Abstract
Bacterial microcompartments (BMCs) are polyhedral organelles found in an increasingly wide variety of bacterial species. These structures, typified by carboxysomes of cyanobacteria and many chemoautotrophs, function to compartmentalize important reaction sequences of metabolic pathways. Unlike their eukaryotic counterparts, which are surrounded by lipid bilayer membranes, these microbial organelles are bounded by a thin protein shell that is assembled from multiple copies of a few different polypeptides. The main shell proteins form hexamers whose edges interact to create the thin sheets that form the facets of the polyhedral BMCs. Each hexamer contains a central pore hypothesized to mediate flux of metabolites into and out of the organelle. Because several distinctly different metabolic processes are found in the various BMCs studied to date, it has been proposed that a common advantage to packaging these pathways within shell-bound compartments is to optimize the concentration of volatile metabolites in the BMC by maintaining an interior pH that is lower than that of the cytoplasm. We have tested this idea by recombinantly fusing a pH-sensitive green fluorescent protein (GFP) to ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), the major enzyme component inside the carboxysome. Our results suggest that the carboxysomal pH is similar to that of its external environment and that the protein shell does not constitute a proton barrier. The explanation for the sundry BMC functions must therefore be sought in the characteristics of the pores that traverse their shells.
Clearly, the subcellular organization of bacteria is much more complex than was once assumed (reviewed in references 17 and 26), and many bacteria are able to compartmentalize metabolic processes into distinct organelles. Among these, the bacterial microcompartments (BMCs) have garnered attention because the genetic potential to form these structures, which consist entirely of protein, is widespread among the bacteria (1). BMCs have been credited with enhancing the activity of the enzyme(s) they contain by providing a unique environment with optimized substrate concentrations or pH, facilitating metabolite channeling, or protecting the cell by sequestering toxic intermediates (1). Based on comparative genomic and biochemical analyses, the interiors of BMCs in various bacteria are populated by different complements of enzymes, suggesting that, collectively, these organelles play a role in a multitude of metabolic pathways. The bounding shells of all BMCs, on the other hand, appear to be built from multimeric assemblies of proteins that belong to the same two families (pfam 00936 and pfam 03319) (7, 11, 12, 29, 30, 32). Despite some structural differences between individual members of the two main shell protein types, the central pores in the pentamers and hexamers formed by these proteins have been implicated in mediating metabolite traffic across the BMC shell and, in some cases, may actively regulate transfer of substrates and products across the shell through a gating mechanism (12, 30). The variations in pore sizes and surface properties between individual BMC shell proteins likely reflect differences in interactions with the metabolites that pass through them and beg the question about functional differences among BMCs and between BMC protein shells and the lipid bilayer-based membranes of eukaryotic organelles.
The carboxysome, the first BMC to be discovered (25), is found in all cyanobacteria and in many nonphotosynthetic chemoautotrophs, exemplified by the aerobic sulfur bacterium Halothiobacillus neapolitanus and its relatives. Its interior is filled with ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) (25), the enzyme that is crucial for carbon assimilation by these bacteria because it catalyzes the fixation of inorganic carbon onto the organic acceptor molecule, ribulose-1,5-bisphosphate. The enzyme is a relatively inefficient catalyst; it has a high Km for its substrate, CO2, a low turnover number, and is also able to fix O2 through a competing, unproductive reaction. The carboxysome compensates for these shortcomings by providing a microcompartment that enhances the catalytic efficiency of RubisCO. Cytosolic bicarbonate is thought to enter the carboxysome freely through the many pores in the shell, but it cannot be used by RubisCO. The action of the carboxysomal carbonic anhydrase that is cosequestered with RubisCO rapidly converts HCO3− to the RubisCO substrate, CO2 (5, 19, 28). The carboxysome shell retards diffusion of CO2 out of the organelle (5) and thereby contributes to the generation of an elevated steady-state concentration of CO2 in the vicinity of the RubisCO active site, a condition that favors the carboxylation reaction. In addition, the carboxysomal shell may reduce the concentration of the competing RubisCO substrate O2 by excluding it from the microcompartment interior (3, 14). Quantitative modeling of CO2 fixation in cyanobacteria supports the role of the shell as a barrier for CO2 diffusion out of the carboxysome (21). Measurements of CO2 hydration rates in purified intact and disrupted carboxysomes suggest that access of CO2 to the interior of the organelle is likewise limited by the shell (5).
The shells of other BMCs have also been proposed to influence the flux of metabolites into and out of the microcompartment interior. The Pdu BMC of Salmonella enterica, which participates in the B12-dependent degradation of propanediol, prevents the toxic intermediate propionaldehyde from diffusing into the cytoplasm (6, 24). The Eut BMC, also found in S. enterica, is thought to encapsulate several enzymes of the ethanolamine utilization pathway, in which acetaldehyde is a central intermediate. The shell of the Eut BMC is also postulated to prevent loss of a crucial metabolite, in this case acetaldehyde, from the interior by diffusion (22). Acetaldehyde, which like propionaldehyde is volatile, is thought to be captured within the Eut BMC not to protect cellular structures from damage but to prevent escape of this important intermediate from the cell (20). In an effort to identify a common mechanism for BMC function, Penrod and Roth (20) made the intriguing suggestion that all BMCs may constitute compartments with an interior pH that is lower than that of the surrounding cytoplasm. Such an environment would promote the conversion of aldehydes in the Pdu and Eut BMCs to less volatile acetals, which are less likely to escape the downstream pathway enzymes localized in the BMC interior. In the carboxysomes of autotrophic bacteria, a lower pH would presumably favor higher concentrations of CO2 by shifting the equilibrium from HCO3− toward CO2.
This unifying model of BMC function necessitates the assumption that the BMC shell resists passage of protons out of the compartment to maintain a pH gradient. Although it is not known whether a thin protein layer can impede proton diffusion, one needs only to look at the mechanism of proton discrimination described for the pores of aquaporins (2, 31) to imagine that the multiple pores in the BMC shell might likewise fulfill the structural requirements of a proton permeability barrier.
An extended discussion of this model at a recent symposium on BMCs (at the 109th General Meeting of the American Society for Microbiology in 2009) prompted us to design a study that directly determined if the BMC shell is able to maintain a lumen pH that is different from that of its surrounding medium.
MATERIALS AND METHODS
Strains and growth conditions.
The Φ(gfp-cbbS)(Hyb)::Kanr mutant was constructed in the Halothiobacillus neapolitanus c2 (ATCC 23641) wild-type background. Cells were cultured in chemostats as described previously (5). Growth curves were established for batch cultures by measuring the optical density at 600 nm.
H. neapolitanus Φ(gfp-cbbS)(Hyb)::Kanr mutant construction.
An in-frame fusion of the genes encoding pH-sensitive GFP (gfp) and the RubisCO small subunit (cbbS) of H. neapolitanus was generated by overlap extension PCR using the following primers: GGATCCCGTTGATCCCTCGTACCACACAACATACTAAGGTGAGTAACCATGAGTAAAGGAGAAGAACTTTTCA, TTTGTATAGTTCATCCATGCCATGTGTAATCCTAGCAGCTGTTACAAACTCAAGAAGGATCATGTGATCTCTC, ACATGGCATGGATGAACTATACAAAATGGCTGAAATGCAGGATTACAAGC, and GGTACCTTAGTTGCCGCGGTAGACCACGAAGGCCAGACCTAAGCTTTGCG (sequence portions in italics denote BamHI and KpnI restriction sites used for cloning purposes; underlined sequences are homologous to the regions that flank the cbbS gene in the H. neapolitanus genome). The resulting fragment was cloned into the pCR-BluntII-TOPO vector (Invitrogen). After excision by digestion with BamHI and KpnI, the insert and a kanamycin resistance cassette (Kanr) with a KpnI and an XhoI site flanking its 5′ and 3′ ends, respectively, were ligated into the BamHI- and XhoI-digested pPROEX-HTb vector (Invitrogen). The excised Φ(gfp-cbbS)::Kanr fragment of the resulting plasmid and the pUC18-cbbLS plasmid (33) were subsequently used to cotransform Escherichia coli DY330 cells (15). Homologous recombination in this strain resulted in the replacement of wild-type cbbS in pUC18-cbbLS by the Φ(gfp-cbbS)::Kanr fragment to yield the plasmid pUC18-cbbL-Φ(gfp-cbbS). Exponentially growing wild-type H. neapolitanus cells were transformed with this construct by electroporation (5). The genotype of the resulting Φ(gfp-cbbS)(Hyb)::Kanr mutant was confirmed by genomic DNA sequencing (University of Maine DNA Sequencing Facility).
Confocal microscopy.
Exponentially growing H. neapolitanus cells were subjected to confocal microscopy with a Zeiss LSM510 Meta confocal microscope equipped with an alpha Plan-Fluar 100×, 1.45-numerical aperture differential interference contrast oil objective lens. GFP fluorescence was excited with a 488-nm laser; emission was detected using a 505 LP filter. Images were analyzed with the Zeiss LSM 510 software version 3.2.
Transmission electron microscopy.
Transmission electron micrographs of purified carboxysomes were captured as described previously (5) and scanned using an Epson Perfection V700 photo flatbed scanner to obtain the final images.
Carboxysome isolation and analysis.
Carboxysomes were purified by sucrose gradient centrifugation as described previously (28). Protein concentrations were estimated with a standard bicinchoninic acid assay (Thermo Scientific). Carboxysomal polypeptides were resolved on a denaturing 10-to-20% SDS-polyacrylamide gradient gel (Bio-Rad) and stained with GelCode Blue (Thermo Scientific). The GFP-CbbS fusion protein in purified mutant carboxysomes was detected by immunoblotting. Blots were probed with a commercially available rabbit anti-GFP polyclonal antibody (Santa Cruz Biotechnology) and goat anti-rabbit, horseradish peroxidase-tagged IgG as secondary antibody; blots were developed with the SuperSignal West Pico chemiluminescence substrate (Thermo Scientific). Immunoreactive bands were visualized using the VersaDoc imaging system (Bio-Rad). Radiometric RubisCO activity assays were conducted in triplicates as described previously (5).
Fluorescence data collection.
Fluorescence excitation spectra were obtained using an Applied Photophysics SX20 stopped-flow spectrophotometer equipped with a 90° fluorescence detector, variable wavelength excitation monochromator, and a 495-nm long-pass cutoff filter. Isolated carboxysomes or bacterial cells in the buffers indicated in the text below and figure legends were loaded into the flow cell, bypassing the stopped-flow circuit with a single-port syringe. Steady-state excitation spectra were collected by scanning the indicated excitation wavelengths and recording the emission intensities.
The pH shift experiments were conducted by placing appropriate concentrations of either intact or freeze/thaw-disrupted carboxysomes (28) in 10 mM Bicine-HCl, 10 mM MgCl2 (pH 7.5) into one syringe of the stopped-flow apparatus. The sample was then rapidly mixed with an equal volume of a dilute solution of either HCl or NaOH that had been previously titrated potentiometrically to result in a final pH of 7.0 or 8.0, respectively, when mixed with the sample buffer. Fluorescence emission intensity was recorded for the indicated time periods automatically when the stopped-flow mechanism was actuated.
RESULTS
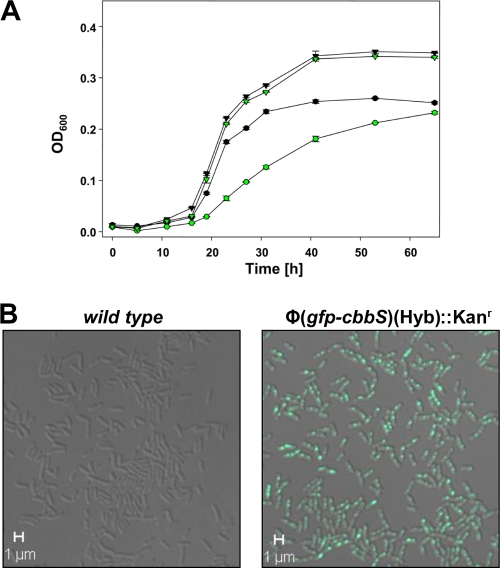
The gene (gfp) encoding a pH-sensitive GFP (pHluorin [13, 16, 18]) was fused in frame to the 5′ end of the carboxysomal RubisCO small subunit gene (cbbS) and used to replace wild-type cbbS in the H. neapolitanus genome via homologous recombination (Fig. 1). Confocal fluorescence microcopy of live, unfixed cultures revealed that the cells of the resulting H. neapolitanus Φ(gfp-cbbS)(Hyb)::Kanr mutant had a normal morphology but contained punctile, highly fluorescent regions (Fig. 2B), consistent with the carboxysome clusters observed in cryo-electron tomograms of cells (9) and in transmission electron micrographs of cell thin sections (27). The mutant exhibited a mild high-CO2-requiring (hcr) phenotype, characterized by slower growth than the wild type at ambient CO2 levels but growth at the same rate as the wild type when the culture was supplemented with 5% CO2 (Fig. 2A).
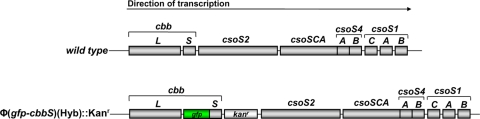
FIG. 1.
Genotype of the cso operon in the wild type and the Φ(gfp-cbbS)(Hyb)::Kanr mutant of Halothiobacillus neapolitanus. Shown are the position and approximate length of the large (cbbL) and small (cbbS) subunit genes of RubisCO, the gene encoding putative structural proteins CsoS2A and CsoS2B, the gene for the shell-associated carboxysomal carbonic anhydrase CsoSCA (csoS3), the two paralogs (csoS4A and csoS4B) encoding pentameric vertex proteins, and the three genes (csoS1A, csoS1B, and csoS1C) encoding the shell proteins that form the facets of the icosahedral carboxysome shell. The gfp-cbbS fusion shown for the mutant gives rise to the pH-sensitive GFP-tagged RubisCO packaged inside the mutant carboxysomes. The kanamycin resistance cassette (Kanr) was included for selection purposes.
FIG. 2.
(A) Growth curves of wild-type (black symbols) and Φ(gfp-cbbS)(Hyb)::Kanr mutant (green symbols) batch cultures of H. neapolitanus. At the ambient CO2 level (circles), growth of the mutant was reduced compared to that of the wild type; growth of the mutant in air supplemented with 5% CO2 (triangles) was indistinguishable from that of the wild type. (B) Composite bright-field and fluorescent confocal images of live wild-type (left) and Φ(gfp-cbbS)(Hyb)::Kanr mutant (right) H. neapolitanus cells. The distribution and position of the fluorescent foci visible in the mutant cells are consistent with carboxysomes and carboxysome clusters documented in the literature (26, 27).
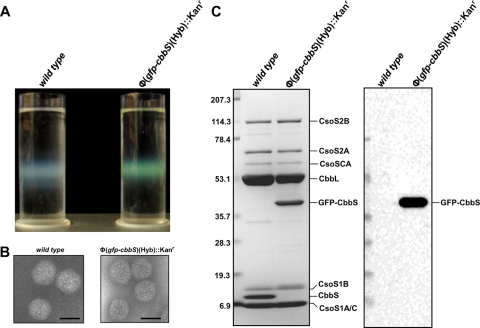
Chemostat-grown mutant cells subjected to the routine carboxysome isolation procedure employed in our laboratory yielded the same heavy carboxysome band normally seen in sucrose density gradients (28). However, in contrast to the whitish-blue, cloudy appearance characteristic of wild-type carboxysomes when present in high concentration, the band of Tyndall-scattering mutant carboxysomes was distinctly fluorescent green (Fig. 3A). Isolated, negatively stained mutant carboxysomes were indistinguishable from their wild-type counterparts in transmission electron micrographs (Fig. 3B). The mutant organelles contained RubisCO holoenzyme molecules with small subunits that consisted entirely of the GFP-CbbS fusion protein; all other protein constituents were present in the proper stoichiometric ratios, as determined by SDS-PAGE and immunoblot analysis (Fig. 3C). The specific activity of RubisCO measured in intact purified mutant carboxysomes was approximately 50% lower than that from wild-type organelles (Table 1) .
FIG. 3.
Characterization of isolated Φ(gfp-cbbS)(Hyb)::Kanr mutant carboxysomes. (A) Wild-type (left) and mutant (right) carboxysomes were purified by centrifugation through a linear sucrose density gradient. Tyndall scattering of the carboxysome band is evident in both gradients, but only Φ(gfp-cbbS)(Hyb)::Kanr mutant carboxysomes are fluorescent green. (B) Transmission electron micrographs of negatively stained wild-type and mutant carboxysomes do not reveal any obvious morphological differences. Bars, 100 nm. (C) Polypeptide composition of purified wild-type and mutant carboxysomes. Polypeptides were separated by SDS-polyacrylamide gel electrophoresis (the stained gel on the left). The GFP-CbbS fusion protein replaces wild-type CbbS in the mutant carboxysomes. The location of GFP was evident after probing an immunoblot (right) with an anti-GFP antibody.
TABLE 1.
Specific activities of RubisCO in different fractions
| Source of RubisCO | Sp act (μmol CO2 fixed/min/mg of protein) |
|
|---|---|---|
| Wild type | Φ(cbbS-gfp) (Hyb)::Kanr | |
| Intact carboxysomes | 1.482 ± 0.006 | 0.736 ± 0.007 |
| Broken carboxysomes | 1.626 ± 0.007 | 0.618 ± 0.014 |
| Shell-enriched fraction | 0.989 ± 0.005 | 0.428 ± 0.002 |
| Freed RubisCO | 2.442 ± 0.037 | 0.011 ± 0.001 |
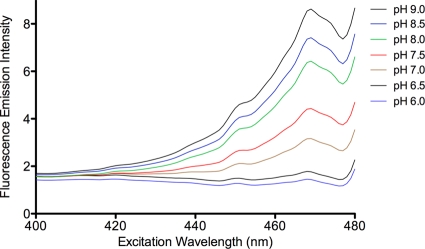
The gfp gene that was used to create the Φ(gfp-cbbS) fusion contains multiple substitutions; these encode a mutant GFP that displays a pH-sensitive excitation peak at 470 nm (16). Indeed, excitation spectra of isolated, intact mutant carboxysomes revealed a major peak at 469 nm when the emission intensity above a 495-nm cutoff was measured. The intensity of emission at this excitation wavelength was maximal at pH 9.0 and decreased as the pH was lowered to 6.0, at which point the peak was barely detectable (Fig. 4). Increasing the pH reversed the loss of fluorescence, as expected (16).
FIG. 4.
Fluorescence excitation spectra of Φ(gfp-cbbS)(Hyb)::Kanr mutant carboxysomes. Equivalent amounts (mg of protein) of purified mutant carboxysomes were suspended in buffers of the pH values indicated in the graph. Fluorescence emission intensity above 495 nm was recorded as excitation wavelengths from 400 to 480 nm were scanned. The scale on the ordinate axis represents voltage output from the spectrofluorometer. A pH-sensitive maximum was observed at 469 nm.
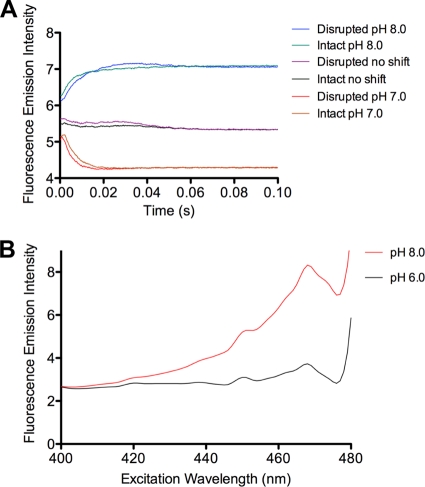
To determine if the intact carboxysome shell could maintain a pH gradient, isolated, intact mutant carboxysomes that had been equilibrated overnight at pH 7.5 in a solution containing 10 mM Bicine-HCl and 10 mM MgCl2 were rapidly mixed with an equal volume of HCl or NaOH solutions that had been electrometrically titrated to produce a final pH of 7.0 or 8.0, respectively, after mixing. Rapid mixing and simultaneous monitoring of fluorescence were carried out in an Applied Photophysics SX 20 stopped-flow spectrophotometer equipped with a fluorescence detector. Changes in fluorescence were observed to occur rapidly, essentially reaching final emission levels after 0.04 to 0.05 s (Fig. 5A). Measurements made after freeze-thaw disruption of the carboxysome shell (28) revealed pH-induced florescence changes practically identical to those observed with intact carboxysomes, suggesting that the intact carboxysome shell is freely permeable to protons.
FIG. 5.
pH-dependent changes of GFP fluorescence in vivo and in vitro. (A) Stopped-flow measurement of fluorescence emitted from intact and disrupted Φ(gfp-cbbS)(Hyb)::Kanr mutant carboxysomes after rapid mixing with HCl or NaOH and a concomitant pH shift from 7.5 to 8.0 or 7.0, respectively, as indicated in the graph. Control (no shift) experiments were performed by mixing the suspended carboxysomes with buffer of an identical pH. (B) Suspensions of H. neapolitanus Φ(gfp-cbbS)(Hyb)::Kanr mutant cells in buffer at pH 6.0 or pH 8.0 containing 10 μM CCCP. Fluorescence emission intensity was recorded at wavelengths above 495 nm while excitation wavelengths were scanned.
The internal pH of mutant carboxysomes was further examined in situ by measuring the fluorescence of Φ(gfp-cbbS)(Hyb)::Kanr H. neapolitanus cells after dissipating the proton gradient across their cytoplasmic membrane (8). Exposure of the cells to the ionophore carbonyl cyanide m-chlorophenylhydrazone (CCCP), which allows the diffusion of protons across the lipid bilayer of the cytoplasmic membrane (10), rapidly equilibrates the cytoplasmic pH to that of the outside medium. However, CCCP should not affect the proton permeability of the carboxysome shell, which consists entirely of protein and does not depend on a lipid bilayer for structural integrity. The internal pH of H. neapolitanus cells suspended in buffer containing CCCP rapidly changed to that of the surrounding buffer, as indicated by a concomitant decrease in GFP fluorescence. If the carboxysome shell were able to maintain a pH gradient, the fluorescence signal generated by GFP within its interior should not change. However, comparison of GFP fluorescence spectra recorded at pH 8.0 and pH 6.0 revealed reduced fluorescence intensity at the lower pH (Fig. 5B). This response is consistent with that displayed by GFP in purified carboxysomes (Fig. 4) and suggests that their shell is freely permeable to protons in vivo and in vitro.
DISCUSSION
Cosequestering metabolically related enzymes into protein-bound microcompartments appears to be a widespread strategy among bacteria (1). In the examples in which a kinetic advantage for the sequestered enzyme has been examined directly, the proteinaceous shell has been proposed to act as a barrier that controls diffusion of volatile metabolites out of the compartment (5, 6, 20, 22, 24). A molecular mechanism that explains the implied barrier properties of the shell has yet to be detailed; however, structural analysis of the major proteins has revealed a physical arrangement in which “edge-on” interactions between hexameric and pentameric oligomers create shells that likely are impervious to the relevant metabolites and intermediates generated within the compartment. The distinctive pores positioned in the center of each hexameric unit provide likely portals for metabolite entry into and exit out of BMCs, and the complex structure of some pores suggests some degree of selectivity for the passage of solutes. The pores formed by CsoS1D from Prochlorococcus marinus MED4 (12) and by EutL from E. coli (23, 30) appear to support some form of active gating. The idea that structural shell elements of BMCs are able to control the passage of protons (20), therefore, is not out of the realm of possibilities. One can envision a mechanism similar to that employed by aquaporin, which allows passage of water molecules through lipid bilayers but blocks proton flux (2).
Considering that a lower internal pH would favor maintaining an optimal concentration of one or more key metabolites inside the Pdu and Eut BMCs as well as the carboxysome, we decided to test whether BMC shells are able to maintain a low pH in the BMC interior. The carboxysome seemed the obvious choice for this study because it has the least complex enzyme content of the three BMCs. Its interior is filled with approximately 270 copies of form I RubisCO; approximately 40 dimers of carbonic anhydrase are tightly associated with the inside of the shell. Our previous study established that CbbL is crucial for packaging of RubisCO into the carboxysome (15) and pointed to cbbS as the target for fusion with gfp. Furthermore, the crystal structure of the carboxysomal RubisCO from H. neapolitanus is known and was able to inform the placement of the pH-sensitive GFP on the N terminus of the small subunit of RubisCO to minimize interference with holoenzyme assembly and function. The fluorescent carboxysomes of the resulting H. neapolitanus mutant, albeit morphologically indistinguishable from wild-type organelles, were compromised in their CO2-fixing ability.
The reduced specific activity of RubisCO in the mutant organelles is likely a reflection of the significant increase in size of the fusion protein, which limits the number of holoenzyme molecules that can be accommodated within the carboxysome interior. The addition of a GFP moiety to CbbS was expected to reduce RubisCO stability and enzymatic performance to some extent, based on previous experience with RubisCO variants (reference 15 and references therein). However, the almost-complete loss of activity of the GFP-RubisCO upon its release from disrupted carboxysomes was surprising, considering that within the carboxysome the mutant enzyme maintains close to half the specific activity of wild-type RubisCO. Clearly, the interior of the carboxysome stabilizes the structure and/or activity of the mutant RubisCO.
A proton concentration that is higher on the inside of the BMC than in the cytoplasm offers a general mechanism underlying the benefit a cell derives from restricted diffusion of intermediates across the BMC boundary (20). However, maintenance of a proton gradient across the BMC shell would require that its thin protein layer, like a lipid bilayer, be impermeable to protons. In view of the results presented here, that possibility seems unlikely. Stopped-flow pH colorimetry experiments performed with purified mutant carboxysomes indicated a rapid equilibration of the carboxysome pH to that of the buffer in which the organelles were suspended. Additionally, consistent results were obtained in suspensions of mutant cells that had been treated with the ionophore CCCP. By comparison, in similar stopped-flow pH decay experiments performed with intact oleate bilayer vesicles, equilibration times approximately 40 times longer were recorded (4). Our study therefore suggests that the carboxysome shell is freely permeable to protons and that no pH gradient exists between the BMC lumen and the cytoplasm.
There is much to learn about the flux of metabolites across the shell of the various BMCs. Structural studies (11, 12, 29, 30, 32) and analytical calculations (32) suggest that the density and geometric arrangement of the pores in the shell permit moderate metabolite exchange between the BMC interior and cytoplasm. The key to BMC function and to any control the shell may assert over solute traffic will be the specific interactions between shell protein structures and the metabolites that pass through the pores of their assemblies.
Acknowledgments
We thank J. M. Henley for providing the gene for the pH-sensitive GFP, pHluorin, B. Kang for assistance with confocal microscopy, K. J. Curry for help with transmission electron microscopy, and C. A. Kerfeld, K. M. Scott, and R. Spreitzer for helpful discussions.
This work was supported by NSF grants MCB 0818680 and MCB 0851070 (to G.C.C. and S.H.).
Footnotes
Published ahead of print on 24 September 2010.
REFERENCES
- 1.Bobik, T. A. 2006. Polyhedral organelles compartmenting bacterial metabolic processes. Appl. Microbiol. Biotechnol. 70:517-525. [DOI] [PubMed] [Google Scholar]
- 2.Burykin, A., and A. Warshel. 2003. What really prevents proton transport through aquaporin? Charge self-energy versus proton wire proposals. Biophys. J. 85:3696-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannon, G. C., C. E. Bradburne, H. C. Aldrich, S. H. Baker, S. Heinhorst, and J. M. Shively. 2001. Microcompartments in prokaryotes: carboxysomes and related polyhedra. Appl. Environ. Microbiol. 67:5351-5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, I. A., and J. W. Szostak. 2004. Membrane growth can generate a transmembrane pH gradient in fatty acid vesicles. Proc. Natl. Acad. Sci. U. S. A. 101:7965-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dou, Z., S. Heinhorst, E. B. Williams, C. D. Murin, J. M. Shively, and G. C. Cannon. 2008. CO2 fixation kinetics of Halothiobacillus neapolitanus mutant carboxysomes lacking carbonic anhydrase suggest the shell acts as a diffusional barrier for CO2. J. Biol. Chem. 283:10377-10384. [DOI] [PubMed] [Google Scholar]
- 6.Havemann, G. D., and T. A. Bobik. 2003. Protein content of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 185:5086-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heldt, D., S. Frank, A. Seyedarabi, D. Ladikis, J. B. Parsons, M. J. Warren, and R. W. Pickersgill. 2009. Structure of a trimeric bacterial microcompartment shell protein, EtuB, associated with ethanol utilization in Clostridium kluyveri. Biochem. J. 423:199-207. [DOI] [PubMed] [Google Scholar]
- 8.Hopfer, U., A. L. Lehninger, and T. E. Thompson. 1968. Protonic conductance across phospholipid bilayer membranes induced by uncoupling agents for oxidative phosphorylation Proc. Natl. Acad. Sci. U. S. A. 59:484-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iancu, C. V., D. M. Morris, Z. Dou, S. Heinhorst, G. C. Cannon, and G. J. Jensen. 2010. Organization, structure, and assembly of α-carboxysomes determined by electron cryotomography of intact cells. J. Mol. Biol. 396:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasianowicz, J., R. Benz, and S. McLaughlin. 1984. The kinetic mechanism by which CCCP (carbonyl cyanidem-chlorophenylhydrazone) transports protons across membranes. J. Membr. Biol. 82:179-190. [DOI] [PubMed] [Google Scholar]
- 11.Kerfeld, C. A., M. R. Sawaya, S. Tanaka, C. V. Nguyen, M. Phillips, M. Beeby, and T. O. Yeates. 2005. Protein structures forming the shell of primitive bacterial organelles. Science 309:936-938. [DOI] [PubMed] [Google Scholar]
- 12.Klein, M. G., P. Zwart, S. C. Bagby, F. Cai, S. W. Chisholm, S. Heinhorst, G. C. Cannon, and C. A. Kerfeld. 2009. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J. Mol. Biol. 392:319-333. [DOI] [PubMed] [Google Scholar]
- 13.Llopis, J., J. M. McCaffery, A. Miyawaki, M. G. Farquhar, and R. Y. Tsien. 1998. Measurement of cytosolic, mitochondrial, and Golgi pH in single living cells with green fluorescent proteins. Proc. Natl. Acad. Sci. U. S. A. 95:6803-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcus, Y., J. A. Berry, and J. Pierce. 1992. Photosynthesis and photorespiration in a mutant of the cyanobacterium Synechocystis PCC 6803 lacking carboxysomes. Planta 187:511-516. [DOI] [PubMed] [Google Scholar]
- 15.Menon, B. B., Z. Dou, S. Heinhorst, J. M. Shively, and G. C. Cannon. 2008. Halothiobacillus neapolitanus carboxysomes sequester heterologous and chimeric RubisCO species. PLoS One 3:e3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miesenboeck, G., D. A. De Angelis, and J. E. Rothman. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394:192-195. [DOI] [PubMed] [Google Scholar]
- 17.Morris, D. M., and G. J. Jensen. 2008. Toward a biomechanical understanding of whole bacterial cells. Annu. Rev. Biochem. 77:583-613. [DOI] [PubMed] [Google Scholar]
- 18.Patterson, G. H., S. M. Knobel, W. D. Sharif, S. R. Kain, and D. W. Piston. 1997. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 73:2782-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peña, K. L., S. E. Castel, C. de Araujo, G. S. Espie, and M. S. Kimber. 2010. Structural basis of the oxidative activation of the carboxysomal γ-carbonic anhydrase, CcmM. Proc. Natl. Acad. Sci. U. S. A. 107:2455-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penrod, J. T., and J. R. Roth. 2006. Conserving a volatile metabolite: a role for carboxysome-like organelles in Salmonella enterica. J. Bacteriol. 188:2865-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reinhold, L., M. Zviman, and A. Kaplan. 1989. A quantitative model for carbon fluxes and photosynthesis in cyanobacteria. Plant Physiol. Biochem. 27:945-954. [Google Scholar]
- 22.Rondon, M., R. Kazmierczak, and J. Escalante-Semerena. 1995. Glutathione is required for maximal transcription of the cobalamin biosynthetic and 1,2-propanediol utilization (cob/pdu) regulon and for the catabolism of ethanolamine, 1,2-propanediol, and propionate in Salmonella typhimurium LT2. J. Bacteriol. 177:5434-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sagermann, M., A. Ohtaki, and K. Nikolakakis. 2009. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc. Natl. Acad. Sci. U. S. A. 106:8883-8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sampson, E. M., and T. A. Bobik. 2008. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J. Bacteriol. 190:2966-2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shively, J. M., F. Ball, D. H. Brown, and R. E. Saunders. 1973. Functional organelles in prokaryotes: polyhedral inclusions (carboxysomes) of Thiobacillus neapolitanus. Science 182:584-586. [DOI] [PubMed] [Google Scholar]
- 26.Shively, J. M., G. C. Cannon, S. Heinhorst, J. A. Fuerst, D. A. Bryant, E. Gantt, J. A. Maupin-Furlow, D. Schueler, F. Pfeiffer, R. Docampo, C. Dahl, J. Preiss, A. Steinbuechel, and B. A. Federici (ed.). 2008. Intracellular structures of prokaryotes: inclusions, compartments and assemblages. Encyclopedia of microbiology, 3rd ed. Elsevier, Amsterdam, Netherlands.
- 27.Shively, J. M., G. L. Decker, and J. W. Greenawalt. 1970. Comparative ultrastructure of the thiobacilli. J. Bacteriol. 101:618-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.So, A. K.-C., G. S. Espie, E. B. Williams, J. M. Shively, S. Heinhorst, and G. C. Cannon. 2004. A novel evolutionary lineage of carbonic anhydrase (epsilon class) is a component of the carboxysome shell. J. Bacteriol. 186:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanaka, S., C. A. Kerfeld, M. R. Sawaya, F. Cai, S. Heinhorst, G. C. Cannon, and T. O. Yeates. 2008. Atomic-level models of the bacterial carboxysome shell. Science 319:1083-1086. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka, S., M. R. Sawaya, and T. O. Yeates. 2010. Structure and mechanisms of a protein-based organelle in Escherichia coli. Science 327:81-84. [DOI] [PubMed] [Google Scholar]
- 31.Tchernov, D., Y. Helman, N. Keren, B. Luz, I. Ohad, L. Reinhold, T. Ogawa, and A. Kaplan. 2001. Passive entry of CO2 and its energy-dependent intracellular conversion to HCO3− in cyanobacteria are driven by a photosystem I-generated ΔμH+. J. Biol. Chem. 276:23450-23455. [DOI] [PubMed] [Google Scholar]
- 32.Tsai, Y., M. R. Sawaya, G. C. Cannon, F. Cai, E. B. Williams, S. Heinhorst, C. A. Kerfeld, and T. O. Yeates. 2007. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 5:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, D., H. M. Ellis, E.-C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]