component of COMC. We further evaluated whether the COMC-associated Pgp3 is trimerized and how the Pgp3 trimer is integrated into the COMC (Fig. 4). The COMC was purified from EB organisms by the use of sequential extractions with 2% (wt/vol) Sarkosyl, and the insoluble fractions generated from each extraction were resolved in a denaturing gel for Western blot detection of Pgp3, MOMP, HSP60, and CPAF (Fig. 4A). A single extraction with Sarkosyl removed most of the chlamydial cytoplasmic HSP60 protein, and additional extractions effectively eliminated all HSP60 from the insoluble fractions. However, both Pgp3 and MOMP were found in abundance in the COMC, confirming that both Pgp3 and MOMP were constitutive components of the COMC. To determine the oligomeric status of the COMC-associated Pgp3, the COMC obtained after 6 extractions with Sarkosyl was reextracted with Sarkosyl, SDS, or PBS alone in the presence or absence of DTT reducing agent and the soluble fractions were collected from these treatments and detected in the native gel-Western blot assay (Fig. 4B). Although additional Sarkosyl extractions performed alone solubilized only minimal amounts of Pgp3 and MOMP oligomers from the COMC, inclusion of DTT dramatically increased the release of both Pgp3 and MOMP. The released Pgp3 was predominantly in the form of trimers, while both monomers and oligomers of MOMP were detected. These observations suggest that disulfide bond-mediated interactions play important roles in maintaining the Pgp3 trimers and MOMP molecules as parts of the COMC. However, the use of DTT alone released neither Pgp3 nor MOMP, suggesting that the disulfide bond interactions are not accessible to DTT in the absence of the nonionic detergents. SDS treatment of the COMC led to the release of monomeric Pgp3 regardless of the presence or absence of DTT, confirming that Pgp3 oligomerization is not dependent on disulfide bond formation. However, disulfide bond interactions may help MOMP oligomerization, since the addition of SDS alone washed off large amounts of oligomerized MOMP whereas the addition of DTT reduced all the oligomers to monomers. Although more studies are required for further teasing out of the relationships of the trimeric Pgp3 molecules with other components of COMC, the experiments described above have revealed that Pgp3 integrates into the outer membrane complexes as trimers and that the integration is likely stabilized, either directly or indirectly, via disulfide bond interactions.
FIG. 4.
Detection of trimeric Pgp3 in chlamydial outer membrane complexes. (A) Lysates of C. trachomatis serovar D-infected HeLa cells (lane 1), purified EBs (lane 2), or insoluble fractions from extraction of purified EBs with Sarkosyl for the number of times indicated on top of the figure (lanes 3 to 6) were subjected to electrophoresis in a 12% denaturing gel. The resolved proteins were detected by Western blot (WB) analysis of Pgp3 (panel a), MOMP (panel b), HSP60 (panel c), and CPAF (panel d) with the corresponding antibodies as listed along the left side of the figure. Note that although the chlamydial cytosolic protein HSP60 was efficiently removed from the insoluble fraction by Sarkosyl extraction, both MOMP and Pgp3 were still associated with the pellet even after six cycles of extraction with Sarkosyl, indicating that Pgp3 is a stable component of the chlamydial outer membrane complex. CPAFc, C-terminal half of CPAF. (B) An insoluble fraction generated by six cycles of Sarkosyl extraction was further extracted with no additives (lanes 1 and 2) or with Sarkosyl (lanes 3 and 4) or 2% SDS (lanes 5 and 6) in the presence (lanes 2, 4, and 6) or absence (lanes 1, 3, and 5) of DTT. The corresponding supernatants were loaded to a 12% native gel for monitoring levels of both Pgp3 and MOMP by Western blot analysis. Note that Sarkosyl treatment in the presence of DTT released significant amounts of both Pgp3 trimers (lane 3 versus lane 4 in panel a) and MOMP (b). Extraction with SDS denatured Pgp3 into monomers with DTT (panel a, lane 6) or without DTT (lane 5). The presence of DTT plus SDS resulted in the conversion of MOMP oligomers into monomers (lane 5 versus lane 6 in panel b).
DISCUSSION
The cryptic plasmid is shared by all C. trachomatis serovars, and plasmid-free C. trachomatis variants are attenuated in inducing pathologies in mouse urogenital tract (7, 32), suggesting that plasmid-encoded or regulated proteins may play important roles in C. trachomatis pathogenesis. Among all the plasmid-encoded and regulated proteins, Pgp3 is the only plasmid-encoded protein that is secreted into the host cell cytosol during chlamydial infection in cell cultures (28 and data not shown). Immunization with expression plasmids encoding Pgp3 induced protective immunity in mouse models (16, 29). Pgp3 is also the most immunodominant antigen during chlamydial infection in humans, and human antibody recognition of Pgp3 is highly dependent on the conformation (9, 30, 45). In the current study, we partially characterized Pgp3 and provided evidence that Pgp3 is produced as a trimer by C. trachomatis during human infection and that Pgp3 trimerization underlies the conformation-dependent recognition by human antibodies. First, Pgp3 purified from a bacterial expression system was identified as a trimer by the use of both native polyacrylamide gel electrophoresis and analytical ultracentrifugation sedimentation velocity analyses. The Pgp3 trimer was resistant to reducing agents and nonionic detergents. Second, both a conformation-dependent MAb and human antibodies detected trimers or higher order of oligomers but not monomers of Pgp3, suggesting that Pgp3 is presented to the human immune system as oligomerized forms during C. trachomatis infection in humans. Third, the endogenous Pgp3 produced in C. trachomatis-infected cells was detected as a trimer regardless of its intracellular locations. Fourth, Pgp3 trimers were detected in cell cultures infected with all C. trachomatis serovars (data not shown), suggesting that Pgp3 trimerization is a common feature of C. trachomatis organisms. Finally, the organism-associated Pgp3 molecules were localized in the outer membrane complex as trimers and disulfide bond formation may not be required for Pgp3 trimerization but can facilitate the integration of Pgp3 trimers into the outer membrane complexes.
Chlamydial proteins such as the secreted serine protease CPAF (24) can oligomerize. CPAF is synthesized as a proenzyme, and homodimerization of CPAF is necessary for acquiring proteolytic activity (17, 18, 24). However, it is not clear why Pgp3 trimerizes. An obvious advantage of trimerization is to increase protein stability. Indeed, trimerized Pgp3 was more than 10-fold more resistant to trypsin digestion (data not shown). Pgp3 trimers were completely resistant to reducing agents such as 2-ME and DTT and to nonionic detergents such as Sarkosyl and Triton. Although the Pgp3 trimers were efficiently stripped from the chlamydial outer membrane complexes by a combination of reducing agents and nonionic detergents, the trimers remained intact. The Pgp3 trimers were also partially resistant to the strongly ionic detergent SDS. The stable Pgp3 trimers were found in both the chlamydial outer membrane complex and host cell cytosol. Thus, whatever functions Pgp3 may have, trimerization appears to be essential for maintaining and promoting its functionality.
Pgp3 contains 4 cysteine residues. Although Pgp3 trimerization may not require disulfide bond formation, disulfide bond interactions may play an important role in stabilizing the Pgp3 trimers in the outer membrane complexes. Consistent with this hypothesis is the observation that brief treatment of Pgp3 trimers with SDS alone led to a band migrating slightly faster than bands representing intact trimers and also to several bands representing Pgp3 with higher orders of oligomerization (Fig. 1D, lane 5). These SDS-accessible cysteine residues may participate in noncovalent interactions that are susceptible to SDS interruption. Once these cysteines are freed by SDS, they may form disulfide bonds within a trimer, leading to the fast-migrating trimer band, or between trimers, causing high-molecular-mass bands in the absence of reducing agents. However, it is not known whether these SDS-accessible cysteines can form disulfide bonds with other outer membrane components, although DTT was required for efficient release of Pgp3 trimers from the outer membrane complexes (Fig. 4B, lane 4), and there are various cysteine-rich proteins (CRP) in the chlamydial outer membrane complex, including CRP10 (also called OmcA), CRP60 (OmcB), and MOMP (OmpA). Disulfide bond interactions between outer membrane components that are independent of Pgp3 can also affect the association of Pgp3 trimers with the outer membrane complexes (OMCs). Regardless of how Pgp3 trimers are anchored in the outer membrane complex, the most important issue is what roles the outer membrane Pgp3 trimers may play in chlamydial biology and pathogenesis. It has been proposed that cysteine-mediated intra- or intermolecular cross-linking in the outer membrane may represent participation in multiple chlamydial processes, including EB attachment and early uptake (1, 10), ATPase activity (33), and EB-RB differentiation (2, 22). Since Pgp3 trimer association with chlamydial OMCs is affected by cysteine cross-linking, it will be interesting to investigate how the Pgp3 trimers may also participate in these diverse processes.
The preferential recognition of Pgp3 trimers by human antibodies and the presence of Pgp3 trimers in the chlamydial outer membrane complexes suggest that human antibodies may be able to neutralize C. trachomatis infectivity. This is because the outer membrane Pgp3 trimers would be an ideal neutralization target for the human anti-Pgp3 trimer antibodies. However, we found that human antibodies that were subjected to affinity purification with Pgp3 trimers were unable to block C. trachomatis infection (data not shown), which is consistent with the observation that the various mouse anti-Pgp3 trimer antibodies also failed to neutralize C. tra chomatis infectivity (data not shown). Thus, despite the robust antibody responses to Pgp3 trimers in humans infected with C. trachomatis (45), the human anti-Pgp3 trimer antibodies may not offer protection against chlamydial infection. This conclusion is supported by the observation that, although the vast majority of the C. trachomatis-infected individuals can develop high titers of anti-Pgp3 trimer antibodies (45), many individuals can be reinfected with C. trachomatis organisms despite the high conservation of Pgp3 among all C. trachomatis serovars. The issue is to determine why human hosts are induced to mount such a robust yet nonprotective antibody response to Pgp3 trimers. We hypothesize that such as a robust anti-Pgp3 trimer antibody response may represent a chlamydial immune evasion mechanism. C. trachomatis may present the highly immunogenic Pgp3 trimers to the host immune system to exhaust the host responsiveness to antigens vital for inducing protective responses. However, immunization with expression plasmid DNA coding for Pgp3 induced protective immunity in mice (29). It would be interesting to investigate the mechanisms of protective immunity in these mice and to test whether the Pgp3 protein produced by the mammalian expression plasmids is trimerized and whether the anti-Pgp3 antibodies in these mice recognize trimeric or monomeric Pgp3.
There are many unanswered questions about Pgp3 despite its potential importance in chlamydial pathogenesis. For example, how is the Pgp3 trimer secreted to the chlamydial outer membrane and further into the host cell cytosol? What is the role of Pgp3 trimers in the outer membrane complexes? Can the Pgp3 trimer function as a molecular switch to regulate outer membrane complex integrity during chlamydial invasion of host cells or RB-EB differentiation? Most importantly, what is the function of the secreted Pgp3 trimers? Since C. trachomatis deficient in plasmid is less virulent and Pgp3 is the only plasmid-encoded protein that is known to secrete into the host cell cytosol, Pgp3 may play an important role in chlamydial pathogenesis.
Acknowledgments
This work was supported in part by grants (to G. Zhong) from the U.S. National Institutes of Health and from the Robert A. Welch Foundation (grant AQ-1399 to P. J. Hart). Support for the Center for Analytical Ultracentrifugation of Macromolecular Assemblies by the Office of the Vice President for Research at the University of Texas Health Science Center is also gratefully acknowledged.
We thank Borries Demeler and Virgil Schirf for help with the analytical ultracentrifugation experiments.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Abromaitis, S., and R. S. Stephens. 2009. Attachment and entry of chlamydia have distinct requirements for host protein disulfide isomerase. PLoS Pathog. 5:e1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allan, I., T. P. Hatch, and J. H. Pearce. 1985. Influence of cysteine deprivation on chlamydial differentiation from reproductive to infective life-cycle forms. J. Gen. Microbiol. 131:3171-3177. [DOI] [PubMed] [Google Scholar]
- 3.Brookes, E., W. Cao, and B. Demeler. 2009. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur. Biophys. J. 39:405-414. [DOI] [PubMed] [Google Scholar]
- 4.Brookes, E., and B. Demeler. 2007. Parsimonious regularization using genetic algorithms applied to the analysis of analytical ultracentrifugation experiments, p. 361-368. In H. Lipson (ed.), GECCO 2007: Genetic and Evolutionary Computation Conference. Association for Computing Machinery, New York, NY.
- 5.Brookes, E., B. Demeler, C. Rosano, and M. Rocco. 2010. The implementation of SOMO (SOlution MOdeller) in the UltraScan analytical ultracentrifugation data analysis suite: enhanced capabilities allow the reliable hydrodynamic modeling of virtually any kind of biomacromolecule. Eur. Biophys. J. 39:423-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson, J. H., W. M. Whitmire, D. D. Crane, L. Wicke, K. Virtaneva, D. E. Sturdevant, J. J. Kupko III, S. F. Porcella, N. Martinez-Orengo, R. A. Heinzen, L. Kari, and H. D. Caldwell. 2008. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect. Immun. 76:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clifton, D. R., K. A. Fields, S. S. Grieshaber, C. A. Dooley, E. R. Fischer, D. J. Mead, R. A. Carabeo, and T. Hackstadt. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U. S. A. 101:10166-10171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comanducci, M., R. Cevenini, A. Moroni, M. M. Giuliani, S. Ricci, V. Scarlato, and G. Ratti. 1993. Expression of a plasmid gene of Chlamydia trachomatis encoding a novel 28 kDa antigen. J. Gen. Microbiol. 139:1083-1092. [DOI] [PubMed] [Google Scholar]
- 10.Conant, C. G., and R. S. Stephens. 2007. Chlamydia attachment to mammalian cells requires protein disulfide isomerase. Cell Microbiol. 9:222-232. [DOI] [PubMed] [Google Scholar]
- 11.Da Ros, C. T., and S. Schmitt Cda. 2008. Global epidemiology of sexually transmitted diseases. Asian J. Androl. 10:110-114. [DOI] [PubMed] [Google Scholar]
- 12.Demeler, B. 2009. UltraScan: A comprehensive data analysis software package for analytical ultracentrifugation experiments. http://www.ultrascan.uthscsa.edu/.
- 13.Demeler, B. 2005. UltraScan: a comprehensive data analysis software package for analytical ultracentrifugation experiments, p. 210-229. In D. Scott, S. Harding, and A. Rowe (ed.), Modern analytical ultracentrifugation: techniques and methods. Royal Society of Chemistry, Cambridge, United Kingdom.
- 14.Demeler, B., and E. Brookes. 2008. Monte Carlo analysis of sedimentation experiments. Colloid Polym. Sci. 286:129-137. [Google Scholar]
- 15.den Hartog, J. E., S. A. Morre, and J. A. Land. 2006. Chlamydia trachomatis-associated tubal factor subfertility: immunogenetic aspects and serological screening. Hum. Reprod. Update 12:719-730. [DOI] [PubMed] [Google Scholar]
- 16.Donati, M., V. Sambri, M. Comanducci, K. Di Leo, E. Storni, L. Giacani, G. Ratti, and R. Cevenini. 2003. DNA immunization with pgp3 gene of Chlamydia trachomatis inhibits the spread of chlamydial infection from the lower to the upper genital tract in C3H/HeN mice. Vaccine 21:1089-1093. [DOI] [PubMed] [Google Scholar]
- 17.Dong, F., M. Pirbhai, Y. Zhong, and G. Zhong. 2004. Cleavage-dependent activation of a chlamydia-secreted protease. Mol. Microbiol. 52:1487-1494. [DOI] [PubMed] [Google Scholar]
- 18.Dong, F., J. Sharma, Y. Xiao, Y. Zhong, and G. Zhong. 2004. Intramolecular dimerization is required for the chlamydia-secreted protease CPAF to degrade host transcriptional factors. Infect. Immun. 72:3869-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engel, J. 2004. Tarp and Arp: how Chlamydia induces its own entry. Proc. Natl. Acad. Sci. U. S. A. 101:9947-9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackstadt, T., E. R. Fischer, M. A. Scidmore, D. D. Rockey, and R. A. Heinzen. 1997. Origins and functions of the chlamydial inclusion. Trends Microbiol. 5:288-293. [DOI] [PubMed] [Google Scholar]
- 22.Hackstadt, T., W. J. Todd, and H. D. Caldwell. 1985. Disulfide-mediated interactions of the chlamydial major outer membrane protein: role in the differentiation of chlamydiae? J. Bacteriol. 161:25-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harding-Esch, E. M., T. Edwards, A. Sillah, I. Sarr, C. H. Roberts, P. Snell, E. Aryee, S. Molina, M. J. Holland, D. C. Mabey, and R. L. Bailey. 2009. Active trachoma and ocular Chlamydia trachomatis infection in two Gambian regions: on course for elimination by 2020? PLoS Negl. Trop. Dis. 3:e573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, Z., Y. Feng, D. Chen, X. Wu, S. Huang, X. Wang, X. Xiao, W. Li, N. Huang, L. Gu, G. Zhong, and J. Chai. 2008. Structural basis for activation and inhibition of the secreted chlamydia protease CPAF. Cell Host Microbe 4:529-542. [DOI] [PubMed] [Google Scholar]
- 25.Hybiske, K., and R. S. Stephens. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U. S. A. 104:11430-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laue, T. M., B. D. Shah, T. M. Ridgeway, and S. L. Pelletier. 1992. Computer-aided interpretation of analytical sedmentation data for proteins, p. 90-125. In S. E. Harding, A. J. Rowe, and J. C. Horton (ed.), Analytical ultracentrifugation in biochemistry and polymer science. Royal Society of Chemistry, Cambridge, United Kingdom.
- 27.Li, D., A. Vaglenov, T. Kim, C. Wang, D. Gao, and B. Kaltenboeck. 2005. High-yield culture and purification of Chlamydiaceae bacteria. J. Microbiol. Methods 61:17-24. [DOI] [PubMed] [Google Scholar]
- 28.Li, Z., D. Chen, Y. Zhong, S. Wang, and G. Zhong. 2008. The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect. Immun. 76:3415-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Z., S. Wang, Y. Wu, G. Zhong, and D. Chen. 2008. Immunization with chlamydial plasmid protein pORF5 DNA vaccine induces protective immunity against genital chlamydial infection in mice. Sci. China C Life Sci. 51:973-980. [DOI] [PubMed] [Google Scholar]
- 30.Li, Z., Y. Zhong, L. Lei, Y. Wu, S. Wang, and G. Zhong. 2008. Antibodies from women urogenitally infected with C. trachomatis predominantly recognized the plasmid protein pgp3 in a conformation-dependent manner. BMC Microbiol. 8:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, X., M. Afrane, D. E. Clemmer, G. Zhong, and D. E. Nelson. 2010. Identification of Chlamydia trachomatis outer membrane complex proteins by differential proteomics. J. Bacteriol. 192:2852-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Connell, C. M., R. R. Ingalls, C. W. Andrews, Jr., A. M. Scurlock, and T. Darville. 2007. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J. Immunol. 179:4027-4034. [DOI] [PubMed] [Google Scholar]
- 33.Peeling, R. W., J. Peeling, and R. C. Brunham. 1989. High-resolution 31P nuclear magnetic resonance study of Chlamydia trachomatis: induction of ATPase activity in elementary bodies. Infect. Immun. 57:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rockey, D. D., J. Wang, L. Lei, and G. Zhong. 2009. Chlamydia vaccine candidates and tools for chlamydial antigen discovery. Expert Rev. Vaccines 8:1365-1377. [DOI] [PubMed] [Google Scholar]
- 35.Seth-Smith, H. M., S. R. Harris, K. Persson, P. Marsh, A. Barron, A. Bignell, C. Bjartling, L. Clark, L. T. Cutcliffe, P. R. Lambden, N. Lennard, S. J. Lockey, M. A. Quail, O. Salim, R. J. Skilton, Y. Wang, M. J. Holland, J. Parkhill, N. R. Thomson, and I. N. Clarke. 2009. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 10:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma, J., A. M. Bosnic, J. M. Piper, and G. Zhong. 2004. Human antibody responses to a Chlamydia-secreted protease factor. Infect. Immun. 72:7164-7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma, J., F. Dong, M. Pirbhai, and G. Zhong. 2005. Inhibition of proteolytic activity of a chlamydial proteasome/protease-like activity factor by antibodies from humans infected with Chlamydia trachomatis. Infect. Immun. 73:4414-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma, J., Y. Zhong, F. Dong, J. M. Piper, G. Wang, and G. Zhong. 2006. Profiling of human antibody responses to Chlamydia trachomatis urogenital tract infection using microplates arrayed with 156 chlamydial fusion proteins. Infect. Immun. 74:1490-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman, K. J., J. R. Daling, A. Stergachis, N. S. Weiss, H. M. Foy, S. P. Wang, and J. T. Grayston. 1990. Sexually transmitted diseases and tubal pregnancy. Sex. Transm. Dis. 17:115-121. [DOI] [PubMed] [Google Scholar]
- 40.Simms, I., H. Ward, I. Martin, S. Alexander, and C. Ison. 2006. Lymphogranuloma venereum in Australia. Sex. Health 3:131-133. [DOI] [PubMed] [Google Scholar]
- 41.Tanzer, R. J., D. Longbottom, and T. P. Hatch. 2001. Identification of polymorphic outer membrane proteins of Chlamydia psittaci 6BC. Infect. Immun. 69:2428-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taylor, H. R., S. L. Johnson, J. Schachter, H. D. Caldwell, and R. A. Prendergast. 1987. Pathogenesis of trachoma: the stimulus for inflammation. J. Immunol. 138:3023-3027. [PubMed] [Google Scholar]
- 43.Thomas, N. S., M. Lusher, C. C. Storey, and I. N. Clarke. 1997. Plasmid diversity in Chlamydia. Microbiology 143(Pt. 6):1847-1854. [DOI] [PubMed] [Google Scholar]
- 44.van Holde, K. E., and W. O. Weischet. 1978. Boundary analysis of sedimentation velocity experiments with monodisperse and paucidisperse solutes. Biopolymers 17:16. [Google Scholar]
- 45.Wang, J., Y. Zhang, C. Lu, L. Lei, P. Yu, and G. Zhong. 2010. A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J. Immunol. 185:1670-1680. [DOI] [PubMed] [Google Scholar]
- 46.Zhong, G. 2009. Killing me softly: chlamydial use of proteolysis for evading host defenses. Trends Microbiol. 17:467-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193:935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]