Abstract
A fitness cost due to imbalanced replichores has been proposed to provoke chromosome rearrangements in Salmonella enterica serovars. To determine the impact of replichore imbalance on fitness, the relative fitness of isogenic Salmonella strains containing transposon-held duplications of various sizes and at various chromosomal locations was determined. Although duplication of certain genes influenced fitness, a replichore imbalance of up to 16° did not affect fitness.
The bacterial chromosome is a dynamic molecule that can undergo various types of rearrangements, including inversions, translocations, and duplications. These rearrangements can alter gene order and change replichore length. Replichores are defined as the halves of the chromosome between the origin of replication and the terminus region in the vicinity of the dif site (4, 6, 10, 15). In most bacteria, both replichores are approximately the same length, with each comprising 180° around the circular chromosome. In this state, the replichores and DNA replication are balanced. Imbalance is introduced when one replichore becomes longer than the other. For example, if the replichores comprise 200° and 160° around the circular chromosome, the replichores are 20° imbalanced. Various studies over the years have investigated the effect of imbalanced DNA replication on fitness in Escherichia coli (8, 9, 16, 17). In a recent analysis, E. coli strains that contained asymmetrical interreplichore inversions were found to have growth defects when the imbalance was at least 50° (8). While these studies have demonstrated that imbalanced replichores can affect fitness, the approaches used to introduce the imbalance either do not occur or are extremely rare in nature.
Duplications play important evolutionary roles because the increase in gene copy number can facilitate adaptation to certain growth conditions (20), as well as being a source for the evolution of new genes (3). Typically duplications occur at frequencies between 10−3 and 10−5 in unselected cultures (2) but are lost at rates of up to 1,000-fold more often if they do not provide a selective advantage (19). Duplications also result in replichore imbalance. However, the effect of duplications on fitness in relation to replichore balance has not been investigated.
A major hurdle in studying the effects of duplications on fitness is that if the duplication is detrimental, haploid revertants will outgrow the parental strain containing the duplication. To circumvent this problem, we used a set of 11 isogenic Salmonella enterica strains with transposon-held duplications (5) (Fig. 1 and Table 1). Culturing these strains in the presence of chloramphenicol selects for the duplication because if the duplication collapses, the transposon is lost and the cells become chloramphenicol sensitive. As the size of the duplications in these strains varies, so does the amount of introduced replichore imbalance, ranging from 5° to 23°. In addition, fitness effects due to the location of the duplication versus the size of the duplication were also discerned, as the collection includes duplications of similar sizes located in different regions of the chromosome.
FIG. 1.
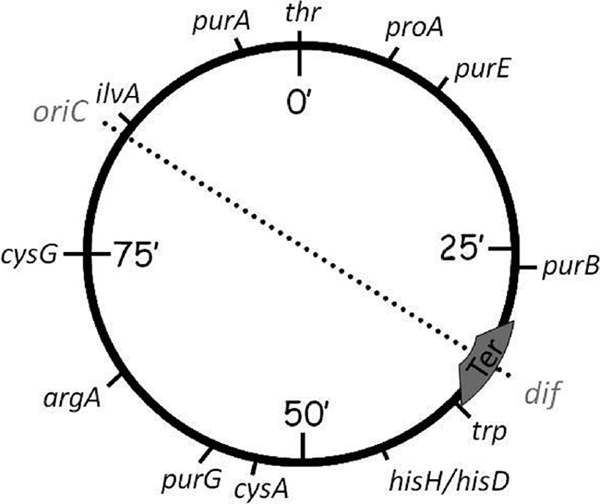
Genetic map of S. enterica serovar Typhimurium LT2 showing genes used as endpoints in constructing transposon-held duplications of the regions between genes. Balanced replichores are indicated by the symmetry of the oriC-Ter axis. Duplications increase the length of one replichore relative to the other, imbalancing axis symmetry.
TABLE 1.
Properties of the S. enterica strains used in this study
| Strain | Alias | Genotype | Duplication location (min) | Duplication size (kbp) | Replichore imbalance (°) | Reference |
|---|---|---|---|---|---|---|
| MST1 | LT2 | Wild type | 1.9 | 11 | ||
| MST3813 | SV4200 | Dup [trp248 MudP hisD9953] | 38-44 | 332.1 | 11.5 | 5 |
| MST3814 | SV3193 | Dup [hisH9962 MudP cysA1586] | 44-53 | 399.7 | 13.7 | 5 |
| MST3815 | SV4015 | Dup [cysA1586 MudP purG2149] | 53-56 | 158.4 | 5.7 | 5 |
| MST3816 | SV4193 | Dup [purG2149 MudP argA9001] | 56-64 | 430.7 | 14.7 | 5 |
| MST3817 | SV4194 | Dup [argA9000 MudP cysG1573] | 64-75 | 484.6 | 16.3 | 5 |
| MST3818 | SV1601 | Dup [cysG1573 MudP ilvA2642] | 75-85 | 486.4 | 16.4 | 5 |
| MST3819 | SV4195 | Dup [ilvA2648 MudP purA1881] | 85-95 | 495.3 | 16.7 | 5 |
| MST3820 | SV4142 | Dup [purA1881 MudP thr469] | 95-0 | 248.7 | 8.8 | 5 |
| MST3821 | SV1604 | Dup [thr469 MudP proA692] | 0-8 | 367.1 | 12.6 | 5 |
| MST3822 | SV1603 | Dup [proA692 MudQ purE2164] | 8-12 | 230.2 | 8.1 | 5 |
| MST3823 | SV1611 | Dup [purE2514 MudP purB1879] | 12-27 | 723.2 | 23.3 | 5 |
| MST1529 | TT11183 | srl-203::Tn10d(Cam) | 1 | |||
| TYT4480 | rrfH::pCE36 | This work | ||||
| MST5198 | rrfH::pCE36 srl-203::Tn10d(Cam) | This work |
The relative fitness of these strains was investigated by measuring growth rates and by assaying growth in a mixed culture with an isogenic competitor strain. Single colony isolates streaked from frozen stocks were used to inoculate broth cultures. Growth was measured in E medium supplemented with 0.2% glucose (minimal medium), Luria-Bertani medium (LB), or LB supplemented with 1× E-salts and 0.2% glucose (LBEDO) (18). Competition assays were done in LB. Media were supplemented with chloramphenicol (20 mg/ml) to maintain the duplications, and the solid media used in competition experiments also contained 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal; 40 mg/ml) to differentiate between the duplication and competitor strains.
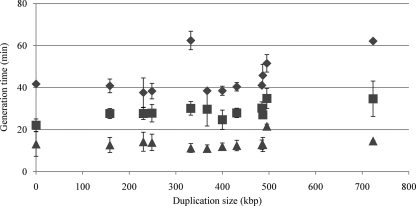
In minimal medium, most of the strains had a generation time of approximately 40 min, except for MST3813, MST3823, and MST3819, which grew somewhat slower (Fig. 2). In LB, the average generation times of all of the duplication-bearing strains were longer than that of the wild type. When grown in LBEDO, all of the strains had generation times of 11 to 15 min. Observed differences in growth rate were not due to lower viability as determined by plate counts. There was no correlation between duplication size and generation time in any of the three media.
FIG. 2.
Generation times of strains as a function of duplication size. Strains were grown in minimal medium (♦), LB (▪), or LBEDO (▴) aerobically at 37°C. Error bars show standard deviations.
Competition indices were determined from mixed cultures with the isogenic competitor strain MST5198 and either the wild-type strain or one of the duplication-bearing strains. The competitor strain contained an integrated plasmid encoding a promoterless lacZ gene (7) driven by the rrnH promoter and Tn10d(Cam) conferring chloramphenicol resistance integrated into the srl locus (Table 1). Strains were grown to saturation overnight, and 10−4 dilutions were used to inoculate mixed cultures. Samples taken at t = 0 (input) and subsequent time points (output) were diluted and spot plated in triplicate. The competition index (C.I.) was calculated as follows: C.I. = (no. output Lac− CFU/no. output Lac+ CFU)/(no. input Lac− CFU/no. input Lac+ CFU).
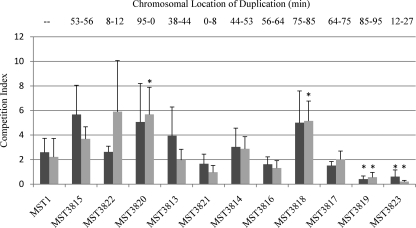
Data are shown for 6 and 12 h, but readings were taken at time points of up to 1 week. There was no correlation between the duplication size and the C.I. (Fig. 3). Most of the duplication-bearing strains competed as well as or slightly better than the wild type against the competitor strain. Two exceptions were MST3819 and MST3823, which both competed significantly more poorly against the competitor strain than did the wild-type strain (Student's t test P < 0.05). While these two strains contain the largest duplications, the size of the duplication in MST3819 was close to the size of the duplications in MST3817 and MST3818. Strains MST3817 and MST3818 were either similar or slightly better competitors than the wild type, indicating that the fitness defect in MST3819 is not due to replichore imbalance.
FIG. 3.
Competition indices of the wild-type and duplication-bearing strains after 6 (▪) and 12 h (░⃞) of growth in mixed cultures with MST5198 (n = 6). Strains are in order of increasing duplication size, with the locations of duplications indicated at the top. Indices statistically significantly different (P < 0.05) from that of the wild-type strain (MST1) are marked (*). Error bars show standard deviations.
To confirm that the duplications in MST3819 and MST3823 were responsible for the observed growth defects, isolates of each strain with collapsed duplications were obtained by growing cultures of each strain without chloramphenicol and then screening for chloramphenicol-sensitive derivatives. The loss of the duplication restored wild-type growth to both strains. The frequencies of duplication collapse in cultures of MST3818 and MST3819 were also compared. Since these two strains have duplications of similar sizes, the collapse frequencies of the duplicated regions, and concomitant loss of chloramphenicol resistance, were expected to be similar. While the reversion frequency of MST3818 cells was 3.5 × 10−3 cells/generation, MST3819 cells reverted with a frequency of 2.8 × 10−2 cells/generation, resulting in 97 to 98% of MST3819 cells being chloramphenicol sensitive and haploid after reaching stationary phase.
In conclusion, the results of this study show that a replichore imbalance of up to 16° introduced by transposon-held duplications in the chromosome does not have a measurable effect on the fitness of S. enterica. The duplicated regions in these strains are larger than the islands acquired via horizontal gene transfer, which have been purported to disrupt replichore balance sufficiently to promote chromosome rearrangements. A duplication that introduced a replichore imbalance of 23° did have a growth defect, but whether the defect is from replichore imbalance or the gene content of the duplication could not be distinguished. This study suggests that the disruption of replichore balance by acquisition of horizontally transferred genes is not the cause of chromosome rearrangements in host-specific S. enterica serovars as hypothesized (12-14).
Acknowledgments
We thank Josep Casadesús for kindly providing the duplication-bearing strains, Moselio Schaechter for valuable discussions and advice, and Michele McKeand for technical assistance.
This research was supported in part by grant U54CA132384 from the National Cancer Institute.
Footnotes
Published ahead of print on 24 September 2010.
REFERENCES
- 1.Altman, E., J. R. Roth, A. Hessel, and K. E. Sanderson. 1996. Transposons currently in use in genetic analysis of Salmonella species, p. 2613-2626. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 2.Anderson, R. P., and J. R. Roth. 1979. Gene duplication in bacteria: alteration of gene dosage by sister-chromosome exchanges. Cold Spring Harb. Symp. Quant. Biol. 43(Pt. 2):1083-1087. [DOI] [PubMed] [Google Scholar]
- 3.Bergthorsson, U., D. I. Andersson, and J. R. Roth. 2007. Ohno's dilemma: evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. U. S. A. 104:17004-17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 5.Camacho, E. M., and J. Casadesús. 2001. Genetic mapping by duplication segregation in Salmonella enterica. Genetics 157:491-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Capiaux, H., F. Cornet, J. Corre, M. I. Guijo, K. Perals, J. E. Rebollo, and J. M. Louarn. 2001. Polarization of the Escherichia coli chromosome. A view from the terminus. Biochimie 83:161-170. [DOI] [PubMed] [Google Scholar]
- 7.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 8.Esnault, E., M. Valens, O. Espeli, and F. Boccard. 2007. Chromosome structuring limits genome plasticity in Escherichia coli. PLoS Genet. 3:e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill, C. W., and B. W. Harnish. 1981. Inversions between ribosomal RNA genes of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 78:7069-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hill, T. M. 1996. Features of the chromosomal terminus region, p. 1602-1614. In F. C. Neidhardt, R. Curtis III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC.
- 11.Lilleengen, K. 1948. Typing of Salmonella typhimurium by means of bacteriophage. Acta Pathol. Microbiol. Scand. Suppl. 77:11-125. [Google Scholar]
- 12.Liu, G. R., W. Q. Liu, R. N. Johnston, K. E. Sanderson, S. X. Li, and S. L. Liu. 2006. Genome plasticity and ori-ter rebalancing in Salmonella typhi. Mol. Biol. Evol. 23:365-371. [DOI] [PubMed] [Google Scholar]
- 13.Liu, G. R., A. Rahn, W. Q. Liu, K. E. Sanderson, R. N. Johnston, and S. L. Liu. 2002. The evolving genome of Salmonella enterica serovar Pullorum. J. Bacteriol. 184:2626-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, S. L., and K. E. Sanderson. 1996. Highly plastic chromosomal organization in Salmonella typhi. Proc. Natl. Acad. Sci. U. S. A. 93:10303-10308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lobry, J. R., and J. M. Louarn. 2003. Polarisation of prokaryotic chromosomes. Curr. Opin. Microbiol. 6:101-108. [DOI] [PubMed] [Google Scholar]
- 16.Louarn, J., J. Patte, and J. M. Louarn. 1982. Suppression of Escherichia coli dnaA46 mutations by integration of plasmid R100.1. derivatives: constraints imposed by the replication terminus. J. Bacteriol. 151:657-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louarn, J. M., J. P. Bouche, F. Legendre, J. Louarn, and J. Patte. 1985. Characterization and properties of very large inversions of the E. coli chromosome along the origin-to-terminus axis. Mol. Gen. Genet. 201:467-476. [DOI] [PubMed] [Google Scholar]
- 18.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria: a laboratory manual. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 19.Reams, A. B., E. Kofoid, M. Savageau, and J. R. Roth. 2010. Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics 184:1077-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonti, R. V., and J. R. Roth. 1989. Role of gene duplications in the adaptation of Salmonella typhimurium to growth on limiting carbon sources. Genetics 123:19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]