Abstract
Methanogens represent some of the most oxygen-sensitive organisms in laboratory culture. Recent studies indicate that they have developed mechanisms to deal with brief oxygen exposure. MsvR is a transcriptional regulator that has a domain architecture unique to a select group of methanogens. Here, runoff in vitro transcription assays were used to demonstrate that MsvR regulates transcription of the divergently transcribed fpaA-rlp-rub operon in Methanothermobacter thermautotrophicus in addition to transcription from its own promoter. The protein products of the fpaA-rlp-rub operon have previously been implicated in oxidative stress responses in M. thermautotrophicus. Additionally, electrophoretic mobility shift assays (EMSAs) and DNase I footprinting were used to confirm a binding site inferred by bioinformatic analysis. Sequence mutations within these binding sites did not significantly alter EMSA shifting patterns on longer templates but did on shorter 50-bp fragments encompassing only the region containing the binding sites. Footprinting confirmed that the regions protected for the longer mutant templates are at different positions within the intergenic region compared to those seen in the intact intergenic region. Oxidized and reduced preparations of MsvR demonstrated different EMSA binding patterns and regions of protection on the intergenic sequence, suggesting that MsvR may play a role in detecting the redox state of the cell.
Biological methane production is limited to a small group of microorganisms in the domain Archaea that are termed methanogens. These organisms are strict anaerobes that can generate methane from H2 and CO2 and/or a limited series of C1 compounds, depending upon the organism (5). Despite a necessity for growth in environments devoid of oxygen, the genomes of methanogens encode various enzymes (e.g., superoxide dismutase and alkyl hydroperoxide reductase) involved in the detoxification of reactive oxygen species. Previous studies have shown peroxide stress has little effect on methane production and overall growth yield in the hydrogenotrophic methanogen Methanothermobacter thermautotrophicus. The same study identified upregulation of the fpaA-rlp-rub operon (mth1350 to mth1352) believed to be involved in the detoxification of reactive oxygen species (10). This operon encodes a flavoprotein (fpaA), a rubrerythrin-like protein (rlp), and a rubredoxin (rub) (10, 19). A homologue of the flavoprotein from the methanogen Methanobrevibacter arboriphilus has been demonstrated to reduce O2 in the presence of H2 and was reclassified as an F420H2 oxidase (24). This lends further evidence to support the role of proteins encoded by this operon in the detoxification of reactive oxygen species.
The archaeal transcription machinery represents a chimeric system with components reminiscent of features found in the other two domains of life, Bacteria and Eukarya. The multisubunit RNA polymerase is similar to the eukaryotic RNA polymerase II complex and, as is the case for the eukaryotic system, is incapable of direct promoter recognition (28). Promoter recognition in Archaea is achieved through use of homologues of the eukaryotic basal transcription factors TATA binding protein (TBP) and TFIIB (TFB in Archaea). Transcription is initiated by TBP binding a TATA box sequence located ∼27 bp upstream of the transcription start site in Archaea followed by TFB binding to a B recognition element (BRE) located just upstream of the TATA box. Once these two factors are bound to the promoter, RNA polymerase is recruited and an open complex is formed (8). Despite features of the eukaryotic basal transcription machinery, the genomes of Archaea encode transcriptional regulators with classical bacterial helix-turn-helix (HTH) DNA binding domains (2). Mechanisms of transcription regulation in Archaea are poorly understood. To date several transcriptional repressors (9, 14, 15) and even fewer activators (11, 16, 21) have been identified and characterized.
Here the previously described M. thermautotrophicus in vitro transcription system (4) is used to demonstrate that a divergently transcribed transcription regulator, MTH1349, regulates the aforementioned oxidative stress response operon. Evidence is provided that this transcriptional regulator is specific to methanogens and that it autoregulates its own expression. The regulator has therefore been named MsvR for methanogen-specific V4R domain-containing regulator. The role of an inverted repeat sequence within the intergenic region is demonstrated through electrophoretic mobility shift assay (EMSA) and DNase I footprinting with native and mutant DNA templates. To identify a mechanism for repression from both the msvR promoter (PmsvR) and the fpaA promoter (PfpaA), order-of-addition in vitro transcription assays were performed. Finally, the potential role of the C-terminal cysteine residues in redox sensing is discussed.
MATERIALS AND METHODS
Reagents.
All chemicals were purchased from Fisher Scientific. FastDigest restriction enzymes and Taq DNA polymerase were purchased from Fermentas. Ligase, DNase I, and Phusion DNA polymerase were purchased from New England Biolabs. Plasmid preps were performed using the Zyppy plasmid miniprep kit (Zymo Research). All plasmids were verified by sequencing at the Oklahoma Medical Research Foundation DNA Sequencing Center.
Sequence analysis.
Analysis of sequences for inverted repeats and palindromes was performed using the Palinsight program (22). Alignments and genome sequences were downloaded and manipulated using the Geneious software program (6).
DNA templates.
A 434-bp DNA fragment containing the msvR-fpaA intergenic region as well as 126 and 185 bp downstream of the msvR and fpaA translation start sites, respectively, was amplified from M. thermautotrophicus genomic DNA using primers LK193 (5′-GACTATTTCCTGGAAGCTGAGTG-3′) and LK194 (5′-GCCATGAGCTCCTGGAAGGTCCCTGG-3′). The PCR product was cloned into pCR2.1 TOPO using a TOPO cloning kit (Invitrogen), and the sequence was confirmed. The resulting plasmid was designated pLK203. The in vitro transcription templates were amplified from pLK203 using the vector-specific M13F and M13R primers, yielding T203, a 620-bp fragment that increases the length of the expected runoff transcripts from both PmsvR and PfpaA. EMSA templates (identified by a GS prefix) were amplified using primer set LK195 (5′-CGGAGAGTATGAGCGACTTGACAGTGCTTTC-3′) and 196 (5′-CCAGTATAATCCATCTGCTATCTTCTCTGCTCTGGCC-3′), generating a 254-bp fragment containing 88 bp of the 5′ end of msvR and 42 bp of the 5′ end of fpaA. T1349 was amplified from pLK203 using primers LK 468 (5′-CCATGGTATTGAAACAGTAATATAGTCC-3′) and M13R. T1350 was amplified using LK467 (5′-CCATGGAACAGAAAAAATTATATAGAAGTTCGTATAG-3′) and M13F. DNase I footprinting templates were amplified using primers M13Fext (with a 5′ 6-carboxyfluorescein [FAM] label) and M13R ext (5′ VIC label) (Applied Biosystems) (16) from the plasmid containing the appropriate template. All PCR products were gel purified and cleaned up with an SV PCR cleanup kit (Promega). Templates representing the mutations listed below in Fig. 3C were generated using the QuikChange site-directed mutagenesis kit (Stratagene). Primers used for site-directed mutagenesis are available upon request. The appropriate templates were amplified from resulting plasmids as done for T203.
Cloning and expression of recombinant MsvR.
The msvR open reading frame was PCR amplified from M. thermautotrophicus genomic DNA and cloned into a vector containing an N-terminal Strep tag. The resulting plasmid was transformed into Escherichia coli Rosetta (Novagen). Expression was induced at an optical density at 600 nm of 0.5 using 0.1 mM isopropyl-β-d-thiogalactopyranoside. Sorbitol was added to a final concentration of 1% (wt/vol), and the cultures were incubated for 16 h at 24°C. Cells were resuspended in NP buffer (50 mM NaH2PO4, 300 mM NaCl; pH 8.0) and lysed by sonication. Protein was purified using Streptactin resin (Qiagen) according to the manufacturer's guidelines. Pure fractions were combined and dialyzed against a protein dilution/storage buffer (PDB; 200 mM KCl, 20 mM Tris [pH 8], 10 mM MgCl2, 50% glycerol). Protein was quantitated using the Quant-iT protein assay kit (Invitrogen). Subsequent protein preparations were calibrated to the original preparation via EMSA titration assays to adjust for differing levels of purity and differences in protein activity from batch to batch. The concentrations listed in the figure legends correspond to the concentration from the purest/most active protein preparation.
Preparation of oxidized and reduced MsvR.
The oxidized and reduced versions of MsvR were prepared per the methods of Bae et al. (3). Briefly, the oxidized version was prepared by incubating 100 μl of a 10 μM protein stock in the presence of 1 mM H2O2 and 10 mM EDTA for 30 min at room temperature. The preparation was then dialyzed against 1 liter of oxygen-saturated PDB (see above). Anaerobic preparations were prepared inside a COY anaerobic glove bag as follows: 100 μl of a 10 μM protein stock was incubated with 10 mM dithiothreitol (DTT) for 30 min at room temperature inside the chamber. The preparation was then dialyzed against 1 liter of nitrogen-saturated PDB. Working stocks were prepared by diluting the protein in oxygen- or nitrogen-saturated PDB.
Runoff in vitro transcription assays.
To generate preinitiation complexes, 10 nM DNA, 80 nM recombinant TBP, 80 nM recombinant TFB, 40 nM RNA polymerase, 20 mM Tris (pH 8), 15 mM MgCl2, 120 mM KCl, 12.5% glycerol, and 5 mM DTT in a 20-μl volume were incubated at 60°C for 15 min. The reaction was chased for 5 min at 60°C with 200 μM ATP, 200 μM CTP, 200 μM GTP, 20 μM UTP, 10 μCi [α-32P]UTP (10 mCi/ml; MP Biomedicals), 10 μg yeast tRNA, and 100 μg heparin to maintain a single round of transcription (9). When included in the reaction mixture, the MsvR concentrations are indicated in the figure legends. Transcription reactions were stopped by the addition of five volumes of 600 mM Tris (pH 8.0), 30 mM EDTA and extracted with an equal volume of phenol-chloroform-isoamyl alcohol (PCI). Reaction mixtures were precipitated by traditional methods, and the pellets were resuspended in 4 μl of formamide loading buffer, heated to 95°C for 2 min, and electrophoresed on a 10% sequencing gel. The TBP, TFB, and M. thermautotrophicus RNA polymerase used in the transcription reactions were purified as previously described (4, 30).
EMSA.
Binding reactions were performed as described above (with PDB replacing TBP, TFB, and RNA polymerase) and incubated at 60°C for 15 min. Reaction mixtures were loaded onto 6% acrylamide gels in 1× Tris-borate buffer and electrophoresed for ∼40 min at 200 V, 20 mA. Gels were stained using SYBR gold and visualized using a Gel-DocXR system (Bio-Rad). Heparin at a final concentration of 125 μg/ml was used to inhibit nonspecific DNA binding (16).
DNase I footprinting.
Binding reactions were set up at room temperature with 10 nM FAM (plus strand)/VIC (minus strand) end-labeled DNA in 20 mM Tris (pH 8), 15 mM MgCl2, 120 mM KCl, 12.5% glycerol, 0.125 mg/ml heparin, with or without 5 mM DTT and 200 nM MsvR (as indicated in the figure legends) in a 20-μl volume for 15 min, followed by addition of 20 μl of a buffer containing 20 mM Tris (pH 8), 15 mM MgCl2, 120 mM KCl, and 5 mM CaCl2. The MsvR/DNA complexes were digested for 1 min with 0.025 units DNase I at room temperature (9). Reactions were stopped by the addition of 40 μl of a solution containing 200 mM NaCl, 30 mM EDTA. Reactions were cleaned up with the SV PCR cleanup kit (Promega) and eluted in 25 μl. The FAM and VIC dyes allowed detection of the fragments with a 3730 DNA analyzer (Applied Biosystems) at the Ohio State University Plant and Microbial Genomics Facility and analysis using Peak Scanner software (Applied Biosystems), and the fragments were aligned to sequences generated using the same primers (31).
RESULTS AND DISCUSSION
MsvR is a transcriptional regulator unique to methanogenic Archaea.
MsvR was identified in the genome of M. thermautotrophicus, where it is located just upstream and divergently transcribed from the fpaA-rlp-rub operon (Fig. 1A). Previous studies had shown that the fpaA-rlp-rub operon is upregulated during peroxide (oxidative) stress (10). In another methanogen, a homologue of the flavoprotein was shown to reduce O2 (24). A search of the Conserved Domain Database (CDD) revealed two major domains within MsvR (17). The N terminus contains a helix-turn-helix DNA binding domain (ArsR family), while the C terminus has a V4R domain (Fig. 1B). Although the function of V4R domains is not well understood (23, 25, 27), a characteristic feature is the presence of five cysteine residues. A BlastP search showed that only members of the Methanobacteriales, Methanomicrobiales, and Methanosarcinales contain proteins with >25% and up to 46% identity with full-length MsvR (1), suggesting that transcriptional regulators with this combined domain architecture are limited to methanogens. Cysteine residues are often used in redox-sensing transcriptional regulators as a means of monitoring the redox state within the cell (3, 12, 13), suggesting that MsvR plays a role in the regulation of the fpaA-rlp-rub operon in response to oxidative stress. The transcript levels for msvR do not change significantly during oxidative stress, suggesting that its activity may be subject to conformational changes and further supporting a role of the C-terminal V4R domain in redox sensing (18, 26).
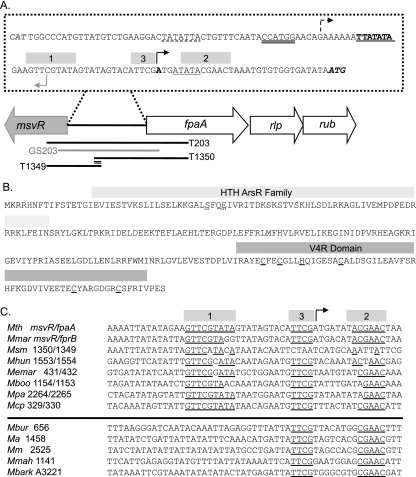
FIG. 1.
(A) Diagram of the M. thermautotrophicus genome region encoding the fpaA-rlp-rub operon (MTH1350 to -1352) and an adjacent transcription regulator (msvR). The sequence of the msvR-fpaA intergenic region is displayed. The PfpaA TATA box is indicated by a solid single underline and bold text, with its corresponding transcription start site identified by a solid black arrow. The PfpaA2 TATA box is represented by a single dashed underline with its putative corresponding transcription start site identified with a dotted arrow. The translation start site for FpaA is shown in italics. The putative PmsvR TATA box is indicated by the thick gray underline, and its putative corresponding transcription start site is identified by a gray arrow. The translation start site for MsvR is shown in gray italics. Potential transcription regulator binding sites are represented by gray boxes. The coverage of templates T203, GS203, T1349, and T1350 is displayed. The overlapping sequence coverage of T1349 and T1350 is double underlined. (B) The amino acid sequence for the putative transcriptional regulator MsvR. Boxes indicate domains identified by the NCBI CDD. The carboxy-terminal amino acid residues that may be important for serving as redox sensors are underlined with a solid black line. N-terminal residues that may be involved in metal binding are represented by a dashed underline. (C) Alignment of the msvR-fpaA intergenic sequence with those of other methanogens (Methanobacteriales and Methanomicrobiales) that have a homologue of the MsvR regulator sharing an intergenic region with a homologue of FpaA. Sequences below the horizontal line are from the promoter regions of msvR genes in members of the Methanosarcinales. The binding regions identified in panel A are displayed here, and conserved nucleotides within these regions are underlined.
An examination of the publicly available genomes of other methanogens and various GenBank sequences showed that this local genomic context is conserved in Methanobrevibacter smithii and Methanothermobacter marburgensis of the Methanobacteriales and Methanospirillum hungatei, “Candidatus Methanoregula boonei,” Methanocorpusculum labreanum Z, Methanosphaerula palustris E1-9c, and Methanoculleus marisnigri of the Methanomicrobiales. Alignment of the intergenic regions of these organisms to the inverted repeats identified by PALINSIGHT (e.g., potential binding sites [Fig. 1A, boxes 1 and 2]) (22) in the intergenic region of M. thermautotrophicus showed sequence conservation (Fig. 1C). The alignment also revealed a conserved region (box 3) between boxes 1 and 2 that overlaps the fpaA-rlp-rub operon transcription start site (Fig. 1C) (19). Additionally, homologues of MsvR were identified in several members of the Methanosarcinales (Methanococcoides burtonii, Methanohalophilus mahii, Methanosarcina acetivorans C2A, Methanosarcina barkeri strain Fusaro, and Methanosarcina mazei Gö1). The MsvR homologue in these organisms is not transcribed divergently from a homologue of FpaA. However, alignment of the msvR promoter regions from these organisms revealed the conservation of boxes 2 and 3 (Fig. 1C). In some but not all of these organisms, a similar sequence pattern could be identified upstream of the FpaA homologues. These sequence patterns were noticeably absent from the promoter regions of genes encoding FpaA homologues in methanogens not containing a homologue of MsvR. Taken together, these results suggest that these sequence boxes play a role in MsvR binding in these regions.
The msvR-fpa-rlp-rub intergenic region contains multiple promoters.
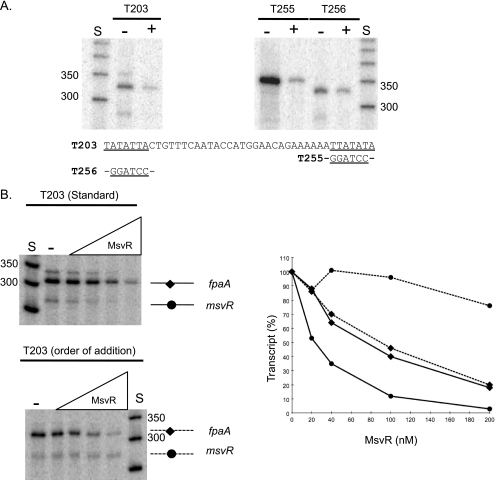
Runoff transcription assays using T203 generated three individual transcripts (Fig. 2A, T203, − lane). Two of the transcripts were of the expected sizes. The dominant transcript (∼311 nucleotides) corresponds to the previously mapped transcription start site for fpaA (19) (Fig. 1A). A smaller transcript of ∼280 nucleotides corresponds to the expected transcript size for msvR. A third unexpected transcript of ∼350 nucleotides was also noted. Previous studies mapping the in vivo transcription start sites did not mention a potential secondary promoter for the fpaA-rlp-rub operon (19). To further map all functional promoters within the intergenic regions, a series of mutations were introduced in mapped and putative TATA boxes for the fpaA-rlp-rub operon. A mutation in the previously mapped PfpaA TATA box, T255 (Fig. 2A), eliminated the 311-nucleotide transcript and resulted in a dominant ∼350-nucleotide transcript. This indicates that, at least in vitro, a secondary fpaA promoter exists (PfpaA2) upstream of PfpaA. A second template, T256, was generated with a mutation in the putative PfpaA2 TATA box. The T256 template generated only two transcripts (∼280 and 311 nucleotides) (Fig. 2A, T256). The larger ∼350-nucleotide transcript was no longer visible, confirming that the mutated region serves as a secondary, less-robust promoter driving transcription of the fpaA-rlp-rub operon in vitro.
FIG. 2.
(A) The fpaA and msvR transcripts generated using the T203 template in the absence (−) or presence (+; 200 nM) of MsvR. Mutations were made in two putative TATA boxes (T255 and T256) for fpaA, and transcripts generated using those templates are displayed (with and without MsvR). (B) The msvR and fpaA transcripts generated from T203 in the presence of increasing concentrations (0 nM, 20 nM, 40 nM, 100 nM, and 200 nM) of MsvR in both standard in vitro transcription assays (solid lines) as well as in an order-of-addition assay (dotted lines), in which the general transcription factors (TBP/TFB) were incubated with the template prior to the addition of MsvR. The amount of transcript produced from promoters in each reaction was quantitated and compared to the respective control reaction (taken as 100% transcript). The data displayed are averages of two independent experiments.
MsvR represses transcription from both promoters.
To determine the role of MsvR in transcription, single-round runoff transcription assays were performed in the absence or presence of MsvR. In the presence of MsvR, transcription was repressed from all three promoters within the intergenic region (Fig. 2A, T203). Similar patterns were observed for the TATA box mutants (T255/T256) (Fig. 2A). Assays using template T203 in the presence of increasing concentrations of MsvR showed enhanced repression of transcription as the concentration of repressor increased (Fig. 2B, standard). The primary fpaA transcript (∼311 nucleotides) was reduced ∼75% in the presence of a 20-fold molar excess of MsvR over the DNA, which corresponds to approximately 10 MsvR homodimers if that is its quaternary state. The secondary ∼350-nucleotide fpaA transcript displays a pattern similar to that of the 311-nucleotide transcript. Notably, repression of the ∼280-nucleotide PmsvR transcript occurs more readily at lower protein concentrations (∼45% at a 2-fold molar excess) than it does for PfpaA (Fig. 2B), indicating a very tight autoregulation. The data clearly indicate that MsvR autorepresses expression from its promoter as well as acting as a transcriptional repressor of PfpaA under reducing conditions present in the transcription assay mixtures. Runoff transcription assays with the M. thermautotrophicus in vitro transcription system are limited to reducing conditions, as both TFB and RNA polymerase activities in vitro are dependent on reducing conditions (4). Therefore, it is not possible to address the effect of oxidized MsvR (MsvRox) on transcription in vitro. To ensure that MsvR in the assay mixtures was indeed in the reduced state, MsvRox was used in binding reaction mixtures containing 5 mM DTT. The shift patterns were identical to those for reduced MsvR (MsvRred), indicating that despite experimental manipulation in the open laboratory atmosphere, the protein was rapidly reduced in the presence of the 5 mM DTT present in the in vitro transcription assay mixtures. The in vitro transcription assay mixtures did not contain nonspecific competitor DNA to prevent nonspecific binding during preinitiation complex formation. EMSAs with MsvR in the presence or absence of nonspecific inhibitors produced similar binding patterns on GS203, indicating that the repression seen in in vitro transcription assays was not the result of nonspecific binding (data not shown).
Mechanism of repression.
To ascertain the mechanism used by MsvR to repress transcription, runoff in vitro transcription assays were performed utilizing specific orders of transcription factor addition. In standard transcription assays all of the proteins (TBP, TFB, RNA polymerase, and MsvR) were incubated with the DNA simultaneously during preinitiation complex formation (Fig. 2B, standard), allowing them to compete equally for binding. Those conditions failed to reveal whether MsvR can still bind and repress transcription if a TBP/TFB/DNA ternary complex has formed. Therefore, transcription assays where TBP and TFB were preincubated with the DNA for 10 min prior to the addition of MsvR and RNA polymerase were performed. Under these conditions repression from PfpaA was similar to that under standard assay conditions, while PmsvR was no longer repressed (Fig. 2B, order of addition). These results indicate that MsvR represses PmsvR through direct abrogation of the TBP/TFB complex. However, continued repression of PfpaA suggests that its transcription is repressed by blocking transcription start site access.
MsvR binds the intergenic region.
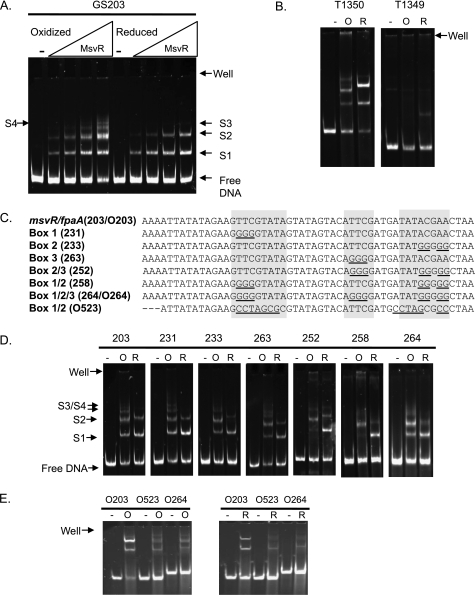
EMSAs were performed to assess the various complexes (species) generated when MsvR binds the intergenic region and whether its oxidation state plays a role (GS203) (Fig. 3A). In order to determine if the oxidation state of MsvR plays a role in DNA binding, both MsvRox and MsvRred preparations were used. The V4R domain of MsvR contains a series of conserved cysteine residues and a histidine at residue 202, which suggests it serves as a redox sensor. This hypothesis was supported by the redox-sensing mechanisms in bacterial regulators. PerR, the transcriptional regulator of oxidative stress in Gram-positive bacteria, utilizes metal-catalyzed oxidation of a histidine residue (13). Conversely, OxyR, a transcriptional regulator of oxidative stress in Gram-negative bacteria, utilizes reversible disulfide bonds (12). Oxidation of the anti-sigma factor RsrA, which involves disulfide bonds and zinc binding, results in induction of the sigma R regulon. This activity involves a CX2CX3H motif in RsrA (3). The histidine at position 202 in MsvR is within such a motif. The presence of features reminiscent of various bacterial redox regulators suggests that it could share a mechanism with one of these regulators or utilize a unique mechanism. In binding reaction mixtures with as little as a 2-fold excess of MsvRox or MsvRred over the DNA concentration, two major shifted species were seen (Fig. 3A, S1 and S2). However, as the concentrations reached a 20-fold excess over the DNA the binding patterns between the two differed. MsvRox produced two additional shifted species (S3 and S4), whereas the S3 shift was at a different position than the S3 for MsvRred (Fig. 3D, 203). S3 was not always present in binding reaction mixtures with MsvRred, suggesting it may be the result of incompletely reduced MsvR. It is possible that MsvR preparations represent a mixed population of protein with cysteine residues in various oxidation states, thereby complicating in vitro analysis of the role of the fully reduced or oxidized states. Even when purified or reduced in anaerobic environments, many of the well-studied redox regulators are easily oxidized after exposure to air (7). What is clear is that in various states of oxidation MsvR exhibits different DNA binding specificities. Throughout multiple EMSA experiments with GS203 and MsvRox or MsvRred preparations, there appeared to be a slight difference in mobility of S2 (Fig. 3, 203). The slight differences in mobility seen for S2 and S3 between MsvRox and MsvRred indicate that these do not represent identical complexes. It is also unclear whether the other shifted species represent identical protein/DNA complexes or if they just have similar molecular weights and therefore migrate to a similar position in the gel. Increasing the ratio of MsvR over DNA to 50:1 resulted in complete shifting of all complexes to the region of the S3/S4 complexes. Even though MsvRox binds to DNA with a slightly different DNA binding pattern than MsvRred, it is still plausible that it no longer functions as a transcriptional repressor. OxyR, the regulator of the oxidative stress response in E. coli, binds to DNA under either oxidizing or reducing conditions with only a slight change in binding patterns, but it functions as an activator in the oxidized conformation and a repressor in the reduced conformation as result of conformational changes in the protein (12, 26).
FIG. 3.
(A) EMSAs with the GS203 template (contains only the intergenic region) in the presence of increasing concentrations (0 nM, 20 nM, 40 nM, 100 nM, and 200 nM) of MsvRox or MsvRred. The binding reaction mixtures contained no DTT. Shifted species are identified as S1, S2, S3, and S4. These species do not necessarily represent the same complex in each reaction but are species with similar overall molecular weights. The gel wells and positions of free DNA are also indicated. (B) EMSAs with T1350 and T1349 in the absence (−) or presence of oxidized (O) or reduced (R) MsvR (200 nM). (C) Mutations generated in each of the three binding boxes as well as combinations of the three binding boxes. (D) EMSAs performed using templates containing various mutations in the binding boxes in the presence of preoxidized or prereduced MsvR (200 nM). A representative EMSA using GS203 is displayed; however, all experiments/gels included binding reactions with GS203 as a positive control to ensure that differences in binding patterns were not due to technical differences between experiments. (E) EMSA with annealed oligonucleotides corresponding to the centralized region containing the three binding boxes identified in panel C, with a 20-fold excess of MsvR. MsvR preparations used are the same as those indicated in panel D.
MsvR binding is sequence specific.
To identify the regions necessary to generate the shifted complexes, two templates were generated from T203 that contained only a 6-bp overlap within the intergenic region (Fig. 1A, double-underlined portion). T1350 included the three binding boxes shown in Fig. 1A and the surrounding promoter region. It was expected that this fragment would fully support MsvR binding. EMSAs performed using these templates confirmed that T1350 supported MsvR binding and that the shifting patterns corresponded to those of GS203. The previously mentioned difference in mobility of the S2 species between MsvR preparations on GS203 is readily visible on T1350. The T1349 template lacks the binding boxes and represents intergenic M. thermautotrophicus DNA sequence. This template was utilized to demonstrate that MsvR binding is specific to the region containing the binding boxes identified in Fig. 1A and does not bind nonspecifically. The T1349 template only generated a faint shift with MsvRred, confirming this (Fig. 3B).
To determine the role of each of the three boxes in MsvR binding to the intergenic region, a series of templates (based on GS203) were made with mutations in one or more of the binding boxes (Fig. 3C). When we used templates T231, T233, and T263 with mutations in only one of the binding boxes, there appeared to be only slight changes in DNA binding for MsvRox, represented by the absence/presence of S4 (Fig. 3D). However, MsvRred binding to these templates showed a reduction in the amount of material in the S2 band with a subsequent increase in the amount in the S1 band. Templates containing mutations in two or more of the binding boxes (T252, T258, and T264) showed similar patterns but did not completely eliminate binding, suggesting other areas within the intergenic regions may also be bound.
To further investigate the role of boxes 1, 2, and 3 in specific binding, a series of ∼50-bp oligonucleotides comprising the regions displayed in Fig. 3C were generated. For O203 a binding pattern similar to that seen for GS203 was observed (Fig. 3E), indicating that this 50-bp region containing the three binding boxes is sufficient to produce the two major shifts seen in Fig. 3A and D. Under oxidizing conditions (Fig. 3E, left) with template O203 a very faint S3 complex was seen. When we used O523 with mutations in box 1 and 2 and O264 with mutations in all three boxes, MsvR binding under either oxidized or reducing conditions was greatly reduced. Binding under oxidizing conditions seemed to be less affected, suggesting that under these conditions the binding specificity is relaxed. It is plausible that in vivo this would be negligible and upon oxidation MsvR loses its affinity for the promoter region, resulting in derepression of the operon. Binding to similar oligonucleotides containing mutations in single binding boxes indicated that the S1 complex is still present but that higher-molecular-weight complexes are absent (data not shown). It is evident that these sequences play a direct role in MsvR binding. Nonetheless, on larger templates these mutations are less detrimental to MsvR binding, suggesting that several lower-affinity binding sites can support binding.
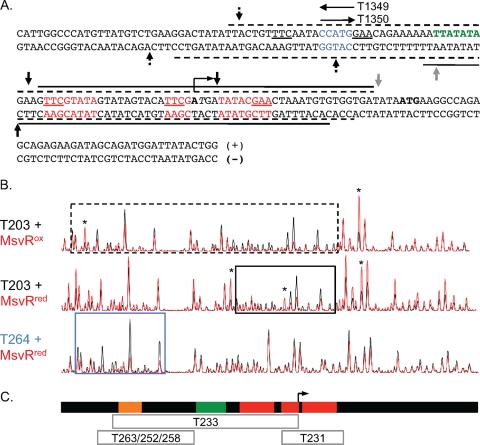
MsvRox and MsvRred have distinguishable footprints.
Since the differences in template binding by MsvRox and MsvRred were subtle, it became important to identify the boundaries of MsvRox and MsvRred binding and to determine whether regions outside the three boxes were bound. Therefore, the T203 template (FAM and VIC labeled) was subjected to DNase I footprinting in the absence or presence of MsvR (200 nM). The resulting fragments were separated by capillary electrophoresis, and peak heights on the chromatograms (with or without MsvR) for each DNA strand were compared to determine regions of protection and hypersensitivity. The data generated from these experiments are depicted in Fig. 4A, and representative chromatograms of MsvRox and MsvRred on the plus strand are shown in Fig. 4B. Variations in the overall intensity of peaks in reactions with and without MsvR are visible in the chromatograms. To compensate for such variations, the presence of hypersensitive and protected sites was confirmed by their appearance across multiple independent experiments. MsvRox binding protects a majority of the intergenic region from DNase I digestion. However, MsvRred binding generates a more defined region of protection encompassing the three binding sites and subsequently blocking the transcription start site for the primary PfpaA (Fig. 4A and B). The area surrounding the PfpaA TATA box is only slightly protected. These binding patterns are consistent with the results obtained from order-of-addition in vitro transcription reactions. The region of protection on the minus strand indicates that the PmsvR TATA box would be blocked. This is consistent with MsvRred no longer repressing transcription from PmsvR when TBP and TFB are allowed to bind first. However, even if TBP and TFB bind first followed by MsvRred, transcription from PfpaA is still repressed. The binding pattern for MsvRred on the plus strand indicates that MsvRred does not completely block the PfpaA TATA box but it does block the transcription start site, thus abrogating RNA polymerase.
FIG. 4.
(A) The msvR and fpaA intergenic region, displayed with corresponding DNase I footprints. The footprints for each strand are displayed with a dashed line for those generated with MsvRox and with a solid line for those generated with MsvRred. The PfpaA TATA box is shown in green, and its transcription start site is identified. The putative binding site sequences identified in Fig. 1 are shown in red. The region of sequence overlap between T1349 and T1350 is shown in blue. Vertical arrows above and below each strand depict hypersensitive sites. Hypersensitive sites present in the oxidized, reduced, or both footprints are depicted by a dotted arrow, solid black arrow, and a solid gray arrow, respectively. (B) Aligned chromatograms of MsvRox and MsvRred footprints on T203 (plus strand) and the MsvRred protection site on T264 (plus strand). The black trace corresponds to reaction mixtures containing only DNA, whereas the red trace corresponds to reaction mixtures containing DNA and MsvR. MsvR protected regions are boxed using the same scheme as for T203 in panel A, and the asterisks depict the hypersensitive sites. The protected region on T264 is boxed in blue. (C) Diagram of MsvR protected regions on fragments containing the mutations identified in Fig. 3C. The diagram is aligned to the chromatograms in panel B. The gray boxes represent the regions protected from DNase I digestion when MsvR is bound to the indicated template. The orange box represents the position of the overlap for templates T1349 and T1350. The red boxes represent binding boxes 1, 2, and 3, and the green box represents the PfpaA TATA box.
Secondary MsvR binding sites.
DNase I footprinting was also performed on mutant templates (Fig. 3C), because EMSAs revealed that binding still occurred on these fragments. Depictions of protected regions are shown for all templates (Fig. 4C), and chromatograms are shown for T203 as well as T264, which has mutations in all three binding boxes (Fig. 4B). On templates T231, T233, and T263 the region of protection for both MsvRox and MsvRred was shifted on each template depending upon the location of the mutation, with T263 showing the most dramatic shift, binding only to the region upstream of the PfpaA TATA box (Fig. 4C, gray boxed areas). This suggested that box 3 might represent a centralized binding box. However, EMSA using O523, which contains mutations in boxes 1 and 2 but not box 3, demonstrated that MsvR could no longer bind even with box 3 intact. On longer templates containing mutations in two out of three of the binding boxes (T252 and T258), regions of protection were only seen upstream of the PfpaA TATA box (Fig. 4C). MsvRred did not protect these regions on the T203 template, suggesting that when its primary binding site is altered it binds to secondary, less-optimal binding sites. Regions protected by MsvR on T264 further supported this. MsvR does indeed provide protection on this template, confirming the EMSA results. This binding occurs upstream of the regions of protection observed for T203 that contained the three binding boxes. Only slight differences were observed in the regions of MsvRox and MsvRred for these templates. Careful examination of these two alternative binding regions revealed the presence of a TTCN9GAA sequence, and the mutants that were constructed (Fig. 3C) included mutations within the TTC or GAA sequence within boxes 1, 2, and 3. Taken together these results suggest that these sequences are likely involved in MsvR binding to T203 and that alternate sites containing similar sequence patterns are utilized in the absence of ideal binding sites. T1349 includes one-half of the TTCN9GAA sequence pattern, which explains the faint shift observed by EMSA when using this template (Fig. 3B). Interestingly, TTC/GAA patterns are seen in the consensus binding site of the archaeal transcription activator Ptr2 (20). It is not uncommon for a transcriptional regulator to bind multiple recognition sequences or sites with limited homology (11, 26, 29).
MsvR as a redox-sensitive archaeal transcription regulator.
All experimentation that utilized MsvRox suggested that it binds both the primary binding boxes illustrated in Fig. 1A as well as a potential secondary binding site just upstream of the PfpaA TATA box. The footprints produced by both preparations suggest they would both function by repressing transcription. However, the bacterial redox-sensing transcriptional regulator, OxyR, binds DNA in both oxidation states, and only slight differences are seen in the footprints. Depending on its oxidation state, OxyR serves as either an activator or repressor. Future studies using MsvR variants with cysteine-to-alanine mutations within the putative V4R domain will be useful in determining the role of the C terminus in dimerization and redox sensing.
Conclusions.
MsvR is a methanogen-specific transcriptional regulator that binds the intergenic region containing its own promoter as well as the promoter of the fpaA-rlp-rub operon. The location of this regulator in relationship to the fpaA-rlp-rub operon indicated it is involved in regulation of gene expression from both promoters. The data presented support this conclusion. MsvRred does tightly autoregulate its own expression by blocking TBP and TFB access to the TATA box/BRE region. Additionally, MsvRred represses transcription from PfpaA (both primary and secondary) by blocking RNA polymerase access to its respective transcription start site. MsvR footprints as well as order-of-addition runoff in vitro transcription assays support this conclusion. Oxidized and reduced preparations of MsvR appear to behave differently in DNA binding assays, indicating that it may be the first redox-sensitive transcriptional regulator described for the archaeal domain.
Acknowledgments
This work was supported by NSF grant MCB003286 and funds provided by the University of Oklahoma.
I thank John N. Reeve and Tyrell Conway for critical review of the manuscript. I also thank Marika Sauter and Jessica Turner for technical contributions and Anne K. Dunn for many helpful discussions.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Aravind, L., and E. V. Koonin. 1999. DNA-binding proteins and evolution of transcription regulation in the Archaea. Nucleic Acids Res. 27:4658-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, J.-B., J.-H. Park, M.-Y. Hahn, M.-S. Kim, and J.-H. Roe. 2004. Redox-dependent changes in RsrA, an anti-sigma factor in Streptomyces coelicolor: zinc release and disulfide bond formation. J. Mol. Biol. 335:425-435. [DOI] [PubMed] [Google Scholar]
- 4.Darcy, T. J., W. Hausner, D. E. Awery, A. M. Edwards, M. Thomm, and J. N. Reeve. 1999. Methanobacterium thermoautotrophicum RNA polymerase and transcription in vitro. J. Bacteriol. 181:4424-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deppenmeier, U. 2002. The unique biochemistry of methanogenesis. Proc. Natl. Acad. Sci. U. S. A. 71:223-283. [DOI] [PubMed] [Google Scholar]
- 6.Drummond, A. J., B. Ashton, M. Cheung, J. Heled, M. Kearse, R. Moir, S. Stones-Havas, T. Thierer, and A. Wilson. 2009. Geneious, version 4.8. Biomatters, Ltd., Auckland, New Zealand.
- 7.Green, J., and M. S. Paget. 2004. Bacterial redox sensors. Nat. Rev. Microbiol. 2:954-966. [DOI] [PubMed] [Google Scholar]
- 8.Hausner, W., J. Wettach, C. Hethke, and M. Thomm. 1996. Two transcription factors related with the eucaryl transcription factors TATA-binding protein and transcription factor IIB direct promoter recognition by an archaeal RNA polymerase. J. Biol. Chem. 270:30144-30148. [DOI] [PubMed] [Google Scholar]
- 9.Karr, E. A., K. Sandman, R. Lurz, and J. N. Reeve. 2008. TrpY Regulation of trpB2 transcription in Methanothermobacter thermautotrophicus. J. Bacteriol. 190:2637-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato, S., T. Kosaka, and K. Watanabe. 2008. Comparative transcriptome analysis of responses of Methanothermobacter thermautotrophicus to different environmental stimuli. Environ. Microbiol. 10:893-905. [DOI] [PubMed] [Google Scholar]
- 11.Kessler, A., G. Sezonov, J. I. Guijarro, N. Desnoues, T. Rose, M. Delepierre, S. D. Bell, and D. Prangishvili. 2006. A novel archaeal regulator protein, Sta1, activates transcription from viral promoters. Nucleic Acids Res. 34:4837-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, C., S. M. Lee, P. Mukhopadhyay, S. J. Kim, S. C. Lee, W.-S. Ahn, M.-H. Yu, G. Storz, and S. E. Ryu. 2004. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat. Struct. Mol. Biol. 11:1179-1185. [DOI] [PubMed] [Google Scholar]
- 13.Lee, J.-W., and J. D. Helmann. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363-367. [DOI] [PubMed] [Google Scholar]
- 14.Lee, S., M. Surma, S. Seitz, W. Hausner, M. Thomm, and W. Boos. 2007. Differential signal transduction via TrmB, a sugar sensing transcriptional repressor of Pyrococcus furiosus. Mol. Microbiol. 64:1499-1505. [DOI] [PubMed] [Google Scholar]
- 15.Lie, T. J., G. E. Wood, and J. A. Leigh. 2006. Regulation of nif expression in Methanococcus maripaludis: roles of the euryarchaeal repressor NrpR, 2-oxoglutarate, and two operators. J. Biol. Chem. 280:5236-5241. [DOI] [PubMed] [Google Scholar]
- 16.Lipscomb, G. L., M. K. Annette, M. C. Darin, J. S. Gerrit, T. Michael, W. W. A. Michael, and A. S. Robert. 2009. SurR: a transcriptional activator and repressor controlling hydrogen and elemental sulphur metabolism in Pyrococcus furiosus. Mol. Microbiol. 71:332-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a Conserved Domain Database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martìnez-Antonio, A., and J. Collado-Vides. 2003. Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6:482-489. [DOI] [PubMed] [Google Scholar]
- 19.Nölling, J., M. Ishii, J. Koch, T. D. Pihl, J. N. Reeve, R. K. Thauer, and R. Hedderich. 1995. Characterization of a 45-kDa flavoprotein and evidence for a rubredoxin, two proteins that could participate in electron transport from H2 to CO2 in methanogenesis in Methanobacterium thermautotrophicum. Eur. J. Biochem. 231:628-638. [DOI] [PubMed] [Google Scholar]
- 20.Ouhammouch, M., R. E. Dewhurst, W. Hausner, M. Thomm, and E. P. Geiduschek. 2003. Activation of archaeal transcription by recruitment of the TATA-binding protein. Proc. Natl. Acad. Sci. U. S. A. 100:5097-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouhammouch, M., and E. P. Geiduschek. 2005. An expanding family of archaeal transcriptional activators. Proc. Natl. Acad. Sci. U. S. A. 102:15423-15428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pareja, E., P. Pareja-Tobes, M. Manrique, E. Pareja-Tobes, J. Bonal, and R. Tobes. 2006. ExtraTrain: a database of extragenic regions and transcriptional information in prokaryotic organisms. BMC Microbiol. 6:29-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podar, A., M. A. Wall, K. S. Makarova, and E. V. Koonin. 2008. The prokaryotic V4R domain is the likely ancestor of a key component of the eukaryotic vesicle transport system. Biol. Direct 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seedorf, H., A. Dreisbach, R. Hedderich, S. Shima, and R. K. Thauer. 2004. F420H2 oxidase (FprA) from Methanobrevibacter arboriphilus, a coenzyme F420-dependent enzyme involved in O2 detoxification. Arch. Microbiol. 182:126-137. [DOI] [PubMed] [Google Scholar]
- 25.Skarfstad, E., E. O'Neill, J. Garmendia, and V. Shingler. 2000. Identification of an effector specificity subregion within the aromatic-responsive regulators DmpR and XylR by DNA shuffling. J. Bacteriol. 182:3008-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storz, G., L. A. Tartaglia, and B. N. Ames. 1990. Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248:189-194. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki, J. Y., and E. B. C. 1995. A prokaryotic origin for light-dependent chlorophyll biosynthesis of plants. Proc. Natl. Acad. Sci. U. S. A. 92:3749-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werner, F., and R. O. J. Weinzierl. 2002. A recombinant RNA polymerase II-like enzyme capable of promoter-specific transcription. Mol. Cell 10:635-646. [DOI] [PubMed] [Google Scholar]
- 29.Wittkopp, P. J. 2010. Variable transcription factor binding: a mechanism of evolutionary change. PLoS Biol. 8:e1000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie, Y., and J. N. Reeve. 2004. Transcription by Methanothermobacter thermautotrophicus RNA polymerase in vitro releases archaeal transcription factor B but not TATA-box binding protein from the template DNA. J. Bacteriol. 186:6306-6310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zianni, M., K. Tessanne, M. Merighi, R. Laguna, and F. R. Tabita. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J. Biomol. Tech. 17:103-113. [PMC free article] [PubMed] [Google Scholar]