Abstract
The peptidoglycan of Selenomonas ruminantium is covalently bound to cadaverine (PG-cadaverine), which likely plays a significant role in maintaining the integrity of the cell surface structure. The outer membrane of this bacterium contains a 45-kDa major protein (Mep45) that is a putative peptidoglycan-associated protein. In this report, we determined the nucleotide sequence of the mep45 gene and investigated the relationship between PG-cadaverine, Mep45, and the cell surface structure. Amino acid sequence analysis showed that Mep45 is comprised of an N-terminal S-layer-homologous (SLH) domain followed by α-helical coiled-coil region and a C-terminal β-strand-rich region. The N-terminal SLH domain was found to be protruding into the periplasmic space and was responsible for binding to peptidoglycan. It was determined that Mep45 binds to the peptidoglycan in a manner dependent on the presence of PG-cadaverine. Electron microscopy revealed that defective PG-cadaverine decreased the structural interactions between peptidoglycan and the outer membrane, consistent with the proposed role for PG-cadaverine. The C-terminal β-strand-rich region of Mep45 was predicted to be a membrane-bound unit of the 14-stranded β-barrel structure. Here we propose that PG-cadaverine possesses functional importance to facilitate the structural linkage between peptidoglycan and the outer membrane via specific interaction with the SLH domain of Mep45.
Polyamines, the ubiquitous polycationic compounds composed of a hydrocarbon backbone with multiple amino groups, exist in all living cells and participate in a wide variety of biological reactions, including DNA, RNA, and protein synthesis (34). However, it has been revealed that some strictly anaerobic eubacteria belonging to the Veillonellaceae family, such as Selenomonas ruminantium, Veillonella alcalescens, Veillonella parvula, and Anaerovibrio lipolyticus, possess polyamines covalently linked to their peptidoglycan (PG) as an essential constituent (8, 16, 17). S. ruminantium possesses a peptidoglycan associated with cadaverine. Cadaverine binds covalently to the α-carboxyl group of the d-glutamic acid residue of peptidoglycan by one of its two amino groups, and the other amino group remains as a free cation (15). In this bacterium, cadaverine is synthesized constitutively from lysine by lysine/ornithine decarboxylase (LDC/ODC [EC 4.1.1.18]), a bifunctional enzyme that decarboxylates both l-lysine and l-ornithine at similar Km and Vmax values (35, 36) and is transferred to a d-glutamic acid residue by a particulate enzyme designated as lipid intermediate:diamine transferase (20). The cadaverine synthesis by LDC/ODC is completely inhibited by dl-α-difluoromethyllysine (DFML) or dl-α-difluoromethylornithine (DFMO), which inhibits the decarboxylating activity toward both l-lysine and l-ornithine (35), and the prevention of the cadaverine synthesis in S. ruminantium was shown to lead to the significant decrease of the amount of the cadaverine covalently linked to peptidoglycan (PG-cadaverine) and result in the growth inhibition (17). Since this inhibitory effect accompanies a drastic morphological change of the cells resulting in an aberrant cell surface structure, PG-cadaverine has been assumed to play a significant role in maintaining the integrity of the cell surface (17).
The cell surface structure of S. ruminantium has a typical Gram-negative three-layer organization, comprising a cytoplasmic membrane, peptidoglycan layer, and outer membrane (18). However, it contains neither the free nor bound form of murein-lipoprotein (19), which plays an important role in the structural linkage between the outer membrane and peptidoglycan, thereby maintaining the structural integrity of the cell surface structures of Gram-negative bacteria (5, 33). The Escherichia coli lpo mutant that lacks murein-lipoprotein becomes hypersensitive to EDTA, resulting in rapid cell lysis upon exposure to EDTA. In contrast, S. ruminantium shows no cell lysis, even in the presence of high concentrations of EDTA, despite the absence of murein-lipoprotein (19). One possible interpretation for these findings was the assumption that PG-cadaverine associates with the structural connection between the outer membrane and peptidoglycan, thereby replacing the function of murein-lipoprotein with an outer membrane component or components. Nevertheless, the factors in the outer membrane interacting with PG-cadaverine have not been identified.
The outer membrane of S. ruminantium contains a 45-kDa major protein (Mep45), which has been proposed to be a peptidoglycan-associating protein (18, 19). Kalmokoff et al. reported that the major outer membrane protein of S. ruminantium OB268, which is similar to Mep45 in size, contains an N-terminal surface-layer homology (SLH) domain (13), a putative functional domain that interacts with cell wall components (27). These findings prompted us to investigate the Mep45 major outer membrane protein of S. ruminantium as a putative outer membrane component interacting with PG-cadaverine. In this report, we characterize the Mep45 protein and its interactions with PG-cadaverine and prove their involvement in the structural linkage between the outer membrane and peptidoglycan.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
S. ruminantium subsp. lactilytica TAM6421 was cultured under anaerobic conditions in Trypticase peptone-yeast extract-glucose (TYG) medium, as described previously (21). The E. coli lpo mutant (37) was cultured in Luria-Bertani (LB) medium supplemented with 0.01% MgSO4·7H2O at 37°C. LB medium was used for culture of E. coli DH5α and BL21(DE3) at 37°C.
Preparation of crude envelopes of S. ruminantium.
Cells were harvested by centrifugation from 200-ml cultures that had reached an optical density at 660 nm (OD660) of 1.0 and resuspended in 10 ml 20 mM sodium-phosphate (NaPi) buffer (pH 7.4). Cells were disrupted by passage through a French pressure cell press at 1,000 lb/in2 (approximately 7.0 × 104 kPa), and unbroken cells were removed by centrifugation at 4,000 × g for 5 min at 4°C. Crude envelopes were pelleted by subsequent centrifugation at 20,000 × g for 20 min at 4°C.
Determination of N-terminal and internal amino acid sequences of Mep45.
The whole proteins of crude envelope preparations of S. ruminantium were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto polyvinylidene difluoride (PVDF) membrane. The protein bands on the membrane were stained with Coomassie brilliant blue (CBB), and the major protein band corresponding to Mep45 was excised and its N-terminal amino acid sequence was analyzed by an automated protein sequencer (ABI491; Applied Biosystems). For sequencing of the internal amino acid sequence, crude envelopes were treated with 100 μg/ml trypsin for the partial digestion of Mep45 that resulted in the formation of a 40-kDa Mep45 fragment that was used for N-terminal sequencing.
Determination of the mep45 nucleotide sequence.
Degenerated oligonucleotide primers were synthesized based on the determined N-terminal (DVPADHW) and internal (AAEFAEE) amino acid sequences of Mep45, namely, 5′-GAYGTICCIGCNGAYCAYTGG-3′ and 5′-TCYTCIGCRAAYTCNGCNGC-3′. These primers were designed to amplify a 209-bp fragment by PCR from the chromosomal DNA of S. ruminantium. Using this 209-bp fragment as a probe, a mep45-coding 4-kbp DNA fragment was identified from PstI-digested chromosomal DNA of S. ruminantium by Southern blot analysis. The 4-kbp fragment was extracted from the agarose gel and used as the template for inverse PCR (36). The amplified fragment was sequenced with the BigDye terminator cycle sequencing ready reaction kit, using an ABI377 DNA sequencer (Applied Biosystems).
Amino acid sequence analysis of Mep45.
Homology analysis was performed using the BLAST algorithm (version 2.2.18) implemented with the EMBL/ GenBank/DDBJ database. A multiple alignment of amino acid sequences was created using the ClustalW program (version 1.83). Prediction of the secondary structure of Mep45 was performed by the PHD method on the PredictProtein server (http://www.predictprotein.org/) (26). Prediction of transmembrane strands and the topology of Mep45 was performed using the PRED-TMBB program (http://bioinfomatics.biol.uoa.gr/PRED-TMBB) (1). Other basic analytical procedures were performed using the GENETYX program (Software Development Co., Tokyo, Japan).
Determination of the protease-accessible site of Mep45.
Intact cells (3 mg wet weight) or crude inside-out envelopes of S. ruminantium, prepared by disruption of the cells with a French pressure cell press (1 mg wet weight), were resuspended in 100 μl 20 mM NaPi buffer containing either 100 μg/ml proteinase K or 100 μg/ml trypsin and incubated at 37°C for 30 min. The reaction was stopped by the addition of phenylmethanesulfonyl fluoride (1 mM final concentration) to the mixture, and the cells and crude inside-out envelopes were then collected by centrifugation at 5,000 × g for 5 min and at 20,000 × g for 20 min, respectively. Total proteins in these preparations were resolved by SDS-PAGE using 12.5% acrylamide gels. The N-terminal amino acid sequences of Mep45 fragments were determined according to the method described above.
Purification of Mep45 and preparation of Mep45 with the SLH domain truncated (ΔSLH-Mep45).
Crude envelopes of S. ruminantium prepared from cells harvested from 200 ml culture at an OD660 of 1.0 were incubated in 8 ml of 1% (wt/vol) Triton X-100 in NaPi buffer for 15 min at 37°C. Mep45 was separated from other membrane proteins as a Triton X-100-resistant precipitate by centrifugation at 20,000 × g for 15 min at 4°C. Mep45 was then extracted from this Triton X-100-insoluble material by incubation in 1.6 ml of 5 M guanidine hydrochloride at 37°C for 30 min. After centrifugation at 20,000 × g for 20 min at room temperature (RT), the supernatant containing Mep45 proteins was obtained and dialyzed against NaPi buffer to remove the guanidine hydrochloride.
Crude envelopes of S. ruminantium were prepared and resuspended in NaPi buffer containing 100 μg/ml of trypsin and incubated at 37°C for 1 h. This enzymatic treatment generates truncation of SLH domain comprising the N-terminal 68 amino acid residues from Mep45. Further purification of ΔSLH-Mep45 was performed according to the method for purification of intact Mep45 as described above.
Construction, expression, and purification of the SLH domain-GST fusion protein (G-SLH).
The DNA fragment encoding the SLH domain of Mep45 (1ASNPF to ALVDK68) was amplified from chromosomal DNA of S. ruminantium using the primers 5′-CGGGATCCGCTAGCAACCCGTTCTCCGATG-3′ (primer I) and 5′-CGGAATTCTCATTTGTCAACCAGAGCCTTGTCC-3′ (primer II), containing BamHI and EcoRI sites, respectively (underlined). The BamH-EcoRI fragment of the amplified product was inserted into the BamHI-EcoRI site of pGEX-4T-1 (GE Healthcare) to obtain the plasmid pGSLH. This plasmid was then introduced into E. coli BL21(DE3) to express the N-terminal glutathione S-transferase (GST)-fused SLH domain (G-SLH) under the control of the tac promoter. E. coli BL21(DE3) harboring pGSLH was grown in 20 ml LB medium containing 100 μg/ml ampicillin at 37°C with shaking, and isopropyl-β-d-thiogalactopyranoside (IPTG; 1 mM final concentration) was added at an OD660 of 0.6. After incubation for an additional 2 h, cells were harvested by centrifugation at 4°C and disrupted by sonication. The cell lysate was centrifuged at 200,000 × g for 1 h at 4°C, and the supernatant was loaded onto a 1-ml GSTap FF column (GE Healthcare) equilibrated with NaPi buffer. After the column had been washed extensively with NaPi buffer, G-SLH was eluted with 50 mM Tris-HCl (pH 8.0) containing 10 mM reduced glutathione. The G-SLH preparation was dialyzed against NaPi buffer at 4°C.
Preparation of peptidoglycan and quantification of PG-cadaverine.
S. ruminantium cells were harvested from 100 ml medium at an OD660 of 1.0 and washed once with NaPi buffer. Cells were suspended in 4 ml of 5% SDS and boiled for 15 min. Insoluble materials were then precipitated by centrifugation at 20,000 × g for 20 min at room temperature (RT) and washed three times with distilled water. Pellets were suspended in 4 ml NaPi buffer containing 100 μg/ml α-amylase and incubated for 12 h at 37°C to degrade contaminating high-molecular-weight (HMW) glycogen. The peptidoglycan preparation was obtained by centrifugation at 20,000 × g for 20 min at RT. Then it was suspended again in 4 ml of 5% SDS, boiled for 15 min, and collected again by centrifugation at 20,000 × g for 20 min. After being washed three times with distilled water, the sample was treated with 1 ml of 10% trichloroacetic acid at 4°C for 30 min. Purified peptidoglycan was collected by centrifugation at 20,000 × g for 20 min at RT and washed extensively with NaPi buffer.
The peptidoglycan of the E. coli lpo mutant was prepared according to the procedure for S. ruminantium peptidoglycan, except for the α-amylase treatment.
The quantification of PG-cadaverine and purified peptidoglycan was performed by first hydrolyzing the sample with 6 M HCl at 110°C for 20 h. The HCl was then removed using an evaporator, and the hydrolysates were dissolved in distilled water. The amounts of cadaverine and d-glutamic acid residue were quantified by high-performance liquid chromatography (HPLC) using TSK-gel Polyaminepak and TSK-gel Aminopak columns (Tosoh, Tokyo, Japan), respectively, by the method described previously (35) with 1 mM cadaverine or glutamic acid as the standard.
Preparation of the peptide moiety of the peptidoglycan (peptide A) and lysozyme-digested peptidoglycan fragment (PG fragment) from S. ruminantium.
Preparation of the peptide moiety of the peptidoglycan [peptide A; l-Ala-d-Glu(cadaverine)-meso-diaminopimelic acid (DAP) -d-Ala] was done according to the method described previously (14). Fifty milligrams of the lyophilized peptidoglycan was digested by 500 μg of Streptomyces albus G-enzymes, which contain glycosidase, N-acetylmuramyl-l-alanine amidase, and endopeptidase (kindly donated by K. Kato, Department of Microbiology and Oral Bacteriology, Osaka University, Osaka, Japan). Digestion was done for 20 h at 37°C in 5 ml of 10 mM NaPi buffer (pH 7.4) containing 50 μl of toluene. Peptide A was purified from the reaction mixture by descending paper chromatography on Whatman 3MM paper at room temperature for 50 h, using the upper phase of n-butanol-acetic acid-water (4:1:5 [vol/vol/vol]) as a solvent. The major band corresponding to peptide A, which was detectable by ninhydrin, was eluted with distilled water and lyophilized. The molecular mass of the purified peptide A preparation was confirmed to correspond to the estimated value (545 Da) by mass spectrometry.
For preparation of lysozyme-digested peptidoglycan fragment (PG fragment), 50 mg of lyophilized peptidoglycan was digested with 100 μg of egg white lysozyme (Wako Pure Chemicals, Japan). Digestion was done in 5 ml of 10 mM NaPi buffer (pH 8.0) for 20 h at 37°C. The reaction mixture was lyophilized and suspended again in 500 μl distilled water. The PG fragment was purified from this suspension by gel filtration chromatography using a G3000SW column (Tosoh, Japan). Only one PG fragment corresponding to a molecular mass of approximately 30 kDa was obtained. The PG fragment was collected and lyophilized.
Peptidoglycan-binding assay.
Fifty microliters of NaPi buffer containing 5 μg protein was mixed with various concentrations of peptidoglycan (0.2 to 2.0 mg wet weight), and allowed to stand for 90 min at RT. The mixture was then centrifuged at 20,000 × g for 20 min at RT to separate the peptidoglycan-associated fractions (precipitates) and non-peptidoglycan-associated fractions (supernatant). The relative amounts of peptidoglycan-associated proteins were quantified by densitometry using the NIH Image program (version 1.63) following immunoblotting and detection with anti-Mep45 antiserum (for Mep45 and ΔSLH-Mep45) and anti-GST antiserum (for G-SLH; GE Healthcare).
Cross-linking experiment.
Purified Mep45 (60 μg/ml) was treated with 0.05% glutaraldehyde or 1 mM BS3 (bis[sulfosuccinimidyl] suberate) (Pierce, Rockford, IL) and incubated at 37°C for 5, 10, or 30 min. The reaction was terminated by the addition of Tris-HCl (pH 7.4; 10 mM final concentration), followed by an additional incubation for 15 min. The resulting Mep45 was analyzed by SDS-PAGE using 7.5% acrylamide gels, and the Mep45 oligomer was detected by immunoblotting using anti-Mep45 antiserum.
Electron microscopy.
S. ruminantium cells were cultivated in 10 ml culture with or without 10 mM DFMO. After incubation for 4 h at 37°C, cells were harvested by centrifugation at 400 × g for 10 min, resuspended in 2% glutaraldehyde, and allowed to stand at RT for 1.5 h. The cells were pelleted by centrifugation at 400 × g for 10 min and then mixed with 2% agar and drawn into the tip of a Pasteur pipette. After the agar solidified, it was extruded from the pipette and washed in 20 mM sodium phosphate buffer (pH 7.4). The agar block was sliced into small cakes and fixed in 1% osmium tetroxide in Veronal buffer for 1.5 h at RT. Agar cakes were then dipped into ethanol at increasing concentrations (50, 60, 70, 80, 90, and 100% for 30 min each) to dehydrate the sample, and then ethanol was replaced by propylene oxide (three times for 30 min). The sample was then embedded in Quetol651 (Nisshin EM Co., Tokyo, Japan) and allowed to stand at 60°C for 20 h for polymerization. Thin sectioning was carried out using diamond knife on an ultramicrotome (Ultracut S; Leica) and mounted onto 150-mesh copper grids, and stained with 4% uranyl acetate and 0.4% lead citrate. Observation was performed using a transmission electron microscope (H-8100; Hitachi Co., Tokyo, Japan) at an accelerating voltage of 100 kV.
Preparation of anti-Mep45 antiserum and immunoblotting.
Anti-Mep45 rabbit antiserum was developed against oligopeptides corresponded to internal amino acid sequence of Mep45 (145DANSNLKSDQGEDS159) that were custom ordered from Operon Biotechnologies (Tokyo, Japan).
Proteins were resolved by SDS-PAGE and electroblotted onto PVDF membrane. Proteins on the membranes were cross-reacted with appropriate antisera, detected using alkaline phosphatase-conjugated anti-rabbit immunoglobulin G (Promega, Madison, WI), and visualized using nitroblue tetrazolium (Wako Pure Chemicals, Japan) and 5-bromo-4-chloro-indolylphosphate (Wako Pure Chemicals, Japan) as substrates.
Chemicals.
dl-α-Difluoromethylornithine (DFMO) was a kind gift from P. Wosten (Wayne State University, United States) and the Merrel Daw Research Institute (Cincinnati, OH).
Nucleotide sequence accession number.
The nucleotide sequence of mep45 has been deposited in the DDBJ/GenBank/EMBL database under accession no. AB252707.
RESULTS
Primary sequence profiles of Mep45.
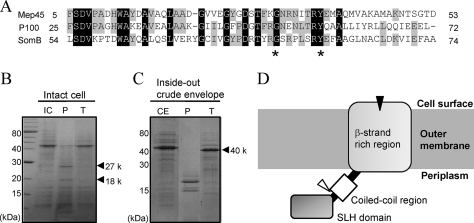
The entire nucleotide sequence of mep45 was 1,299 bp long, encoding a protein of 432 amino acid (aa) residues, and it contained a typical prokaryote-type signal peptide located at the N-terminal 23 aa residues. The molecular mass of mature Mep45 with 409 amino acid residues was calculated as 45,540 Da, which was consistent with an estimated size from the mobility on SDS-PAGE. The deduced amino acid sequence revealed that the N-terminal region of mature Mep45 showed significant homology to the SLH domain of envelope proteins from bacteria belonging to the Deinococcus-Thermus group and cyanobacteria, such as P100 from Thermus thermophilus (60% identity) (4) and SomB from Synechococcus sp. strain PCC6301 (53% identity) (9), respectively. The SLH-bearing proteins are essentially classified into three groups: the S-layer proteins mostly studied in Gram-positive bacteria, the extracellular enzymes and proteins, and the outer membrane proteins from Gram-negative bacteria (3). The SLH domain of the outer membrane proteins possesses the following specific features: it exists in a single copy at the N terminal, and it contains particularly conserved Gly33 (relative position 33) and Tyr40 residues (3). Mep45 is classified as an SLH-bearing outer membrane protein and possesses a single copy of the SLH domain with the typical Gly and Tyr residues, which are designated in Fig. 1A. Another region of Mep45 showed significant homology to the OmpM1 outer membrane protein of Mitsuokella multacida (62% identity) (12).
FIG. 1.
Amino acid sequence profiles of Mep45 and determination of the protease-accessible site. (A) Amino acid sequence alignment of the N-terminal SLH domain of Mep45 with the corresponding region of P100 and SomB. Asterisks denote highly and specifically conserved residues found in SLH-bearing outer membrane proteins. (B and C) SDS-PAGE of proteinase K or trypsin treatment of intact cells (B) and crude envelopes (C) of S. ruminantium. Intact cells (3 mg) or crude envelopes (1 mg) were incubated with 100 μg/ml proteinase K or trypsin at 37°C for 30 min. Gels were stained with Coomassie brilliant blue. The bands designated by the arrowheads are the fragments of Mep45 produced by the protease treatment. Abbreviations: IC, intact cells; CE, crude envelopes; P, proteinase K-treated sample; T, trypsin-treated sample. (D) Schematic representation of the topology of Mep45. The locations of proteolytic sites of proteinase K and trypsin are indicated as black and open triangles, respectively. The N-terminal SLH domain protrudes into the periplasmic space, and the C-terminal β-strands span the outer membrane, except for at least one region exposed to the external cell surface, which is the protease-sensitive site.
On the basis of the homology search and secondary structure prediction, the domain arrangement of Mep45 was suggested to be comprised of (i) a single copy of the N-terminus SLH domain, (ii) an α-helical stretch adjacent to the SLH domain with a high propensity for coiled-coil formation, and (iii) a C-terminal β-strand-rich region. This domain arrangement is common to the SLH-bearing outer membrane proteins found in the bacteria belonging to the Deinococcus-Thermus group and cyanobacteria, including P100 and SomB. The C-terminal β-strand-rich regions of these proteins are proposed to form a membrane-bound β-barrel structure, and the pore-forming activity of this region was experimentally demonstrated in SomB (10). Hence, Mep45 is supposed to construct the membrane-bound unit within its C-terminal β-strand-rich region, and the N-terminal SLH domain of this protein likely acts as a modular domain appended to this region (Fig. 1D).
The N-terminal SLH domain is exposed to periplasmic space.
To clarify the fundamental topology profile of Mep45 in the outer membrane, we investigated the cell-surface- or periplasm-exposed regions of Mep45 by determining the protease-accessible site. To determine the cell-surface-exposed region, intact cells of S. ruminantium were treated with proteinase K or trypsin and the digestion pattern was analyzed by SDS-PAGE (Fig. 1B). Proteinase K digested Mep45 into two major fragments of 27 and 18 kDa, whereas trypsin showed no digestion. The N-terminal amino acid sequence of the 27-kDa fragment corresponded to the N-terminal sequence of mature Mep45, indicating that the N terminus of Mep45 was masked from proteinase K digestion at the cell surface. Conversely, the N-terminal sequence of the 18-kDa fragment possessed the internal sequence of Mep45 (238GRHDD242) located in the C-terminal β-strand-rich region. This suggests that this proteolytic site is exposed to the cell surface.
Similarly, to assess the periplasm-exposed region of Mep45, the crude envelope fractions of S. ruminantium were investigated by treatment with the same proteases (Fig. 1C). Trypsin digestion of Mep45 produced a major fragment of 40 kDa, and its N-terminal sequence corresponded to the internal sequence (70LAAEF75). This proteolytic site was located at the junction between the SLH domain and coiled-coil region, indicating that the N-terminal SLH domain before 70L is exposed to the periplasmic space. The other portion of Mep45 was resistant to trypsin, despite evenly distributed lysine and arginine residues. Proteinase K digestion of Mep45 produced a predominant band of 20 kDa, but the internal sequence could not be determined because it contained several peptides.
Taken together, these observations suggest that the N-terminal SLH domain of Mep45 protruded into the periplasmic space and the C-terminal β-strand-rich region possesses at least one site that is exposed to the cell surface (Fig. 1D).
Mep45 interacts with peptidoglycan in a manner dependent on cadaverine covalently linked to peptidoglycan.
The peptidoglycan-binding ability of Mep45 was investigated by mixing with purified peptidoglycan from wild-type S. ruminantium, and the peptidoglycan-associated and non-peptidoglycan-associated fractions were separated by centrifugation after incubation for 90 min. The presence of Mep45 in each fraction was detected by Western immunoblotting (Fig. 2A). When Mep45 was mixed with peptidoglycan from S. ruminantium, it precipitated as a peptidoglycan-associated form, representing the peptidoglycan-binding ability of Mep45. However, this interaction was decreased when peptidoglycan was prepared from S. ruminantium grown in medium containing DFMO (data not shown). This finding suggests the importance of peptidoglycan-bound cadaverine (PG-cadaverine) for the interaction between Mep45 and peptidoglycan.
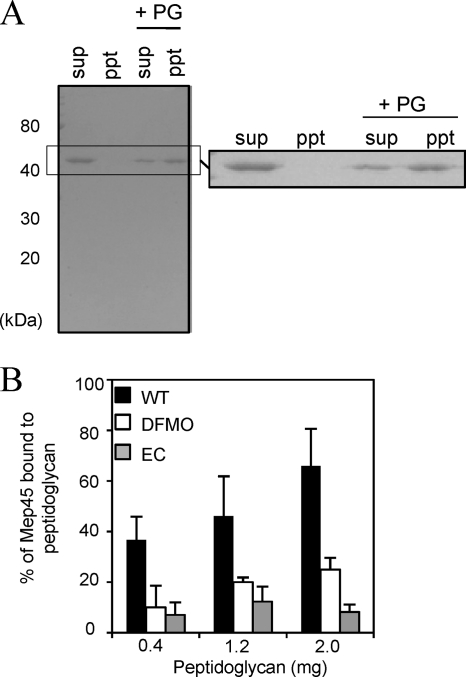
FIG. 2.
In vitro peptidoglycan-binding assay. (A) Western immunoblot of Mep45 binding to wild-type S. ruminantium peptidoglycan. Solubilized Mep45 (3 μg) was mixed with or without 1 mg of purified peptidoglycan (PG). The peptidoglycan-associated Mep45 (ppt) and non-peptidoglycan-associated Mep45 in the supernatant fraction (sup) were detected by using anti-Mep45 antiserum. (B) Effect of the PG-cadaverine on the interaction between Mep45 and peptidoglycan. Relative amounts of peptidoglycan-associated Mep45 were quantified densitometrically following immunoblotting. Solubilized Mep45 (5 μg) was incubated with 0.4, 1.2, or 2.0 mg of purified peptidoglycan from (i) wild-type S. ruminantium cells (WT [closed bars]), (ii) S. ruminantium cells cultivated in the presence of 10 mM DFMO (open bars), and (iii) the E. coli lpo mutant (EC [shaded bars]). The values represent the ratio of the amount of Mep45 after binding to peptidoglycan over the initial amount (mean ± standard deviation [SD] of triplicate experiments).
To confirm whether PG-cadaverine is involved in the interaction between Mep45 and peptidoglycan, the peptidoglycan-binding assay was performed using the following three types of peptidoglycan preparations: (i) peptidoglycan from S. ruminantium that possesses d-glutamic acid residues saturated 100% with covalently linked cadaverine (wild type), (ii) peptidoglycan from S. ruminantium cells cultivated in the presence of 10 mM DFMO that possesses peptidoglycans containing covalently linked cadaverine in approximately 35% of d-glutamic acid residues (deficient type), and (iii) peptidoglycan from the E. coli lpo mutant containing neither cadaverine nor murein-lipoprotein (null type). The assay was performed on 0.4, 1.2, or 2.0 mg of each peptidoglycan, and the relative amounts of peptidoglycan-associated Mep45 were compared (Fig. 2B). It was determined that less Mep45 was captured by the cadaverine-deficient peptidoglycan or the peptidoglycan from the E. coli lpo mutant than wild-type S. ruminantium peptidoglycan. Approximately 70% of Mep45 added to the reaction mixture was captured in the peptidoglycan fraction by the addition of 2 mg of wild-type peptidoglycan. In contrast, approximately 25% and 10% of Mep45 was captured by the cadaverine-deficient peptidoglycan and the peptidoglycan from the E. coli lpo mutant, respectively. Addition of free cadaverine to the reaction mixture had no effect on the ability to bind to peptidoglycan (data not shown), indicating that only the bound form but not the free form of cadaverine had an effect on the binding ability of Mep45. These results strongly suggest that the interaction between Mep45 and peptidoglycan was highly dependent on the presence of PG-cadaverine.
Mep45 interacts with peptidoglycan through its N-terminal SLH domain.
The SLH domain of Mep45 was determined to protrude inward into the periplasmic space, and hence interaction with peptidoglycan is likely. To investigate the function of the SLH domain of Mep45, Mep45 with the SLH domain truncated (ΔSLH-Mep45) was prepared and analyzed for peptidoglycan-binding ability using the peptidoglycan from wild-type S. ruminantium (Fig. 3A and B). It was found that the ΔSLH-Mep45 showed drastically decreased binding to the peptidoglycan compared to the intact Mep45. Approximately 15% of ΔSLH-Mep45 was captured by the addition of 2 mg of peptidoglycan. The basal level of ΔSLH-Mep45 bound to peptidoglycan did not increase as the amount of peptidoglycan increased; therefore, anything detected here is nonspecific. This result suggests that the SLH domain of Mep45 is responsible for the ability to bind to peptidoglycan.
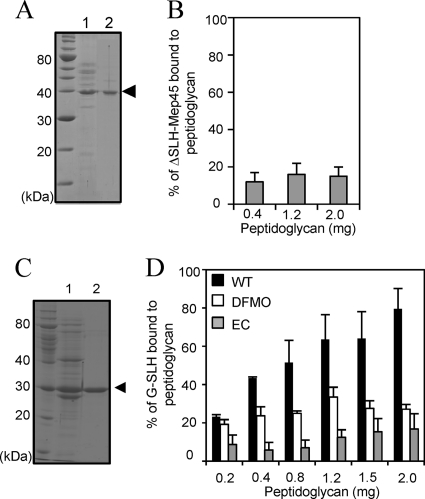
FIG. 3.
Functional analysis of the SLH domain of Mep45. (A) Preparation of Mep45 with the SLH domain truncated (ΔSLH-Mep45). Lane 1, trypsin-digested crude envelopes; lane 2, purified ΔSLH-Mep45 preparation. The arrowhead indicates the ΔSLH-Mep45 bands. (B) Peptidoglycan-binding assay of ΔSLH-Mep45. Solubilized ΔSLH-Mep45 (100 μg/ml) was incubated with 0.4, 1.2, or 2.0 mg of peptidoglycan from wild-type S. ruminantium cells. (C) Preparation of recombinant SLH domain fused with glutathione S-transferase (G-SLH). Lane 1, cell extracts of E. coli BL21(DE3) expressing G-SLH; lane 2, purified G-SLH. The arrowhead indicates the G-SLH band. (D) Peptidoglycan-binding assay of G-SLH of (i) wild-type S. ruminantium cells (WT [closed bars]), (ii) S. ruminantium cells cultured in the presence of 10 mM DFMO (open bars), and (iii) the E. coli lpo mutant (EC [shaded bars]). In panels B and D, anti-glutathione S-transferase antiserum was used to detect G-SLH and relative amounts of peptidoglycan-associated G-SLH were represented as the amount relative to initial amount of G-SLH added to the reaction mixture (mean ± SD of triplicate experiments).
To examine the peptidoglycan-binding property of the SLH domain, the recombinant SLH domain was fused to glutathione S-transferase (G-SLH) and the peptidoglycan-binding assay was performed using three types of peptidoglycan with different cadaverine substitutions, as described above (Fig. 3C and D). It was determined that G-SLH showed the ability to bind to the peptidoglycan of wild-type S. ruminantium in a dose-dependent manner. In contrast, the relative amount of peptidoglycan-associated G-SLH was observed to be drastically decreased for cadaverine-deficient peptidoglycan and the peptidoglycan from the E. coli lpo mutant.
These results indicate that G-SLH associates with the peptidoglycan in a PG-cadaverine-dependent manner, providing evidence that the SLH domain acts as the functional domain of Mep45 for interacting with the peptidoglycan of wild-type S. ruminantium.
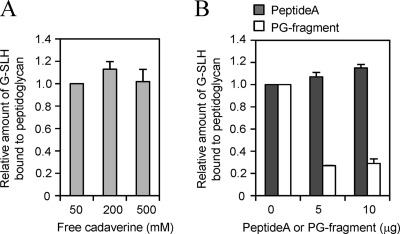
The entire structure of the peptidoglycan is required for the interaction with the SLH domain.
The SLH domain of Mep45 was revealed to bind to the peptidoglycan in a manner dependent on the presence of PG-cadaverine. However, it remained unclear whether it binds directly to the PG-cadaverine, since the exogenous free cadaverine did not inhibit the binding (Fig. 4). To clarify the structural constituent of the peptidoglycan involved in the interaction with SLH domain, we prepared the peptide moiety of the peptidoglycan [peptide A; l-Ala-d-Glu(cadaverine)-mesoDAP-d-Ala] and lysozyme-digested peptidoglycan fragment (PG fragment). We examined the inhibitory effect of these two samples on the interaction between the SLH domain and intact peptidoglycan (Fig. 4). The peptide A or PG fragment was exogenously added to the reaction mixture containing G-SLH and the intact peptidoglycan, and the binding ability of G-SLH was examined. As a result, the PG fragment inhibited the binding of G-SLH to the intact peptidoglycan, but peptide A did not. This indicates that peptide A itself is not the binding ligand for the SLH domain, and the entire structure of the peptidoglycan, especially the presence of saccharide moiety, is required for the interaction. Thus, we conclude that the PG-cadaverine itself does not bind immediately to the SLH domain but mediates the interaction between the SLH domain and the peptidoglycan in an indirect way.
FIG. 4.
The structural constituent of the peptidoglycan required for interaction with SLH domain. The assay for G-SLH binding to peptidoglycan was performed in the presence of free cadaverine (A) or peptide A or the PG fragment (B). The free cadaverine, peptide A, or PG fragment was exogenously added to the reaction mixture containing G-SLH and 1 mg of peptidoglycan from wild-type S. ruminantium, and the relative amount of G-SLH bound to the peptidoglycan was measured.
PG-cadaverine-deficient S. ruminantium cells display the decreased number of linkages between the outer membrane and peptidoglycan.
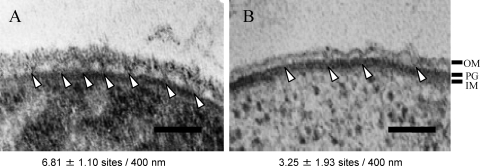
Specific interactions between Mep45 and PG-cadaverine are expected to serve as the structural linkage between peptidoglycan and outer membrane. To this end, we examined the cell-surface structure of wild-type S. ruminantium cells and PG-cadaverine-deficient cells (Fig. 5). The cell surface of wild-type cells showed a typical Gram-negative-type three-layer structure comprised of an inner membrane, peptidoglycan, and an outer membrane with a ruffled structure, as previously reported (19). As shown in Fig. 5A, the outer membrane and peptidoglycan layers are apparently connected each other at several points on the cell surface. The number of connection sites per 400-nm cell surface was 6.81 ± 1.10. In contrast, this linkage was largely absent from the cell surface of cells cultivated in the presence of 10 mM DFMO, which results in a 65% reduction of PG-cadaverine (Fig. 5B). The number of connection sites of this cell was decreased to 3.25 ± 1.93, displaying approximately 55% reduction compared to the wild-type cell. These observations are consistent with the notion that PG-cadaverine is involved in the structural linkage between the outer membrane and peptidoglycan.
FIG. 5.
Electron micrograph of cell surface structures of wild-type S. ruminantium cells (A) and PG-cadaverine-deficient cells (B). Arrowheads indicate the sites where the outer membrane (OM) and peptidoglycan are connected. IM, inner membrane. The numbers of the connection sites per 400 nm of cell surface were counted from 40 cells each. The difference between the wild-type cell and PG-cadaverine-deficient cell was significant (P < 0.05 according to Student's t test). Bar, 100 nm.
Mep45 forms an unusual high-molecular-weight complex.
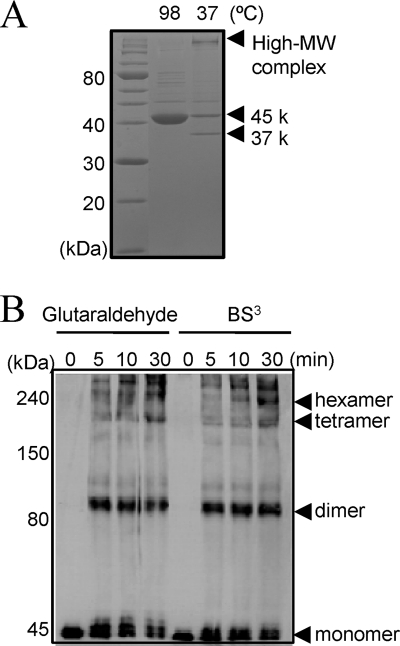
From previous research, Mep45 was shown to form SDS-resistant oligomers in the outer membrane (19). The significant stability in SDS is one of the typical features of porins in general, whose oligomer does not dissociate unless heated at high temperatures in 2% SDS (11, 24). Without heat denaturation, the oligomeric form of porin is able to be clearly distinguished by SDS-PAGE as the protein band of the oligomer, which migrates to the appropriate position representing the molecular weight of the oligomer. To examine the structural profiles, purified Mep45 was analyzed under heated and unheated conditions by SDS-PAGE (Fig. 6A). The heat-denatured Mep45 was observed as a single protein band of 45 kDa on SDS-PAGE. However, when Mep45 was analyzed under the unheated condition, the protein band remained in the well of the SDS-PAGE gel, implying the formation of a high-molecular-weight (HMW) complex of Mep45 oligomers (Fig. 6A). In addition, a faster-migrating band of 37 kDa was observed under the unheated condition. This likely represents a typical feature of β-barrel proteins generally referred to as “heat modifiability,” in which the faster-migrating band represented by the unheated sample is considered to be caused by the incomplete denaturing of the protein (22, 23). The presence of the 37-kDa protein band of Mep45 suggests the formation of the β-barrel structures, consistent with predicted structural analyses based on the primary sequence analysis of Mep45.
FIG. 6.
Structural analysis of Mep45. (A) SDS-PAGE analysis of Mep45 under heated or unheated conditions. 45 k and 37 k, 45 kDa and 37 kDa, respectively. (B) Subunit structure of Mep45 determined by cross-linking experiment using glutaraldehyde or BS3. Mep45 oligomers were detected by immunoblotting using anti-Mep45 antiserum.
To analyze the subunit structure, the purified Mep45 preparation was treated with 0.05% glutaraldehyde or 1 mM BS3 (bis[sulfosuccinimydyl] suberate) and the cross-linked oligomer was analyzed by Western immunoblotting using anti-Mep45 antiserum following SDS-PAGE (Fig. 6B). It was determined that the dimer, tetramer, and hexamer forms of Mep45 were detected, indicating that dimeric Mep45 molecules existed as a structural subunit involved in the formation of the HMW Mep45 complex.
DISCUSSION
On the basis of the observations shown here, Mep45 is considered to play the role of an anchor protein that connects the outer membrane and peptidoglycan. Based on the primary sequence analysis, Mep45 was suggested to be comprised of essentially two distinct domains, the N-terminal SLH domain and the C-terminal β-strand-rich region (Fig. 1). The N-terminal SLH domain of Mep45 was determined to protrude into the periplasmic space, acting as the functional domain for interacting with peptidoglycan (Fig. 1 and 3). On the other hand, the C-terminal β-strand-rich region is supposed to be the membrane-bound unit (Fig. 1).
The interaction between Mep45 and peptidoglycan occurred in a manner dependent on the presence of PG-cadaverine (Fig. 2 and 3), suggesting the necessity of PG-cadaverine for the structural linkage between the outer membrane and peptidoglycan in this bacterium. The electron micrographs of cell surface structure strongly support this, in which the number of arch-like outer membrane structures connecting with peptidoglycan was apparently decreased according to the deficiency of the amount of PG-cadaverine (Fig. 5).
The PG-cadaverine-dependent interaction between the SLH domain and peptidoglycan displays the novel binding mechanism between them, while a number of SLH domains studied so far have been shown to bind to the secondary cell wall polymers (27). However, the PG-cadaverine itself does not serve as the binding ligand for the SLH domain, since it was determined that the entire structure of the peptidoglycan, especially the presence of the saccharide moiety, was required for the binding of the SLH domain (Fig. 4). Thus, we conclude that PG-cadaverine plays a functional role for mediating the interaction between the SLH domain and the peptidoglycan in an indirect way. Further analysis is desired to clarify the binding mechanism(s).
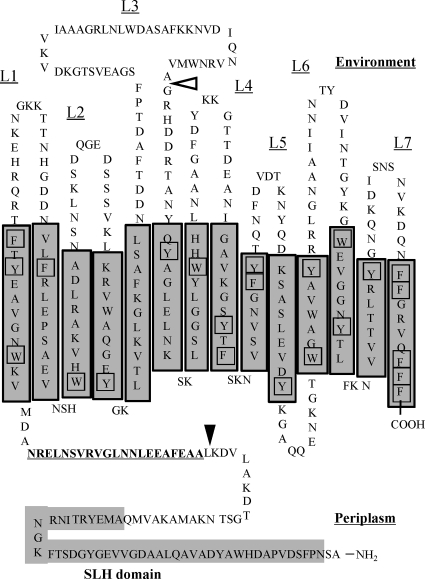
The C-terminal β-strand-rich region of Mep45 is considered to form β-barrel structure. Based on the analysis of the transmembrane strand using the PRED-TMBB program (1), the C-terminal region of Mep45 is predicted to be a 14-stranded β-barrel (Fig. 7). This topology model is supported by the following features which are typical for β-barrel proteins: (i) short periplasmic loops with long variable external loops, (ii) the preferential location of aromatic residues at the near ends of the β-strands contacting the membrane surface, and (iii) the presence of a C-terminal phenylalanine residue typical for β-barrel proteins important for correct folding (28, 32). The large extracellular loop domain (L3) is shown to contain the region accessible by proteinase K (Fig. 1B), and hence this region is externally exposed from the β-barrel. Similarly, the large loop-like L3 domain is also proposed in the predicted structure of OmpM1 of M. multacida (12), but its functional importance remains to be elucidated. In addition, the biochemical features of Mep45 described below are consistent with those of β-barrel proteins. (i) As shown in Fig. 1, the β-barrel-rich region was shown to be trypsin resistant despite having evenly distributed lysine and arginine residues in its primary sequence. High resistance to tryptic digestion is one of the typical features of β-barrel proteins and is often used as a criterion for monitoring correct folding of the β-barrel structure (32). (ii) Purified Mep45 showed heat modifiability on SDS-PAGE that also represents a typical feature of β-barrel proteins (Fig. 6).
FIG. 7.
Proposed model of Mep45 in the outer membrane. Shown are PRED-TMBB program predictions of transmembrane strands and the topology of Mep45. Transmembrane strands are shaded. The amino acid sequence with predicted high propensity to form an α-helical coiled-coil structure is underlined. Aromatic residues located near the ends of the transmembrane strand contacting the membrane surfaces are boxed. Proteinase K- and trypsin-sensitive cleavage sites are indicated as open and closed triangles, respectively.
On the other hand, Mep45 possesses some features different from general β-barrel proteins like porins. Mep45 forms far larger high-molecular-weight complexes than the oligomeric form of general porins. The Mep45 protein is not readily solubilized by detergents but can be extracted with guanidine hydrochloride, which is a chaotropic reagent commonly used to solubilize S-layer proteins, ubiquitous cell-surface proteins among Gram-positive bacteria (27). Kalmokoff et al. previously reported the presence of the major outer membrane protein, which is similar to Mep45 in size, constructing an S-layer-like crystalline array on the cell surface of S. ruminantium OB268, and characterized it as the regularly ordered outer membrane protein representing a kind of primitive intermediate between S-layers and Gram-negative outer membrane proteins (13). Therefore, Mep45 in our strain may also share several features of S-layer proteins.
We examined the number of Mep45 molecule per single cell by immunoblot analysis using the purified Mep45 as a standard and calculated the number of Mep45 molecules in 1 μm2 of cell surface according to Smit et al. (30; data not shown). The estimated number of Mep45 molecules was 1.78 × 105 molecules/1 μm2 of cell surface, which means that approximately 80 to 100% of the cell surface is expected to be covered by the Mep45 molecules based on the hypothesis that Mep45 is equivalent in size to the β-barrel proteins such as OmpF and α-hemolysin, whose structures have been determined to be 16- and 14-stranded β-barrels, respectively, as resolved by X-ray crystallography (29). Thus, it appears reasonable that Mep45 covers a large part of the cell surface and plays the role of the attachment system anchoring the outer membrane to the peptidoglycan in this bacterium.
The mechanism of structural linkages that occur between the outer membrane and peptidoglycan in S. ruminantium proposed here is very different from that of conventional Gram-negative bacteria possessing the murein-lipoprotein and Tol-Pal system (5, 6, 33). The Tol-Pal system is a highly conserved multiprotein apparatus shared by Gram-negative bacteria, comprised of the TolA inner membrane protein, TolQ, TolR, the TolB periplasmic protein, and the Pal peptidoglycan-associated protein. Mutation of the genes encoding these proteins leads to functional and morphological alterations of the envelope, such as hypersensitivity to external damaging agents, formation of membrane blebs, and leakage of periplasmic components. Genome sequence data from S. ruminantium (unpublished) suggest that none of the Tol-Pal system genes are conserved in this organism. However, phenotypic defects observed in E. coli mutants with mutations in murein-lipoprotein or the Tol-Pal system have not been detected in S. ruminantium. These facts suggest that the structural linkage between the outer membrane and peptidoglycan in S. ruminantium, comprised of Mep45 and PG-cadaverine, seems to act as a unique anchoring system able to replace the function of the murein-lipoprotein and Tol-Pal system. However, it remains unknown whether the defects in cell morphology induced by DFMO (17) are just a consequence of the lack of PG-cadaverine or are related to other inhibitory effects occurring in concert with this induction.
Based on gene homology analyses, Mep45-like proteins were suggested to be conserved among the bacteria in the Veillonellaceae family. Since these bacteria also possess polyamines covalently linked to peptidoglycan (8, 16), specific interaction between Mep45-like outer membrane proteins and peptidoglycan-linked polyamines is likely to occur. Hence, this mechanism of structural linkage between the outer membrane and peptidoglycan would be widely distributed among the Veillonellaceae. In addition, it is worth emphasizing that Mep45 is closely related to the SLH-bearing outer membrane proteins found in the Deinococcus-Thermus group bacteria and cyanobacteria (Fig. 1). The relationship between the SLH-bearing outer membrane protein and cell wall was examined in P100 from T. thermophilus, which was shown to interact with the secondary cell wall polymer through its SLH domain and was proposed to serve as an anchoring protein (2). In case of cyanobacteria, there have been no reports about the interaction between such proteins and peptidoglycan, but the covalent binding of N-acetylputrescine to the peptidoglycan has been described for the chloroplasts of Cyanophora paradoxa (25), implying the possible occurrence of the polyamine-containing peptidoglycan in cyanobacteria and binding mechanism within its SLH-bearing outer membrane protein. Therefore, among these bacteria, SLH-bearing outer membrane proteins possibly play the common functional role involved in the anchoring system of the outer membrane to peptidoglycan, through the interaction between the SLH domain and the cell wall components. Phylogenetic analyses have determined that bacteria from the Veillonellaceae family, Deinococcus-Thermus group, and cyanobacteria are far distant from conventional Gram-negative bacteria (7, 31), implying that the Gram-negative-type cell surface structures of these bacteria might be maintained by mechanisms different from those of conventional Gram-negative bacteria.
Acknowledgments
We are grateful to K. Itoh, H. Ishida, and S. Ohwada for technical assistance with electron microscopy and T. Yamada for mass spectrometry measurements.
Footnotes
Published ahead of print on 17 September 2010.
REFERENCES
- 1.Bagos, B. G., T. D. Liakopoulos, I. C. Spyropoulos, and S. J. Hamodrakas. 2004. PRED-TMBB: a web server for predicting the topology of β-barrel outer membrane proteins. Nucleic Acids Res. 32:W400-W404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cava, F., M. A. dePedro, H. Schwarz, A. Henne, and J. Berenguer. 2004. Binding to pyruvylated compounds as an ancestral mechanism to anchor the outer envelope in primitive bacteria. Mol. Microbiol. 52:677-690. [DOI] [PubMed] [Google Scholar]
- 3.Engelhardt, H., and J. Peters. 1998. Structural research on surface layers: a focus on stability, surface layer homology domains, and surface layer-cell wall interactions. J. Struct. Biol. 124:276-302. [DOI] [PubMed] [Google Scholar]
- 4.Faraldo, M. M., M. A. dePedro, and J. Berenguer. 1992. Sequence of S-layer gene of Thermus thermophilus HB8 and functionality of its promoter in Escherichia coli. J. Bacteriol. 174:7458-7462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fung, J., T. J. MacAlister, and L. I. Rothfield. 1978. Role of murein lipoprotein in morphogenesis of the bacterial division septum: phenotypic similarity of lkyD and lpo. J. Bacteriol. 133:1467-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Godlewska, R., K. Wiśniewska, Z. Pietras, and E. K. Jagusztyn-Krynicka. 2009. Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol. Lett. 298:1-11. [DOI] [PubMed] [Google Scholar]
- 7.Gupta, R. S. 1998. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62:1435-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamana, K. 1999. Distribution of cell wall-linked polyamines within the Gram-negative anaerobes of the subbranch Sporomusa belonging phylogenetically to Gram-positive taxa. Microbios 100:145-157. [Google Scholar]
- 9.Hansel, A., F. Pattus, U. J. Jürgens, and M. H. Tadros. 1998. Cloning and characterization of the genes coding for two porins in the unicellular cyanobacterium Synechococcus PCC 6301. Biochim. Biophys. Acta 1399:31-39. [DOI] [PubMed] [Google Scholar]
- 10.Hansel, A., and M. H. Tadros. 1998b. Characterization of two pore-forming proteins isolated from the outer membrane of Synechococcus PCC 6301. Curr. Microbiol. 36:321-326. [DOI] [PubMed] [Google Scholar]
- 11.Hasegawa, Y., H. Yamada, and S. Mizushima. 1976. Interaction of outer membrane proteins 0-8 and 0-9 with peptidoglycan sacculus of Escherichia coli K-12. J. Biochem. 80:1401-1409. [DOI] [PubMed] [Google Scholar]
- 12.Kalmokoff, M. L., J. W. Austin, T. D. Cyr, M. A. Hefford, R. M. Teather, and L. B. Selinger. 2009. Physical and genetic characterization of an outer-membrane protein (OmpM1) containing an N-terminal S-layer-like homology domain from the phylogenetically Gram-positive gut anaerobe Mitsuokella multacida. Anaerobe 15:74-81. [DOI] [PubMed] [Google Scholar]
- 13.Kalmokoff, M. L., J. W. Austin, M. F. Whitford, and R. M. Teather. 2000. Characterization of a major envelope protein from the rumen anaerobe Selenomonas ruminantium OB268. Can. J. Microbiol. 46:295-303. [DOI] [PubMed] [Google Scholar]
- 14.Kamio, Y., Y. Itoh, and Y. Terawaki. 1981. Chemical structure of peptidoglycan in Selenomonas ruminantium: cadaverine links covalently to the d-glutamic acid residue of peptidoglycan. J. Bacteriol. 146:49-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamio, Y., Y. Itoh, Y. Terawaki, and T. Kusano. 1981. Cadaverine is covalently linked to peptidoglycan in Selenomonas ruminantium. J. Bacteriol. 145:122-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamio, Y., and K. Nakamura. 1987. Putrescine and cadaverine are constituents of peptidoglycan in Veillonella alcalescens and Veillonella parvula. J. Bacteriol. 169:2881-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamio, Y., H. Pösö, Y. Terawaki, and L. Paulin. 1986. Cadaverine covalently linked to a peptidoglycan is an essential constituent of the peptidoglycan necessary for the normal growth in Selenomonas ruminantium. J. Biol. Chem. 261:6585-6589. [PubMed] [Google Scholar]
- 18.Kamio, Y., and H. Takahashi. 1980. Isolation and characterization of outer and inner membrane of Selenomonas ruminantium lipid compositions. J. Bacteriol. 141:888-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamio, Y., and H. Takahashi. 1980. Outer membrane proteins and cell surface structure of Selenomonas ruminantium. J. Bacteriol. 141:899-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamio, Y., Y. Terawaki, and K. Izaki. 1982. Biosynthesis of cadaverine-containing peptidoglycan in Selenomonas ruminantium. J. Biol. Chem. 257:3326-3333. [PubMed] [Google Scholar]
- 21.Kanegasaki, S., and H. Takahashi. 1967. Function of growth factors for rumen microorganisms. I. Nutritional characteristics of Selenomonas ruminantium. J. Bacteriol. 93:456-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mogensen, J. E., and D. E. Otzen. 2005. Interactions between folding factors and bacterial outer membrane proteins. Mol. Microbiol. 57:326-346. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura, K., and S. Mizushima. 1976. Effects of heating in dodecyl sulfate solution on the conformation and electrophoretic mobility of isolated major outer membrane proteins from Escherichia coli K-12. J. Biochem. 80:1411-1422. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido, H., and M. Vaara. 1985. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 49:1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfanzagl, B., A. Zenker, E. Pittenauer, G. Allmaier, J. Martinez-Torrecuadrada, E. R. Schmid, M. A. dePedro, and W. Löffelhardt. 1996. Primary structure of cyanelle peptidoglycan of Cyanophora paradoxa: a prokaryotic cell wall as part of an organelle envelope. J. Bacteriol. 178:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rost, B., and C. Sander. 1993. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc. Natl. Acad. Sci. U. S. A. 90:7558-7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sára, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulz, G. E. 1994. Structure-function relationships in porins as derived from a 1.8 Å resolution crystal structure, p. 343-362. In J. M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier, Amsterdam, Netherlands.
- 29.Schulz, G. E. 2002. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta 1565:308-317. [DOI] [PubMed] [Google Scholar]
- 30.Smit, J., Y. Kamio, and H. Nikaido. 1975. Outer membrane of Salmonella typhimurium: chemical analysis and freeze-fracture studies with lipopolysaccharide mutants. J. Bacteriol. 124:942-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stackebrandt, E., H. Pohla, R. Kroppenstedt, H. Hans, and C. R. Woese. 1985. 16S rRNA analysis of Sporomusa, Selenomonas, and Megasphaera: on the phylogenetic origin of Gram-positive eubacteria. Arch. Microbiol. 143:270-276. [Google Scholar]
- 32.Struyvé, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki, H., Y. Nishimura, S. Yasuda, A. Nishimura, M. Yamada, and Y. Hirota. 1978. Murein-lipoprotein of Escherichia coli: a protein involved in the stabilization of bacterial cell envelope. Mol. Gen. Genet. 167:1-9. [DOI] [PubMed] [Google Scholar]
- 34.Tabor, C. W., and H. Tabor. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takatsuka, Y., M. Onoda, T. Sugiyama, K. Muramoto, T. Tomita, and Y. Kamio. 1999. Novel characteristics of Selenomonas ruminantium lysine decarboxylase capable of decarboxylating both L-lysine and L-ornithine. Biosci. Biotechnol. Biochem. 63:1063-1069. [DOI] [PubMed] [Google Scholar]
- 36.Takatsuka, Y., Y. Yamaguchi, M. Ono, and Y. Kamio. 2000. Gene cloning and molecular characterization of lysine decarboxylase from Selenomonas ruminantium delineate its evolutionary relationship to ornithine decarboxylase from eukaryotes. J. Bacteriol. 182:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yem, D. W., and H. C. Wu. 1978. Physiological characterization of an Escherichia coli mutant altered in the structure of murein lipoprotein. J. Bacteriol. 133:1419-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]