Abstract
PilO is an oligosaccharyl transferase (OTase) that catalyzes the O-glycosylation of Pseudomonas aeruginosa 1244 pilin by adding a single O-antigen repeating unit to the β carbon of the C-terminal residue (a serine). While PilO has an absolute requirement for Ser/Thr at this position, it is unclear if this enzyme must recognize other pilin features. To test this, pilin constructs containing peptide extensions terminating with serine were tested for the ability to support glycosylation. It was found that a 15-residue peptide, which had been modeled on the C-proximal region of strain 1244 pilin, served as a PilO substrate when it was expressed on either group II or group III pilins. In addition, adding a 3-residue extension culminating in serine to the C terminus of a group III pilin supported PilO activity. A protein fusion composed of strain 1244 pilin linked at its C terminus with Escherichia coli alkaline phosphatase (which, in turn, contained the above-mentioned 15 amino acids at its C terminus) was glycosylated by PilO. E. coli alkaline phosphatase lacking the pilin membrane anchor and containing the 15-residue peptide was also glycosylated by PilO. Addition of the 3-residue extension did not allow glycosylation of either of these constructs. Site-directed mutagenesis of strain 1244 pilin residues of the C-proximal region common to the group I proteins showed that this structure was not required for glycosylation. These experiments indicate that pilin common sequence is not required for glycosylation and show that nonpilin protein can be engineered to be a PilO substrate.
Colonization and dissemination of the opportunistic pathogen Pseudomonas aeruginosa rely to a large extent on the ability of this organism to produce functional type IV pili (26). These protein fibers, which radiate from the cell pole, are adhesion factors (51), mediate a form of surface translocation referred to as twitching motility (10, 37), and are important in biofilm formation (39). The pili of this organism are primarily composed of a monomeric subunit called pilin (PilA). Type IV pili can be differentiated into two classes (a or b) on the basis of the PilA sequence and structure (23). Although they display considerable sequence variation, the majority of the type IVa pilins of P. aeruginosa can be placed into one of three groups on the basis of primary structure and antigenicity, as well as by the presence of auxiliary pilin genes found immediately downstream from pilA (8, 33). We previously determined that pilin from P. aeruginosa 1244, which belongs to group I (8), contained an O-antigen repeating unit covalently attached to the β-hydroxyl group of a serine residing at the C terminus of this protein (7). While the specific physiological role of the pilin glycan in this organism is not clear, the presence of this saccharide influences pilus hydrophobicity and has a pronounced effect on virulence, as determined in a mouse respiratory model (47). The metabolic origin of the pilin saccharide is the O-antigen biosynthetic pathway (14), and its attachment is catalyzed by an oligosaccharyl transferase (OTase) called PilO (6). Specific regions of this cytoplasmic membrane protein necessary for glycosylation activity have been identified (42). Topological studies of PilO have shown that these regions face the periplasm, suggesting that pilin glycosylation takes place in this chamber (42). Here the glycan substrate is the O-antigen repeating unit covalently linked to the undecaprenol carrier lipid.
PilO has a very relaxed glycan substrate specificity, as indicated by the evidence that it is able to utilize a number of structurally dissimilar O-antigen repeating units as substrate (14), and requires only features of the reducing end sugar to carry out pilin glycosylation (28). WaaL, the enzyme that transfers polymerized O antigen to core lipid A, from Escherichia coli also has a similar broad glycan specificity (19). Recent studies (18) provided evidence that PglL, an OTase of Neisseria meningitidis, recognized only the carrier lipid and was able to attach a variety of saccharides to the pilin of this organism. Although the glycan specificity of PilO is relaxed, this enzyme will not attach other carrier lipid-bound saccharides, such as the peptidoglycan subunit or polymerized O-antigen repeating unit, to pilin. This is indicated by the absence of pilins with increased mass in O-antigen-negative mutants or the production of multiple pilin sizes in the wild-type strain (6).
In vivo analysis of mutagenized P. aeruginosa 1244 pilin showed that the C-terminal serine of this protein was a major pilin glycosylation recognition feature of PilO (27). In addition, modification (substitution of the C-terminal amino acid with a 3-residue sequence terminating in serine) of a group II pilin allowed PilO-dependent attachment of the O-antigen repeating unit (27). While these results suggested that the preponderance of pilin structural information was not required for glycosylation, it was not clear whether regions common among the P. aeruginosa pilins were needed. In the present study three types of experiments were carried out in order to answer this question. First, the glycosylation site was extended away from the pilin surface with the addition of a 15-residue peptide which terminates with serine. Second, an engineered periplasmic protein containing the glycosylation residue at its C terminus was fused with pilin and tested for PilO activity. Finally, this periplasmic protein containing no pilin common region was constructed and tested. Evidence presented in this paper suggests that PilO requires only the glycosylation target residue.
The work presented also indicated that, in addition to pilins, nonpilin protein free in the periplasm or anchored to the cytoplasmic membrane could be engineered so as to serve as a PilO substrate. These results suggest that a wide range of pilins and nonpilin proteins can be engineered to serve as substrate for glycosylation, a finding that would potentially have practical value, particularly in the area of vaccine construction. In addition to elucidating the protein specificity of the PilO system, the present work determined that the peptide extension used can supply functional epitope information to the modified protein, in addition to providing a site for glycosylation. Altogether, the results presented suggest that engineering of pilins and nonpilin proteins for the biological generation of protein-peptide-saccharide constructs is a potentially important strategy in vaccine design.
MATERIALS AND METHODS
Culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Cultures were grown at 37°C on solid or liquid (with agitation at 250 rpm) medium. Bacteria were grown on LB medium for general culturing, on CAYE medium (2% agar, 0.75% Casamino Acids, 0.15% yeast extract) for production of pili, and on Trypticase soy broth for expression of PhoA fusions. The concentrations of antibiotics used in the selective media were as follows: ampicillin at 50 μg/ml for Escherichia coli, carbenicillin at 250 μg/ml for P. aeruginosa, kanamycin at 35 μg/ml for E. coli and 100 μg/ml for P. aeruginosa, and tetracycline at 100 μg/ml for P. aeruginosa. When added, isopropyl-β-d-thiogalactopyranoside (IPTG) was present in a final concentration of either 0.1 mM (E. coli) or 5.0 mM (P. aeruginosa).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| P. aeruginosa | ||
| PA1244 | Wild-type strain, pilin group I | 44 |
| PA1244N3 | Interruption of sigma factor rpoN with tetracycline cassette, Tcr | 43 |
| PA103 | Wild-type strain, pilin group II | 34 |
| PA103 | Interruption of wzy gene with a gentamicin cassette (aacC1), GmrwzyPaO11::aacC1 | 13 |
| PA683 | Wild-type strain, pilin group III | This study |
| E. coli | ||
| XL-Gold | endA1 supE44 thi-1 recA1 gryA96 relA1 lacHte | Stratagene |
| HB101 | supE44 hsdS20 recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 4 |
| Plasmids | ||
| pHK734 | pTrc99a with E. coli phoA | 31 |
| pCR2.1-TOPO | 3.9-kb cloning vector, Apr | InVitrogen |
| pECAP | pMMB66EH with pHK734 phoA | This study |
| pECAPaas | pMMB66EH with mutated E. coli phoA that had an AAS stop extension added to the C-terminal lysine residue | This study |
| pECAPpep | pECAP with DNA coding for C-terminal TAWKPNYAPANAPKS stop | This study |
| pMMB66EH | 8,807-bp broad-host-range expression vector, IPTG inducible tac promoter, Ampr Cbr | 20 |
| pUCP26PilO | pUCP26 with pilO under the control of tac promoter, Tcr | 27 |
| pPAC24 | pMMB66EH with 1244 pilA | 6 |
| pPAC46 | pMMB66EH with 1244 pilAO | 6 |
| pUC46 | pilAO in pUC19 | 9 |
| pW136A | pPAC46 with pilA codon for W136 converted to A | This study |
| pK137A | pPAC46 with pilA codon for K137 converted to A | This study |
| pN139A | pPAC46 with pilA codon for N139 converted to A | This study |
| pY140A | pPAC46 with pilA codon for Y140 converted to A | This study |
| pP142A | pPAC46 with pilA codon for P142 converted to A | This study |
| pN144A | pPAC46 with pilA codon for N144 converted to A | This study |
| pP146A | pPAC46 with pilA codon for P146 converted to A | This study |
| pK147A | pPAC46 with pilA codon for K147 converted to A | This study |
| pS148A | pPAC46 with pilA codon for S148 converted to A | 9 |
| pRMCD28 | Apr, PhoA fusion vector | 12 |
| pPilAPhoA | pRMCD28 with 1244 pilA | This study |
| pPilAP | pMMB66EH with 1244 pilA-E. coli phoA fusion | This study |
| pPilAPaas | pPilAP with DNA coding for C-terminal AAS stop | This study |
| pPilAPpep | pPilAP with DNA coding for C-terminal TAWKPNYAPANAPKS stop | This study |
| pSAD100 | pUC18 with PA103 pilA | 27 |
| pSD5 | pMMB66EH with PA103 pilA | 27 |
| pMAM | pSD5 with DNA coding for C-terminal TAWKPNYAPANAPKS stop | This study |
| pPAL100 | pMal-cRI containing DNA coding for a malE-pilO gene fusion | 42 |
| pUCP24 | Broad-host-range cloning vector, pUC18 derived, Gmr | 50 |
| pUCP24PilO | pUCP24 with pilO under control of tac promoter | This study |
| p683PilA | pMMB66EH with PA683 pilA | This study |
| p683aas | p683pilA with DNA coding for C-terminal AAS stop | This study |
| p683pep | p683pilA with DNA coding for C-terminal TAWKPNYAPANAPKS stop | This study |
Plasmid construction.
Details of plasmid construction, including the oligonucleotide primers used, are presented in the supplemental material.
Cellular alkaline phosphatase determination.
Cellular alkaline phosphatase determination employed the protocol of Daniels et al. (12). Culture absorbance at 600 nm was determined using a Spectronic 20 spectrometer. Absorbances at 415 and 595 nm were measured using a Bio-Rad model 3550 microplate reader. PhoA enzyme units were calculated using a standard equation (36).
Protein sample preparation.
Strains expressing pilin constructs were grown on solid media in the presence of necessary antibiotics. Cell extracts were prepared as described previously (8).
For PilAP, PilAPI, and PilAPII, broth starter cultures of P. aeruginosa PA103 wzyPaO11::aacC1 (where wzyPaO11 is wzy of P. aeruginosa serotype O11) containing the plasmids of interest were grown overnight. Seven milliliters of the starter cultures was used to inoculate 300 ml of the same medium containing 5.0 mM IPTG. These cultures were grown overnight and centrifuged at 5,000 rpm for 30 min in a GSA rotor. The pellet was resuspended in 100 ml of 0.05 M Tris HCl, pH 7.3, and centrifuged at 5,000 rpm for 30 min in a GSA rotor. These cells were resuspended in 10 ml of lysis buffer (10 mM phosphate, 30 mM NaCl, 0.25% Tween 20, 10 mM β-mercaptoethanol, 10 mM EDTA, 10 mM EGTA) and subjected to sonication on ice with three 15-s bursts using a Biosonik III sonicator (Bronwill Scientific) and then centrifuged at 10,000 rpm for 30 min in an SS-34 rotor. The supernatant was subjected to centrifugation at 30,000 rpm for 1 h in an SW60Ti rotor, after which the pellets were rinsed with 5 ml of ice-cold phosphate-buffered saline. This pellet was dissolved with 200 μl of lysis buffer. The protein concentration was determined using the bicinchoninic acid assay (Pierce).
In order to analyze ECAP, ECAPI, and ECAPII, broth starter cultures of P. aeruginosa PA103 wzyPaO11::aacC1 containing the plasmids of interest and 5 mM IPTG were grown overnight as described above. The periplasmic fraction of these cells was isolated using the protocol of Poole and Hancock (41). This material was centrifuged at 35,000 rpm for 1.5 h in a SW-60 Ti rotor, and the supernatant was dialyzed overnight against 2 liters of 0.05 M Tris HCl, pH 7.3. The sample was lyophilized and dissolved with 1 ml of deionized water. This solution was then fractionated by ammonium sulfate precipitation (17), followed by gel filtration by fast-performance liquid chromatography using a Superose 12 column. Fractions containing PhoA were determined using the alkaline phosphatase assay of Garen and Levinthal (21).
Mass spectrometric analysis.
Analysis of ECAP and ECAPI was carried out on GluC-digested samples. Thirty micrograms of purified protein in 50 μl of 0.1 M ammonium bicarbonate was treated according to the manufacturer's directions. In brief, the protein was reduced with the addition of dithiothreitol to 10 mM, followed by alkylation with the addition of iodoacetamide to 20 mM. This material was digested with 1.5 μg GluC (Thermo Scientific) for 22 h at 37°C. The peptide fragments produced were separated by capillary C18 high-pressure liquid chromatography using a ThermoElectron Surveyor liquid chromatograph with a Micro AS autosampler. Direct analysis of the column effluent was carried out using a ThermoElectron LCQ Deca XP Plus quadrupolar ion trap mass spectrometer utilizing a nanospray ionization source. This instrument was set up to perform both mass spectrometry (MS) and MS/MS analysis. The computer algorithm controlling these experiments selected the three largest ions from an MS spectrum for MS/MS analysis in triplicate. This process was repeated throughout the chromatographic separation. MS and MS/MS data analysis employed the SEQUEST program, in which the data obtained were compared with those from an in silico digestion of Escherichia coli alkaline phosphatase. Pilin mass analysis was carried out by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS using a PerSeptive Biosystems Voyager STR with DE and a high-m/z detector.
Electrophoresis and Western blot methods.
The electrophoresis and Western blot methods described previously (42) were followed. Polyacrylamide gels with 16% T and 3% C were used when PilA constructs were analyzed. Polyacrylamide gels with 10% T and 3% C were used when PhoA, PilAPhoA, and derivatives of these proteins were examined. All primary antibodies were mouse IgG. A monoclonal antibody (Sigma-Aldrich Co.) was used to detect PhoA. A P. aeruginosa IATS (International Antigenic Typing Scheme) (35) serotype O11-specific monoclonal antibody (ERFA Biotech) was used to detect proteins bearing this saccharide. Monoclonal antibody 11.14 (46) was used to detect proteins bearing the O7 repeating unit. Monoclonal antibody 5.44 (8), which was specific for the α-β loop region, or monoclonal antibody 6.45 (8), which was specific for the disulfide loop region, was used to detect P. aeruginosa strain 1244-specific pilin epitopes. Monoclonal antibody 2.97 (46) was used to recognize strain PA103 pilin. Secondary antibodies (anti-mouse IgG) were either phosphatase (Kirkegaard & Perry Laboratories) or Alexa Fluor (Sigma-Aldrich) labeled. The latter label was detected using a Molecular Dynamics 595 fluorimager, as described previously (42). This protocol was also used to quantitate PhoA fusions. Here, protein samples were separated by polyacrylamide gel electrophoresis using 1-mm 10% T and 3% C gels. The separated proteins were electroblotted for 23 min to 0.45-μm-pore-size nitrocellulose paper at 100 V using a 25 mM Tris HCl-192 mM glycine-20% methanol buffer. Under these conditions, staining with Coomassie brilliant blue showed that no protein with the apparent molecular weight of the fusion remained in the gel. After the gels were blocked and the Alexa Fluor secondary antibody was applied, relative fluorescence levels were determined using the ImageQuant program. Utilizing a double layer of membrane showed that no fusion passed through the first nitrocellulose paper.
Site-directed mutagenesis.
Mutagenesis of DNA coding for the P. aeruginosa 1244 pilin disulfide loop residues was carried out as described in the supplemental material. The plasmid encoding the protein to be tested also contained a functional pilO gene. The mutagenized pilA was expressed in P. aeruginosa 1244N3, where the glycosylation state was indicated by a shift in apparent molecular weight, as determined by Western blotting using a pilin-specific monoclonal antibody.
Generation of polyclonal anti-683 pilin serum.
The protocol used for the generation of polyclonal anti-683 pilin serum was approved by Duquesne University's Institutional Animal Care and Use Committee (IACUC). All animal storage and experimentation took place at the USDA-approved Duquesne University Animal Care Facility. Preparation of strain 683 pili followed a standard methodology (29). A total of 6 female BALB/c mice (Hilltop Lab Animals, Scottdale, PA), approximately 6 to 8 weeks old and weighing 25 g, were immunized using a protocol described previously (29).
Twitching motility.
The twitching motility protocol was carried out as described previously (7).
Homology modeling.
The E. coli PhoA molecular structure was obtained from the Protein Data Bank (http://www.rcsb.org/pdb/explore/explore.do?structureId=1ALK). PhoA homology models of the PhoA constructs were generated using 3D-PSSM software online (http://www.sbg.bio.ic.ac.uk/3dpssm/index2.html). The tertiary homology structural output was analyzed using the DS Viewer Pro program (version 6.0) for surface-exposed C-terminal residues and surface charge.
RESULTS
Glycosylation of engineered pilins.
We have previously shown that the P. aeruginosa PA103 pilin, a protein that is normally not glycosylated and that is a member of the group II family, could be engineered to act as a glycosylation substrate by substituting three residues (AAS) for the C-terminal amino acid (27). These and other experiments indicated the importance of the C-terminal serine but did not eliminate the possibility that PilO required other pilin components that are common to both proteins. Representatives of the three major pilin groups of P. aeruginosa show strong sequence identity over the N-proximal region of this protein and, to a lesser extent, in the C-proximal region (Fig. 1), a characteristic shared with other type IV pilins (25). Experiments were carried out to determine whether the glycosylation site could function if it was extended a significant distance from the protein surface or if its close proximity with the pilin common regions was required. To do this, the pilA gene of strain PA103 was extended so as to code for 15 more residues. The sequence chosen (Fig. 2 A) was based on the strain 1244 C-proximal region (TAWKPNYAPANCPKS). The cysteine of this sequence was replaced with an alanine in order to avoid potential incorrect disulfide formation. This sequence was selected in part because it is a component of a stable protein, which suggested that the fusion might be resistant to degradation. It was also picked because it is a known B-cell epitope and is the target of characterized monoclonal antibodies (8, 9), suggesting that fusions containing this structure could be detected by immunoblotting. A group II pilin was chosen as the platform over the 1244 protein to avoid potential DNA excision problems which could occur with a repetitive contiguous sequence.
FIG. 1.
Comparison of the P. aeruginosa 683 PilA primary structure with pilin group I, II, and III representatives. Sequence identity is indicated by gray highlighting. The alignment of the three representative pilin groups is based on the common sequence of the N-proximal region and the disulfide loop region. Cysteines forming the disulfide loop are in white with a black background. Identity between PA14 and 683 pilin residues is indicated by a colon. The GenBank accession numbers for strains 1244, PA103, and PA14 are CAA58768.1, P08015.1, and ABJ13792.1, respectively.
FIG. 2.
Features of the fusions employed in this study. (A) Parent and modified pilins; (B) pilin-alkaline phosphatase constructs (modified and unmodified); (C) parent and modified alkaline phosphatase. The cartoons show the relative sizes and orientations of construct components (gray boxes, PilA; white boxes, PhoA) as well as the modification site (black boxes). The protein product name is listed in the first column of each panel. The plasmid containing the gene expressing this protein is in the second. The third column shows essential features of the protein construct. Here, the C-terminal additions are in white with a black background. The PilA-PhoA junctions are shown in black with a gray background.
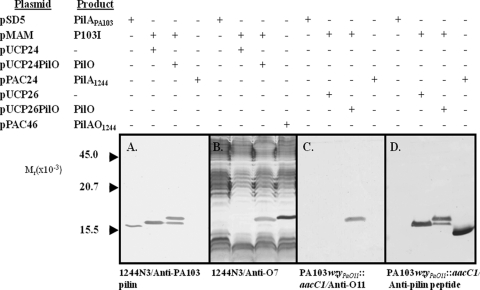
This protein construct, which was coded for by pMAM and which was termed P103I (Fig. 2A), was assayed by Western blotting in a PilAO-negative (PilAO−) background using a PA103 pilin-specific monoclonal antibody as the probe (Fig. 3 A). PilO was delivered from a second plasmid. The pilin produced in the absence of PilO had an increased size, as would be expected from the presence of the added peptide. The stability of this protein construct was indicated by the absence of pilin degradation products. Pilin generated in the presence of a functional pilO gene produced an antigen species of even greater apparent molecular weight, suggesting that this protein had been modified by PilO. A blot produced in an identical manner was probed with an antibody specific for the O-antigen repeating unit of the host strain (Fig. 3B). While the lipopolysaccharide ladder was seen in all cases, the presence of PilO gave an additional band which was at an identical position as the putatively modified P103I, suggesting that this protein contains bound O-antigen repeating unit. In order to confirm this, pMAM was expressed in P. aeruginosa PA103wzyPaO11::aacC1, which produced an antigenically dissimilar pilin and carrier-lipid-bound O-antigen repeating unit (but no polymerized O antigen). This system allowed Western blot detection of the pilin glycan with an O-antigen-specific antibody without visual interference from the lipopolysaccharide ladder. Here a reaction was seen only when pMAM was expressed in the presence of a functional pilO gene (Fig. 3C). In addition, the apparent molecular weight of this antigen matched that of the putative glycosylated construct seen in Fig. 3A and B. The presence of the peptide extension in both the putative glycosylated and the nonglycosylated constructs was confirmed by Western blotting (Fig. 3D) using monoclonal antibody 6.45, which has previously been determined to be specific for the C-terminal region of strain 1244 pilin (8). The control for this response is the reaction of monoclonal antibody 6.45 with nonglycosylated 1244 pilin produced by pPAC24 (Fig. 3D). A twitching assay showed that while cloned PA103 pilA expressed in a PilAO− mutant of strain 1244 supported motility, expression of pMAM did not (results not shown). In addition, electrophoretic analysis of the supernatant of sheared cells showed the absence of pilin protein (results not shown), indicating that the construct is not assembled into a pilus. Altogether, these results suggest that moving the glycosylation site away from the pilin surface does not interfere with PilO action and is evidence that the pilin common regions are not necessary for glycosylation.
FIG. 3.
Western blot of natural and modified PilA of Pseudomonas aeruginosa PA103 in the presence and absence of PilO. Antibodies used were monoclonal antibody 2.97, which is specific for strain P. aeruginosa PA103 pilin (A); monoclonal antibody 11.14, which is specific for the O antigen of P. aeruginosa IATS serotype O7 (B); monoclonal antibody O11, which is specific for the O antigen of P. aeruginosa IATS serotype O11 (C); and monoclonal antibody 6.45, which is specific for the disulfide loop region of strain 1244 pilin (D). The plasmids of panels A and B were expressed in P. aeruginosa 1244N3. The plasmids of panels C and D were expressed in P. aeruginosa PA103wzyPaO11::aacC1. Glycosylated and nonglycosylated 1244 pilin was produced from pPAC46 and pPAC24, respectively.
The P. aeruginosa group III pilins are the third most common type produced by this organism (33). Nucleotide sequencing of the pilA of P. aeruginosa 683 revealed that the pilin coded for by this gene was nearly identical to the group III pilin of strain PA14 and that these pilins, while they share core sequence features, were considerably larger than members of the other two groups (Fig. 1). In addition, accessory gene tfpY, typically found immediately downstream of the group III pilA (33), was present in this position in strain 683 (results not shown). Published results have shown that PA14 pilin is not glycosylated (2). We determined, using MALDI-TOF MS, that the strain 683 pilin isolated had a mass of 17,475 kDa, which agreed well with the predicted value (17,453 kDa), further suggesting that this protein was also not modified.
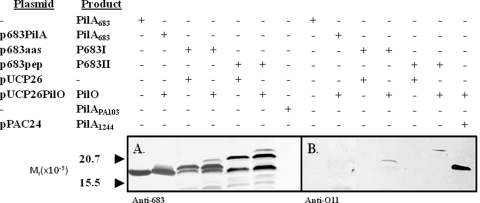
Experiments were carried out to determine (i) if the strain 683 pilin could be engineered to be a PilO substrate and (ii), if so, whether the glycosylation site could be functional if it is extended away from the protein. In order to do this, two modified pilins were constructed. The first, which was analogous to a group II construct that was shown to support glycosylation (27), was p683aas (Fig. 2A). This plasmid produced P683I, which was strain 683 pilin with an AAS extension at its C terminus. The second, p683pep, coded for P683II, which contained the C-terminal 15-residue epitope described above (Fig. 2A). Expression of these constructs in P. aeruginosa PA103wzyPaO11::aacC1 was analyzed by Western blotting using the procedure described above and employing either a 683 pilin-specific polyclonal antibody or the O-antigen-specific monoclonal antibody as probe. Figure 4 A shows that both p683aas and p683pep, expressed in the absence of a functional pilO gene, produced antigen of the expected increased apparent molecular weight as well as a form matching unmodified pilin. In the presence of functional pilO, a larger form yet was seen with both constructs, suggesting that these pilins had undergone posttranslational modification. To test this, a blot run with identical strain 683 pilin samples was probed with the O-antigen-specific monoclonal antibody. Here it can be seen (Fig. 4B) that a band of the same apparent molecular weight as the largest pilin structure detected by the anti-683 pilin serum was produced only in the presence of functional pilO. pPAC24 expressed in the presence of pPUCP26 produced the glycosylated pilin control of Fig. 4B. A twitching assay of these constructs showed that while p683PilA supported motility when it was expressed in a PilA− P. aeruginosa strain, neither of the plasmids producing modified pilins did so (results not shown). Further, only trace amounts of pili were present in the supernatants of sheared cells (results not shown). Overall, these results indicate that group III pilin, in spite of size and sequence differences, can be engineered to function as a glycosylation substrate for PilO by adding either a short peptide extension or the 15-residue structure. Further, moving the glycosylation site away from the pilin surface, as with P103I, supports PilO function, which suggests that the pilin common sequences are not needed for modification.
FIG. 4.
Western blot of natural and modified P. aeruginosa 683 PilA in the presence or absence of PilO. The antibodies used were a polyclonal preparation raised against pure 683 pilin (A) or a monoclonal antibody against IATS serotype O11 (B). All plasmids were expressed in P. aeruginosa PA103wzyPaO11::aacC1. Strain 683 pilin and strain PA103 pilin were supplied as purified proteins. pPAC24 contains a functional pilA gene but no pilO.
Glycosylation of engineered nonpilin protein.
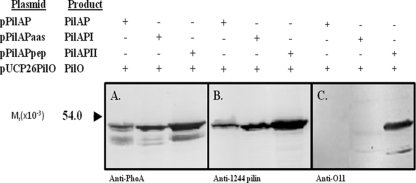
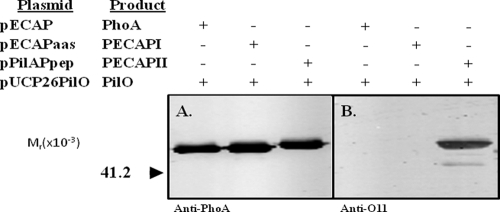
To further test the PilO substrate requirements, experiments were carried out to determine whether nonpilin protein could be engineered to function as a glycosylation target. In the first of two strategies, we constructed a protein fusion that targeted the glycosylation site to the periplasmic side of the cytoplasmic membrane, the location of PilO's catalytic region and glycan substrates (42). To accomplish this, the C-terminal residue of P. aeruginosa 1244 PilA was fused with the N terminus of E. coli PhoA lacking a leader sequence, thus producing PilAP (Fig. 2B). PhoA from Escherichia coli was chosen because it shared with strain 1244 pilin surface characteristics that were necessary for glycosylation (27). Specifically, it has a C-proximal region that extends away from the protein surface and a net positive charge in this region (results not shown). PilAP was engineered so as to have a C-terminal AAS (PilAPI, coded by pPilAPaas) or the pilin peptide described above (PilAPII, coded by pPilAPpep) at its C terminus (Fig. 2B). High levels of alkaline phosphatase (grown under conditions which suppress expression of the chromosomal phoA gene) were produced by P. aeruginosa strains expressing these plasmids, indicating that the fusion was active and that the PilA component of the construct was able to target the PhoA portion to the periplasm of this organism (Table 2). The membrane fraction of P. aeruginosa PA103wzyPaO11::aacC1 expressing these constructs (along with a plasmid producing functional PilO) was analyzed by Western blotting using an anti-PhoA serum as a probe. The results (Fig. 5 A) showed antigens of the expected size and indicated that, although degradation was seen, the fusion targeted as anticipated. A Western blot of the periplasmic fraction of these strains using the same probe showed antigens of lower molecular weight, suggesting that the fusions were subject to degradation, resulting in the release of PhoA (results not shown). The presence of the PilA component of this fusion was confirmed by Western blotting (Fig. 5B) using a monoclonal antibody specific for a central region of the strain 1244 pilin primary structure. These samples were also assayed by Western blotting using the O-antigen-specific monoclonal antibody as a probe in order to test for the presence of O-antigen repeating unit (Fig. 5C). Before this step was carried out, the membrane fractions were normalized after determination of relative antigen levels using a quantitative Western blot. Figure 5C shows that neither the control construct nor the fusion containing the AAS extension produced a reaction, indicating that these proteins were not suitable glycosylation substrates for PilO. However, fusion PilAPII produced a strong response, suggesting that, as above, the added peptide allowed this protein to function as a PilO substrate. Altogether, these results suggest that a nonpilin protein anchored to the outer surface of the cytoplasmic membrane can be engineered to act as substrate for this enzyme. This is further evidence that the pilin common region is not required for PilO activity. The second strategy employed aimed to determine whether a nonpilin periplasmic protein which was not fused to pilin (and not anchored in the cytoplasmic membrane) could serve as a glycosylation substrate. Here, E. coli PhoA (PECAP, coded for by pECAP), a protein that normally targets to the periplasm, was engineered so as to have a C-terminal AAS (PECAPI, coded by pECAPaas) or the pilin peptide described above (PECAPII, coded by pECAPpep) at its C terminus (Fig. 2C). To test for protein glycosylation, plasmids coding for these constructs were expressed in P. aeruginosa PA103wzyPaO11::aacC1, which, as described above, allowed Western blot assay of glycosylated protein without lipopolysaccharide interference. Here, PilO was supplied from a plasmid carrying a functional pilO gene. Table 2 shows that high levels of alkaline phosphatase activity were produced under growth conditions suppressing expression of the chromosomal phoA, indicating that the PhoA constructs were active and targeted correctly. These proteins were purified from the periplasmic fraction using salt fractionation and gel filtration, which resulted in preparations containing only minor contamination, as determined by polyacrylamide gel electrophoresis followed by protein staining with Coomassie brilliant blue (results not shown). Aliquots of this material containing equivalent amounts of protein were then subjected to Western blot analysis using an anti-PhoA antibody as a probe. Here, antigens of the anticipated size were seen for each sample (Fig. 6 A). When an identical blot was probed with the O-antigen-specific monoclonal antibody, a strong reaction was seen with PECAPII, indicating that, as with PilAPII, this construct was a glycosylation substrate (Fig. 6B). No reaction was seen with the PECAPI construct. In order to clarify whether the engineered portion of PECAPI was intact and a good potential substrate, this construct, as well as the control protein, was digested with GluC and subjected to liquid chromatography followed by mass spectrometry (MS and MS/MS). An intact C-terminal fragment (DLFYTMKAALGLK) of the control protein was detected with a cross-correlation score of 2.65. While this fragment was absent in PECAPI, another fragment (DLFYTMKAALGLKAAS) which was not present in the control digestion and which had a cross-correlation score of 3.38 was seen. These results indicated that this protein contains the potential glycosylation site and that the absence of modification indicated that PilO was not able to recognize this structure.
TABLE 2.
Alkaline phosphatase levels
| Plasmida | Alkaline phosphatase activityb |
|---|---|
| pPAC24 | 0.0 |
| pPilAP | 11,507.1 |
| pPilAPaas | 11,836.4 |
| pPilAPpep | 10,413.1 |
| pECAP | 15,629.1 |
| pECAPaas | 20,569.1 |
| pECAPpep | 9,675.1 |
Plasmids were expressed in P. aeruginosa PA103 wzyPaO11::aacC1/pUCP26PilO.
Activity is given in standard units (36).
FIG. 5.
Western blot of natural and modified PilA-PhoA fusions in the presence of PilO. The antibodies used were a monoclonal antibody specific for E. coli PhoA (A); monoclonal antibody 5.44, which was specific for a P. aeruginosa 1244 pilin epitope (B); and monoclonal antibody O11, which was specific for P. aeruginosa IATS serotype O11 (C). All plasmids were expressed in P. aeruginosa PA103wzyPaO11::aacC1 containing pUCP26PilO. See the legend to Fig. 2 for construct details.
FIG. 6.
Western blot of natural and modified E. coli PhoA produced in the presence of PilO. The antibodies used were a monoclonal antibody specific for E. coli PhoA (A) and monoclonal antibody O11, which was specific for P. aeruginosa IATS serotype O11 (B). All plasmids were expressed in P. aeruginosa PA103wzyPaO11::aacC1 containing pUCP26PilO. See the legend to Fig. 2 for construct details.
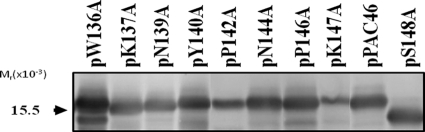
The finding that PECAPII is glycosylated while PECAPI is not raises the possibility that PilO recognizes a portion of the group I disulfide loop region, in addition to the C-terminal residue. This is unlikely, since we have previously shown that mutant 1244 pilin lacking the entire disulfide loop is glycosylated (27). In order to reexamine this point, eight of the conserved group I disulfide loop and tail residues (see Fig. S1 in the supplemental material) found in this portion of P. aeruginosa 1244 PilA were replaced with alanines and tested for their ability to support glycosylation. Figure 7 shows that this treatment did not influence the ability of PilO to glycosylate pilin. These results confirm the findings of previous studies (27) and suggest that, other than the C-terminal serine/threonine, no pilin components are required for protein glycosylation by PilO. Altogether, the results presented here provide evidence that the pilin constant region is not required for glycosylation and suggest that the potential for the use of PilO in glycoengineering is considerably broadened with the finding that nonpilin proteins can be employed as substrates.
FIG. 7.
Glycosylation of mutant P. aeruginosa 1244 pilins. Cloned pilA from this organism was subjected to site-directed mutagenesis, resulting in clones which produced the pilins seen. The primary antibody employed was monoclonal antibody 5.44. Pilin produced from pPAC46 was the glycosylated control, while pilin from pS148A, which has been shown to be nonglycosylated (9), was the negative control.
DISCUSSION
The type IV pilins make up a protein family that is characterized by common sequence and structural features (11). While previous work (27) has shown that PilO requires an unblocked serine or threonine occupying the pilin C-terminal position, shared pilin components are also potential recognition features for pilin glycosylation. The evidence offered in the present paper suggests that this is not the case. Here it was shown that extension of the PilO site away from the pilin surface by 15 residues does not prevent glycosylation. In addition, insertion of a protein between the pilin C terminus and this peptide extension likewise results in active protein glycosylation. Finally, attaching this peptide modification to the carboxyl end of a native periplasmic protein also results in efficient production of a stably glycosylated product. The peptide extension used in each of these experiments was designed from the C-proximal portion of the disulfide loop and the pilin tail region of strain 1244 pilin, which left open the possibility that PilO recognized portions of the peptide used for extension of the glycosylation site. The finding that these residues, as present in native pilin, were not required for PilO activity suggested that this region was not a determinant in glycosylation. Overall, the results presented here suggest that PilO recognizes only the C-terminal serine. While the C-terminal residue is crucial to PilO activity, other factors are also important. Previous studies showed that a minimum distance from the pilin surface as well as a compatible surface charge is required for activity (27). Interference of enzyme activity by protein surface properties may be responsible for the finding that while attachment of both the AAS and the peptide extensions to the pilins allowed glycosylation, the shorter structure did not support modification of either the PilAPI or the PECAPI fusions. These results may be due to steric hindrance caused by protein surface structures or to a charge effect caused by the presence of the required metal or the noncovalent attachment of substrate (or product) to the PhoA catalytic site (32). The extension of the glycosylation site well away from the protein surface may have allowed modification of PilAPII and PECAPII. A longer C-terminal extension may produce a more efficient interaction between PilO and P683I and P683II. However, the inefficient glycosylation of both P683I and P683II may also reflect an interference caused by the bulky disulfide region of this protein or a construct instability (since glycosylation was not improved using P683II) introduced by modification at the C terminus. The physical environment of the glycosylation site and the composition of any peptide extension will be important considerations in the future design of both pilin and nonpilin constructs.
PglO, an OTase, of Neisseria gonorrhoeae and related species was found to be able to glycosylate a number of cellular proteins, in addition to pilin (5, 49). While the simple protein substrate requirements of PilO and the ready availability of its O-antigen substrate raise the possibility that this enzyme has other protein substrates as well, the absence of reactive antigens seen when pilO was expressed in a Wzy-negative strain (Fig. 2C) suggests that this is not the case. One reason might be that while PglO is able to modify internal serines (1), PilO is able to glycosylate only a C-terminal serine or threonine (9, 27). Thus, the PilO protein substrate requirements are, while simple, likely found naturally on very few proteins. Another is that the oligosaccharides utilized by PilO vary in charge, size, and hydrophobicity, depending on the lipopolysaccharide serotype. The glycan substrate for the general glycosylation systems of Neisseria and Campylobacter are structurally less variable (22, 48). While this variable modification is clearly tolerated on the surface of a structural protein such as pilin, as indicated by the finding of a variety of pilin-associated O antigens among P. aeruginosa strains (unpublished observations), the presence of this diversity of O-antigen repeating units could have a differentially negative effect on the activity of an enzyme or chaperone protein. Overall, these results point to dissimilarity in protein substrate specificities between PglO and PilO, which suggests that there is a difference in the physiological role of protein glycosylation in the organisms that produce these enzymes.
The ability of PilO to attach heterologous O-antigen repeating units to pilins (14) suggests the possibility of constructing a vaccine that could provide both pilin and O-antigen-specific protection. Both of these structures are strongly immunogenic and have been considered vaccine candidates (16). Immunization with either pili (38) or O antigen (15, 40, 52) from P. aeruginosa has been shown to be protective, as determined in murine models. We have shown that the P. aeruginosa strain 1244 pilin-bound O-antigen repeating unit is immunogenic (9) and that this structure provides O-antigen-specific protection (29). A T-cell-dependent response against this saccharide is indicated by the strong IgG reaction seen (29). This vaccine target is not limited to the lipopolysaccharide of P. aeruginosa, since the minimal PilO glycan requirement is found in the O-antigen repeating units of many bacterial pathogens (14, 28). In addition, the evidence presented here that PilO requires no pilin component other than the C-terminal residue suggests that structurally different pilins from a variety of pathogens could be engineered to act as glycosylation substrates. Candidates for research in this area would be the type IVb pilins (23), including the structurally unusual Flp fimbriae (3), as well as the chaperone-assisted pili (45).
The present work demonstrates that it is possible to insert a peptide epitope between pilin and the associated O-antigen repeating unit. This addition allows the incorporation of a distinctly different vaccine constituent that could target a pathogen component structurally unrelated to pilin or the O antigen. For example, it may be possible to construct a single vaccine that could protect against colonization and dissemination (pilin response), have a neutralizing effect (from a peptide based on a toxin epitope), and stimulate opsonization (due to production of O-antigen-specific antibodies). Using a similar line of reasoning, a tripartite construct could be made that was composed of a nonpilin protein platform to which a glycosylated peptide was attached. Again, this structure could potentially stimulate three separate types of response. The glycosylation of PECAPII suggests that other free periplasmic proteins can be engineered for such activity, while modification of the PilAPII fusion indicates that membrane-anchored nonpilin proteins can serve as substrates for PilO. It will be of particular interest to determine whether transiently periplasmic species such as proteins transported by the type II or type IV secretion systems can also be engineered for O-antigen glycosylation.
Delivery of multiple forms of the same immunogen can produce a suppressed response against each component. The pilins of Dichelobacter nodosus (30) and the O antigen of P. aeruginosa (24) are examples of this phenomenon. A further disadvantage of numerous forms of a single immunogen is that many preparations are required to produce a single vaccine cocktail. These problems could potentially be avoided by incorporating antigenically unrelated epitopes in the same vaccine structure using the tripartite strategy described above. Cocktails of these preparations incorporating only the most common immunogen forms could potentially avoid the immunogenic suppression response. The targeting of three separate epitopes could compensate for incomplete immunogen coverage. Studies are required to determine the relative immunogenicity and protection afforded by such preparations.
Supplementary Material
Acknowledgments
This study was supported by a grant (AI054929) from the NIH to P.C.
Mass spectrometric analysis of peptide fragments was carried out at the University of Pittsburgh Genomics and Proteomics Core Laboratories. Pilin mass analysis was performed at the Mellon Institute Center for Molecular Analysis, Carnegie Mellon University.
Footnotes
Published ahead of print on 10 September 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aas, F. E., W. Egge-Jacobsen, H. C. Winther-Larsen, C. Lovold, P. G. Hitchen, A. Dell, and M. Koomey. 2006. Neisseria gonorrhoeae type IV pili undergo multisite hierarchical modifications with phosphoethanolamine and phosphocholine requiring an enzyme structurally related to lipopolysaccharide phosphoethanolamine transferases. J. Biol. Chem. 281:27712-27723. [DOI] [PubMed] [Google Scholar]
- 2.Asikyan, M. L., J. V. Kus, and L. L. Burrows. 2008. Novel proteins that modulate type IV pilus retraction dynamics in Pseudomonas aeruginosa. J. Bacteriol. 190:7022-7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernard, C. S., C. Bordi, E. Termine, A. Filloux, and S. de Bentzmann. 2009. Organization and PprB-dependent control of the Pseudomonas aeruginosa tad locus, involved in Flp pilus biology. J. Bacteriol. 191:1961-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolivar, F., and K. Backman. 1979. Plasmids of Escherichia coli as cloning vectors. Methods Enzymol. 68:245-267. [DOI] [PubMed] [Google Scholar]
- 5.Borud, B., F. E. Aas, A. Vik, H. C. Winther-Larsen, W. Egge-Jacobsen, and M. Koomey. 2010. Genetic, structural, and antigenic analyses of glycan diversity in the O-linked protein glycosylation systems of human Neisseria species. J. Bacteriol. 192:2816-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castric, P. 1995. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology 141:1247-1254. [DOI] [PubMed] [Google Scholar]
- 7.Castric, P., F. J. Cassels, and R. W. Carlson. 2001. Structural characterization of the Pseudomonas aeruginosa 1244 pilin glycan. J. Biol. Chem. 276:26479-26485. [DOI] [PubMed] [Google Scholar]
- 8.Castric, P. A., and C. D. Deal. 1994. Differentiation of Pseudomonas aeruginosa pili based on sequence and B-cell epitope analyses. Infect. Immun. 62:371-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comer, J. E., M. A. Marshall, V. J. Blanch, C. D. Deal, and P. Castric. 2002. Identification of the Pseudomonas aeruginosa 1244 pilin glycosylation site. Infect. Immun. 70:2837-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Craig, L., M. E. Pique, and J. Tainer. 2004. Type IV pilus structure and bacterial pathogenicity. Nat. Rev. Microbiol. 2:363-378. [DOI] [PubMed] [Google Scholar]
- 12.Daniels, C., C. Vindurampulle, and R. Morona. 1998. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy). Mol. Microbiol. 28:1211-1222. [DOI] [PubMed] [Google Scholar]
- 13.Dean, C. R., C. V. Franklund, J. D. Retief, M. J. Coyne, K. Hatano, D. Evans, G. B. Pier, and J. B. Goldberg. 1999. Characterization of the serogroup O11 O-antigen locus of Pseudomonas aeruginosa PA103. J. Bacteriol. 181:4275-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiGiandomenico, A., M. J. Matewish, A. Bisaillon, J. R. Stehle, J. S. Lam, and P. Castric. 2002. Glycosylation of Pseudomonas aeruginosa 1244 pilin: specificity of glycan substrate. Mol. Microbiol. 46:519-530. [DOI] [PubMed] [Google Scholar]
- 15.DiGiandomenico, A., J. Rao, K. Archer, T. S. Saidi, J. Gardner, A. N. Neely, G. B. Pier, and J. B. Goldberg. 2007. Intranasal immunization with heterologously expressed polysaccharide protects against multiple Pseudomonas aeruginosa infections. Proc. Natl. Acad. Sci. U. S. A. 104:4624-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doring, G., and G. B. Pier. 2008. Vaccines and immunotherapy against Pseudomonas aeruginosa. Vaccine 26:1011-1024. [DOI] [PubMed] [Google Scholar]
- 17.Englard, S., and S. Seifter. 1990. Precipitation techniques, p. 285-300. In M. P. Deutscher (ed.), Guide to protein purification, vol. 182. Academic Press, Inc., San Diego, CA. [DOI] [PubMed] [Google Scholar]
- 18.Faridmoayer, A., M. A. Fentabil, M. F. Haurat, W. Yi, R. Woodward, P. G. Wang, and M. F. Feldman. 2008. Extreme substrate promiscuity of the Neisseria oligosaccharyl transferase involved in protein O-glycosylation. J. Biol. Chem. 283:34596-34604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldman, M. F., C. L. Marolda, M. A. Monteiro, M. B. Perry, A. J. Parodi, and M. A. Valvano. 1999. The activity of a putative polyisopronol-linked sugar translocase (Wzx) involved in Escherichia coli O antigen assembly is independent of the chemical structure of the O repeat. J. Biol. Chem. 274:35129-35138. [DOI] [PubMed] [Google Scholar]
- 20.Furste, J. P., W. Pansegrau, F. R. Blocker, P. Scholz, M. Bagdasarian, and E. Lanka. 1986. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 48:119-131. [DOI] [PubMed] [Google Scholar]
- 21.Garen, A., and C. Levinthal. 1960. A fine structure genetic and chemical study of the enzyme alkaline phosphatase of Escherichia coli. Biochim. Biophys. Acta 38:470-483. [DOI] [PubMed] [Google Scholar]
- 22.Guerry, P., and C. M. Szymanski. 2008. Campylobacter sugars sticking out. Trends Microbiol. 16:428-435. [DOI] [PubMed] [Google Scholar]
- 23.Hansen, J. K., and K. T. Forest. 2006. Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion. J. Mol. Microbiol. Biotechnol. 11:192-207. [DOI] [PubMed] [Google Scholar]
- 24.Hatano, K., and G. B. Pier. 1998. Complex serology and Immune response of mice to variant high-molecular-weight O polysaccharides isolated from Pseudomonas aeruginosa serogroup O2 strains. Infect. Immun. 66:3719-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazes, B., P. A. Sastry, K. Hayakawa, R. J. Read, and R. T. Irvin. 2000. Crystal structure of Pseudomonas aeruginosa PAK pilin suggests a main-chain-dominated mode of receptor binding. J. Mol. Biol. 299:1005-1017. [DOI] [PubMed] [Google Scholar]
- 26.Heiniger, R. W., H. C. Winther-Larsen, R. J. Pickles, M. Koomey, and M. C. Wolfgang. 2010. Infection of human mucosal tissue by Pseudomonas aeruginosa requires sequential and mutually dependent virulence factors and novel pilus-associated adhesin. Cell. Microbiol. 12:1158-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horzempa, J., J. E. Comer, S. A. Davis, and P. Castric. 2006. Glycosylaton substrate specificity of Pseudomonas aeruginosa 1244 pilin. J. Biol. Chem. 281:1128-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horzempa, J., R. D. Dean, J. B. Goldberg, and P. Castric. 2006. Pseudomonas aeruginosa 1244 pilin glycosylation: glycan substrate recognition. J. Bacteriol. 188:4244-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horzempa, J., T. K. Held, A. S. Cross, D. Furst, M. Qutyan, A. N. Neely, and P. Castric. 2008. Immunization with a Pseudomonas aeruginosa 1244 pilin provides O-antigen-specific protection. Clin. Vaccine Immunol. 15:590-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt, J. D., D. C. Jackson, L. E. Brown, P. R. Wood, and D. J. Stewart. 1994. Antigenic competition in a multivalent foot rot vaccine. Vaccine 12:457-464. [DOI] [PubMed] [Google Scholar]
- 31.Kadokura, H., and J. Beckwith. 2009. Detecting folding intermediates of a protein as it passes through the bacterial translocation channel. Cell 138:1164-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim, E. E., and H. W. Wyckoff. 1991. Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J. Mol. Biol. 218:449-464. [DOI] [PubMed] [Google Scholar]
- 33.Kus, J. V., E. Tullis, D. G. Cvitkovitch, and L. L. Burrows. 2004. Significant differences in type IV pilin allele distribution among Pseudomonas aeruginosa isolates from cystic fibrosis (CF) versus non-CF patients. Microbiology 150:1315-1326. [DOI] [PubMed] [Google Scholar]
- 34.Liu, P. V. 1973. Exotoxins of Pseudomonas aeruginosa. I. Factors that influence the production of exotoxin A. J. Infect. Dis. 128:506-513. [DOI] [PubMed] [Google Scholar]
- 35.Liu, P. V., H. Matsumoto, H. Kusama, and T. Bergan. 1983. Survey of heat-stable, major somatic antigens of Pseudomonas aeruginosa. Int. J. Syst. Bacteriol. 33:256-264. [Google Scholar]
- 36.Manoil, C. 1991. Analysis of membrane protein topology using alkaline phosphatase and beta-galactosidase gene fusions. Methods Cell Biol. 35:61-75. [DOI] [PubMed] [Google Scholar]
- 37.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 38.Ohama, M., K. Hiramatsu, Y. Miyajima, K. Kishi, M. Nasu, and J. Kadota. 2006. Intratracheal immunization with pili protein protects against mortality associated with Pseudomonas aeruginosa pneumonia in mice. FEMS Immunol. Med. Microbiol. 47:107-115. [DOI] [PubMed] [Google Scholar]
- 39.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 40.Pier, G. B., H. F. Sidberry, and J. C. Sadoff. 1978. Protective immunity induced in mice by immunization with high-molecular-weight polysaccharide from Pseudomonas aeruginosa. Infect. Immun. 22:919-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poole, K., and R. A. Hancock. 1984. Phosphate transport in Pseudomonas aeruginosa. Involvement of a periplasmic phosphate-binding protein. Eur. J. Biochem. 144:607-612. [DOI] [PubMed] [Google Scholar]
- 42.Qutyan, M., M. Paliotti, and P. Castric. 2007. PilO of Pseudomonas aeruginosa 1244: subcellular location and domain assignment. Mol. Microbiol. 66:1444-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramphal, R., L. Koo, K. S. Ishimoto, P. A. Totten, J. C. Lara, and S. Lory. 1991. Adhesion of Pseudomonas aeruginosa pilin-deficient mutants to mucin. Infect. Immun. 59:1307-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramphal, R., J. Sadoff, M. Pyle, and J. D. Silipigni. 1984. Role of pili in the adherence of Pseudomonas aeruginosa to injured tracheal epithelium. Infect. Immun. 44:38-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruer, S., S. Stender, A. Filloux, and S. de Bentzmann. 2007. Assembly of fimbrial structures in Pseudomonas aeruginosa: functionality and specificity of chaperone-usher machineries. J. Bacteriol. 189:3547-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadoff, J. C., D. C. Wright, S. Futrovsky, H. Sidberry, H. Collins, and B. Kaufmann. 1985. Characterization of mouse monoclonal antibodies directed against Pseudomonas aeruginosa lipopolysaccharides. Antibiot. Chemother. 36:134-146. [DOI] [PubMed] [Google Scholar]
- 47.Smedley, J. G., III, E. Jewell, J. Roguskie, J. Horzempa, A. Seyboldt, D. B. Stolz, and P. Castric. 2005. Influence of pilin glycosylation on Pseudomonas aeruginosa 1244 pilus function. J. Bacteriol. 73:7922-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szymanski, C. M., and B. W. Wren. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 3:225-237. [DOI] [PubMed] [Google Scholar]
- 49.Vik, A., F. E. Aas, J. H. Anonsen, S. Bisborough, A. Schneider, W. Egge-Jacobsen, and M. Koomey. 2009. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. U. S. A. 106:4447-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West, S. E. H., H. P. Schweizer, C. Dall, A. K. Sample, and L. J. Runyen-Janecky. 1994. Construction of improved Escherichia coli-Pseudomonas shuttle vectors derived from pUC18/19 and the sequence of the region required for their replication in Pseudomonas aeruginosa. Gene 128:81-86. [DOI] [PubMed] [Google Scholar]
- 51.Wood, D. E., D. C. Strauss, W. G. Johanson, V. K. Berry, and J. A. Bass. 1980. Role of pili in adherence of Pseudomonas aeruginosa to mammalian buccal epithelial cells. Infect. Immun. 29:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zuercher, A. W., M. P. Horn, H. Wu, Z. Song, C. J. Bundgaard, H. K. Johansen, N. Hoiby, P. Marcus, and A. B. Lang. 2006. Intranasal immunization with conjugate vaccine protects mice from systemic and respiratory tract infection with Pseudomonas aeruginosa. Vaccine 24:4333-4342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.