Abstract
Colicins are bacterial antibiotic toxins produced by Escherichia coli cells and are active against E. coli and closely related strains. To penetrate the target cell, colicins bind to an outer membrane receptor at the cell surface and then translocate their N-terminal domain through the outer membrane and the periplasm. Once fully translocated, the N-terminal domain triggers entry of the catalytic C-terminal domain by an unknown process. Colicin K uses the Tsx nucleoside-specific receptor for binding at the cell surface, the OmpA protein for translocation through the outer membrane, and the TolABQR proteins for the transit through the periplasm. Here, we initiated studies to understand how the colicin K N-terminal domain (KT) interacts with the components of its transit machine in the periplasm. We first produced KT fused to a signal sequence for periplasm targeting. Upon production of KT in wild-type strains, cells became partly resistant to Tol-dependent colicins and sensitive to detergent, released periplasmic proteins, and outer membrane vesicles, suggesting that KT interacts with and titrates components of its import machine. Using a combination of in vivo coimmunoprecipitations and in vitro pulldown experiments, we demonstrated that KT interacts with the TolA, TolB, and TolR proteins. For the first time, we also identified an interaction between the TolQ protein and a colicin translocation domain.
Colicins are bacterial toxins produced by Escherichia coli strains and are active against E. coli or related strains (17). These bacterial antibiotic toxins play an important role in the E. coli colonization of environmental niches, including the mammal gastrointestinal tract (25, 32, 49, 50). The classification of colicins is based on differences in the mechanisms of action, such as pore formation (colicins A, B, E1, K, Ia, N, 5, etc.), degradation of nucleic acids (including DNases [colicins E2, E7, and E9], 16S RNases [colicins E3, E4, and E6], or tRNases [colicins D and E5]), or degradation of lipid II (colicin M) (17, 34). Colicins are also categorized depending on their import machines: colicins using the Tol proteins are classified as group A (colicins A, E1 to E9, K, N, etc.), whereas colicins using the ExbBD-TonB proteins are classified as group B (colicins B, D, Ia, M, 5, etc.). However, the transport across the periplasm is only one of the three steps of the mechanism of action. Colicins bind to an outer membrane receptor and are translocated through the outer membrane and the periplasm (14, 35, 55, 56). Finally, the C-terminal domain (responsible for the activity) is translocated to its final destination (inner membrane or cytoplasm) depending on its mechanism of action. Colicins are divided into three different structural and functional domains that correspond to the three steps of the mechanism of action: the N-terminal domain is required for translocation, the central domain is involved in receptor binding, and the C-terminal domain carries the activity (4, 5). During the translocation step, the N-terminal domain of the colicin interacts with components of the import machine: colicins A, E1, and N interact with the TolA protein; colicins A, E3, E7, and E9 interact with the TolB protein; and colicins A and E3 interact with TolR (6, 12, 13, 15, 21, 23, 26, 27, 30, 39, 48, 54). In some cases, the domains of the Tol proteins involved in colicin binding have been identified. Reciprocally, the regions of colicins in interaction with the Tol proteins have been delineated. In colicin A, the TolA binding sequence (ABS) is contained within residues 37 to 98 (13, 30), in which a SYNT motif (residues 57 to 60) has been shown to be essential for TolA binding (18, 46). The TolB box and the TolR binding sequences have also been identified in colicin A (27, 30). The TolB box is well conserved within TolB-dependent colicins, including colicins A and E2 to E9, and is composed of residues DG[T,S]GWSSE (12, 13). These residues form a loop penetrating within the TolB beta-propeller (39, 57), mimicking the TolB-Pal interaction (9, 10). Interestingly, the Tol-dependent, pore-forming colicin K does not possess a TolB box (see Fig. 1A), raising the hypothesis that its translocation might be TolB independent or that colicin K interacts with TolB differently than do other TolB-dependent colicins. In this study, we tested the Tol requirements for colicin K translocation and showed that colicin K requires the TolA, TolB, TolQ, and TolR proteins. Production of the N-terminal domain of colicin K in the periplasm of wild-type (WT) cells induces specific tol defects and tolerance to Tol-dependent colicins and bacteriophage, suggesting that the colicin K N-terminal domain binds and titrates the Tol proteins. Further in vivo coimmunoprecipitation and in vitro pulldown experiments demonstrated interactions between the colicin K N-terminal domain and the TolA, TolB, and TolR proteins. For the first time, we also identified an interaction between a colicin translocation domain and the fourth component of the Tol complex, the TolQ protein.
FIG. 1.
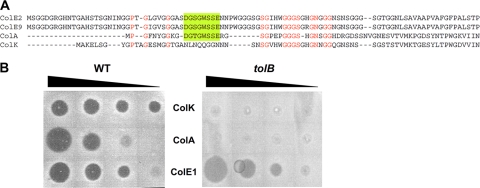
In the absence of an identifiable TolB-binding sequence, colicin K translocation is TolB dependent. (A) Sequence alignment of colicin K and three TolB-dependent colicins (A, E2, and E9). Conserved residues are indicated by red letters. The characterized TolB binding sequence is indicated by the green box (defined in references 12 and 27). (B) Colicin spot assays using serial dilutions of colicins A (TolB dependent), E1 (TolB independent), and K on a wild-type (WT) strain and its tolB derivative (from left to right, 100, 10, 1, and 0.1 ng of colicins have been spotted, respectively).
MATERIALS AND METHODS
Bacterial strains, growth conditions and chemicals.
Bacterial strains used in this study are Escherichia coli K-12 derivatives. C600 and its tolQRA and tolQRABPal derivatives (16) have been used for fractionation experiments. The GM1 (F+), 1292, and W3110 wild-type strains have been used, as well as their tolQ (TPS13 [52]), tolR (TPS300 [52]), tolA (JC7782 [7]), tolB (JC898 [7]), pal (pal892 [7]), and tonB (KP1344 [33]) derivatives. Routinely, bacterial strains were maintained on Luria-Bertani (LB) plates and were grown aerobically in LB medium at 37°C. When required, media have been supplemented with ampicillin (100 μg/ml), chloramphenicol (40 μg/ml), or kanamycin (50 μg/ml). Sodium deoxycholate (DOC), paraformaldehyde, 5-bromo-4-chloro-3-indolylphosphate, nitroblue tetrazolium, Torula yeast RNA, and isopropyl-β-d-thiogalactopyranoside (IPTG) were purchased from Sigma-Aldrich.
Plasmid construction.
Plasmid pKTFLAG (called pKT here) was obtained using a double PCR technique (3, 53) using the Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA). The sequence encoding the N-terminal domain of colicin K (cka-N) was amplified by PCR using oligonucleotides 5′-GGTTTCGCTACCGTAGCGCAGGCCGCTATGGCTAAAGAACTAAGTGGA and 5′-GCTCAGCCGGATCCAAGCTTGGAATTCTTATTTATCATACTCATCTTTATAATCAGTTTCATGTTCAGCGACCTC and the colicinogenic plasmid pColK235 (49) as template. The oligonucleotides allow the insertion of a sequence encoding a FLAG epitope upstream of the stop codon (underlined in the sequence above), and extensions annealing on plasmid pIN-III-ompA2 (24) (italicized in the sequences above). The PCR products were then used as oligonucleotides for a second PCR using the target plasmid pIN-III-ompA2 as template. The pQ8R plasmid, which encodes TolQ fused at its C terminus with 8 histidine residues, was constructed by a recombinant PCR technique (1) using pQR (tolQR genes under the control of the T7 promoter [22]) and oligonucleotides 5′-CATTAACTCGAGGAGGTTTACCATGGCCAGAGCGCGTGGAC and 5′-TAAACCTCCTCGAGTTAATGGTGATGATGATGATGATGATGGGCGCCCTTGTTGCTCTCGCTAACGG (8-histidine coding sequence underlined). Plasmid pOK-QHA, allowing the production of a TolQ protein carrying a C-terminal hemagglutinin (HA) epitope, has been constructed similarly by a recombinant PCR technique allowing the deletion of the tolR gene and using pOK-QHAR as template and oligonucleotides 5′-GGAGGATACCCATACGACGTCCCAGACTACGCTTAAACATCTGCGTTTCCCTTGCTT and 5′-AGCGTAGTCTGGGACGTCGTATGGGTATCCTCCCCCCTTGTTGCTCTCGCT AACGGT. TolQ cysteine substitutions were introduced into pOK-QHA by site-directed mutagenesis using complementary oligonucleotides carrying the mutation to be introduced (oligonucleotides are available on request). All the constructions have been verified by restriction, protein production, and DNA sequencing. KTFLAG (called KT here) expression has been induced using 100 μM IPTG on liquid cultures and 10 μM IPTG on plates throughout the study.
Outer membrane stability assays.
Outer membrane stability has been assessed by measuring cell susceptibilities to various detergents and by measuring the level of periplasmic leakage. Detergent susceptibility was estimated on sodium deoxycholate (DOC)-containing plates (1% final concentration), by spotting normalized serial dilutions of the strain to be tested. For the quantitative plate assay, strains to be tested were grown aerobically in LB medium. At an optical density at 600 nm (OD600) of 0.8, cultures were then diluted in LB medium to a concentration of ∼2,000 bacteria/ml. One hundred microliters (≈200 bacteria) was then plated on LB agar plates or LB agar plates supplemented with 1% DOC. After overnight incubation at 37°C, CFU were counted, and the survival (number of CFU on DOC plates relative to that on the LB control plate) was calculated and expressed as a percentage of the value of the WT strain carrying the empty vector. For liquid broth assay, the strains to be tested were grown aerobically in LB medium to an OD600 of ∼0.8. Cultures were then diluted to an OD600 of 0.05 in LB medium containing 0, 0.5%, 1%, or 2% SDS in presence or absence of 100 μM IPTG and incubated at 37°C. The OD600 was measured after 120 min. The OD600 values were plotted against the SDS concentration, and the 50% lethal dose (LD50) was calculated as the concentration of SDS required to cause a 50% decrease of turbidity compared to growth in LB plain medium. Periplasmic leakage was estimated on RNA-containing plates, as previously described (19). Briefly, cells were plated on LB agar supplemented with 1% RNA (type VI from Torula yeast [Sigma]). After overnight growth, RNA was precipitated with cold 10% trichloroacetic acid, and the leakage of the periplasmic RNase I enzyme was detected by identification of clear zones surrounding colonies. Electron microscopy analyses were performed as previously described (7).
Colicin susceptibility assays.
For the plate assay, colicin susceptibility was verified by the presence of halos on a lawn of the strain to be tested as previously described (30). Data are reported as the maximal 10-fold serial dilution of the colicin stock (1 mg/ml; 10−1, corresponding to a concentration of 0.1 mg/ml) sufficient to inhibit growth. Liquid broth assays were performed as previously described (30, 46): strains to be tested were grown aerobically overnight in LB medium. Cultures were then diluted to an OD600 of 0.05 in LB medium containing serial dilutions of colicins and incubated at 37°C. The OD600 was measured after 120 min, and the percentage of survival (relative to that in LB medium without colicin) was plotted against colicin concentration. The 50% lethal dose (concentration of colicin required to inhibit 50% of bacterial growth after 120 min) was then determined from the graph.
Filamentous bacteriophage infection.
Cell susceptibility to filamentous bacteriophage infection was determined using the tetracycline-resistant Ff phage fd-Tc as previously described (46). Briefly, F+ strains to be tested were grown to exponential growth phase. fd-Tc phage was mixed with ∼500 cells at a multiplicity of 10:1 (phage/bacterium ratio) and incubated for 30 min at 25°C without shaking and then for 45 min at 37°C with shaking before being plated on LB agar medium. Infection frequencies are reported as the number of CFU of infected bacteria (selected on ampicillin-plus-tetracycline plates) compared to the number of CFU of total bacteria (selected on ampicillin plates).
Cell fractionation.
Cell fractionation was performed as previously described (2). The membrane pellet was resuspended in 1 ml of 1 M carbonate sodium (Na2CO3) and incubated on a rotating wheel for 30 min at room temperature. Ultracentrifugation at 90,000 × g for 40 min then separated the integral membrane fraction (pellet) from the membrane-associated fraction (supernatant). The periplasmic, cytoplasmic, and membrane-associated fractions were precipitated with 15% trichloroacetic acid and resuspended in loading buffer prior to analyses by SDS-PAGE and immunoblotting.
Coimmunoprecipitation.
Coimmunoprecipitation experiments were performed essentially as previously described (19). Exponentially growing cells (2 × 109) were harvested, washed with 20 ml of 10 mM sodium phosphate buffer (NaPi, pH 6.8), and resuspended in NaPi buffer supplemented with 1% formaldehyde. After incubation at room temperature for 20 min, the cross-linking reaction was stopped by the addition of 0.3 M Tris-HCl, pH 6.8, and the cells were washed once in 20 mM Tris-HCl (pH 6.8). The cell pellet was then subjected to solubilization for 30 min at 37°C in TES (10 mM Tris-HCl, pH 7.5, 5 mM EDTA, 1% SDS) in the presence of protease inhibitors (Complete; Roche), and diluted 15-fold in TNE (10 mM Tris-HCl, pH 7.5, 5 mM EDTA, 150 mM NaCl) supplemented with 1% Triton X-100. After incubation for 2 h at room temperature with vigorous shaking, the extract was centrifuged for 15 min at 18,000 × g to remove unsolubilized material. Supernatants were then incubated overnight at 4°C with antibodies coupled to either protein A-Sepharose CL-4B beads (Pharmacia), protein G-agarose beads (Roche), or anti-FLAG M2-agarose beads (Sigma-Aldrich). Beads were then washed twice with TNE supplemented with 1% Triton X-100, once in TNE supplemented with 0.1% Triton X-100 and 0.1% Tween, and once in TNE supplemented with 0.1% Triton X-100. The immunoprecipitated material was heated in loading buffer prior to analyses by SDS-PAGE and immunoblotting.
Protein purification.
Histidine-tagged variants of the C-terminal domain (6HisTolA3 and TolA36His) of the TolA protein, of the full-length TolB protein (6HisTolB), of the unacylated Pal protein (Pal6His), and of the periplasmic domain of the TolR (TolR2-36His) and TonB (TonB2-36His) proteins have been purified as previously reported (21, 23, 31, 54). Purification of TolQ8His from E. coli BL21(DE3) omp8 cells (47) carrying the plasmid pQ8HR was done as follows: expression of tolQ8His was induced for 3 h at 30°C with 0.5 mM IPTG and the bacterial cell pellet was resuspended in 20 mM Tris-HCl, pH 8.0, 100 mM NaCl (TN buffer) in the presence of protease inhibitors (Complete-EDTA free; Roche) and DNase. After sonication, membranes were pelleted and membrane proteins were extracted overnight at 4°C under vigorous agitation in the same buffer supplemented with 1% dodecyl-β-d-maltoside (DDM; Fluka). After removal of unsolubilized material by centrifugation, the 8-histidine-tagged TolQ protein was purified by ion metal affinity chromatography (nickel resin; Qiagen) in TN buffer supplemented with 0.1% DDM.
Copurification.
Copurification experiments were performed by ion metal affinity chromatography. Twenty micrograms of the His-tagged protein (6HisTolA3, TolA36His, 6HisTolB, Pal6His, TolR2-36His, TolQ8His, and TonB2-36His) was immobilized on 100 μl of cobalt beads (or nickel for TolQ8His; nickel and cobalt affinity resins from Qiagen) for 40 min. Beads were washed once with the TN buffer and incubated with periplasmic extracts from C600tolQRABPal cells producing KT-FLAG (obtained from 2 × 1010 cells by using the fractionation protocol). Because the FLAG epitope-tagged colicin K N-terminal domain adsorbs on cobalt and nickel beads, 10 mM imidazole was used during the binding and washing steps to limit KT nonspecific binding. After 2 h of incubation at room temperature, beads were washed twice with TN buffer supplemented with 10 mM imidazole and once with 50 mM imidazole and resuspended in loading buffer prior to analyses by SDS-PAGE and immunoblotting. For pulldown experiments with the TolQ8His protein, buffers were supplemented with 0.1% DDM.
Miscellaneous methods.
For detection by immunostaining, proteins were transferred onto nitrocellulose membranes, and blots were probed with the indicated antibodies and either secondary goat anti-rabbit, anti-mouse, or anti-rat antibodies coupled to alkaline phosphatase (Millipore) and developed using 5-bromo-4-chloro-3-indolylphosphate and nitroblue tetrazolium. Anti-FLAG M2 and anti-maltose binding protein (anti-MBP; MalE) antibodies were purchased from Sigma-Aldrich, and anti-HA 3F10 was purchased from Roche. Anti-EF-Tu and anti-AcrA were kind gifts from Vincent Géli and Muriel Masi, respectively. Other polyclonal antibodies are from our laboratory collection and have been described elsewhere (anti-TolA [22], anti-TolB [12], anti-TolR [31], and anti-Pal [16]).
RESULTS AND DISCUSSION
Colicin K was described in the 1970s as a bacterial toxin using the Tol system for its import into Escherichia coli cells (20, 41, 42). Since then, it has been thought that colicin K uses the products of the tolQ, tolR, tolA, and tolB genes to enter into bacteria. However, Luria reported confusion between colicins A and K over a 20-year period because of the distribution of an incorrect producer plasmid (40). Sequence alignments with colicin A show that the TolA and TolR binding sequences (ABS and RBS, respectively) are present whereas the previously identified TolB binding box (BBS), characterized by a DG[T,S]GWSSE motif, is not present (Fig. 1A). The confusion in the early days of colicin K studies and the absence of an identifiable BBS raised the question whether colicin K penetration requires the periplasmic TolB protein. To definitively answer the question, we performed colicin spot assays, which demonstrated that TolB is necessary for colicin K import (Fig. 1B). This result suggests that if an interaction with TolB occurs, the colicin K region responsible for TolB binding is different from those of other group A colicins.
Production and periplasmic location of the N-terminal domain of colicin K.
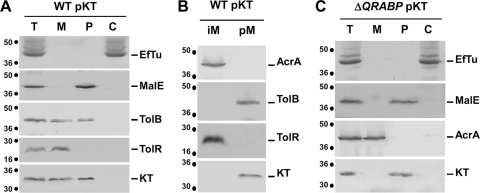
To study the transit step of colicin K translocation, a DNA fragment encoding the N-terminal domain of colicin K was cloned into the pIN-III-ompA2 vector (24), downstream of the OmpA cleavable sequence signal, allowing its IPTG-inducible expression and export to the periplasm. This approach has been widely used to study Tol-colicin or TonB-colicin interactions (12, 13, 29, 30, 46; for a review, see reference 11). The N-terminal domain of colicin K corresponds to residues 1 to 254, for which the boundaries have been defined by sequence comparisons with the Tol- and OmpA-dependent, Tsx-independent colicin S4 (similar N-terminal domains but different central domains [43]) and the Tol- and OmpA-independent, Tsx-dependent colicins 5 and 10 (similar central domains but different translocation domains [44, 45]) (see Fig. S1 in the supplemental material). For immunology-based experiments, a FLAG epitope was fused at the C terminus of the colicin K domain. Pilot experiments showed that the FLAG-tagged colicin K translocation domain (KTFLAG; here called KT) was produced with an expected molecular mass of ∼30 kDa. Fractionation studies confirmed that this domain is correctly exported to the periplasm (Fig. 2A); however, when produced in WT cells, a portion of KT cofractionated with the membrane fraction (Fig. 2A). To test whether this may reflect affinity of the N-terminal domain of colicin K for the membranes (i.e., through protein-lipid contacts) or with subunits anchored in the membrane (i.e., through protein-protein contacts), we treated the membrane fraction with sodium carbonate to disturb the protein-protein interactions. Results displayed in Fig. 2B showed that KT was released from the membrane fraction upon treatment with Na2CO3, suggesting that KT is peripherally associated with the membrane fraction. One may hypothesize that KT is retained through interaction with membrane-associated subunits of its translocation machine, the Tol proteins. Indeed, KT cofractionated with the periplasm fraction in tolQRABPal cells (Fig. 2C), further suggesting that interaction(s) with Tol subunit(s) tethers KT to the membrane fraction.
FIG. 2.
The colicin K N-terminal domain is tethered to the membrane fraction through interaction with Tol components. Total (T) wild-type (WT) (A and B) and tolQRABPal (ΔQRABP) (C) cells producing KT were subjected to fractionation to separate the periplasm (P), cytoplasm (C), and membrane (M) fractions. Membrane proteins from the M fraction of panel A were separated into integral membrane proteins (iM) and peripherally associated membrane proteins (pM) by Na2CO3 treatment (B). Samples were loaded onto a 12.5% acrylamide SDS-PAGE gel and subjected to immunodetection with antibodies raised against the cytoplasmic EF-Tu, the periplasmic MalE, the peripherally membrane-associated TolB, the integral membrane TolR and AcrA proteins, and the FLAG epitope carried by the colicin K translocation domain (KT). Molecular mass markers (in kDa) are indicated on the left of each panel.
Periplasmic expression of the colicin K N-terminal domain induces tol phenotypes.
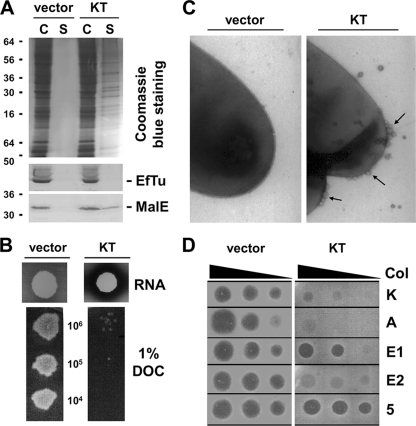
It has been previously shown that production of the N-terminal domain of colicin A or E3 (12, 13) or of the minor capsid protein g3p of filamentous bacteriophage f1 (8, 46) into the periplasm of WT cells induces tol phenotypes, as does the periplasmic production of the TolA C-terminal domain (37) or the TolR central domain (31), probably through titration of Tol subunits and/or prevention of Tol complex formation. These tol phenotypes include (i) the release of periplasmic proteins and of outer membrane vesicles (OMV), (ii) the increased susceptibility to toxic compounds, and (iii) a specific tolerance to group A colicins (7, 36, 38) or to filamentous bacteriophage infection (46). To test whether the production of KT into the periplasm of WT cells induces the tol syndromes, we performed phenotypic analyses. During the fractionation protocol, a portion of the periplasmic MalE protein—but not the EF-Tu cytoplasmic elongation factor—was found in the supernatant fraction of GM1 pKT cells but not in GM1 pIN-III-ompA2 culture supernatants (Fig. 3A), suggesting that periplasmic release occurs upon KT production. The leakiness of the GM1 pKT strain relative to that of the GM1 strain carrying the empty vector pIN-III-ompA2 was confirmed on RNA-containing plates: the release of the periplasmic RNase I led to a degradation of RNA contained in the medium (Fig. 3B). The susceptibility to toxic compounds was tested on deoxycholate (DOC)-containing plates. Figure 3B shows that GM1 pIN-III-ompA2 cells were resistant to 1% deoxycholate whereas GM1 pKT cells grew only faintly on these plates. Quantitative measurements on DOC plates or in SDS-containing liquid broth confirmed these data (see Fig. S2A in the supplemental material; Table 1). Electron microscopy experiments further demonstrated formation of OMV at the cell surface of GM1 pKT cells (Fig. 3C). Finally, colicin susceptibility experiments showed that GM1 pIN-III-ompA2 cells were killed by group A and group B colicins while GM1 pKT cells displayed a specific tolerance to group A but not group B colicins (Fig. 3D). Here again, quantitative measurements in liquid broth containing various concentrations of colicin confirmed these results and further showed that GM1 pKT cells displayed ∼10,000-fold protection against colicins K, A, and E2; 100-fold protection against colicin E1; and no protection against the group B colicins Ia and 5 relative to control cells (see Fig. S2B; Table 2 ). Similarly, the production of KT induced a 20-fold decrease of fd filamentous bacteriophage infection frequencies (Tables 1 and 2). Similar effects have been reported upon periplasmic production of colicin A or of the minor capsid protein g3p (13, 46). Overall, the results for the phenotypic consequence of KT production demonstrate that KT induces characteristic tol membrane defects and colicin and bacteriophage tolerance.
FIG. 3.
Periplasmic production of KT induces tol phenotype. Comparison between wild-type cells carrying the empty vector pIN-III-ompA2 (vector) or pKT (KT) for periplasm content release (A) (C, cell pellet; S, culture supernatant); RNase I release (B, upper panels); deoxycholate (DOC) susceptibility (B, lower panels) (from top to bottom, 106, 105, and 104 cells were spotted, respectively); release of outer membrane vesicles (C) (arrows indicate vesicles); and susceptibility to colicins K, A, E1, E2, and 5 (D) (from left to right, 100, 10, and 1 ng of colicins have been spotted, respectively). Quantitative survival measurements in the presence of various concentrations of sodium dodecyl sulfate and colicins are shown in Fig. S2 in the supplemental material and reported in Table 1.
TABLE 1.
Phenotypic consequences of the periplasmic production of the colicin K translocation domain
| Strain and IPTG inductiona | Outer membrane stability |
Infection by: |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DOCb | SDSc | RNased | Colicine |
fd bacteriophagef | ||||||
| ColK | ColA | ColE1 | ColE2 | ColIa | Col5 | |||||
| WT pIN-III-ompA2 | ||||||||||
| Noninduced | 94.0 | 2 | − | 0.07 | 0.07 | 0.07 | 0.04 | 0.05 | 0.04 | 2.8 × 10−1 |
| Induced | 92.0 | 2 | − | 0.07 | 0.07 | 0.07 | 0.04 | 0.05 | 0.04 | 3.1 × 10−1 |
| WT pKT | ||||||||||
| Noninduced | 3.0 | 0.35 | + | >500 | >500 | 7 | 500 | 0.05 | 0.04 | 2.1 × 10−2 |
| Induced | 1.3 | 0.35 | + | >500 | >500 | 8 | 500 | 0.05 | 0.04 | 1.5 × 10−2 |
Induction with 10 mM IPTG.
Percent survival on 1% sodium deoxycholate (DOC) plates.
Percent sodium dodecyl sulfate (SDS) concentration required to inhibit 50% of bacterial growth after 120 min (50% lethal dose) (from Fig. S2A in the supplemental material).
Presence (+) or absence (−) of the halo surrounding the bacterial spot on RNA plates.
Colicin concentration (ng/ml) required to inhibit 50% of bacterial growth after 120 min (50% lethal dose) (from Fig. S2B).
Infection frequency, i.e., number of infected cells per total cells.
TABLE 2.
Protection by the KT domain against exogenous colicins or bacteriophage
| Colicin or bacteriophage | Protectiona |
|---|---|
| ColK | >7,000 |
| ColA | >7,000 |
| ColE1 | 100 |
| ColE2 | 12,500 |
| ColIa | 1 |
| Col5 | 1 |
| fd bacteriophage | 20 |
Protection by the KT domain against exogenous colicins or bacteriophage is reported as the ratio of 50% lethal dose of colicin or bacteriophage for WT pKT cells to 50% lethal dose of colicin or bacteriophage for WT pIN-III-ompA2 cells (Table 1).
The colicin K N-terminal domain interacts with the Tol proteins.
The data from the fractionation studies (Fig. 2) and phenotypic analyses (Fig. 3) converge to suggest the hypothesis that, as colicins A, E1, and E3 and the bacteriophage minor capsid protein g3p N-terminal domains do (6, 12, 13, 46), the colicin K N-terminal domain interacts with components of its import machinery.
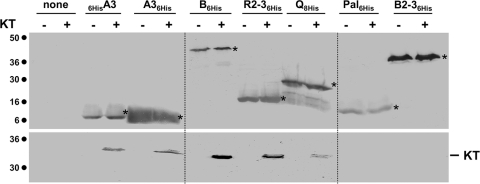
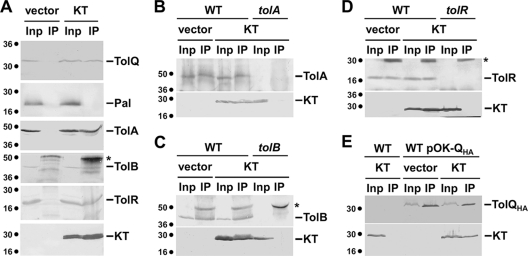
To test whether KT makes contacts with the Tol proteins, we performed in vivo and in vitro protein-protein interaction experiments. We first performed affinity pulldown experiments using purified proteins or domains. His-tagged variants of TolB (6HisTolB), of the C-terminal domain of TolA (TolA36His and 6HisTolA3), of the periplasmic domains of TolR and of TonB (TolR2-36His and 6HisTonB2-3, respectively), of the unacylated Pal protein (Pal6His), and of the full-length TolQ protein (TolQ8His) were purified to homogeneity. Purified proteins were immobilized on cobalt beads, which were then mixed with periplasm extracts from GM1 pIN-III-ompA2 or GM1 pKT cells. As shown in Fig. 4, KT copurified with 6HisTolA3, TolA36His, 6HisTolB, and TolR2-36His and to a lesser extent with TolQ8His, but not with 6HisTonB2-3 or Pal6His.
FIG. 4.
KT interacts in vitro with TolA, TolB, TolQ, and TolR. Copurification experiments using histidine-tagged purified proteins immobilized on ion metal affinity chromatography columns and periplasmic extracts of tolQRABPal cells carrying the empty vector pIN-III-ompA2 (−) or pKT (+). The immobilized protein is indicated (A3, C-terminal domain of the TolA protein; B, full-length TolB protein; R2-3, periplasmic domain of the TolR protein; Q, full-length TolQ protein; Pal, Pal lipoprotein devoid of its acylated N-terminal cysteine residue; B2-3, periplasmic domain of the TonB protein; none, no protein immobilized). Immunodetected His-tagged proteins are indicated by asterisks. Molecular mass markers (in kDa) are indicated on the left.
To confirm these results, we next assayed for formation of immunoprecipitable KT-Tol complexes in GM1 cells producing KT. As shown in Fig. 5A, anti-FLAG antibody precipitated KT and the TolA, TolB, and TolR proteins from extracts of GM1 pKT cells. However, the anti-FLAG antibody did not precipitate the Pal lipoprotein from GM1 pKT extracts. In control experiments, the anti-FLAG antibody did not precipitate the Tol proteins from GM1 pIN-III-ompA2 cell extracts. The interaction of colicin N-terminal domains with the TolQ protein has never been tested due to the lack of specific anti-TolQ antibodies. We thus constructed the pOK-QHA plasmid, allowing the production of a TolQ protein fused to a C-terminal hemagglutinin (HA) epitope tag to which monoclonal antibodies are commercially available. The TolQHA protein is functional as tested by complementation of the TPS13 (tolQ) mutant for cell envelope defects and colicin sensitivity (data not shown). Interestingly, the anti-FLAG antibody precipitated both KT and the TolQHA protein from TPS13 pOK-QHA pKT cell extracts but not from TPS13 pOK-QHA pIN-III-ompA2 extracts (Fig. 3A). We then performed reciprocal immunoprecipitation experiments using specific anti-Tol antibodies (Fig. 5B to E). KT and the corresponding proteins were precipitated from GM1 pKT cell extracts with the anti-TolA (Fig. 5B), -TolB (Fig. 5C), and -TolR (Fig. 5D) antibodies but not with the anti-Pal antibodies (data not shown). Similarly, the anti-HA antibody precipitated TolQHA and KT from TPS13 pOK-QHA pKT cell extracts (Fig. 5E). Overall, results displayed in Fig. 5 showed that KT interacts with the TolA, TolB, TolQ, and TolR proteins but not with the Pal lipoprotein. Since TolQ interacts with several Tol subunits, including TolR and TolA (22, 31), our results cannot rule out the hypothesis that TolQ indirectly interacts with KT. We thus performed coimmunoprecipitation experiments from tolQRA pOK-QHA pKT cell extracts and obtained identical results (data not shown), demonstrating that the TolQ-KT interaction is likely direct. As a further control, we tested whether a FLAG-tagged unrelated protein (SciSp; the periplasmic domain of a type VI secretion-associated protein [3]) interacted with the Tol proteins by coimmunoprecipitation. We did not observe interaction between SciSp-FLAG and any of the Tol proteins (data not shown), demonstrating that the FLAG epitope is not responsible for artifactual interactions.
FIG. 5.
KT interacts in vivo with TolA, TolB, TolQ, and TolR. Detergent-solubilized extracts of wild-type cells producing TolQHA and carrying the empty vector pIN-III-ompA2 (vector) or pKT (KT) producing the FLAG epitope-tagged colicin K N-terminal domain were subjected to immunoprecipitation using the anti-FLAG antibody (A). The total solubilized material (input [Inp]) and the precipitated material (IP) were loaded on a 12.5% acrylamide SDS-PAGE gel and immunodetected with the anti-HA and anti-FLAG monoclonal antibodies and with the anti-Pal, -TolA, -TolB, and -TolR polyclonal antibodies. Detergent-solubilized extracts of the same strains were subjected to immunoprecipitation using anti-TolA (B), anti-TolB (C), anti-TolR (D), and anti-HA (E) (precipitation of the HA epitope-tagged TolQ protein) antibodies. The corresponding mutant strain carrying the KT-producing vector has been used as an additional control for specificity. Immunodetected proteins are indicated on the right. Asterisks indicate heavy or light chains of immunoglobulins. Molecular mass markers (in kDa) are indicated on the left.
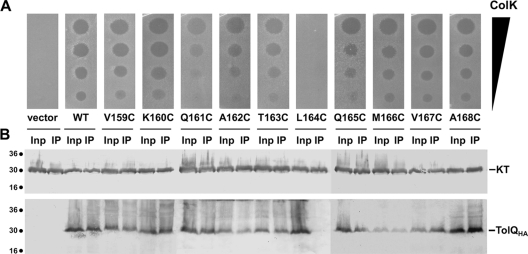
The results of the TolQ-KT interaction suggested that the colicin K N-terminal domain should make contacts with periplasmic regions of the TolQ subunit. TolQ is a three-transmembrane segment (three-TMS)-spanning protein with a 17-amino-acid N-terminal periplasmic tail, a large cytoplasmic domain between TMS1 and TMS2, and a short periplasmic loop connecting TMS2 and TMS3 (residues 159 to 168). We therefore engineered TolQ cysteine substitutions at each position of the periplasmic loop (V159C to A168C). The WT TolQHA protein and each of the mutant derivatives accumulated at comparable levels (data not shown). We then tested these mutant proteins for their ability (i) to restore colicin K susceptibility and (ii) to interact with KT. As shown in Fig. 6, the L164C mutation affected the TolQ-KT interaction and led to susceptibility to colicin K. A less dramatic effect was shown for the Q161C mutation on colicin susceptibility, but this was not accompanied by a defect in the coimmunoprecipitable TolQQ161C-KT complex. This result suggests that the TolQ periplasmic loop participates in colicin K interaction. Because TolQL164C interacts with TolA and TolR (data not shown), this result further confirms that TolQ makes direct contacts with KT, independently of TolR and TolA.
FIG. 6.
The TolQ periplasmic loop L164 residue is required for TolQ-KT interaction. (A) Colicin spot assays using serial dilutions of colicin K (from top to bottom, 1,000, 100, 10, and 1 ng of colicins have been spotted) on tolQ cells producing HA epitope-tagged TolQ (WT) or cysteine variants (V159C to A168C). (B) Detergent-solubilized extracts of tolQ cells coproducing TolQHA or its Cys derivatives and the FLAG epitope-tagged colicin K N-terminal domain were subjected to immunoprecipitation using the anti-FLAG antibody. The total solubilized material (input [Inp]) and the precipitated material (IP) were loaded on a 12.5% acrylamide SDS-PAGE gel and immunodetected with the anti-HA (TolQHA) and anti-FLAG (KT) monoclonal antibodies. Immunodetected proteins are indicated on the right. Molecular mass markers (in kDa) are indicated on the left.
Concluding remarks.
In this study, we first clarified the genetic requirements for colicin K uptake and showed that colicin K requires the products of the tolA, tolB, tolQ, and tolR genes. We then used a combination of fractionation, in vivo coimmunoprecipitation, and in vitro pulldown experiments coupled to phenotypic analyses to characterize colicin K import. Overall, our results demonstrate that the colicin K N-terminal domain interacts with components of its import machinery, including the TolB and TolQ proteins. An important novelty raised by this study is the first evidence for complex formation between a colicin translocation domain and the TolQ protein. This represents a novel interaction involving a colicin and the fourth component of the import machine. This has been made possible by the construction of a plasmid encoding a tagged, functional, and immunoprecipitable TolQ protein. Thus, colicin K interacts with four proteins in the periplasm of E. coli: TolA, TolB, TolQ, and TolR. One of the important questions to address is to determine whether these interactions occur simultaneously or whether there are sequential and hierarchical contacts of the colicin N-terminal domain with periplasmic partners. Interestingly, the three-TMS TolQ protein interacts with the colicin K N-terminal domain targeted into the periplasm, suggesting that a periplasmic portion of TolQ is involved in the interaction. TolQ possesses a periplasmic N-terminal 17-residue extension and a 10-residue periplasmic loop connecting TMS2 and -3, which are therefore interesting candidates for the colicin K binding site. Our work showed that mutation of the TolQ periplasmic residue L164 abolishes colicin K binding and activity. These data suggest that the periplasmic loop is involved in TolQ-KT interaction, but further studies need to determine whether this loop corresponds to the colicin K binding site or whether other TolQ regions are required for an efficient and functional interaction. Interestingly, only a few residues of the periplasmic loop, including L164, are conserved within TolQ proteins (19). Current work in our laboratory is dedicated to delineating regions within the colicin K and TolQ proteins involved in complex formation. Also, despite the colicin K requirements for TolB, colicin K lacks the characteristic TolB box (DG[T,S]GWSSE). The absence of an identifiable TolB box in the colicin K sequence raises the question of how colicin K contacts the TolB protein. Colicins have developed important variations in the uptake mechanism strategy, and colicin K-TolB binding might be an additional example highlighting the important diversity of colicin modes of penetration. Cocrystals between the TolB protein and peptides corresponding to the colicin E9 TolB box or colicin A N-terminal fragment have been obtained, showing that the TolB binding peptides form a penetrating hairpin inside the beta-propeller of the TolB protein (39, 57); however, the modes of interaction of the colicins A and E9 peptides with TolB are distinct, demonstrating that similar TolB binding boxes might interact differently with TolB (57). More generally, distinct modes of interaction have been described for colicins A, E1, and N binding to TolA (28, 51), which may explain why the KT periplasmic production led to different effects on colicin A, E1, or E2 susceptibility. This diversity in the transport mechanisms engendered by colicins (variations in the receptors, the translocators, the import components, and the modes of action) explains how colicin-producing strains are so powerful in colonizing various niches.
Supplementary Material
Acknowledgments
We thank Alain Bernadac for electron microscopy experiments; Daniel Baty and Stéphane Bonetto for the gift of the fd-Tc bacteriophage preparation; Danièle Cavard for the pColK235 plasmid; Marie Guérin for construction of the SciSp-producing plasmid; Emilie Goemaere for TolQ cysteine substitutions; Vincent Géli and Muriel Masi for antibodies; Stéphanie Pommier, Laure Journet, and Anne Walburger for discussions and critical reading of the manuscript; and the members of the Bouveret, Sturgis, and Lloubès research groups for helpful suggestions. We thank the members of the A.B.-A. committee meeting for discussions and Gérard Manvuossicon for encouragements.
Work in this group is supported by grants from the Agence Nationale de la Recherche (SODATOL [ANR-07-BLAN-67] and PEPGLYCOL [ANR-07-MIME-020]) to R.L. and encouraged by the Institut National des Sciences Biologiques of the Centre National de la Recherche Scientifique through a PEPS grant (Projet Exploratoire—Premier Soutien) to E.C. A.B.-A. is a recipient of a doctoral fellowship from the French Ministry of Education and Research.
Footnotes
Published ahead of print on 24 September 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ansaldi, M., M. Lepelletier, and V. Méjean. 1996. Site-specific mutagenesis by using an accurate recombinant polymerase chain reaction method. Anal. Biochem. 234:110-111. [DOI] [PubMed] [Google Scholar]
- 2.Aschtgen, M. S., C. S. Bernard, S. de Bentzmann, R. Lloubès, and E. Cascales. 2008. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 190:7523-7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aschtgen, M. S., M. Gavioli, A. Dessen, R. Lloubès, and E. Cascales. 2010. The SciZ protein anchors the enteroaggregative Escherichia coli type VI secretion system to the cell wall. Mol. Microbiol. 75:886-899. [DOI] [PubMed] [Google Scholar]
- 4.Baty, D., M. Frenette, R. Lloubès, V. Geli, S. P. Howard, F. Pattus, and C. Lazdunski. 1988. Functional domains of colicin A. Mol. Microbiol. 2:807-811. [DOI] [PubMed] [Google Scholar]
- 5.Bénédetti, H., M. Frenette, D. Baty, M. Knibiehler, F. Pattus, and C. Lazdunski. 1991. Individual domains of colicins confer specificity in colicin uptake, in pore-properties and in immunity requirement. J. Mol. Biol. 217:429-439. [DOI] [PubMed] [Google Scholar]
- 6.Bénédetti, H., C. Lazdunski, and R. Lloubès. 1991. Protein import into Escherichia coli: colicins A and E1 interact with a component of their translocation system. EMBO J. 10:1989-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernadac, A., M. Gavioli, J. C. Lazzaroni, S. Raina, and R. Lloubès. 1998. Escherichia coli tol-pal mutants form outer membrane vesicles. J. Bacteriol. 180:4872-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke, J. D., P. Model, and N. D. Zinder. 1982. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol. Gen. Genet. 186:185-192. [DOI] [PubMed] [Google Scholar]
- 9.Bonsor, D. A., I. Grishkovskaya, E. J. Dodson, and C. Kleanthous. 2007. Molecular mimicry enables competitive recruitment by a natively disordered protein. J. Am. Chem. Soc. 129:4800-4807. [DOI] [PubMed] [Google Scholar]
- 10.Bonsor, D. A., O. Hecht, M. Vankemmelbeke, A. Sharma, A. M. Krachler, N. G. Housden, K. J. Lilly, R. James, G. R. Moore, and C. Kleanthous. 2009. Allosteric beta-propeller signalling in TolB and its manipulation by translocating colicins. EMBO J. 28:2846-2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouveret, E., L. Journet, A. Walburger, E. Cascales, H. Bénédetti, and R. Lloubès. 2002. Analysis of the Escherichia coli Tol-Pal and TonB systems by periplasmic production of Tol, TonB, colicin, or phage capsid soluble domains. Biochimie 84:413-421. [DOI] [PubMed] [Google Scholar]
- 12.Bouveret, E., A. Rigal, C. Lazdunski, and H. Bénédetti. 1997. The N-terminal domain of colicin E3 interacts with TolB which is involved in the colicin translocation step. Mol. Microbiol. 23:909-920. [DOI] [PubMed] [Google Scholar]
- 13.Bouveret, E., A. Rigal, C. Lazdunski, and H. Bénédetti. 1998. Distinct regions of the colicin A translocation domain are involved in the interaction with TolA and TolB proteins upon import into Escherichia coli. Mol. Microbiol. 27:143-157. [DOI] [PubMed] [Google Scholar]
- 14.Cao, Z., and P. E. Klebba. 2002. Mechanism of colicin binding and transport through outer membrane porins. Biochimie 84:399-412. [DOI] [PubMed] [Google Scholar]
- 15.Carr, S., C. N. Penfold, V. Bamford, R. James, and A. M. Hemmings. 2000. The structure of TolB, an essential component of the Tol-dependent translocation system, and its protein-protein interaction with the translocation domain of colicin E9. Structure 8:57-66. [DOI] [PubMed] [Google Scholar]
- 16.Cascales, E., A. Bernadac, M. Gavioli, J. C. Lazzaroni, and R. Lloubès. 2002. Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J. Bacteriol. 184:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cascales, E., S. K. Buchanan, D. Duché, C. Kleanthous, R. Lloubès, K. Postle, M. Riley, S. Slatin, and D. Cavard. 2007. Colicin biology. Microbiol. Mol. Biol. Rev. 71:158-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cascales, E., and R. Lloubès. 2004. Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol. Microbiol. 51:873-885. [DOI] [PubMed] [Google Scholar]
- 19.Cascales, E., R. Lloubès, and J. N. Sturgis. 2001. The TolQ-TolR proteins energize TolA and share homologies with the flagellar motor proteins MotA-MotB. Mol. Microbiol. 42:795-807. [DOI] [PubMed] [Google Scholar]
- 20.Davies, J. K., and P. Reeves. 1975. Genetic of resistance to colicins in Escherichia coli K-12: cross-resistance among colicins of group A. J. Bacteriol. 123:102-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deprez, C., L. Blanchard, F. Guerlesquin, M. Gavioli, J. P. Simorre, C. Lazdunski, D. Marion, and R. Lloubès. 2002. Macromolecular import into Escherichia coli: the TolA C-terminal domain changes conformation when interacting with the colicin A toxin. Biochemistry 41:2589-2598. [DOI] [PubMed] [Google Scholar]
- 22.Dérouiche, R., H. Bénédetti, J. C. Lazzaroni, C. Lazdunski, and R. Lloubès. 1995. Protein complex within Escherichia coli inner membrane. TolA N-terminal domain interacts with TolQ and TolR proteins. J. Biol. Chem. 270:11078-11084. [DOI] [PubMed] [Google Scholar]
- 23.Dérouiche, R., G. Zeder-Lutz, H. Bénédetti, M. Gavioli, A. Rigal, C. Lazdunski, and R. Lloubès. 1997. Binding of colicins A and E1 to purified TolA domains. Microbiology 143:3185-3192. [DOI] [PubMed] [Google Scholar]
- 24.Ghrayeb, J., H. Kimura, M. Takahara, H. Hsiung, Y. Masui, and M. Inouye. 1984. Secretion cloning vectors in Escherichia coli. EMBO J. 3:2437-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gillor, O., I. Giladi, and M. A. Riley. 2009. Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol. 9:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gokce, I., E. M. Raggett, Q. Hong, R. Virden, A. Cooper, and J. H. Lakey. 2000. The TolA-recognition site of colicin N. ITC, SPR and stopped-flow fluorescence define a crucial 27-residue segment. J. Mol. Biol. 304:621-632. [DOI] [PubMed] [Google Scholar]
- 27.Hands, S. L., L. E. Holland, M. Vankemmelbeke, L. Fraser, C. J. Macdonald, G. R. Moore, R. James, and C. N. Penfold. 2005. Interaction of TolB with the translocation domain of colicin E9 requires an extended TolB box. J. Bacteriol. 187:6733-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hecht, O., H. Ridley, J. H. Lakey, and G. R. Moore. 2009. A common interaction for the entry of colicin N and filamentous phage into Escherichia coli. J. Mol. Biol. 388:880-893. [DOI] [PubMed] [Google Scholar]
- 29.Howard, S. P., C. Herrmann, C. W. Stratilo, and V. Braun. 2001. In vivo synthesis of the periplasmic domain of TonB inhibits transport through the FecA and FhuA iron siderophore transporters of Escherichia coli. J. Bacteriol. 183:5885-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Journet, L., E. Bouveret, A. Rigal, R. Lloubès, C. Lazdunski, and H. Bénédetti. 2001. Import of colicins across the outer membrane of Escherichia coli involves multiple protein interactions in the periplasm. Mol. Microbiol. 42:331-344. [DOI] [PubMed] [Google Scholar]
- 31.Journet, L., A. Rigal, C. Lazdunski, and H. Bénédetti. 1999. Role of TolR N-terminal, central, and C-terminal domains in dimerization and interaction with TolA and TolQ. J. Bacteriol. 181:4476-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkup, B. C., and M. A. Riley. 2004. Antibiotic-mediated antagonism leads to a bacterial game of rock-paper-scissors in vivo. Nature 428:412-414. [DOI] [PubMed] [Google Scholar]
- 33.Larsen, R. A., M. G. Thomas, and K. Postle. 1999. Protonmotive force, ExbB and ligand-bound FepA drive conformational changes in TonB. Mol. Microbiol. 31:1809-1824. [DOI] [PubMed] [Google Scholar]
- 34.Lazdunski, C. J., E. Bouveret, A. Rigal, L. Journet, R. Lloubès, and H. Bénédetti. 1998. Colicin import into Escherichia coli cells. J. Bacteriol. 180:4993-5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazzaroni, J. C., J. F. Dubuisson, and A. Vianney. 2002. The Tol proteins of Escherichia coli and their involvement in the translocation of group A colicins. Biochimie 84:391-397. [DOI] [PubMed] [Google Scholar]
- 36.Lazzaroni, J. C., P. Germon, M. C. Ray, and A. Vianney. 1999. The Tol proteins of Escherichia coli and their involvement in the uptake of biomolecules and outer membrane stability. FEMS Microbiol. Lett. 177:191-197. [DOI] [PubMed] [Google Scholar]
- 37.Levengood-Freyermuth, S. K., E. M. Click, and R. E. Webster. 1993. Role of the carboxyl-terminal domain of TolA in protein import and integrity of the outer membrane. J. Bacteriol. 175:222-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lloubès, R., E. Cascales, A. Walburger, E. Bouveret, C. Lazdunski, A. Bernadac, and L. Journet. 2001. The Tol-Pal proteins of the Escherichia coli cell envelope: an energized system required for outer membrane integrity? Res. Microbiol. 152:523-529. [DOI] [PubMed] [Google Scholar]
- 39.Loftus, S. R., D. Walker, M. J. Maté, D. A. Bonsor, R. James, G. R. Moore, and C. Kleanthous. 2006. Competitive recruitment of the periplasmic translocation portal TolB by a natively disordered domain of colicin E9. Proc. Natl. Acad. Sci. U. S. A. 103:12353-12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luria, S. E. 1982. The mistaken identity of colicin A. J. Bacteriol. 149:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagel de Zwaig, R., and S. E. Luria. 1967. Genetics and physiology of colicin-tolerant mutants of Escherichia coli. J. Bacteriol. 94:1112-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nomura, M., and C. Witten. 1967. Interaction of colicins with bacterial cells. 3. Colicin-tolerant mutations in Escherichia coli. J. Bacteriol. 94:1093-1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pilsl, H., D. Smajs, and V. Braun. 1999. Characterization of colicin S4 and its receptor, OmpW, a minor protein of the Escherichia coli outer membrane. J. Bacteriol. 181:3578-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pilsl, H., and V. Braun. 1995. Novel colicin 10: assignment of four domains to TonB- and TolC-dependent uptake via the Tsx receptor and to pore formation. Mol. Microbiol. 16:57-67. [DOI] [PubMed] [Google Scholar]
- 45.Pilsl, H., and V. Braun. 1995. Strong function-related homology between the pore-forming colicins K and 5. J. Bacteriol. 177:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pommier, S., M. Gavioli, E. Cascales, and R. Lloubès. 2005. Tol-dependent macromolecule import through the Escherichia coli cell envelope requires the presence of an exposed TolA binding motif. J. Bacteriol. 187:7526-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prilipov, A., P. S. Phale, P. Van Gelder, J. P. Rosenbusch, and R. Koebnik. 1998. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol. Lett. 163:65-72. [DOI] [PubMed] [Google Scholar]
- 48.Raggett, E. M., G. Bainbridge, L. J. Evans, A. Cooper, and J. H. Lakey. 1998. Discovery of critical TolA-binding residues in the bactericidal toxin colicin N: a biophysical approach. Mol. Microbiol. 28:1335-1343. [DOI] [PubMed] [Google Scholar]
- 49.Rijavec, M., M. Budic, P. Mrak, M. Müller-Premru, Z. Podlesek, and D. Zgur-Bertok. 2007. Prevalence of ColE1-like plasmids and colicin K production among uropathogenic Escherichia coli strains and quantification of inhibitory activity of colicin K. Appl. Environ. Microbiol. 73:1029-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riley, M. A., and J. E. Wertz. 2002. Bacteriocins: evolution, ecology, and application. Annu. Rev. Microbiol. 56:117-137. [DOI] [PubMed] [Google Scholar]
- 51.Schendel, S. L., E. M. Click, R. E. Webster, and W. A. Cramer. 1997. The TolA protein interacts with colicin E1 differently than with other group A colicins. J. Bacteriol. 179:3683-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun, T. P., and R. E. Webster. 1987. Nucleotide sequence of a gene cluster involved in entry of E colicins and single-stranded DNA of infecting filamentous bacteriophages into Escherichia coli. J. Bacteriol. 169:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van den Ent, F., and J. Löwe. 2006. RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 67:67-74. [DOI] [PubMed] [Google Scholar]
- 54.Walburger, A., C. Lazdunski, and Y. Corda. 2002. The Tol/Pal system function requires an interaction between the C-terminal domain of TolA and the N-terminal domain of TolB. Mol. Microbiol. 44:695-708. [DOI] [PubMed] [Google Scholar]
- 55.Yamashita, E., M. V. Zhalnina, S. D. Zakharov, O. Sharma, and W. A. Cramer. 2008. Crystal structures of the OmpF porin: function in a colicin translocon. EMBO J. 27:2171-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zakharov, S. D., and W. A. Cramer. 2004. On the mechanism and pathway of colicin import across the E. coli outer membrane. Front. Biosci. 9:1311-1317. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, Y., C. Li, M. N. Vankemmelbeke, P. Bardelang, M. Paoli, C. N. Penfold, and R. James. 2010. The crystal structure of the TolB box of colicin A in complex with TolB reveals important differences in the recruitment of the common TolB translocation portal used by group A colicins. Mol. Microbiol. 75:623-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.