Abstract
The ethanolamine-utilizing bacterial microcompartment (Eut-BMC) of Escherichia coli is a polyhedral organelle that harbors specific enzymes for the catabolic degradation of ethanolamine. The compartment is composed of a proteinaceous shell structure that maintains a highly specialized environment for the biochemical reactions inside. Recent structural investigations have revealed hexagonal assemblies of shell proteins that form a tightly packed two-dimensional lattice that is likely to function as a selectively permeable protein membrane, wherein small channels are thought to permit controlled exchange of specific solutes. Here, we show with two nonisomorphous crystal structures that EutM also forms a two-dimensional protein membrane. As its architecture is highly similar to the membrane structure of EutL, it is likely that the structure represents a physiologically relevant form. Thus far, of all Eut proteins, only EutM and EutL have been shown to form such proteinaceous membranes. Despite their similar architectures, however, both proteins exhibit dramatically different pore structures. In contrast to EutL, the pore of EutM appears to be positively charged, indicating specificity for different solutes. Furthermore, we also show that the central pore structure of the EutL shell protein can be triggered to open specifically upon exposure to zinc ions, suggesting a specific gating mechanism.
Bacterial microcompartments are subcellular organelles that are found in many prokaryotic organisms (10, 32). In contrast to the lipidic vesicles of many eukaryotic cells, these enclosures are entirely composed of proteins. Recent imaging by electron microscopy revealed capsid-like particles obeying 2-, 3- and 5-fold symmetries that suggest icosahedral symmetry (4, 13, 27). Shell proteins are thought to form a tightly sealed membrane structure that separates the lumen from the cytosol. Similar to the lipidic membranes of vesicles, these proteinaceous membranes have been suggested to provide a selectively permeable solute barrier, wherein specific pores maintain an optimal biochemical environment for the catabolic reactions inside (25).
The ethanolamine-utilizing bacterial microcompartment (Eut-BMC) enables some bacteria to survive on ethanolamine as the sole source for carbon, nitrogen, and energy (25). It is encoded by a 17-gene-containing operon, and homologues of its genes have been identified in Escherichia coli, Salmonella enterica serovar Typhimurium, Mycobacterium tuberculosis, and Clostridium kluyveri among other prokaryotic pathogens (22). Largely based on sequence comparisons, the compartment's outer shell was proposed to be composed of five different shell proteins: Eut-K, -L, -M, -N, and -S, all of which are fairly small proteins that typically consist of about 100 amino acids. Only EutL is about twice the size, with 216 amino acids as a result of two tandemly duplicated shell protein domains (26).
To date, little is known about the composition, architecture, and function of bacterial microcompartments. Recent structural investigations of BMC particles and individual shell proteins, however, have contributed greatly to a basic understanding of BMC architecture. Electron microscopy, for example, has revealed polyhedral shell structures that are composed of a thin layer of proteins. Intriguingly, crystallizations revealed that some shell proteins also assemble into tightly packed two-dimensional arrays that may resemble the facets of the compartments (28). Within an array, these proteins typically assembled into hexamers or trimers (in the case of tandem domain proteins) that exhibited a distinct hexagonal shape. As this geometry was suggested to be of fundamental importance to the microcompartment architecture, we will here refer to it as a “tile” or “tile structure.” While it has not yet been proven directly that the assembly of proteins in the crystals is identical to that of the BMC, their almost seamless two-dimensional packing has been suggested to be of physiological relevance as it could provide an efficient barrier to prevent leakage of toxic by-products into the cytoplasm (4, 25). Overall, however, it is not understood how the various shell proteins assemble to form the polyhedral structure while maintaining an efficiently tight seal. In particular, the interactions among the shell proteins and their arrangements within facets, edges, and vertices have remained elusive.
In the study presented here, we demonstrate for the first time that the shell protein EutM is also able to form tightly packed two-dimensional arrays. With two independently determined crystal structures, we show that its protein array closely resembled that of EutL and other carboxysomal proteins. As a result, we hypothesize that this assembly represents a physiologically relevant form. Both crystal forms also revealed the C-terminal tail of the protein, which is proposed to serve as a potential interaction site with other factors.
Furthermore, we show that the pore structure of EutL can be triggered to open upon exposure to specific solutes. A first structure of EutL was previously determined in our laboratory, and it revealed three water-filled pores per tile (26). Interestingly, its structure consisted of two tandemly repeated shell protein domains, which assembled into an almost perfectly shaped hexagonal structure. This architectural feature was recently also found in shell proteins of other microcompartments (11, 20). Each of the pores of an EutL tile was coated with acidic residues, which indicated a possible pathway for positively charged molecules such as ethanolamine. Inspection of the structure also suggested specific metal binding sites on its surface. In order to verify this idea, we performed systematic soaking studies of the crystals with selected divalent metals. Surprisingly, we found that zinc ions bound to the protein specifically not at the suspected sites but at different sites that caused a dramatic opening of a central pore. This unprecedented observation of a specifically triggered pore opening is consistent with another previous observation (30) and may point to a mechanism for regulation of permeability.
MATERIALS AND METHODS
Cloning.
The genes for Eut-K, -M, -N, and -S were cloned by PCR directly from the E. coli genome. The resulting products were cloned into a pET101 vector (Invitrogen) according to the company's protocol and were subsequently transformed into chemically competent TOP10 cells (Invitrogen). Plasmids of clones that were positively identified by PCR were transformed into BL21-AI cells for overexpression. Fifty milliliters of LB medium overnight culture was used to inoculate 2 liters of LB medium. Cells were induced with 1 mM isopropyl-β-d-thiogalactopyranoside at an optical density at 600 nm (OD600) of 0.6. Induction was carried out for 3 h before centrifugation at 5,000 rpm in a Beckman J1 rotor. The resulting cell paste was subjected to lysis by French press in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF). Following a high-speed centrifugation at 12,000 rpm (Beckmann JA20 rotor), the protein was isolated from the supernatant by standard nickel affinity chromatography (Novagen).
Crystallography.
Crystals of EutL were obtained as described previously (24). In order to enable soaking of various metal compounds, crystals were washed repeatedly in deionized water for at least 5 min. Soaking was carried out with 1 M ZnCl2, CoCl2, MgCl2, and NiCl2, respectively, for about 1 min prior to flash-freezing and data collection. Control data sets were obtained with crystals that were either extensively washed with deionized water or washed and soaked with 10 mM ethanolamine. Data collections and refinements from at least three different crystals of each soak were obtained. Crystal form I of EutM was crystallized with 100 mM sodium dihydrogen phosphate, 100 mM potassium dihydrogen phosphate, 100 mM MES (morpholineethanesulfonic acid) buffer (pH 6.5), and 2 M sodium chloride. Crystals typically appeared after 48 h. Attempts to cocrystallize EutM and EutL were carried out with a 1:1 mixture of both proteins at a 1 mg/ml concentration each. As each of the proteins crystallized under nearly identical conditions individually, a refined matrix screen around these conditions was performed with both proteins. Diffraction-quality crystals were obtained after 4 to 6 days. EutM crystals of form II were obtained with 100 mM sodium acetate (pH 4.6) and 2 M ammonium sulfate. Synchrotron data collections were carried out at SSRL beamline 7-1 equipped with a Quantum ADSC Q315R charge-coupled device (CCD) detector. Additional diffraction data were collected on a Rigaku FRE generator equipped with a Raxis 4++ imaging plate and with a Saturn-92 CCD area detector. Integration and scaling were performed with the programs MOSFLM and XDS (9, 15, 16). Determination of the EutM structures was carried out by molecular replacement with the program MOLREP, using the carboxysomal protein CcmK2 (Protein Data Bank identification no. [PDB ID] 2A1B; residues 2 to 90) as a search model. Programs of CNS version 1.2 and REFMAC (5, 9) were used to refine the structure. Model building was carried out with programs COOT (9) and O. Maskless averaging of the electron density was attempted to improve the densities with the programs of the RAVE package (21) as well as with CNS. Figures 1 to 4 and 6 were prepared with the program PyMol (DeLano, http://www.pymol.org).
FIG. 1.
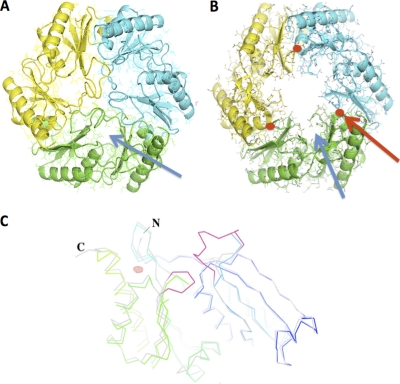
(A to C) Full-atom representation of a EutL trimer (tile) superimposed onto a ribbon diagram. (A) “Closed” conformation; (b) “open” conformation. The backbone atoms of individual monomers are colored in blue, green, and yellow, respectively. The solvent channel of the zinc-free protein is highlighted by the blue arrow. The positions of the zinc atoms are shown in red (red arrow). (C) Ribbon diagram of EutL closed (light gray) and EutL open (rainbow colored with N terminus in blue and C terminus in green) superimposed. The two loop structures that underwent dramatic conformational changes are highlighted in red. In this crystal form, two molecules are contained within the asymmetric unit. The Fo-Fc electron density of the zinc was calculated at a resolution of 2.7 Å (6.0 σ above the mean) and is shown in red.
Determination of protein mass from crystals.
In order to identify the protein species in the crystals, mass determinations were carried out with a PerSeptive Voyager DE matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometer (Perkin-Elmer). Individual crystals were washed extensively in deionized water prior to application onto the specimen holder in the presence of sinapinic acid (in freshly prepared aqueous 30% acetonitrile and 0.1% trifluoroacetic acid). Individual crystals, typically of size 0.2 by 0.2 by 0.02 mm, were selected via the spectrometer's microscope for UV laser-induced ionization. This way, multiple mass determinations of proteins from different individual crystals were obtained. In order to confirm the success of the washing procedure, mass determinations of the washing solutions were confirmed to be devoid of any residual protein in all cases.
RESULTS
Structure of EutL.
Crystallizations and refinements were carried out as described previously (24). The zinc-free structure of EutL (PDB ID 3GFH) was used as an isomorphous replacement model for the structural determination. Structures of the protein that were obtained from water-soaked crystals or from crystals soaked with 10 mM ethanolamine exhibited no conformational changes. While the structures of the NiCl2−, CoCl2−, and MgCl2− soaked crystals also revealed no significant structural changes, all independently collected data sets from crystals that were soaked with ZnCl2 showed dramatic conformational rearrangements (Fig. 1). The best of these, EutL-Zn3, was refined to an R value of 22% and an Rfree value of 26% at a 2.65-Å resolution (Table 1). Calculation of 2Fo-Fc and Fo-Fc electron density maps revealed that both molecules of the asymmetric unit have largely remained the same compared to the search molecule (Fig. 1C). However, dramatic rearrangements of the loops between residues 70 to 79 and 181 to 183 were observed. In both of the molecules of the asymmetric unit, the corresponding density was entirely absent due to thermal or static disorder. Furthermore, the β-strands that connect the loops between residues 176 and 180 and 65 and 69 exhibited altered positions.
TABLE 1.
Crystallographic data and refinement statisticsa
| Data set | Result for: |
||
|---|---|---|---|
| EutL (zinc) | EutM form I | EutM form II | |
| X-ray source (SSRL) | BL 7-1 | BL 7-1 | BL 7-1 |
| Cell dimensions (Å) | a = 67.42, α = 90 | a = 70.0, α = 90 | a = 69.16, α = 90 |
| b = 67.42, β = 90 | b = 149.25, β = 120.1 | b = 69.16, β = 90 | |
| c = 79.60, γ = 120 | c = 70.1, γ = 90 | c = 28.95, γ = 120 | |
| Resolution (Å) (high-resolution bin) | 58.42-2.65 (2.75-2.65) | 21.97-2.7 (2.8-2.7) | 17.29-2.0 (2.1-1.9) |
| Wavelength (Å) | 0.97946 | 0.94946 | 0.97946 |
| Space group | P3 | P21 | P6 |
| No. of reflections (unique) | 11,250 (847) | 56,948 (5418) | 4,477 (737) |
| No. of reflections (observed) | 31,465 (2383) | 117,751 (10775) | 49,593 (8935) |
| R-merge | 0.117 (0.363) | 0.03 (0.084) | 0.077 (0.229) |
| I/σ〈I〉 | 3.9 (1.9) | 19.3 (9.0) | 21.45 (12.1) |
| Completeness (%) | 98.9 (98.3) | 84.1 (77.9) | 81.1 (98) |
| Redundancy | 2.8 (2.8) | 2.1 (1.9) | 11.1 (12.1) |
| Rwork/Rfree | 0.22/0.261 | 0.24/0.33 | 0.224/0.28 |
| RMSD bond angles | 1.97 | 1.99 | 1.99 |
| RMSD bond lengths | 0.022 | 0.022 | 0.021 |
| Ramchandran statistics (%) | |||
| Core | 70.9 | 81.0 | 88.6 |
| Allowed | 28.5 | 19.9 | 11.4 |
| General | 0.6 | 0.1 | 0.0 |
| Disallowed | 0.0 | 0.0 | 0.0 |
| PDB ID | 3MPV | 3MPW | 3MPY |
Rsym = Σ|I − 〈I〉|/Σ〈I〉, where I is the observed intensity and 〈I〉 is the statistically weighted average intensity of multiple symmetry related observation. R-factors: R = Σ‖Fcalc − |Fobs‖/Σ|Fobs|, where Fcalc and Fobs are the calculated and observed structure factors respectively. Rfree (4a) was calculated by the same formula with 7 % of randomly selected reflections. The data sets for EutL and EutM form II exhibited twinning, and refinements were carried out with the program REFMAC. The partial respective twin fractions for EutL (zinc) were 0.56 for twin operator h, k, l and for h, k, −l, 0.44 and those for EutM form I were for h, k, l 0.815 and −0.185 for −h −k, k, −l. As described earlier (8), the degree of twinning varied among different crystals in both cases. Ramachandran statistics were obtained with the program PROCHECK (22a). RMSD, root mean square deviation from ideal geometry. The structure of EutM form I contains 12 EutM chains in the asymmetric unit, and it was collected to relatively low resolution. The six disordered C-terminal tails and His tags may have contributed to the relatively high Rfree value of this structure. As described in the text, data sets from all soaked crystals were obtained in triplicate and occasionally measured at other X-ray sources.
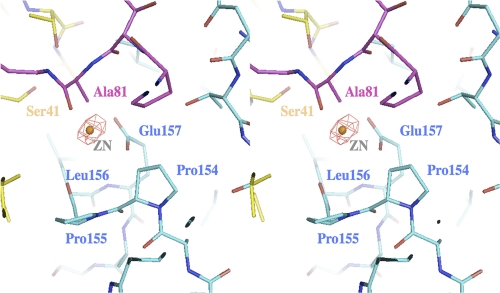
Two major positive electron densities were observed in Fo-Fc deletion maps. With 7 σ above the mean, these spherical densities were interpreted as zinc ions. The first site was found on the 3-fold axis and was coordinated by three Arg97. The second site was located at the amino-terminal end of helix 5 (residues 155 to 169) (Fig. 2). In an unusual constellation, the carboxyl of Glu157 and the carbonyl of Pro155 participated in the coordination of the zinc that resulted in the capping of the helix. Zinc-mediated helix capping that involves backbone carbonyls has been reported previously and support this interpretation (23). Interestingly, superposition of the zinc-free structure of EutL showed that the position of Ala81 would directly interfere with the position of the zinc. As Ala81 is part of the loop structure that occludes the center of the EutL tile, the binding of zinc may have triggered the substantial conformational rearrangements, causing it to open up (Fig. 2). With an apparent diameter of approximately 12 Å, this new opening was about 5 to 6 times wider than the previously determined pore. As the loops, however, were still present in the channel, albeit not visible, the average diameter was likely to be smaller. In the following discussion, we will refer to the former structure (3GFH) as the “closed” form (despite the presence of three small channels) and the zinc structure as the “open” form.
FIG. 2.
Stereo diagram of the structural environment of the zinc binding site at the interface between two adjacent monomers (highlighted in blue and in yellow). Superimposed onto this structure is the loop conformation (residues 78 to 82 (purple)) of EutL in the absence of zinc. The Fo-Fc electron density was calculated same way as in Fig. 1C. The density was observed at the N-terminal site of helix 4 involving the carboxylate of Glu157 and the backbone oxygen of Pro155. Systematic soaks of the crystals with either water, buffers at various pH values, or other metals revealed that only the addition of zinc chloride resulted in a dramatic opening of the channel. Fo (zinc)-Fo (native) electron densities calculated in the absence of any solvent molecules resulted in similarly strong positive densities at this location.
Structure of EutM.
In crystal form II, molecular replacement was performed with a single peptide chain of CcmK2 that yielded a single clear solution with an initial R value of 41%. In crystal form I, the search was performed with the refined EutM hexamer of crystal form II and resulted in two clear positions within the asymmetric unit (starting R factor of 45%). In both crystal forms, therefore, the proteins formed a hexameric tile structure that was common to shell proteins of other microcompartments (18).
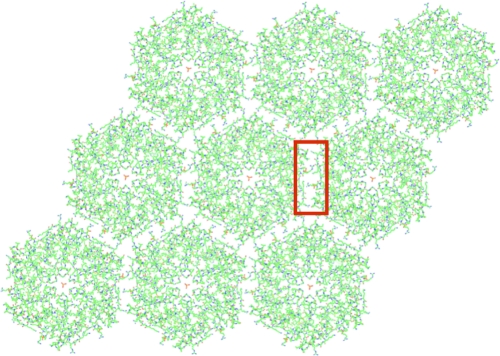
Comparison of the crystal structures of EutM and EutL revealed further striking similarities. In both cases, the protein tiles assembled into a two-dimensional array that extended throughout the crystal (Fig. 3). In comparison to the EutL array, the distance between the tiles was somewhat increased by approximately 3 Å. Whereas in crystal form II a sulfate ion was observed on the 2-fold axis between adjacent tiles, two phosphate ions were positioned off the 2-fold axis in crystal form I. The identity of this ion was inferred from the positively charged environment and the strength of the density. The observation of sulfate ions between the contacting tiles of crystal form II was also found previously in the carboxysomal shell protein CcmK1 (29). Despite the, thus far, unprecedented finding of two differently positioned phosphate ions along the tile's edge in crystal form I, however, the packing of the EutM tiles remained indistinguishable from that of crystal form II.
FIG. 3.
Crystal packing of the EutM monomers into a 2D proteinaceous membrane. Shown is the crystal lattice of crystal form I. Despite the different crystallization conditions and packing interactions, the lateral packing interactions of the two-dimensional lattice of crystal forms I and II remained the same. Also visible is the central phosphate ion trapped in the pore of each tile. The honeycomb-patterned tile packing is thereby similar to the one observed in EutL and other carboxysomal crystal structures, suggesting a common assembly principle. The corresponding interactions between the tiles (within the red rectangle) are highlighted in Fig. 6.
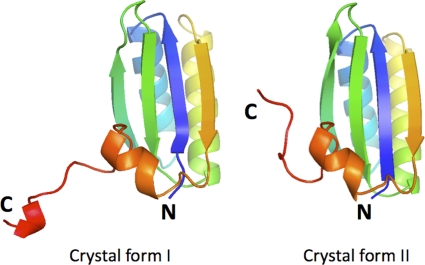
Also consistent with previous observations (29) was the presence of a single phosphate ion (in crystal form I) or sulfate ion (in crystal form II) in the central pore of each tile, which was coordinated to the backbone amide of Gly38. The two EutM structures thus differed majorly only in the way the membranes were stacked on top of each other in the crystal. Whereas in crystal form II, the arrays were stacked on top of each other in a unidirectional fashion, in crystal form I, the arrays were sandwiched on top of each other face-to-face. Interestingly, in crystal form I, the C-terminal extension was revealed to interact with the extensions of the other, non-crystallographically related molecule of the asymmetric unit (Fig. 4). Most likely due to disorder, only three out of the six tails were observed within a single tile. The interactions of the extended tail in this crystal form thus produced a larger distance between the protein arrays in the crystal and hence the much larger unit cell dimension along b. Overall, the electron density of the extensions appeared weak with higher-than-average B values. In crystal form II, on the other hand, this eight-residue long tail extended toward the center of the concave side of the tile and was not involved in any crystal contacts.
FIG. 4.
Ribbon diagram of the respective monomers of EutM in crystal forms I and II. The rainbow coloring is shown with blue at the N terminus and with red at the C terminus. In both cases, the last seven residues adopt a fundamentally different conformation. In crystal form I, however, its structure resembled the corresponding peptide conformation of CcmK2. In all cases, the structure of the His tag was not visible.
Structure of the EutM pore.
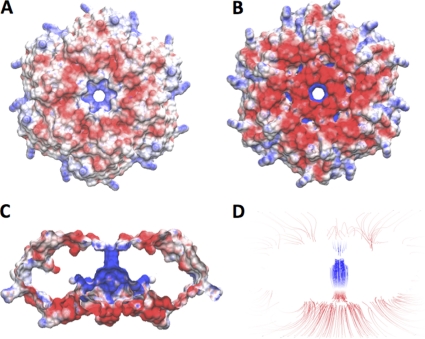
The hexameric arrangement of the proteins exhibits a single central pore. At its bottleneck, which is defined by the distance between the two opposing amide nitrogens of Gly38, the pore measured approximately 8.6 Å in diameter. On either side, the pore was preceded by funnel-shaped entrances, which were decorated with a ring of positively charged residues. Electrostatic surface calculations with the programs APBS (2) and VMD (12) demonstrated a significantly positively charged surface (Fig. 5). The observation of negatively charged ions such as sulfate or phosphate of the respective crystallization buffers in the pore's center is thus consistent with this finding. Overall, the size and the electrostatic environment of the pore dramatically contrasted the pore structure of the EutL tile, which exhibits three strongly acidic channels in the “closed” form.
FIG. 5.
(A to D) Surface representation of the electrostatic charge distribution of a EutM tile of crystal form II. The convex site of the tile (A) exhibits a moderately negatively charged crown (red) with a strongly positively charge in the center leading to the channel (blue). A somewhat stronger negative charge distribution was evident on the concave site of the tile (B), which is largely caused by the C-terminal extension of this crystal form. The strongest charge distribution was observed within the channel (C). The high concentration of positive field lines within the channel is shown in panel D. The positive charge of the channel is likely to provide access for negatively charged solutes to enter or leave the BMC. Electrostatic calculations and surface representations were performed with APBS (2) and VMD (12).
Interactions within the proteinaceous membranes.
Due to the highly similar sizes and shapes of the EutM and EutL tiles, we suspected that both array-forming proteins EutL and EutM might also cross-interact to form a heterogeneous protein membrane. In order to substantiate this idea, we identified the major contacts within the two-dimensional arrays to assess the feasibility for such cross-interactions. In both cases (i.e., within the EutL and EutM arrays), the lateral crystal contacts were dominated by hydrophobic interactions of arginines or lysines, which associated in an antiparallel fashion. In EutM, however, the orientations of these side chains was somewhat angled with respect to the tile edge (Fig. 6).
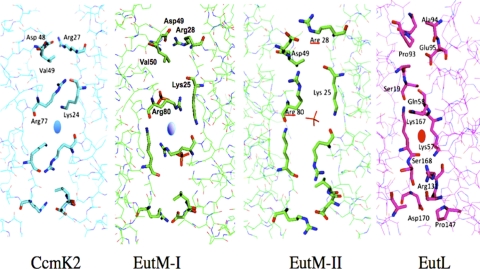
FIG. 6.
Congruent tiling of the protein hexagons within each two-dimensional lattice. The corresponding regions of the proteins CcmK2, EutM (form I and form II) and EutL are shown, with the contacting residues highlighted as a “stick” representation instead of as a “wire.” The respective anions are also shown as “sticks.” The crystal contacts of both CcmK2 and EutM (both crystal forms) are maintained by specific residues that are arranged in a 2-fold symmetrical manner (with the 2-fold axis shown as a blue oval). In crystal form II, a sulfate ion sits on the 2-fold axis. EutL also exhibits large residue side chains such as arginines and lysines in the interface, but at different relative positions, suggesting that the crystal contacts of EutL and EutM may be incompatible for mixed tiling. Furthermore, due to the imperfect sequence repeat of both domains within EutL, only a pseudo-2-fold rotation of the contacting residues is observed (the pseudo-axis is shown as a red oval).
As can also be seen in this figure, the relative positionings of the contacting residues along the respective tile edges did not match. While in EutL, Lys57 and Lys167 interacted across the pseudo-2-fold axis, the hydrophobic interaction between Lys24 and Arg77 was offset from the 2-fold axis in EutM. Moreover, an ionic interaction between Arg28 and Asp49 was observed in EutM that was missing in EutL. Possible implications for this mismatch of interactions are discussed below.
As both proteins were shown to crystallize under almost identical conditions into proteinaceous arrays individually, it was hypothesized that they might also do so in combination. Crystallization trials were therefore screened around the conditions that were previously reported (24) with a 1:1 mixture of both proteins as well as with customized refined matrix screens. Typically, crystals were observed after about 4 days and continued to grow to diffraction quality for about a week. The crystals also displayed a hexagonal morphology, which was almost identical in size and shape to that of the EutM crystals. Using synchrotron radiation, data sets of several crystals were collected to about a 2.2-Å resolution. Determinations of the structures, however, revealed no electron densities that could be attributed to the presence of both proteins within a single crystal. These results were also confirmed by single-crystal mass spectrometry. Numerous larger and smaller crystals were washed extensively with deionized water (EutL and EutM crystals remaining stable under these conditions) to remove all noncrystalline protein. Individual crystals were then mixed with sinapinic acid and subjected to MALDI-TOF mass spectrometry. In all cases, only the mass of EutM was uniquely identified, with no indication of the presence of EutL (see Fig. S1 in the supplemental material).
DISCUSSION
One of the most fascinating aspects of bacterial microcompartments is the formation of a semipermeable membrane structure (8). Similarly to the membranes of lipidic vesicles, these proteinaceous membranes are thought to provide an efficient solute barrier in order to maintain a specific reaction environment within the BMC (25). As the entire compartment is likely to be sealed with respect to specific solutes, the interfaces of contacting edges and vertices are likely to be sealed as well. Consequently, the polyhedral symmetry of the compartment and its semipermeable properties may impose specific architectural requirements on the structural units that compose its shell.
A first compelling model of a carboxysomal microcompartment was put forward by the Yeates laboratory (28). The authors suggested that the triangular facets of the icosahedral compartment are composed of hexagonally shaped tile units and that the 5-fold symmetric vertices may be sealed by specific pentameric shell proteins (7). At present, however, little is known about the specific interactions between the various shell proteins within a compartment. In particular, the composition and stoichiometry of the microcompartment's facets are not understood. Thus far, proteomic interaction databases such as Int-Act or DIP (1, 6, 19) have revealed only a single potential interaction between EutL and EutK. This method, however, has failed to identify other interactions most likely due to the nonstringent growth condition of the tested bacteria.
EutL and EutM exhibit substantially different pore structures that suggest different specific functions. Earlier investigations of EutL have revealed three small acidic channels within a tile that were interpreted as possible pathways for small positively charged solutes to enter the compartment. The observation of the dramatic pore opening in response to zinc binding was surprising as it changed this structure entirely. Interestingly, none of the other metals tested (including the mercury ions that were used to determine the structure) (26) resulted in similar conformational changes.
In contrast to the findings presented here, another recent study of EutL also revealed “closed” and “open” conformations, which, however, were obtained among crystals that were grown under identical conditions (30). At this point, it remains unclear as to why some crystals exhibited only the “open” form and others only the “closed” form of the protein. With the structures of EutL described here, only the zinc-bound form of the protein was repeatedly found to reveal a central opening. The proximity of the zinc binding sites to the flexible loop regions and the resultant conformational changes of the peptide backbone adjacent to the binding site suggest that zinc binding may have stabilized an “open” conformation specifically. Moreover, comparison of both “open” structures revealed significant differences in the surroundings of the gating loop regions that were attributed to the presence of the zinc.
At this point, it is not clear if (or why) zinc may be of any physiological importance to the function of the microcompartment. It seems reasonable to speculate, however, that a continuously flexible (or even open) conformation will not be advantageous for the maintenance of a specialized environment within the BMC. The previously observed “open” conformation (30), as well as the structure presented here, however, may demonstrate an inherent potential of the two loop structures of EutL to rearrange. The zinc-induced opening of the pores may therefore be indicative of another specific, yet-to-be-identified solute (or protein) that may open the pores in a controlled fashion.
As can be seen in Fig. 5 and Fig. S2 in the supplemental material, the electrostatic potential of EutL and EutM increases dramatically toward the center of the pore. Even though the field calculations were performed in the absence of water and other solutes, the strong potential inside the pores may indicate that it may not only filter specific charges and sizes of solutes through the membrane but may also help accumulate and hold specific solutes in proximity to the pore to facilitate passive transport. Given the positive charge of ethanolamine, it seems unlikely that the overall positive charge of the EutM channel is involved in the permeation of this compound. Rather, other reaction species or catabolic by-products such as aldehydes or carbon dioxide are more likely to pass through this channel (25).
Structural investigations by electron microscopy revealed that the carboxysomal shell is rather thin, with a depth of only about 30 to 40 Å (27), which approximately corresponds to the width of a single layer of shell proteins. Furthermore, high-resolution imagery revealed periodicities within the shell structure that closely resembled the spacing found in the 2D arrays of shell protein crystals (17) suggesting that they provide a structural basis. It is thus most intriguing that EutL and EutM formed similarly packed 2D arrays that matched this spacing. In both cases, the assembly of the shell proteins within the 2D arrays fit the geometrical definition of tiling, whereby equally sized and shaped plane figures fill a plane with no overlaps and no gaps. Based on this tile congruence (i.e., the same shapes and sizes of the tiles), heterogeneous assembly of tiles from different shell proteins into a tightly packed array seems possible. This concept of mixed tiling would allow differently functional proteins to be integrated into the compartment's shell structure without necessarily jeopardizing its seal. One of the challenges of research is therefore to elucidate the specific interactions between the different tile species in order to establish the composition of the facets and to identify the mechanisms of assembly.
The two crystal structures of EutM presented here represent the first examples of this protein to assemble into a nearly solid proteinaceous membrane. A recent structural determination of this protein by Tanaka et al. (30) also reported its hexameric tile structure. Despite the similar structure, however, these tiles did not assemble into a two-dimensional array, and its C-terminal tail structure was not resolved. The observations made here suggest, therefore, that EutM and EutL are likely to be major components of the compartment's shell.
Given the differently charged structures of the pores, it appears likely that the pores of EutM and EutL occupy different lateral positions in the compartment's shell. This structural difference of the proteins thus raises the question of whether the membrane of a single facet is predominantly composed of a single species of EutL (or EutM) or a mixture of both. In order to probe for this possibility, we took advantage of the fact that each of the proteins, EutL and EutM, crystallized readily under nearly identical conditions. Our observation, however, that the protein mixture did not cocrystallize suggests that the hexagon edges might be incompatible for mixed congruent tiling. Consistent with this observation, native gel electrophoresis as well as size exclusion chromatography of a EutL-EutM mixture also did not reveal any interactions between the two proteins (data not shown).
Analysis of the respective lateral crystal contacts of EutL and EutM tiles, also, did not support a mixed-tiling model that includes both proteins. Figure 6 shows the crystal contacts along a single edge of a tile. Whereas the interactions across the EutM tiles were 2-fold symmetric, the interactions across the EutL tiles were pseudosymmetric due to the imperfect repeat of its sequence (26). In both cases, the interactions were maintained by salt bridges and by hydrophobic interactions of large side chains such as lysine and arginine. Another significant difference was the relative spacing of the interacting residues along the tile's edge. As can be seen in Fig. 6, the positioning of the long aliphatic lysine residues of EutL was near the center of each edge. In EutM, however, the long antiparallel Arg7 and Lys24 side chains form similarly structured hydrophobic interactions—but which are offset from the 2-fold axis. In other words, if an interaction between a EutM tile edge and a EutL tile were modeled by superposition, Lys24 of EutM would be forced interact with Ser19 of EutL and Asp48 of EutM would be placed in proximity to Glu95 of EutL—all of which would result in weak or unfavorable interactions.
An alternative possibility would be to arrange the faces of the EutL and EutM tiles in opposite orientations: e.g., the concave side of EutL alternates with the convex side of EutM. Such an unusual arrangement has been observed only once, thus far, with the carboxysomal protein CcmK4 (18). In addition to the resulting relatively large void spaces between nonidentical tiles, which were also present in the modeled EutL-EutM interface, this model would also project weak interactions (as judged by the contacting residues and surface areas). It is therefore questionable if such an arrangement is of biological relevance.
The cocrystallization experiments as well as the model-building studies may therefore suggest that, despite their similar size and shape, the protein tiles may only assemble into a proteinaceous membrane if they exhbit highly similar and compatible contact surfaces.
These findings may thus have direct implications on the current model of the Eut compartment's shell structure. Given the lack of affinity, however, it is doubtful that a single facet is majorly composed of an interlaced mixture of the two proteins. Rather, other proteins may be required to integrate both proteins into the compartment's shell. Based on the overall similarities, however, one may speculate about whether CcmK2 could possibly replace EutM within the microcompartment's structure—or alternatively—whether both of these proteins can interact directly. As the tile dimensions and the contacting tile residues are highly conserved, the engineering of an artificial protein membrane that is composed of EutM and CcmK2 may be feasible.
Recent electron microscopic images of carboxysomes have also revealed close association of large proteins with the compartment's shell structure, such as ribulose bisphosphate carboxylase/oxygenase (14). Similarly, ethanolamine ammonia lyase or other proteins may thus also have a direct role in the assembly of the Eut facets. At this point, however, it is not clear whether these proteins are surface associated or whether they are integrated into the structure of the protein membrane. Given the very tight packing between the tiles in the crystal, the latter possibility may not be feasible. The identification of additional specific interactions is thus of great importance.
A sequence alignment between EutM and CcmK2 suggests only weak homology between the C-terminal residues 91 to 97. In the structure of the CcmK2, this tail formed an extended conformation before ending in a short α-helix. Similarly, the 10-residue tail structure of EutM (crystal form I) was also folded into a strand-helix conformation. In crystal form II, on the other hand, the same sequence adopted a mostly extended conformation (Fig. 4). Interestingly, in CcmK2, this tail structure was found to cause dimerization of the protein tiles in the crystal and in solution (29). Based on this finding, it was hypothesized that the tail serves as an interaction site for other factors. Given these additional structural similarities, EutM may represent the Eut counterpart of the caboxysomal CcmK2 protein.
Overall, a number of studies have now confirmed the formation of proteinaceous membrane structures of individual shell proteins (18, 26, 28, 30, 31). In all cases, the membranes are composed of hexagonal protein tiles that serve as fundamental building blocks. As the tiles can easily be envisioned to assemble into triangular facets of different sizes, they may also provide a simple explanation for the different sizes of microcompartments (3, 14, 27, 32).
The two crystal structures of EutM presented here demonstrated the intrinsic potential of these shell proteins to assemble into such a structure autonomously. Despite the highly similar structures and shapes of the EutL and EutM tiles, unproductive and competing interactions between different tile species may be prevented due to the specificity of contacting residues along the tile edges. The study presented here may thus point to a Lego-like assembly principle of bacterial microcompartments: a common underlying shape of fundamental building blocks that exhibit specific interactions between the tiles to ensure productive assembly. Further studies on tile interactions will be pivotal to the understanding of structure and function of this supramolecular assembly.
Supplementary Material
Acknowledgments
We are grateful for the expert help we received from Lisa Dunn as well as from P. Dunten, C. Smith, and T. Doukov of the Stanford Synchrotron Research Laboratory. Moreover, we express our sincere thanks to Chris Mohr and Steve Jordan at AMGEN (Thousand Oaks, CA) for the possibility to collect additional X-ray data. We thank Herbert Waite and Dong Soo Hwang for help with the MALDI-TOF mass determinations.
This work was in part supported by the International Training Program (ITP) from the Japanese Society for the Promotion of Science (JSPS).
Footnotes
Published ahead of print on 17 September 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arifuzzaman, M., M. Maeda, A. Itoh, K. Nishikata, C. Takita, R. Saito, T. Ara, K. Nakahigashi, H. C. Huang, A. Hirai, K. Tsuzuki, S. Nakamura, M. Altaf-Ul-Amin, T. Oshima, T. Baba, N. Yamamoto, T. Kawamura, T. Ioka-Nakamichi, M. Kitagawa, M. Tomita, S. Kanaya, C. Wada, and H. Mori. 2006. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 16:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, N. A., D. Sept, S. Joseph, M. J. Holst, and J. A. McCammon. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037-10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobik, T. A. 2006. Polyhedral organelles compartmenting bacterial metabolic processes. Appl. Microbiol. Biotechnol. 70:517-525. [DOI] [PubMed] [Google Scholar]
- 4.Brinsmade, S. R., T. Paldon, and J. C. Escalante-Semerena. 2005. Minimal functions and physiological conditions required for growth of Salmonella enterica on ethanolamine in the absence of the metabolosome. J. Bacteriol. 187:8039-8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Brunger, A. T. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472-475. [DOI] [PubMed] [Google Scholar]
- 5.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 6.Butland, G., J. M. Peregrin-Alvarez, J. Li, W. Yang, X. Yang, V. Canadien, A. Starostine, D. Richards, B. Beattie, N. Krogan, M. Davey, J. Parkinson, J. Greenblatt, and A. Emili. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531-537. [DOI] [PubMed] [Google Scholar]
- 7.Cai, F., B. B. Menon, G. C. Cannon, K. J. Curry, J. M. Shively, and S. Heinhorst. 2009. The pentameric vertex proteins are necessary for the icosahedral carboxysome shell to function as a CO2 leakage barrier. PLoS One 4:e7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon, G. C., C. E. Bradburne, H. C. Aldrich, S. H. Baker, S. Heinhorst, and J. M. Shively. 2001. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl. Environ. Microbiol. 67:5351-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CCP4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 10.Cheng, S., Y. Liu, C. S. Crowley, T. O. Yeates, and T. A. Bobik. 2008. Bacterial microcompartments: their properties and paradoxes. Bioessays 30:1084-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heldt, D., S. Frank, A. Seyedarabi, D. Ladikis, J. B. Parsons, M. J. Warren, and R. W. Pickersgill. 2009. Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochem. J. 423:199-207. [DOI] [PubMed] [Google Scholar]
- 12.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33-38. [DOI] [PubMed] [Google Scholar]
- 13.Iancu, C. V., H. J. Ding, D. M. Morris, D. P. Dias, A. D. Gonzales, A. Martino, and G. J. Jensen. 2007. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J. Mol. Biol. 372:764-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iancu, C. V., D. M. Morris, Z. Dou, S. Heinhorst, G. C. Cannon, and G. J. Jensen. 2010. Organization, structure, and assembly of alpha-carboxysomes determined by electron cryotomography of intact cells. J. Mol. Biol. 396:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabsch, W. 1993. Automatic processing of diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26:795-800. [Google Scholar]
- 16.Kabsch, W. 1988. Evaluation of single-crystal diffraction data from a position-sensitive detector. J. Appl. Crystallogr. 21:916-924. [Google Scholar]
- 17.Kaneko, Y., R. Danev, K. Nagayama, and H. Nakamoto. 2006. Intact carboxysomes in a cyanobacterial cell visualized by Hilbert differential contrast transmission electron microscopy. J. Bacteriol. 188:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerfeld, C. A., M. R. Sawaya, S. Tanaka, C. V. Nguyen, M. Phillips, M. Beeby, and T. O. Yeates. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936-938. [DOI] [PubMed] [Google Scholar]
- 19.Kerrien, S., Y. Alam-Faruque, B. Aranda, I. Bancarz, A. Bridge, C. Derow, E. Dimmer, M. Feuermann, A. Friedrichsen, R. Huntley, C. Kohler, J. Khadake, C. Leroy, A. Liban, C. Lieftink, L. Montecchi-Palazzi, S. Orchard, J. Risse, K. Robbe, B. Roechert, D. Thorneycroft, Y. Zhang, R. Apweiler, and H. Hermjakob. 2007. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 35:D561-D565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein, M. G., P. Zwart, S. C. Bagby, F. Cai, S. W. Chisholm, S. Heinhorst, G. C. Cannon, and C. A. Kerfeld. 2009. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J. Mol. Biol. 392:319-333. [DOI] [PubMed] [Google Scholar]
- 21.Kleywegt, G. J., and T. A. Jones. 1999. Software for handling macromolecular envelopes. Acta Crystallogr. D Biol. Crystallogr. 55:941-944. [DOI] [PubMed] [Google Scholar]
- 22.Kofoid, E., C. Rappleye, I. Stojiljkovic, and J. Roth. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 23.Liu, J., J. Dai, and M. Lu. 2003. Zinc-mediated helix capping in a triple-helical protein. Biochemistry 42:5657-5664. [DOI] [PubMed] [Google Scholar]
- 24.Nikolakakis, K., A. Ohtaki, K. Newton, A. Chworos, and M. Sagermann. 2009. Preliminary structural investigations of the Eut-L shell protein of the ethanolamine ammonia-lyase metabolosome of Escherichia coli. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 65:128-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Penrod, J. T., and J. R. Roth. 2006. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J. Bacteriol. 188:2865-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sagermann, M., A. Ohtaki, and K. Nikolakakis. 2009. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc. Natl. Acad. Sci. U. S. A. 106:8883-8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmid, M. F., A. M. Paredes, H. A. Khant, F. Soyer, H. C. Aldrich, W. Chiu, and J. M. Shively. 2006. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J. Mol. Biol. 364:526-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka, S., C. A. Kerfeld, M. R. Sawaya, F. Cai, S. Heinhorst, G. C. Cannon, and T. O. Yeates. 2008. Atomic-level models of the bacterial carboxysome shell. Science 319:1083-1086. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, S., M. R. Sawaya, M. Phillips, and T. O. Yeates. 2009. Insights from multiple structures of the shell proteins from the beta-carboxysome. Protein Sci. 18:108-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka, S., M. R. Sawaya, and T. O. Yeates. 2010. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327:81-84. [DOI] [PubMed] [Google Scholar]
- 31.Tsai, Y., M. R. Sawaya, G. C. Cannon, F. Cai, E. B. Williams, S. Heinhorst, C. A. Kerfeld, and T. O. Yeates. 2007. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 5:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeates, T. O., C. A. Kerfeld, S. Heinhorst, G. C. Cannon, and J. M. Shively. 2008. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6:681-691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.