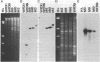
Abstract
Progress in archaebacterial molecular biology requires tools for genetic analysis. We describe vectors that can be selected and maintained in either Halobacterium volcanii or Escherichia coli. A genetic determinant for resistance to the 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitor mevinolin was isolated by "shotgun cloning" into a derivative of the endogenous H. volcanii plasmid pHV2, to form pWL2, which transforms sensitive H. volcanii to mevinolin resistance at high frequency. The resistance determinant, portions of pHV2, and an ampicillin- and tetracycline-resistance-conferring pBR322 derivative, pAT153, were ligated together to form the shuttle vectors pWL101 and pWL102. We describe conditions for the use of these vectors and provide preliminary definition of regions essential for drug resistance and for plasmid replication and maintenance.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts A. W., Chen J., Kuron G., Hunt V., Huff J., Hoffman C., Rothrock J., Lopez M., Joshua H., Harris E. Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3957–3961. doi: 10.1073/pnas.77.7.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertani G., Baresi L. Genetic transformation in the methanogen Methanococcus voltae PS. J Bacteriol. 1987 Jun;169(6):2730–2738. doi: 10.1128/jb.169.6.2730-2738.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera J. A., Bolds J., Shields P. E., Havel C. M., Watson J. A. Isoprenoid synthesis in Halobacterium halobium. Modulation of 3-hydroxy-3-methylglutaryl coenzyme a concentration in response to mevalonate availability. J Biol Chem. 1986 Mar 15;261(8):3578–3583. [PubMed] [Google Scholar]
- Charlebois R. L., Lam W. L., Cline S. W., Doolittle W. F. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8530–8534. doi: 10.1073/pnas.84.23.8530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline S. W., Doolittle W. F. Efficient transfection of the archaebacterium Halobacterium halobium. J Bacteriol. 1987 Mar;169(3):1341–1344. doi: 10.1128/jb.169.3.1341-1344.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett N. R., DasSarma S. Characterization of the small endogenous plasmid of Halobacterium strain SB3 and its use in transformation of H. halobium. Can J Microbiol. 1989 Jan;35(1):86–91. doi: 10.1139/m89-013. [DOI] [PubMed] [Google Scholar]
- Hofman J. D., Schalkwyk L. C., Doolittle W. F. ISH51: a large, degenerate family of insertion sequence-like elements in the genome of the archaebacterium, Halobacterium volcanii. Nucleic Acids Res. 1986 Sep 11;14(17):6983–7000. doi: 10.1093/nar/14.17.6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagramanova V. K., Derckacheva N. I., Mankin A. S. The complete nucleotide sequence of the arcaebacterial plasmid pHSB from Halobacterium, strain SB3. Nucleic Acids Res. 1988 May 11;16(9):4158–4158. doi: 10.1093/nar/16.9.4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe R. J., Harriss J. V., Sharp Z. D., Douglas M. G. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene. 1983 Nov;25(2-3):333–341. doi: 10.1016/0378-1119(83)90238-x. [DOI] [PubMed] [Google Scholar]
- Mevarech M., Werczberger R. Genetic transfer in Halobacterium volcanii. J Bacteriol. 1985 Apr;162(1):461–462. doi: 10.1128/jb.162.1.461-462.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer F., Weidinger G., Goebel W. Characterization of plasmids in halobacteria. J Bacteriol. 1981 Jan;145(1):369–374. doi: 10.1128/jb.145.1.369-374.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomm M., Sherf B. A., Reeve J. N. RNA polymerase-binding and transcription initiation sites upstream of the methyl reductase operon of Methanococcus vannielii. J Bacteriol. 1988 Apr;170(4):1958–1961. doi: 10.1128/jb.170.4.1958-1961.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomm M., Wich G. An archaebacterial promoter element for stable RNA genes with homology to the TATA box of higher eukaryotes. Nucleic Acids Res. 1988 Jan 11;16(1):151–163. doi: 10.1093/nar/16.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Walseth T. F., Johnson R. A. The enzymatic preparation of [alpha-(32)P]nucleoside triphosphates, cyclic [32P] AMP, and cyclic [32P] GMP. Biochim Biophys Acta. 1979 Mar 28;562(1):11–31. doi: 10.1016/0005-2787(79)90122-9. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrell V. E., Nagle D. P., Jr, McCarthy D., Eisenbraun A. Genetic transformation system in the archaebacterium Methanobacterium thermoautotrophicum Marburg. J Bacteriol. 1988 Feb;170(2):653–656. doi: 10.1128/jb.170.2.653-656.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]