Abstract
Complex gene regulatory circuits exhibit emergent properties that are difficult to predict from the behavior of the components. One such property is the stability of regulatory states. Here we analyze the stability of the lysogenic state of phage λ. In this state, the virus maintains a stable association with the host, and the lytic functions of the virus are repressed by the viral CI repressor. This state readily switches to the lytic pathway when the host SOS system is induced. A low level of SOS-dependent switching occurs without an overt stimulus. We found that the intrinsic rate of switching to the lytic pathway, measured in a host lacking the SOS response, was almost undetectably low, probably less than 10−8/generation. We surmise that this low rate has not been selected directly during evolution but results from optimizing the rate of switching in a wild-type host over the natural range of SOS-inducing conditions. We also analyzed a mutant, λprm240, in which the promoter controlling CI expression was weakened, rendering lysogens unstable. Strikingly, the intrinsic stability of λprm240 lysogens depended markedly on the growth conditions; lysogens grown in minimal medium were nearly stable but switched at high rates when grown in rich medium. These effects on stability likely reflect corresponding effects on the strength of the prm240 promoter, measured in an uncoupled assay system. Several derivatives of λprm240 with altered stabilities were characterized. This mutant and its derivatives afford a model system for further analysis of stability.
Gene regulation often involves complex interlocking circuits whose parts work together to produce systems with particular properties. Most current descriptions of complex gene regulatory circuits are essentially qualitative and represent the causal connections among the components in a wiring diagram. However, complex circuits have “emergent” or systems properties that arise from the interactions of the components. The properties of the parts cannot readily predict these systems properties, and while the wiring diagram may predict the presence of systems properties, it generally does not predict the quantitative behavior of the system. Hence, a combination of quantitative modeling and experimental analysis of systems behavior is needed to provide a deeper understanding of emergent properties.
One important systems property is stability—the ability of a system in a particular regulatory state to maintain that state in the face of noise, such as chance fluctuations in the concentrations of regulatory molecules. A stable system rarely changes state; an unstable one switches readily. In metazoans, many regulatory states, once chosen, are stabilized by mechanisms involving chromatin structure and are essentially irreversible, leading to stable specification of cell type. In prokaryotes, by contrast, most regulatory states appear to be reversible, in keeping with the organism's responsiveness to its environment, and the stability of regulatory states depends on the ongoing function of the regulatory molecules themselves. It is not feasible to predict the stability of a circuit from its wiring diagram, since this lacks details such as the strength of interaction parameters. In addition, state switching is believed in the system we study to involve stochastic features of the system that occur in a subpopulation of the cells (5); such features are not included in a wiring diagram.
In this work, we have analyzed stability in the well-studied phage λ system. Like most viruses, λ can grow and produce new virions, following a developmental pathway termed the lytic pathway. Infected cells express a set of lytic genes, resulting in DNA replication, virion production, and cell lysis after ∼60 min. The λ regulatory circuitry can also exist in a highly stable regulatory state, the lysogenic state, in which the viral genome is inserted into the genome of the bacterial host and phage lytic genes are repressed by the action of the CI repressor. This state is highly stable in the absence of perturbations, but lysogenic cells can undergo an epigenetic switch to the lytic pathway. When the host SOS system is induced by DNA damage or treatments that inhibit DNA replication, the host RecA protein is activated to a form that stimulates specific cleavage and inactivation of CI (34, 36). Accordingly, the λ system is balanced such that, during normal growth, the lysogenic state is almost completely stable, but it can be almost completely destabilized by an active mechanism for inactivation of CI.
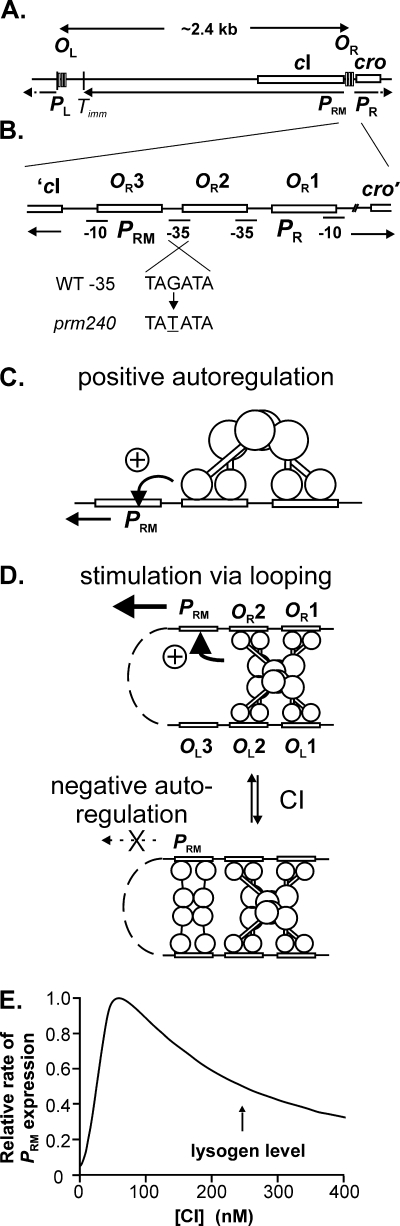
One contribution to this wide range in stability is the pattern of CI regulation in the lysogenic state. The cI gene is expressed under the control of the PRM promoter (Fig. 1A and B), which is regulated in a complex manner in response to CI levels. PRM is intrinsically a very weak promoter but is subject to two forms of positive autoregulation. At low to moderate CI levels (Fig. 1C), CI stimulates PRM ∼10-fold by binding to OR2 and contacting RNA polymerase. Binding to OR1 and OR2 is cooperative; occupancy by CI changes over a narrow range of CI concentrations. PRM can be further stimulated about 2-fold when a CI-mediated loop is formed between OL and OR (Fig. 1D), provided that CI is not bound to OR3 (1, 2). At higher CI levels, CI negatively autoregulates PRM by binding to OR3 (Fig. 1D), and this binding is favored at lysogenic CI levels by cooperativity due to looping (17). As a result, the rate of CI expression varies markedly in response to CI levels (Fig. 1E). An additional regulatory control is that, at low CI levels, expression from the PR promoter (Fig. 1A) leads to production of another repressor, Cro protein. Cro then represses PRM by binding to OR3. This double-negative effect is not essential for switching, but it apparently acts in cells that are “on the edge” to drive them toward switching (3, 54).
FIG. 1.
Maps of λ, functions of CI, and regulation of CI expression. (A) Map of the immunity region of λ, to scale. PL and PR transcripts continue beyond the region shown. (B) Expanded map, to scale, of the OR region, showing the location of the CI and Cro binding sites OR1, OR2, and OR3, the location of the PRM and PR promoters, and the sequence change in prm240. (C) Positive autoregulation of PRM by CI bound at OR2; CI binds cooperatively to OR1 and OR2. Both subunits of a dimer contact a subunit in the other dimer (59), as schematized. (D) Positive and negative autoregulation of PRM by CI-mediated looping. Details of protein-protein interactions in looped forms are not known. (E) Expression of PRM as a function of CI. This curve is the output of a stochastic simulation (J. W. Little and A. P. Arkin, unpublished data), based on a mechanistic model that includes stimulation of PRM by looping. It is meant to illustrate the response of PRM to CI levels, not to represent exact values. It is qualitatively similar to one measured (17) with a reporter construct that did not allow stimulation of PRM by looping and to a curve calculated (1) from a model that includes this feature.
Negative autoregulation contributes to the intrinsic stability of the lysogenic state (17). PRM expression increases if the CI level falls, counterbalancing the depletion (9). However, if CI levels fall below a certain level (the peak in Fig. 1E), negative autoregulation no longer occurs. At still lower CI levels, positive feedback begins to fail; this helps drive the system toward switching (19), and synthesis of Cro helps make the switch irreversible. Hence, as CI levels fall to a critical value, which we shall term the “switching threshold,” switching becomes increasingly likely. The value for the switching threshold is not known, but measurements of CI activity after SOS induction suggest that it is in the range of 10 to 20% of the lysogen level of CI. This value represents only 25 to 50 monomers of CI in the cell. At such a low value, stochastic events almost certainly play a major role in determining whether switching will occur. Accordingly, two cells with the same history and amount of CI may follow different fates, and the switching threshold should not have a discrete value but a narrow range.
A lysogenic cell that switches to the lytic state after SOS induction produces ∼100 virions and releases them into the medium upon lysis. Hence, measurement of free phage in cultures provides a simple assay for switching and an estimate of its rate. Even in an untreated wild-type host lysogenic for λ, the lysogenic state breaks down at an easily detectable level, yielding free phage; about 1 cell in 105 switches state (37). This process is termed “spontaneous induction,” since it occurs without an overt stimulus and yet depends on the SOS regulatory system, as shown by two lines of evidence. First, host recA mutants cannot support cleavage of CI, and recA mutant lysogens yield few free phage (10). Second, cI ind mutations make CI resistant to cleavage; λind mutant lysogens also yield few free phage (23, 44). A likely explanation for the SOS dependence of free phage release is that cells often undergo sporadic DNA damage, which can induce the SOS system (14). Single-cell experiments show that in a small fraction of untreated cells, the SOS system is induced (40, 47), presumably due to sporadic damage, and it is plausible that in a smaller fraction this induction is severe and prolonged enough to result in prophage induction. These genetic data indicate that spontaneous SOS induction limits the stability of the lysogenic state.
Here we analyze the intrinsic stability of the lysogenic state in the absence of the SOS system. Spontaneous switching of this state presumably follows a pathway similar to that occurring after prophage induction, with the major difference that the latter process includes a mechanism for active CI degradation, whereas spontaneous switching presumably occurs as a consequence of random fluctuations in CI levels. To determine the intrinsic stability of the lysogenic state, we have analyzed stability in a recA mutant host, addressing several related questions. First, how stable is this regulatory circuit? As we show, it is extremely stable. Second, what mechanisms contribute to this high degree of stability? Third, is stability dependent on conditions? Insight into the latter question came from analysis of a mutant, λprm240, whose lysogenic state was only marginally stable. Finally, what is the level of CI at the switching threshold?
MATERIALS AND METHODS
Media, chemicals, and reagents.
LB and M9 minimal media are as described previously (43). LBGM, LBMM, and TMG are as described previously (37). Restriction enzymes and DNA ligase were from New England Biolabs or Fermentas Inc. IPTG and XGal were from GoldBioTech. Pfu Turbo DNA polymerase for site-directed mutagenesis (22) was from Stratagene; GoTaq master mix (Promega) was used for routine PCR. Polymyxin B was from Sigma. BugBuster HT and rLysozyme were from Novagen.
Bacterial and phage strains.
Many of the strains used are listed in Table 1. Construction of phage strains is described in the supplemental material. All bacterial strains were derivatives of Escherichia coli K-12. Bacterial strains not listed include those lysogenized with reporter gene fusions and several lysogens not analyzed in detail. All λ derivatives carried the bor::kan substitution (37), which is not listed. Phage strains isolated as free phage from JL5904 are listed in Table S3 in the supplemental material.
TABLE 1.
Strains employed
| Strain, phage, or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Bacterial strains | ||
| JL2497 | N99 lacZΔM15/F′ lacIqlacZΔM15::Tn9; used as wild type | 37 |
| JL5016 | JL5902 (λJL615) | This worka |
| JL5024 | JL6112 resistant to λ | This workc |
| JL5902 | JL2497 Δ(srl-recA)306::Tn10 | 37 |
| JL5904b | JL5902 (λJL163) | 37 |
| JL5906b | JL5902 (λJL169) | 37 |
| JL5908b | JL5902 (λJL172) | 37 |
| JL5924b | JL5902 (λJL175) | 37 |
| JL5932b | JL2497 (λJL163) | 37 |
| JL6093 | JL5904 resistant to λ | This workc |
| JL6112 | JL5902 (λJL240) | This worka |
| JL6142 | JL2497 Δ(lacIPOZYA) F− | 4 |
| JL6799 | JL2497 (λJL240) | This worka |
| JL6839 | JL2497 (λJL615) | This worka |
| JL6994 | JL6142/pJWL615/pJWL486 | 41 |
| JL6995 | JL6142/pJWL615/pA3B2 | 41 |
| JL7198 | JL468/pJWL1063 | This work |
| Phage strains | ||
| λJL163 | λ+bor::kan, used as wild type | 37 |
| λJL169 | λ OR121 | 37 |
| λJL172 | λ OR3′23′ | 37 |
| λJL175 | λ OR323 | 37 |
| λJL176 | λ v3 (mutation in OR1) | This workd |
| λJL188 | λ OR323 derivative with OR1 sequence TATCCCTTGCGGTAATAe | 37 |
| λJL240 | λprm240 | This work |
| λJL291 | λ OR2: TAACACCGTCCGTGTTG | This workf |
| λJL293 | λ OR2: TAACACCATGCGTGTTG | This workf |
| λJL343 | λ OR1: TAACTCTGGCGGTGATA | This workf |
| λJL465 | λprm240 OR2− | This workd |
| λJL466 | λprm240 OR1− | This workd |
| λJL473 | λprm240 OL3-4 | This workd |
| λJL615 | λprm240 cro-z8 | This workd |
| λJL815 | λ cI D38N | 41 |
| λJL1387 | λprm240 OR3-r1 | This workd |
| M13 KO7 | Helper phage for packaging pJWL1063 | 62 |
| Plasmids | ||
| pA3B2 | Δ35 lacP::cI; provides low level of CI; Cmr; vector pACYC184 | 63 |
| pGB2 | Spcr; compatible with pACYC184 and ColE1-derived plasmids | 12 |
| pJWL334 | Modified polylinker; high copy no.; vector pBS(−) | 42 |
| pJWL486 | Vector control for pA3B2; vector pACYC184 | 42 |
| pJWL615 | lacIq; vector pGB2 | 42 |
| pJWL1063 | lacP::cI oriM13; vector pBR322 | This workd |
| pRS414, pRS591 | Vectors for making lacZ protein fusions; vector pBR322 | 56 |
| pRS1274 | Vector for making lacZ operon fusions (described as pRS528 in reference 56); vector pBR322 | 56 |
Lysogens of λJL240, λJL473, λJL615, and λJL1387 were isolated by plating phage on tryptone plates at 30°C, followed by selection for lysogens by streaking on tryptone plates with kanamycin at 30°C and a single lysogen test (see Materials and Methods). Lysogens of λJL465 and λJL466 were isolated in the same way but on M9 glucose plates at 30°C.
Lysogen was used and described in reference 37 but not identified by strain number.
JL6093 and JL5024 were isolated from JL5904 and JL6112, respectively, by selection for resistance to λvir (from D. Mount). These strains cannot adsorb λ.
Isolated as described in the supplemental material.
The altered OR1 sequence in λJL188 was derived (37) from λJL175, which contains three changes (underlined) in OR1, by a fourth mutation, shown in bold, changing G to A. This latter mutation probably weakens binding of Cro to the operator, based on the way it was isolated, and probably CI binding as well; it also likely weakens PR, since it changes the first base of the −10 region away from consensus. This phage exhibited the unstable lysogen phenotype in the ΔrecA host, with a growth defect on tryptone plates more severe than that of λprm240 lysogens at 37°C. Colony size on minimal plates was like that of the wild type at both temperatures.
Isolated as described in the text and in Table S3 in the supplemental material. The mutated bases are underlined; the corresponding WT bases for the mutations in λJL291, λJL293, and λJL343 are G, G, and C, respectively. The OR1 mutation was reported to weaken CI binding by only ∼0.6 kcal/mol (52), a small increase whose effect should be even less due to pairwise cooperative DNA binding. Hence this mutation might have little effect on stability. The effect of the OR2 mutations on CI binding has not been tested. These mutants were isolated in screens that did not involve the use of mitomycin C.
Plasmids.
Many of the plasmids used are listed in Table 1; others are listed in the supplemental material, which describes most of the constructions as well.
Phage methodology.
General phage methodology was as described previously (37). The test for single lysogens was as described previously (48). It was modified for use in strains with an unstable lysogenic state, as follows. Strain JL7198 was infected with M13 KO7 as described previously (62), and the resulting lysate, containing a preponderance of packaged pJWL1063, was sterilized by heating at 65°C for 20 min. It was then used to transduce this plasmid into the strain being tested. Cells were grown to ∼108/ml, infected with the lysate, and shaken 30 min. To select for cells bearing pJWL1063, cells were spread on ampicillin plates for isolation of single colonies, or ampicillin was added to 100 μg/ml and cells were grown overnight. The test for single lysogens (48) was carried out on colonies or overnight cultures as described. High levels of CI made by pJWL1063 prevent spontaneous phage induction, which interferes with this test. Use of this method requires that the host carry an F plasmid, as JL5902 and JL2497 do.
Single-lysogen test of λprm240 lysogens.
λprm240 was plated on JL2497 (grown on LBMM) at 30°C on M9 glucose plates. Single plaques were streaked on M9 glucose-Kan plates at 30°C. Nineteen single colonies were purified under the same conditions; colonies were then streaked onto tryptone-Kan plates at 37°C to determine colony phenotype and separately transformed with pJWL1063; transformed (AmpR) colonies were subjected to the single-lysogen test. For each of the three isolates giving medium-sized colonies with a few small ones, 13 transformants were tested, and all were multiple lysogens. The remaining 16 colonies formed small colonies and were single lysogens.
Isolation of free phage.
For free phage from lysogens of λJL163, strain JL5904 was used for cultures grown in LBGM or M9 glucose medium; for growth in M9 glycerol or M9 succinate, strain JL6093 was used to prevent adsorption of free phage. Colonies of JL5904 or JL6093 were formed on a tryptone plate at 37°C and inoculated into LBGM. Cultures were grown at 37°C to 5 × 107 to 10 × 107 cells/ml and chilled; cells were removed by centrifugation, and the supernatant fluid was treated with chloroform. Chloroform was removed by adding fluid to a Falcon polystyrene tube, which adsorbs CHCl3, and aliquots were plated as indicated. To distinguish λprm240 from the wild type, free phage were plated on JL2497 on plates containing mitomycin C at 0.25 μg/ml in the bottom agar; this low level of mitomycin C allows the indicator to survive. Lysogens of relatively stable strains were grown and treated as described above; lysogens of phages carrying prm240 were streaked onto M9 glucose or tryptone plates and incubated at 30°C to avoid selective pressure, followed by growth under the conditions indicated.
Sequencing of free phage.
DNA sequencing was carried out as described previously (37). Templates were made by PCR of cored plaques, using the appropriate primers. Six isolates (four from cells grown in LBGM and two from cells grown in M9 succinate) for which no mutation was found in the cI-cro interval were sequenced in the intervals lying between the following coordinates in the λ genome (GenBank accession number J02459.1): left of sib to right of xis, 27451 to 29250; middle of gam to right of cIII, 32880 to 33550; left of N to first part of O, 34500 to 39170; no mutations were found in these intervals.
β-Galactosidase assays.
β-Galactosidase assays were done as described previously (41), except that assays were done at 37°C to increase the sensitivity. Accordingly, values for PRM are higher than reported (41, 42).
Medium-shift experiments.
Cells were grown with aeration in M9 glucose medium at 30°C to 108 cells/ml. Cultures were diluted at least 20-fold into LBGM at 37°C and shaken at 37°C. At intervals, the optical density at 590 nm (OD590) was read, and samples were taken for measurement of free phage: an aliquot was centrifuged 1 min at ≥10,000 × g, and the supernatant fluid was treated with chloroform. In some experiments, aliquots were also taken to measure CI levels: a portion (generally 2 optical density units [ODU]) of the culture was chilled rapidly to 0°C, followed by centrifugation 1 min at ≥10,000 × g; pellets were resuspended in 1 ml 10 mM Tris-HCl (pH 8)-10 mM MgSO4 and centrifuged 1 min at ≥10,000 × g; pellets were frozen at −20°C for processing as indicated below. In some experiments, several different dilutions of the initial culture were made to allow OD590 measurement over a longer time frame.
Measurement of CI levels.
CI levels were determined using Western blots, using a rabbit polyclonal antibody. Cells isolated as described above were lysed using BugBuster HT (50 μl per ODU of cells) containing rLysozyme. Protein concentrations were measured using the Bio-Rad protein assay reagent. Samples for SDS gel electrophoresis contained a constant amount of protein per well; in some cases, extracts were diluted with an extract from a nonlysogenic strain grown under the same conditions as the sample being assayed. JL6112 extracts were compared with JL5904 extracts diluted with JL5902 extract. Following electrophoresis, protein was transferred to a nitrocellulose membrane (Schleicher and Schuell BA83), blocked, and exposed to rabbit polyclonal antibody against CI, followed by goat anti-rabbit secondary antibody conjugated with IRDye 800CW (LI-COR). Dried membranes were scanned using the LI-COR Odyssey infrared imaging system. A gel mobility shift assay, done as described previously (17), also gave results consistent with the those of the Western blotting experiment for strain JL6112 grown in LB medium (not shown).
RESULTS
Stability of the lysogenic state.
To measure the frequency with which the lysogenic state switches to the lytic state, we grew cultures of the ΔrecA (λ+) strain JL5904 to mid-exponential phase, removed the cells by centrifugation, and characterized phage in the supernatant fluid. Some phage form clear plaques and carry cI mutations; others form turbid plaques, because lysogens can form in the center of the plaque. We analyzed free phage forming turbid plaques. The prophage carried a marker for kanamycin resistance, allowing selection for lysogens by growth on kanamycin plates. Adsorption of released phage to the cells was prevented by growth in media containing glucose, which represses the LamB receptor to which λ binds (29, 61).
We reported (37) that such cultures contain ∼500 free phage/ml that form turbid plaques. Others have obtained similar results (5, 10, 51). We later found that this value is variable (see below). In addition, two lines of evidence showed that almost all free phage forming turbid plaques were not wild-type λ, but mutants. First, lysogens arising in most turbid plaques formed a range of colony sizes, while lysogens of λ+ formed uniformly large colonies. For reasons given below, we term this the “unstable lysogen phenotype.” Second, and strikingly, sequencing of the cI-cro interval from many isolates with this phenotype showed that all had the same mutation in the PRM promoter, which we term prm240 (Fig. 1B). This mutant was isolated from all cultures analyzed; it is described below (“Analysis of λprm240”). We conclude that most of the free phage forming turbid plaques are mutants, a conclusion also reached on the basis of other evidence by others (7).
To measure the titer of wild-type free phage, we used a screen to discriminate phage with prm240 from wild-type phage. Phage were plated on a recA+ indicator in the presence of the SOS-inducing agent mitomycin C; λ+ formed faintly turbid plaques, while λprm240 formed clear plaques. Without mitomycin C, the titer of turbid plaques varied substantially, as expected if most arose by mutation (Table 2). With mitomycin C, most plaques were clear. Phage from turbid plaques were isolated, and the cI gene and the OR region were sequenced (Table 2). For isolates with no mutations, the OL region was also sequenced. In total, 42 isolates with no mutations in these intervals were isolated from 12 ml of culture supernatant.
TABLE 2.
Analysis of free phage from a ΔrecA (λ+) mutant lysogena
| Culture | No. of free phage/0.6 ml without mitomycin C |
No. of turbid plaques/2 ml with mitomycin C | Sequences of turbid plaque isolates | |
|---|---|---|---|---|
| Clear | Turbid | |||
| A | 57 | 44 | 10 | 10 WT |
| B | 102 | 326 | 9 | 4 WT, 5 cI S149F |
| C | 104 | 29 | 5 | 5 WT |
| D | 62 | 34 | 13 | 13 WT |
| E | 90 | 107 | 15 | 10 WT, 5 cI S149F |
| F | 62 | 73 | 0 | 0 |
| Totals | 52/12 ml | 42 WT, 10 cI S149F/12 ml | ||
Free phage from six independent 50-ml cultures were isolated as described in Materials and Methods and plated on JL2497. Plaques formed without mitomycin C were scored for clear or turbid phenotype. Since most of the phage forming turbid plaques carry prm240, the abundance of phage with this allele is about the same as those forming clear plaques due to mutation of the cI gene, indicating that prm240 is a mutational hot spot. High titers of turbid plaque formers in culture B, and perhaps E and F, probably resulted from “jackpots,” in which a prm240 mutant arose early in the culture, as likely also occurred previously (37). For plaques formed with mitomycin C (0.25 μg/ml) in the bottom agar, only turbid plaques were scored. Sequences of the cI-cro interval and the OL region were determined. λcI S149F showed the unstable lysogen phenotype. The S149F allele changes the catalytic serine residue involved in CI autocleavage (58); plaques of this phage, which are less turbid than the wild type without mitomycin C, remain turbid in its presence since CI cleavage cannot occur. The mutant protein is somewhat defective for repressor function (63), probably leading to frequent switching. Other cI ind mutants would also form turbid plaques, but perhaps few such mutants are also leaky for repressor function.
Other screens also yielded isolates for which no mutations were found. For several of these we sequenced other intervals involved in gene regulation (see Materials and Methods), and found no mutations, suggesting that they are indeed wild type. We conclude that wild-type free phage are present at roughly 2 to 5 phage/ml culture. We calculate (see Discussion) that the frequency of switching is probably <10−8 per cell per generation.
We speculated that a wild-type lysogen might be less stable during growth in minimal medium; slowly growing cells are smaller, so that stochastic effects on cI expression or function might be more frequent. To test this idea, we grew JL5904 or JL6093 in minimal medium with glucose, glycerol, or succinate as the carbon source. Contrary to our expectation, the free phage titer was very low. This resulted in part because λ virions are somewhat unstable in minimal medium, with a half-life of 30 to 60 min (not shown). Despite this complication, we infer that the low titer also reflected a low rate of free phage production. Few of the isolates conferred the unstable lysogen phenotype. They included cI mutants (see Table S3 in the supplemental material) and an OR1 mutant, as well as several for which no mutations could be found. The instability of λ virions prevents a quantitative estimate of the titer of wild-type phages, but we may conclude that growth in minimal medium does not markedly destabilize the lysogenic state.
Analysis of mutant free phage and existing mutants affecting stability.
In addition to wild-type phage, in the above screens we recovered free phage with mutations in cI or in OR (Table 1; see Table S3 in the supplemental material). We analyzed free phage levels from lysogens of these mutants. Surprisingly, lysogens of λJL291, λJL293, and λJL343, with changes in OR2 or OR1, did not release markedly elevated levels of free phage (data not shown). It is uncertain whether the level is higher than that of the wild type and unclear why these mutants were initially recovered as free phage. Several mutants with changes in cI gave a range of phenotypes, and lysogens in JL5902 had a wide range of stabilities (see Table S3 in the supplemental material).
We also analyzed several existing mutants expected to have a lower level of CI and/or a higher switching threshold, and found (Table 3) that each had a substantially higher switching rate than the wild type. The first has an OR1 mutation, v3, which weakens binding of CI to OR1 but has almost no effect on Cro binding (52, 60). We attribute the higher level of free phage to the expectation that, in a v3 lysogen, the occupancy of OR1 and OR2 by CI would be lower than in a λ+ lysogen and hence that its switching threshold would be elevated. The second mutant carries the cI D38N mutation, which nearly abolishes positive autoregulation of cI from PRM (31, 41) and should reduce the level of CI; it also markedly destabilized the lysogenic state.
TABLE 3.
Free phage levels in unstable lysogensa
| Phage | Mutation | Free phage level in recA lysogen (phage/ml) |
|---|---|---|
| λJL176b | OR1 v3 | 1,000 |
| λJL815c | cI D38N | 4 × 105 |
| λJL169d | OR121 | 6,000 |
| λJL172d | OR3′23′ | 500 |
| λJL175d | OR323 | 105 |
| λJL163 | None (WT) | (2 to 5) |
Cells were grown and free phage were recovered as described in Materials and Methods. The titer of turbid plaques is indicated. The value for the WT, from Table 2, is that of WT free phage only and is given for comparison.
Lysogens of λv3 formed colonies of normal size. The value given for free phage titer was about the same in six independent cultures in the same experiment. Of 24 plaques tested, all gave uniformly large Kanr colonies. Of six isolates sequenced in the OR region, all retained v3 and had no other changes.
Of seven free phage sequenced in the cI-cro interval, all had the cI D38N mutation and OR+.
Free phage forming turbid plaques from the λOR323 lysogen were probably of that genotype, since they were present at high levels and formed the small plaques characteristic of λOR323. Lysogens of λOR121 and λOR3′23′ yielded the given titer of free phage forming turbid plaques in each of three independent cultures. We sequenced six isolates of each; in each case, all six had the parental sequence in the cI-cro interval; in the OL region, all were wild type, except that one derivative of λOR3′23′ had a mutation in OL1 that should weaken CI binding substantially.
In the third type of mutant tested, OR1 and/or OR3 are mutated so that both have the same sequence (37). In λOR121, OR3 is changed to the sequence of OR1; in λOR323, OR1 is changed to OR3; and in λOR3′23′, both sites are changed to a hybrid site termed OR3′. Consistent with previous findings (37), the present, more-detailed analysis showed that all three mutants were less stable than the wild type. We ascribe this to the reduced level of CI found in mutant lysogens (37). We previously suggested that the CI level is reduced because CI occupancy patterns at OR are altered (37). Subsequent work suggests that these changes might also affect looping between OL and OR (1, 17), giving aberrant looped forms that are not generally favored in the wild type. The effect of this is hard to predict: it might reduce CI levels if it prevents looping-mediated activation of PRM (1), this activation might instead be more efficient, or changes in looped forms might affect the cooperative interactions among CI dimers that lead to negative autoregulation.
In sum, our evidence indicates that the stability of the lysogenic state can be reduced by mutations, either in cis-acting sites or in CI itself, that weaken the ability of CI to repress the lytic promoters and/or to regulate CI expression. Presumably, the proper choice of mutations in cI, in the operators, or in PRM could afford any desired level of stability. We turn next to a mutant that lies at the opposite end of the stability spectrum from the wild type.
Analysis of λprm240.
The following evidence indicates that lysogens of λprm240 are barely stable and readily switch their regulatory state from lysogenic to lytic. As noted above, λprm240 showed the “unstable lysogen phenotype”; among lysogens arising in a plaque, most formed small colonies, while a small fraction made larger colonies. Since prm240 is a down-mutation in PRM (see below), we surmised that cells in small colonies made a low level of CI and often switched to the lytic state, reducing the number of progeny cells, while cells in the larger colonies made more CI and switched less frequently. We found that lysogens forming small colonies carried a single copy of the prophage, while those making large colonies carried multiple tandem prophages (see Materials and Methods), and presumably made more CI due to gene dosage. Further analysis used single lysogens.
Remarkably, growth conditions influenced the severity of the unstable lysogen phenotype. Colony sizes of recA+ (λprm240) and ΔrecA (λprm240) lysogens were much closer to those of wild-type lysogens at 30°C than at 37°C, suggesting that the lysogenic state was more stable at 30°C. On minimal medium, colony sizes of λprm240 and wild-type lysogens were the same, suggesting that λprm240 lysogens were relatively stable in minimal medium. The mechanistic basis for these conditional responses is unclear.
These responses to growth conditions were not specific to prm240. Other PRM mutants also show the unstable lysogen phenotype (42). We tested six such isolates on the ΔrecA host. Colonies of lysogens were the same size as wild-type colonies on minimal plates. On tryptone plates, most were like the wild type at 30°C, while MD17 and MD12 were slightly smaller; at 37°C, colony sizes were, in rank order, wild type (WT) > MD12 > MD13 > λprm240 ≈ MD8 > MD17 ≥ MD11 ≈ MD47. Hence, the detailed response to growth conditions may depend on the properties of the given promoter.
We analyzed further the stability of λprm240 lysogens in a range of growth media, at both 30°C and 37°C. Few free phage were released in minimal medium (Table 4), consistent with the colony phenotype and the paucity of λprm240 isolates from JL5904 grown in minimal medium. At higher growth rates, titers of free phage increased markedly. At the highest growth rate, in LB medium at 37°C, cultures grew slowly (see also below) and released very high levels of free phage. We conclude that growth at higher rates markedly destabilizes λprm240 lysogens, as does growth at higher temperatures to a lesser extent.
TABLE 4.
Free phage titer of λprm240 lysogena
| Medium | Temp (°C) | Doubling timeb for λJL163 lysogen (min) | Free phage/cellb for λ prm240 lysogen | 37°C/30°Cc |
|---|---|---|---|---|
| M9 glucose | 30 | 160 | 5.4 × 10−6 | 11 |
| 37 | 110 | 5.9 × 10−5 | ||
| M9 glucose + | 30 | 127 | 0.012 | 18 |
| Casamino Acids | 37 | 73 | 0.21 | |
| Tryptone broth | 30 | 80 | 0.08 | 16 |
| 37 | 54 | 1.3 | ||
| LB | 30 | 72 | 1.8 | 10 |
| 37 | 44 | 17 |
Strain JL6112 was grown overnight in tryptone broth at 30°C and then grown as follows. For growth in M9 glucose with or without Casamino Acids, an aliquot was grown exponentially for many generations in M9 glucose at 30°C, followed by exponential growth for at least five generations under the indicated conditions. For growth in tryptone or LB, an aliquot was diluted into tryptone and grown for three generations at 30°C, followed by exponential growth for at least five generations in the indicated conditions and sampling for free phage as described in Materials and Methods.
For calculation of doubling time and phage/cell, cell growth and cell numbers were determined on cultures of JL5904 growing in parallel. Titers given for minimal medium are likely underestimated by a factor of 2 to 5, since λ virions were somewhat unstable in minimal medium (see text). To test for readsorption of free phage in tryptone broth (which lacks glucose), in a separate experiment strain JL6112 and its λ-resistant derivative JL5024 were grown in parallel in tryptone broth, at both 30 and 37°C. At each temperature, free phage levels for both cultures were the same within experimental error. Hence, readsorption is not significant under our conditions.
Ratio of titer of free phage at 37°C to that of free phage at 30°C.
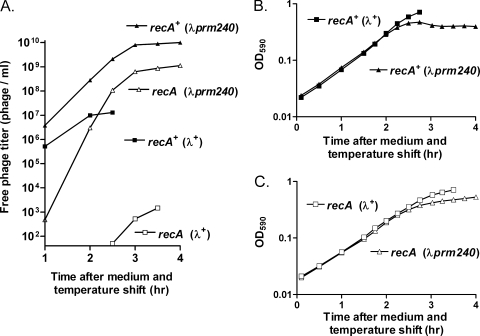
To assess the time course with which the lysogenic state was destabilized after a shift from minimal to rich medium, we followed growth and phage production of recA+ and ΔrecA λ+ and λprm240 lysogens after a switch from the most stable to the least stable growth condition. After growth in minimal medium at 30°C, cells were transferred to LB medium at 37°C. In both host strains, λprm240 lysogens grew like the wild-type control for about 2 h, followed by growth at a lower rate (Fig. 2B and C). Levels of free phage underwent a progressive and enormous increase with time, reaching a high value at about 3 h, after which the level of free phage increased more slowly as cell growth continued (Fig. 2A).
FIG. 2.
Destabilization of λprm240 lysogens after medium and temperature shift. Cultures were grown and sampled as described in Materials and Methods. All measurements were from the same experiment and are separated for clarity. Strains were JL5932 and JL6799 (recA+), and JL5904 and JL6112 (ΔrecA). (A) Free phage levels for all strains. Note the logarithmic scale. (B and C) Growth curves for recA+ (B) and recA mutant (C) lysogens. OD590, optical density at 590 nm. Cell debris from lysis of λprm240 lysogens made a minor contribution to the OD590 at later times.
The magnitude of these effects was greater in the recA+ host than in its ΔrecA counterpart. The recA+ (λprm240) culture underwent partial lysis, followed by an essentially flat growth curve (Fig. 2B). Evidently the likelihood of switching was about the same as that of dividing. Net growth of the recA mutant (λprm240) culture slowed, presumably because a substantial fraction of the cells in the culture switched and lysed. Free phage levels in the recA+ culture were ∼10-fold higher than in its recA mutant counterpart. The more frequent switching in recA+ cells may result in part from the presence of activated RecA in a fraction of the cells in the culture; the greater yield of free phage may result both from higher switching rates and the generally healthier state of recA+ cells.
We reasoned that the CI levels in the λprm240 lysogen grown under a range of conditions could provide insight into the level of CI at the switching threshold for λprm240—that is, the level of CI at which switching becomes likely (see introduction). We compared CI levels in the λ+ and λprm240 lysogens in cultures grown in tryptone broth at 30° and in LB medium at 37°C using Western blotting to quantify CI levels; the respective values were about 30% and 20%. The precision of this assay was not sufficient to allow a detailed correlation between CI levels and switching rates under various growth conditions. We conclude that the switching threshold for λprm240 is about 25% of the wild-type level of CI (see Discussion).
Mutations affecting stability of λprm240.
We analyzed the effects of mutations that stabilized or further destabilized λprm240. We first compared ΔrecA λprm240 lysogens with those of λJL465 and λJL466 (with additional mutations in OR2 and OR1, respectively). Under all conditions tested (30 and 37°C in M9 medium and in tryptone broth), titers for λJL465 and λJL466 were elevated about 5- and 10-fold, respectively, relative to those for λprm240 (data not shown). In tryptone broth at 37°C, growth of λJL465 and λJL466 lysogens nearly ceased, as seen in LB medium for the recA+ (λprm240) lysogen (Fig. 2B), suggesting that the likelihood of switching was about equal to that of cell division. We conclude that the OR2 and OR1 mutations in these isolates destabilize λprm240, presumably by weakening binding of CI to these sites. The finding that lysogens of the single OR1 and OR2 mutants still had very low levels of free phage (see above) further emphasizes the extreme stability of the wild type.
We next removed or weakened negative autoregulation by combining prm240 with either of two changes. The first derivative, λJL473, carried the r1 mutation in OR3; this mutation eliminates negative autoregulation at CI levels found in a WT lysogen (17). The second derivative, λJL1387, carried OL3-4, with four mutations in OL3, abolishing CI binding to OL3 (18). This change also reduces negative autoregulation (Fig. 1D). We expected with either mutant that, if negative autoregulation was substantial in a λprm240 lysogen, then relieving it would lead to higher CI levels in the prm240 OR3-r1 and prm240 OL3 mutants, markedly stabilizing the lysogen. We found that yields of free phage after extended growth in LB medium at 30 or 37°C were reduced 5 to 10-fold from that seen with λprm240 (data not shown), suggesting that the rate of switching is reduced by 5- to 10-fold in the double mutants. This modest effect suggests that negative autoregulation was scarcely operative during growth in LB medium. Consistent with these reduced switching rates, growth of lysogens in LB medium was only slightly slower than that of the λ+ lysogen (data not shown), in contrast to the marked reduction of growth of the λprm240 lysogen (Fig. 2).
It is not clear what the course of events is during the switching process of λprm240. Presumably an early event is transient expression of the lytic promoters PL and PR. An early product of PR expression is the Cro protein, which can act to repress PRM. We reasoned that, if Cro plays an essential role in switching, then removing Cro should stabilize the mutant lysogen. Complete removal of Cro by mutation is lethal for lytic growth (20). However, if PRM repression plays an important role in the switching process, then reducing the level of Cro by mutation might act to stabilize λprm240.
We tested this expectation with the use of a mutation termed cro-z8, which changes the 5′-UTR of the cro mRNA and gives a reduced level of Cro (46). We (J. W. Little and K. Newell, unpublished data) reisolated this mutation as a suppressor of a lysogenization defect in a mutant phage (see supplemental material), a property suggesting that it has a functional defect in Cro function. As judged by its effect on expression of a PR::lacZ protein fusion, cro-z8 reduced Cro expression to ∼25 to 30% of the wild-type level (Table 5). Hence, the cro-z8 mutation might be expected to stabilize λprm240. Comparison of λprm240 and λprm240 cro-z8 lysogens grown under the same conditions as used for the reporter assays (Table 5) showed that, although cro-z8 did stabilize the lysogen, the effect was rather modest. As the λprm240 lysogen became more unstable, the effect of cro-z8 became smaller. These data suggest that Cro may play a minor role in certain switching events for λprm240 (see Discussion).
TABLE 5.
Effect of cro-z8 mutation on PR expression and on stability of λprm240 under various growth conditions
| Medium | Growth temp (°C) | β-Galactosidase activitiesa |
Phage yield (phage/cell)b |
||||
|---|---|---|---|---|---|---|---|
| PR+ | PR+cro-z8 | Ratio | λprm240 | λprm240 cro-z8 | Ratio | ||
| Tryptone | 30 | 2,304 | 714 | 0.31 | 0.08 | 0.008 | 10 |
| 37 | 3,869 | 1,268 | 0.33 | 1.3 | 0.2 | 6.5 | |
| LB | 30 | 2,203 | 730 | 0.33 | 1.8 | 0.37 | 4.8 |
| 37 | 3,491 | 872 | 0.25 | 17 | 7 | 2.3 | |
For the β-galactosidase assays, reporters carried the prm240 allele and either the wild-type or cro-z8 mutant allele; the presence of the prm240 allele made little or no difference in the activity of PR under these conditions (not shown). We surmise that the effect of the cro-z8 mutation differed from the lower value (11%) found when Cro was expressed from the lac promoter on a plasmid (46) due to differences in the 5′ leader of the mRNA. Since PR was more active at 37°C than at 30°C, both in tryptone broth and in LB, direct comparisons between results at different temperatures are not meaningful, since the level of Cro should also be somewhat higher in the cro-z8 lysogen at 37°C.
Properties of the prm240 promoter.
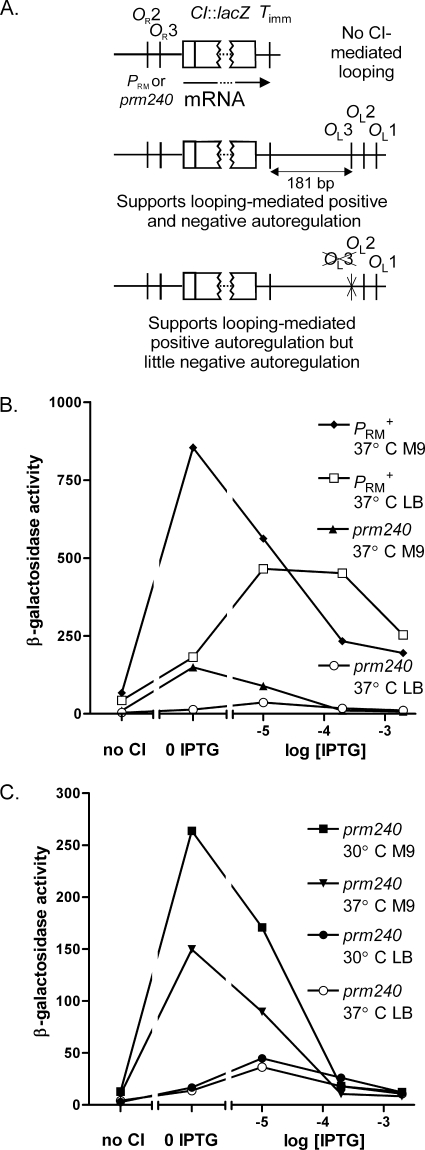
We assessed the strength and regulation of the prm240 promoter, using a reporter gene in an uncoupled assay system. The PRM::lacZ protein fusions carry either PRM+ or prm240 and various alleles of the OL region distal to the lacZ reporter gene (Fig. 3A and supplemental material). The first construct (Fig. 3A, middle) carries OL and a region lying upstream of OL3 that supports (1) the stimulatory effect of looping and should mimic the situation in the prophage. We tested its response to graded levels of CI, supplied from a lacP::cI fusion on a plasmid in the presence of graded levels of IPTG. This plasmid produces some CI in the absence of IPTG. A control strain lacked the CI-producing plasmid. Assays were carried out both in M9 minimal medium and in LB medium at 30°C and at 37°C. Unexpectedly, the shapes of the dose-response curves differed between those for M9 medium and LB medium. This resulted, at least in part, because the levels of CI were much greater in reporter strains grown in M9 medium than in those in LB medium (data not shown; see Fig. 3 legend). Hence, comparisons between the two growth media at a given IPTG level are not meaningful, but comparisons between the WT and prm240 promoters under a given condition are valid.
FIG. 3.
Reporter constructs and promoter activity for PRM+ and prm240. (A) Structure of reporter constructs (not to scale). The three constructs shown (see the supplemental material) encode an mRNA that is terminated at Timm; hence, all three mRNAs are identical and their levels of expression are directly comparable (1). A second version of each construct contained the r1 allele in OR3, which weakens CI binding (see text). Maps are not to scale. (B and C) Reporter assays. Cells were grown at the temperature and in the medium indicated in the figure. Two reporter constructs were used, one with PRM+ and the other with prm240 (see text and panel A, middle construct). For each construct and growth condition, two strains were used, one lacking CI and one carrying a plasmid with a weak lacP::cI fusion. The latter strain was grown with no IPTG or with the indicated levels of IPTG, providing various levels of CI. Data for PRM+ at 30°C were obtained in separate experiments and are shown in Fig. S3 in the supplemental material. (B). Expression of PRM+ and prm240 at 37°C. (C). Data for prm240 fusions, with an expanded scale. As judged by a gel shift assay (not shown), CI levels in the reporter strain grown in M9 medium at 30°C and in LB medium at 37°C with 2 mM IPTG were roughly 3 times and 1 time, respectively, the level in JL5932 grown in parallel; in LB medium at 37°C, the CI levels in cells grown at 0.01 and 0.2 mM IPTG were roughly 0.1 and 0.5, respectively, of the lysogen level, bracketing the value (∼0.2 the lysogen level) seen in JL6112 under these growth conditions. To compare the rates of expression with the levels seen in wild-type and λprm240 lysogens, we compared the rate of PRM+ expression at the wild-type lysogen level (panel B), at 2 mM IPTG, with the rate of prm240 expression at the CI level in a λprm240 lysogen. These values were 250 and ∼40 units, respectively; hence, the value for prm240 is 15 to 20% of the value for PRM+, similar to the ratio of observed CI levels in lysogens.
Reporter assays (Fig. 3 and Fig. S3 in the supplemental material) showed several notable features. First, prm240 was somewhat weaker at 37°C than at 30°C (Fig. 3C), suggesting that the CI level would be lower at 37°C, consistent with the higher rate of phage production at 37°C. Wild-type PRM was less responsive to the temperature, showing somewhat higher expression levels at 30°C than at 37°C only at low levels of CI, at which positive autoregulation should be occurring (see Fig. S3 in the supplemental material).
Second, prm240 was markedly weaker in LB medium than in M9 medium (Fig. 3), both at 30°C and at 37°C. The WT promoter was less affected by the change in growth medium (Fig. S3 in the supplemental material). Though the mechanistic basis for the effects of temperature and growth rate on the strength of prm240 is not understood, these effects correlate well with the rates of λprm240 phage production in response to these changes in conditions.
Third, prm240 was much weaker than WT under all conditions. In M9 medium at 30°C, its strength was about 20% of that of wild-type PRM. In LB medium, it was much weaker yet. In LB medium, the observed values fit reasonably well with the observed levels of CI in the λprm240 lysogen (see Fig. 3 legend).
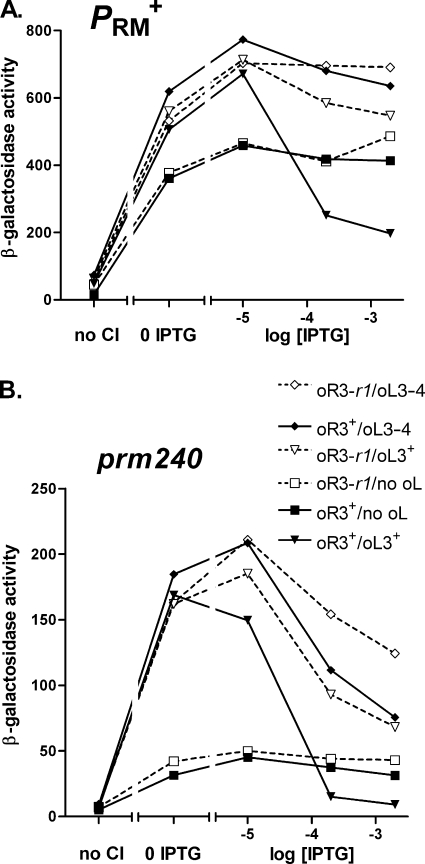
We next tested whether regulation of prm240 resembled that of PRM+. As shown above (Fig. 3), CI stimulates prm240, as with the wild type. To test if prm240 was subject to negative autoregulation and to stimulation by looping, we used reporter constructs (Fig. 3A, top and bottom) with two other alleles of the OL region—one which has only the Timm terminator and which cannot support looping, and one which has the OL region but with four mutations in OL3, abolishing CI binding to that site. For each allele of OL, reporters with OR3+ and the OR3-r1 mutation (to reduce negative autoregulation) were made; the assay was done in M9 medium at 30°C to maximize the activity of prm240.
In strains with PRM+, the presence of OL stimulated expression at low CI levels (Fig. 4A) and led to repression at high levels. Repression was largely prevented in the presence of both of the OR3-r1 and OL3 mutations and weakened by each mutation individually. This pattern, and the magnitude of the stimulation, was expected from previous work (1, 17), although this work did not use an uncoupled assay system in conjunction with the region upstream of OL as used here.
FIG. 4.
Positive and negative autoregulation of PRM+ and prm240. Cells were grown at 30°C in M9 glucose; experiments were otherwise as in Fig. 3. Data for PRM+ (A) and prm240 (B) were obtained in separate experiments. For each promoter, six reporter strains were used, which differed in the alleles of OR3 and OL present, as indicated. Symbols are the same for both panels.
Strikingly, prm240 was stimulated by looping to a substantially greater degree (Fig. 4B) than was PRM+. A large stimulation was also seen in cells grown in LB medium (data not shown), but its magnitude was hard to assess due to the low value in the absence of looping. At higher CI levels, negative autoregulation was observed and was more complete than with WT PRM. This effect was less pronounced when OR3 or OL3 was mutated and still less so when both were mutated, as seen with the WT (1), but some residual repression was seen. Hence, though regulation of prm240 is qualitatively similar to that of PRM+, quantitative differences are seen (see Discussion).
DISCUSSION
Estimate of switching rate for the WT.
We found a low but detectable number of wild-type free phage in cultures of ΔrecA (λ+) lysogens. There are several difficulties in estimating the rate of switching to the lytic pathway. First, recA mutant cultures include a sizable fraction of dead cells (11); it is unknown if a cell that cannot form a colony can switch and/or support phage growth. Presumably the number of cells able to switch lies between the viable cell titers measured for this strain and for its recA+ counterpart, a value ∼2-fold higher in our strains. Second, the burst size of cells that have switched is not known. Possibly it is lower than observed after UV irradiation or infection, since residual CI may partially repress the lytic promoters (54). However, we have found (unpublished data) that in JL5932, the recA+ counterpart of JL5904, the burst size of cells given a low UV dose is substantially larger than reported (54), suggesting that the effect of residual CI is not large in our strains, at least after UV induction. Third, the burst size likely varies widely after individual switching events. Wide variability in individual burst sizes has been observed with lytic phages after single (16) or multiple (30) infection and with λ after multiple infection (33); since our estimates involved only a few switching events, this adds another level of uncertainty. Finally, the switching rate is so low that probably only a few events occurred in the cultures we studied, adding some statistical uncertainty.
With these caveats, we can make a rough estimate of switching rates, if we assume that the number of cells that can switch equals the number of viable cells and that the average burst size is 100 phage per switching event. We observed ∼4 free wild-type phage/ml culture at 5 × 107 viable cells/ml, or ∼2 switching events in a 50-ml culture. These phage represent the progeny of cells that had switched to the lytic state prior to 1 h before, a time at which the cultures contained ∼109 cells. Accordingly, of 109 cells arising by cell division about two had switched, i.e., there were ∼2 × 10−9 switching events per cell division. Given all the uncertainties, a conservative estimate is that the switching rate is probably <10−8 per cell division.
This number is so low that its exact value is not very meaningful. It is lower than the mutation rate for an average gene. It is unclear whether wild-type λ was released by cells that were wild type except for the recA mutation, since we cannot recover these cells. A few mutants with an elevated but still low switching rate are known (51). Perhaps host mutants exist with a greatly enhanced switching rate but have not been analyzed because they are lethal. We conclude that the lysogenic state is extremely stable and that the rate of switching is so low that a precise measurement would be difficult.
Evolution of stability.
Why does λ have such a stable lysogenic state? Evolution almost certainly did not operate on recA mutant lysogens. In a recA+ host, RecA-dependent spontaneous switching occurs at a much higher rate (see introduction), so there is not an obvious need for the lysogenic state to have such a high intrinsic stability.
We surmise that the stability is tuned to maximize the overall reproductive rate at which phage genomes increase in the environment, whether as free phage or as prophages. The level of SOS activation varies markedly; in addition to the low level of spontaneous switching, the SOS system gives efficient induction after a large dose of DNA damage and an intermediate rate of phage production (the “subinduced” state) in response to low doses of DNA damage, whether applied in a single dose by UV (8) or by chronic treatment with agents such as mitomycin C (see reference 35).
We do not study λ or its host in their natural environments (27, 53), and it is unclear how the level of SOS activation varies in such environments. Since the colon is almost anaerobic (28), there might be less oxidative damage to DNA and less sporadic activation of the SOS system. Conversely, the presence of DNA-damaging agents from the diet (e.g., aflatoxins) or produced by other gut flora might increase the level of SOS activation on occasion. In sum, we do not know how to “integrate” the selective pressures, or the level of SOS induction, operating in the likely range of natural environments, as would be needed to estimate how changes in stability of the circuit would influence the overall rate of phage genome production.
At the mechanistic level, mutations can modulate both the intrinsic stability of the circuitry (see Results) and the rate of CI cleavage (13, 24, 25) (our unpublished data). Different combinations of these two features would likely give essentially the same rate of phage production over the naturally occurring range of SOS-inducing treatments. In particular, a circuit that is less stable might be combined with a lower rate of CI cleavage. Nonetheless, several wild phage isolates with λ immunity specificity have the same regulatory circuitry in the immunity region as λ (15), suggesting that covariation in stability and cleavage rate is not frequent in extant λ immunity phages.
Selection might also operate on the host. Many lambdoid phages carry genes that confer a selective advantage to the host and express these genes, either in the lysogenic state (lysogenic conversion) or after spontaneous induction (38). Lambdoid phages can be considered a single species, with assortment of functional modules on an evolutionary time scale (32). Hence, the immunity region of λ, which is also carried on other phages (15), is likely associated at times with genes that confer a selective advantage on the host. This would select for a highly stable lysogenic state.
Switching threshold.
We define the switching threshold as that level of CI at which switching becomes likely. Our data suggest that its value for λprm240 is roughly 25% of the level of CI in a wild-type λ lysogen, a value measured at ∼250 monomers of CI per cell growing in rich medium (49). That is, the switching threshold for this mutant is in the range of ∼60 molecules per cell. This value is rough, because it represents the average of cells in the population, and generally the cells with the lowest values are most likely to switch. It is known (7) that in the wild type the level of CI varies substantially, and it is reasonable to expect at least this much variation for a λprm240 lysogen, particularly since negative autoregulation would not act to counteract fluctuations (9, 17).
The value for wild-type λ is almost certainly less than this, since with its stronger PRM it can “fight back” to replenish CI more effectively than λprm240 can do. Previous work (8) led to an estimate of 10% of the lysogen level for the switching threshold, based on the amount of CI DNA binding activity found at 30 min after UV, but interpretation of these data involves several assumptions, including knowing the time that cells switch and assuming that all cells switch at the same time.
With this small number of molecules, stochastic events play a large role in determining the fate of the cell (39, 50). That is, there is not a discrete number at which switching invariably occurs; instead, at any given number of CI molecules, there is a certain probability of switching, a probability that increases as the number of CI molecules decreases or the cell size increases. Specific pathways are suggested by the properties of mutants we studied, as we now discuss.
Mechanisms of switching.
Though switching of λprm240 lysogens likely follows a pathway generally similar to that for prophage induction (see introduction), the low level of CI and the lack of an active mechanism (cleavage) for its removal suggest two additional mechanisms, both involving stochastic effects, that may play a role in some λprm240 switching events. First, cell division may reduce abruptly the level of CI in some cells. CI that is free in solution or bound nonspecifically to DNA should partition between progeny cells according to a binomial distribution (50). A cell receiving substantially less than half the CI might lose positive autoregulation, leading soon to switching. This mechanism could not operate during SOS-mediated prophage induction; cell division is blocked during that process by the SulA protein, which is expressed at high levels by the SOS system.
Second, DNA replication may also play a role in switching of λprm240. A cell growing in rich medium usually has four copies of the region where λ integrates (45); hence a lysogen has four prophages, each with six CI binding sites. If four of these sites on each prophage (all but OR3 and OL3) are occupied by CI dimers, 32 CI monomers, roughly half the number present at the switching threshold, would be bound specifically as dimers. When these prophages are replicated, two events might contribute to switching by derepressing the lytic promoters. When the replication fork passes the prophage, the number of binding sites doubles. Since initiations at oriC are somewhat synchronized (57), replication of the prophage may occur at about the same time on all four chromosomes, and there may not be enough CI to bind fully all the operators. In addition, bound CI will dissociate from the DNA, at least transiently, during passage of the replication fork, possibly leading to some expression of lytic promoters before CI can rebind. Hence, near the switching threshold the mechanics of DNA replication may favor switching, particularly in combination with the potential for unequal CI partitioning upon cell division as discussed above. Again, this mechanism would likely not operate after DNA damage, since the forks are stalled at lesions and would traverse the prophage only in a small fraction of cells. In our view, both the above events (binomial partitioning and effects of DNA replication) should be included in stochastic models of cellular processes.
The effects of secondary mutations on the stability of λprm240 help provide insight into the mechanisms of switching. Mutations in OR1 and OR2 destabilized λprm240, an effect likely resulting from derepression of the lytic promoters at somewhat higher levels of CI; that is, the switching threshold was probably higher. In contrast, mutations eliminating negative autoregulation stabilized λprm240. They presumably led to somewhat higher CI levels, making expression of lytic promoters somewhat less likely.
More puzzling is the finding that cro-z8, a mutation leading to lower levels of Cro protein, reduced only modestly the switching rate of λprm240. Two lines of evidence suggested that this mutation would have a large stabilizing effect. First, it was isolated (see Fig. S2 in the supplemental material) from a mutant phage that could not form stable lysogens, acting as a suppressor that allowed lysogenization. Second, λcI Y210N, with a mutation that eliminates cooperative CI binding, cannot lysogenize (6); we found here that the cI Y210N cro-z8 double mutant formed turbid plaques and displayed the unstable lysogen phenotype, so that cro-z8 suppressed the lysogenization defect. Hence, in both cases, cro-z8 shifts the balance toward the lysogenic state.
We suggest several possibilities for the modest effect of cro-z8 on the stability of λprm240. Perhaps the cro-z8 mutation provides adequate levels of Cro to give some repression of PRM. Alternatively, Cro-mediated repression of PRM might not be important for switching of λprm240, in apparent contrast to the effects of cro mutations in the two contexts just mentioned. One likely difference is that these phages carried PRM+, while cells containing prm240 should have far lower levels of CI. Some switching events might occur as a result of stochastic processes based on cell division or DNA replication, and Cro-mediated repression of PRM might not be important in these cases. The finding (Table 5) that cro-z8 has a smaller effect at higher rates of switching is consistent with this suggestion. A final possibility is that, despite its low average expression rate, cro-z8 may allow wild-type levels of Cro in a small fraction of cells, making enough Cro to support switching in those cells. Perhaps the promoter is expressed in bursts, as observed for other promoters (26), or the secondary structure of the mutant mRNA may allow it to have switch-like behavior, alternating between translationally active and inactive forms.
Properties of the prm240 promoter.
We found that prm240 was regulated in response to CI similarly to PRM+ (Fig. 3 and 4), with two quantitative differences. First, the degree of stimulation by looping was greater. This finding suggests that, whatever the mechanism by which looping stimulates PRM, it is substantially more effective with prm240, and further analysis of prm240 may help determine this mechanism. Second, prm240 was more sensitive to negative autoregulation than WT PRM (Fig. 4). This pattern suggests that CI can bind more effectively after initiation of transcription on the mutant promoter than on its wild-type counterpart. A plausible mechanism is that CI can compete more successfully with RNA polymerase for binding to the weaker promoter; it is known that the primary effect of prm116, another PRM down-mutation affecting this same position in the −35 region, is to weaken RNA polymerase binding (55).
Substantial repression of prm240 was observed (Fig. 4) in the presence of the OR3-r1 mutation, which weakens CI binding to OR3 ∼100-fold (17, 52). We expect a less severe effect on CI binding to the mutated site in the context of a dodecamer, since multiple weak interactions in the complex will partially compensate (21). Even with the OR3-r1 OL3 template, with no specific binding to OL3, some repression remained, suggesting that a repressive complex could form in the absence of specific CI-OL3 interactions; again, multiple cooperative interactions among the components, and residual nonspecific binding of CI to OL3 or a nearby sequence, might make this possible. Hence, the finding that no repression is observed on this template with WT PRM implies that RNA polymerase is bound nearly all the time to WT PRM; otherwise, the same repressive complex could form and confer some repression.
Properties of λprm240.
The stability of λprm240 lysogens was markedly responsive to temperature and growth rate. Several findings suggest that these responses reflect a more general aspect of cell and/or viral physiology. First, other PRM mutations conferred similar behavior (see Results). Second, another mutant, λJL188 (see Table 1), also exhibited the unstable lysogen phenotype. It has four mutations in the OR1 site, which should affect the binding of CI and Cro and the strength of the PR promoter. Hence, these conditional responses can arise from changes in other cis-acting sites. Third, free phage levels made by a recA mutant (λOR323) lysogen were ∼200-fold lower when cells were grown in minimal medium (unpublished data), though the actual difference is likely less than this, due to instability of λ virions in this medium (see Results). Finally, preliminary data suggest that the set point (see reference 42) for UV induction of a recA+ (λOR323) lysogen is markedly higher in minimal medium than in LB medium. In sum, the responses of λprm240 to growth conditions are not specific to the mutation in that phage. We speculate that the λ circuitry has evolved to fine-tune the probability of induction in response to the growth conditions.
Why does prm240 appear so frequently? We found it in every culture examined. It is likely a hot spot for mutation. In addition, λprm240 lysogens, once arisen, have another property that should contribute to the abundance of free mutant phage. If we consider the relationship between the switching rate of various mutants and the resulting titer of phage, intuitively a mutant with an intermediate switching rate should yield the highest titer of phage. If the rate is zero (as is nearly so with WT), no free phage should result. If a mutant always switches as a result of a newly arising cI mutation, the cell would give no progeny, and one might obtain ∼5 to 10 bursts in the lysate, yielding ∼500 to 1,000 phage. For lysogens with switching rates between these extreme values, some mutant cells would switch and some would divide to give two daughter cells. Hence, the number of cells capable of releasing free phage would continue to increase, as would the number of free phage. This increase should occur in the presence of a large excess of JL5904, just as is observed with the λprm240 lysogen by itself (Fig. 2). Hence, there should be an optimal switching rate giving the highest titer of free phage.
We verified this expectation by simulating the relationship between switching rate and total phage titer, as described in the supplemental material. This simulation also showed that the optimum switching rate depended on the length of time the culture was grown, becoming progressively lower with longer growth times (see Fig. S4 in the supplemental material). Hence, it is plausible that a mutant could exist with an optimum switching rate. Though it is unclear whether prm240 is such a mutant, the use of λprm240 derivatives with altered degrees of stability should enable tests of this expectation.
Supplementary Material
Acknowledgments
We are grateful to Meadow Anderson, Adam Arkin, John Clark, Carol Dieckmann, Telsa Mittelmeier, and Bruce Patterson for helpful discussions; to Andrew Capaldi, Carol Dieckmann, Don Court, Ian Dodd, Gary Gussin, Telsa Mittelmeier, and Lynn Thomason for comments on the manuscript; and to Meadow Anderson and Haw Yang for communicating results before publication.
This work was supported by grant GM24178 from the National Institutes of Health.
Footnotes
Published ahead of print on 24 September 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anderson, L. M., and H. Yang. 2008. DNA looping can enhance lysogenic CI transcription in phage lambda. Proc. Natl. Acad. Sci. U. S. A. 105:5827-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, L. M., and H. Yang. 2008. A simplified model for lysogenic regulation through DNA looping. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2008:607-610. [DOI] [PubMed] [Google Scholar]
- 3.Atsumi, S., and J. W. Little. 2006. Role of the lytic repressor in prophage induction of phage λ as analyzed by a module-replacement approach. Proc. Natl. Acad. Sci. U. S. A. 103:4558-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atsumi, S., and J. W. Little. 2004. Regulatory circuit design and evolution using phage λ. Genes Dev. 18:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aurell, E., S. Brown, J. Johanson, and K. Sneppen. 2002. Stability puzzles in phage λ. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 65:051914. [DOI] [PubMed] [Google Scholar]
- 6.Babić, A. C., and J. W. Little. 2007. Cooperative DNA binding by CI repressor is dispensable in a phage λ variant. Proc. Natl. Acad. Sci. U. S. A. 104:17741-17746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baek, K., S. Svenningsen, H. Eisen, K. Sneppen, and S. Brown. 2003. Single-cell analysis of λ immunity regulation. J. Mol. Biol. 334:363-372. [DOI] [PubMed] [Google Scholar]
- 8.Bailone, A., A. Levine, and R. Devoret. 1979. Inactivation of prophage λ repressor in vivo. J. Mol. Biol. 131:553-572. [DOI] [PubMed] [Google Scholar]
- 9.Becskei, A., and L. Serrano. 2000. Engineering stability in gene networks by autoregulation. Nature 405:590-593. [DOI] [PubMed] [Google Scholar]
- 10.Brooks, K., and A. J. Clark. 1967. Behavior of λ bacteriophage in a recombination deficient strain of E. coli. Virology 1:283-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Capaldo-Kimball, F., and S. D. Barbour. 1971. Involvement of recombination genes in growth and viability of Escherichia coli K-12. J. Bacteriol. 106:204-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Churchward, G., D. Belin, and Y. Nagamine. 1984. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31:165-171. [DOI] [PubMed] [Google Scholar]
- 13.Cohen, S., B. J. Knoll, J. W. Little, and D. W. Mount. 1981. Preferential cleavage of phage λ repressor monomers by recA protease. Nature 294:182-184. [DOI] [PubMed] [Google Scholar]
- 14.Cox, M. M., M. F. Goodman, K. N. Kreuzer, D. J. Sherratt, S. J. Sandler, and K. J. Marians. 2000. The importance of repairing stalled replication forks. Nature 404:37-41. [DOI] [PubMed] [Google Scholar]
- 15.Degnan, P. H., C. B. Michalowski, A. C. Babić, M. H. J. Cordes, and J. W. Little. 2007. Conservation and diversity in the immunity regions of wild phages with the immunity specificity of phage λ. Mol. Microbiol. 64:232-244. [DOI] [PubMed] [Google Scholar]
- 16.Delbrück, M. 1945. The burst size distribution in the growth of bacterial viruses (bacteriophages). J. Bacteriol. 50:131-135. [DOI] [PubMed] [Google Scholar]
- 17.Dodd, I. B., A. J. Perkins, D. Tsemitsidis, and J. B. Egan. 2001. Octamerization of λ CI repressor is needed for effective repression of PRM and efficient switching from lysogeny. Genes Dev. 15:3013-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodd, I. B., K. E. Shearwin, A. J. Perkins, T. Burr, A. Hochschild, and J. B. Egan. 2004. Cooperativity in long-range gene regulation by the λ CI repressor. Genes Dev. 18:344-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrell, J. E., Jr. 2002. Self-perpetuating states in signal transduction: positive feedback, double-negative feedback and bistability. Curr. Opin. Cell Biol. 14:140-148. [DOI] [PubMed] [Google Scholar]
- 20.Folkmanis, A., W. Maltzman, P. Mellon, A. Skalka, and H. Echols. 1977. The essential role of the cro gene in lytic development by bacteriophage λ. Virology 81:352-362. [DOI] [PubMed] [Google Scholar]
- 21.Frankel, A. D., and P. S. Kim. 1991. Modular structure of transcription factors: implications for gene regulation. Cell 65:717-719. [DOI] [PubMed] [Google Scholar]
- 22.Giese, K. C., C. B. Michalowski, and J. W. Little. 2008. RecA-dependent cleavage of LexA dimers. J. Mol. Biol. 377:148-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gimble, F. S., and R. T. Sauer. 1985. Mutations in bacteriophage λ repressor that prevent RecA-mediated cleavage. J. Bacteriol. 162:147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gimble, F. S., and R. T. Sauer. 1986. λ repressor inactivation: properties of purified Ind− proteins in the autodigestion and RecA-mediated cleavage reactions. J. Mol. Biol. 192:39-47. [DOI] [PubMed] [Google Scholar]
- 25.Gimble, F. S., and R. T. Sauer. 1989. λ repressor mutants that are better substrates for RecA-mediated cleavage. J. Mol. Biol. 206:29-39. [DOI] [PubMed] [Google Scholar]
- 26.Golding, I., J. Paulsson, S. M. Zawilski, and E. C. Cox. 2005. Real-time kinetics of gene activity in individual bacteria. Cell 123:1025-1036. [DOI] [PubMed] [Google Scholar]
- 27.Hartl, D. L., and D. E. Dykhuizen. 1984. The population genetics of Escherichia coli. Annu. Rev. Genet. 18:31-68. [DOI] [PubMed] [Google Scholar]
- 28.He, G., R. A. Shankar, M. Chzhan, A. Samouilov, P. Kuppusamy, and J. L. Zweier. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. U. S. A. 96:4586-4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hendrix, R. W., and R. L. Duda. 1992. Bacteriophage λPaPa: not the mother of all λ phages. Science 258:1145-1148. [DOI] [PubMed] [Google Scholar]
- 30.Hershey, A. D., and R. Rotman. 1949. Genetic recombination between host-range and plaque-type mutants of bacteriophage in single bacterial cells. Genetics 34:44-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain, D., B. E. Nickels, A. Hochschild, and S. A. Darst. 2004. Structure of a ternary transcription activation complex. Mol. Cell 13:45-53. [DOI] [PubMed] [Google Scholar]
- 32.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 33.Kaiser, A. D. 1955. A genetic study of the temperate coliphage λ. Virology 1:424-443. [DOI] [PubMed] [Google Scholar]
- 34.Little, J. W. 1993. LexA cleavage and other self-processing reactions. J. Bacteriol. 175:4943-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Little, J. W. 1983. The SOS regulatory system: control of its state by the level of RecA protease. J. Mol. Biol. 167:791-808. [DOI] [PubMed] [Google Scholar]
- 36.Little, J. W., and D. W. Mount. 1982. The SOS regulatory system of Escherichia coli. Cell 29:11-22. [DOI] [PubMed] [Google Scholar]
- 37.Little, J. W., D. P. Shepley, and D. W. Wert. 1999. Robustness of a gene regulatory circuit. EMBO J. 18:4299-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livny, J., and D. I. Friedman. 2004. Characterizing spontaneous induction of Stx encoding phages using a selectable reporter system. Mol. Microbiol. 51:1691-1704. [DOI] [PubMed] [Google Scholar]
- 39.McAdams, H. H., and A. Arkin. 1997. Stochastic mechanisms in gene expression. Proc. Natl. Acad. Sci. U. S. A. 94:814-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCool, J. D., E. Long, J. F. Petrosino, H. A. Sandler, S. M. Rosenberg, and S. J. Sandler. 2004. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol. Microbiol. 53:1343-1357. [DOI] [PubMed] [Google Scholar]
- 41.Michalowski, C. B., and J. W. Little. 2005. Positive autoregulation of cI is a dispensable feature of the phage λ gene regulatory circuitry. J. Bacteriol. 187:6430-6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michalowski, C. B., M. D. Short, and J. W. Little. 2004. Sequence tolerance of the phage λ PRM promoter: implications for evolution of gene regulatory circuitry. J. Bacteriol. 186:7988-7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 44.Mount, D. W. 1976. A method for the isolation of phage mutants altered in their response to lysogenic induction. Mol. Gen. Genet. 145:165-167. [DOI] [PubMed] [Google Scholar]
- 45.Nielsen, H. J., B. Youngren, F. G. Hansen, and S. Austin. 2007. Dynamics of Escherichia coli chromosome segregation during multifork replication. J. Bacteriol. 189:8660-8666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pakula, A. A., V. B. Young, and R. T. Sauer. 1986. Bacteriophage lambda cro mutations: effects on activity and intracellular degradation. Proc. Natl. Acad. Sci. U. S. A. 83:8829-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pennington, J. M., and S. M. Rosenberg. 2007. Spontaneous DNA breakage in single living Escherichia coli cells. Nat. Genet. 39:797-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Powell, B. S., M. P. Rivas, D. L. Court, Y. Nakamura, and C. L. Turnbough, Jr. 1994. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 22:5765-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reichardt, L., and A. D. Kaiser. 1971. Control of λ repressor synthesis. Proc. Natl. Acad. Sci. U. S. A. 68:2185-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenfeld, N., J. W. Young, U. Alon, P. S. Swain, and M. B. Elowitz. 2005. Gene regulation at the single-cell level. Science 307:1962-1965. [DOI] [PubMed] [Google Scholar]
- 51.Rozanov, D. V., R. D'Ari, and S. P. Sineoky. 1998. RecA-independent pathways of lambdoid prophage induction in Escherichia coli. J. Bacteriol. 180:6306-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarai, A., and Y. Takeda. 1989. λ repressor recognizes the approximately 2-fold symmetric half-operator sequences asymmetrically. Proc. Natl. Acad. Sci. U. S. A. 86:6513-6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Savageau, M. A. 1983. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am. Nat. 122:732-744. [Google Scholar]
- 54.Schubert, R. A., I. B. Dodd, J. B. Egan, and K. E. Shearwin. 2007. Cro's role in the CI-Cro bistable switch is critical for λ's transition from lysogeny to lytic development. Genes Dev. 21:2461-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shih, M. C., and G. N. Gussin. 1983. Mutations affecting two different steps in transcription initiation at the phage λ PRM promoter. Proc. Natl. Acad. Sci. U. S. A. 80:496-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 57.Skarstad, K., E. Boye, and H. B. Steen. 1986. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 5:1711-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slilaty, S. N., and J. W. Little. 1987. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc. Natl. Acad. Sci. U. S. A. 84:3987-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stayrook, S., P. Jaru-Ampornpan, J. Ni, A. Hochschild, and M. Lewis. 2008. Crystal structure of the λ repressor and a model for pairwise cooperative operator binding. Nature 452:1022-1025. [DOI] [PubMed] [Google Scholar]
- 60.Takeda, Y., A. Sarai, and V. M. Rivera. 1989. Analysis of the sequence-specific interactions between Cro repressor and operator DNA by systematic base substitution experiments. Proc. Natl. Acad. Sci. U. S. A. 86:439-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thirion, J. P., and M. Hofnung. 1972. On some genetic aspects of phage λ resistance in E. coli K12. Genetics 71:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vieira, J., and J. Messing. 1987. Production of single-stranded plasmid DNA. Methods Enzymol. 153:3-11. [DOI] [PubMed] [Google Scholar]
- 63.Whipple, F. W., N. H. Kuldell, L. A. Cheatham, and A. Hochschild. 1994. Specificity determinants for the interaction of λ repressor and P22 repressor dimers. Genes Dev. 8:1212-1223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.