Abstract
Two-component systems (TCS) and phosphorelay systems are mechanisms used by bacteria and fungi to quickly adapt to environmental changes to produce proteins necessary for survival in new environments. Bacterial pathogens use TCS and phosphorelay systems to regulate genes necessary to establish infection within their hosts, including type III secretion systems (T3SS). The Yersinia enterocolitica ysa T3SS is activated in response to NaCl by YsrS and YsrR, a putative hybrid sensor kinase and a response regulator, respectively. Hybrid TCS consist of a sensor kinase that typically has three well-conserved sites of phosphorylation: autophosphorylation site H1, D1 within a receiver domain, and H2 in the histidine phosphotransferase (HPt) domain. From H2, the phosphoryl group is transferred to D2 on the response regulator. A curious feature of YsrS is that it lacks the terminal HPt domain. We report here the identification of the HPt-containing protein (YsrT) that provides this activity for the Ysr system. YsrT is an 82-residue protein predicted to be cytosolic and α-helical in nature and is encoded by a gene adjacent to ysrS. To demonstrate predicted functions of YsrRST as a phosphorelay system, we introduced alanine substitutions at H1, D1, H2, and D2 and tested the mutant proteins for the ability to activate a ysaE-lacZ reporter. As expected, substitutions at H1, H2, and D2 resulted in a loss of activation of ysaE expression. This indicates an interruption of normal protein function, most likely from loss of phosphorylation. A similar result was expected for D1; however, an intriguing “constitutive on” phenotype was observed. In addition, the unusual feature of a separate HPt domain led us to compare the sequences surrounding the ysrS-ysrT junction in several Yersinia strains. In every strain examined, ysrT is a separate gene, leading to speculation that there is a functional advantage to YsrT being an independent protein.
Yersinia enterocolitica is a human pathogen that causes several gastrointestinal conditions, with symptoms that include fever, vomiting, and diarrhea. In an otherwise healthy host, the infection is usually self-limiting, lasting 1 to 2 weeks (44). However, individuals with high blood iron levels or with compromised immune systems can develop a systemic Y. enterocolitica infection that is often fatal (8). Pigs are a major reservoir of some biotypes of pathogenic Y. enterocolitica (28), and while it is often isolated from the tonsils and intestinal tract, Y. enterocolitica does not cause obvious disease in swine (10). The bacteria are shed into the environment via feces or can contaminate pork products during processing, both of which can lead to contamination of human food and water supplies. In addition, Y. enterocolitica is frequently found during surveys of food-borne pathogens in milk supplies (22). The prevalence of Y. enterocolitica in contaminated meat and milk products may be enhanced by its ability to grow at cool temperatures, even as low as 0°C (3).
Based on biochemical properties, Y. enterocolitica strains are subdivided into several biovars. Interestingly, there is a considerable range in the severity of disease caused by the different biovars. Biovar 1B strains are the most highly pathogenic for humans, and 1B is the only biovar that is lethal in a mouse model of yersiniosis (37, 38, and references therein). Not surprisingly, these isolates contain virulence factors not found in other biovars (19, 41). Among these is the chromosomally encoded Ysa type III secretion system (T3SS). In mouse infection studies, deletion of apparatus genes (ysa) or individual effector genes (ysp) results in a modest attenuation only at 24 h postinfection (27, 45) and a 10-fold increase in the 50% lethal dose, both following oral inoculation (17). Thus, the role of the Ysa T3SS in pathogenicity is subtle in the mouse model, leading to speculation that the secreted effectors (called Ysps) are important for gastroenteritis but not for systemic infection (45). In vitro, secretion of Ysps through the Ysa T3SS apparatus is only detected when the bacteria are cultured at 26°C and in the presence of a high concentration of salt (e.g., NaCl) (17, 50). Subsequent studies provided evidence that the genes encoding both the apparatus and effector proteins are upregulated by growth in a high concentration of salt, indicating that the salt dependence is at the level of transcription (45-47).
As Y. enterocolitica exists as both a commensal and a pathogenic organism and has the capacity to grow in a wide range of temperatures, the expression of many genes must be tightly regulated to ensure survival under these often stressful conditions. One mechanism widely used by prokaryotes to respond to changes in their environment is signal transduction via two-component systems (TCS) (49). In the canonical TCS, a membrane-bound sensor histidine kinase (HK) autophosphorylates at a conserved histidine (His) residue in response to an environmental cue. The phosphoryl group is then transferred to a conserved aspartate (Asp) residue in the receiver domain of a response regulator (RR) protein. RR proteins also contain an effector domain, which is often a DNA-binding moiety; phosphorylation of the receiver domain activates the effector domain to bind DNA and alter gene transcription (13). A common variation of the canonical TCS is a phosphorelay (2). Phosphorelays involve two or more proteins and three phosphoryl group transfer events (His→Asp→His→Asp). In one subclass of the phosphorelay group, a hybrid sensor kinase contains not only an HK domain but also receiver (REC) and histidine phosphotransferase (HPt) domains, and the first two phosphotransfer events are within the sensor protein. The final phosphotransfer event is to the RR protein. Additionally, variations on this theme have been reported where three or four proteins are required to provide the domains necessary for the phosphorelay (5, 12, 36, 39).
Studies conducted in our laboratory and others showed that the putative TCS proteins YsrR and YsrS are required for expression of the ysa apparatus genes and specifically of the ysaE promoter (Fig. 1 A) (30, 45-47). Deletion of either gene results in a lack of NaCl-induced activation of the ysa genes. Furthermore, the level of ysaE expression in a ΔysrS mutant strain is the same as that in a wild-type strain grown in the absence of NaCl, thus implying that YsrS responds to the NaCl (46). One conundrum of this model is that YsrS appears to belong to the hybrid class of sensor kinases but lacks the terminal HPt domain presumably required to transfer the phosphate to YsrR. In this study, we report the identification of a gene encoding this HPt function and demonstrate genetically that the Ysr proteins do indeed conduct a phosphorelay cascade that leads to activation of the ysaE promoter and, hence, the ysa and ysp genes.
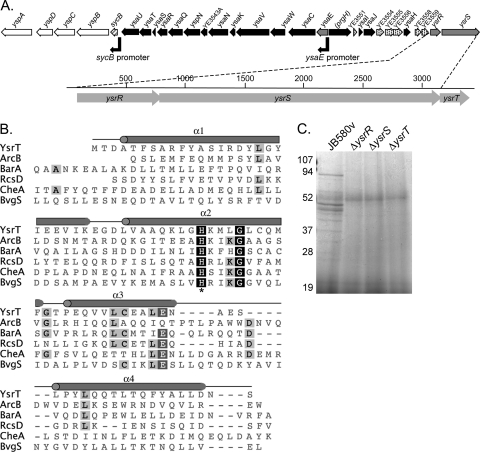
FIG. 1.
Identification of YsrT. (A) Genomic organization of the ysa locus, zooming in on the ysrRST operon. Black boxes, apparatus genes; white boxes, secreted proteins; gray boxes, transcriptional regulators; hashed, chaperone/regulator; speckled, unknown. (B) Alignment of YsrT (YE3561a) with defined HPt domains. These domains were defined in other organisms, and the sequences given here are for the Y. enterocolitica homologs (accession numbers are given in Materials and Methods). The predicted α-helices are designated with gray barrels, and the conserved histidine is marked with an asterisk. (C) Secretion of Ysps into culture supernatants at 26°C. Proteins were prepared as described in Materials and Methods. Supernatants equivalent to an OD of 2.5 were separated by SDS-PAGE and stained with Coomassie blue. Values to the left are the sizes of protein standards in kDa.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this work are listed in Table 1 and described below. All cultures of Escherichia coli strains were grown in LB (1% tryptone, 0.5% yeast extract, 170 mM NaCl; Difco) at 37°C. Cultures of Y. enterocolitica were grown at 26°C in LB, L broth (1% tryptone, 0.5% yeast extract, 0 mM NaCl), or L broth with 290 mM NaCl (referred to as LB-290). For preparation of secreted proteins, brain heart infusion (BHI; Difco) and BHI with 490 mM NaCl were used. Antibiotics were added as needed at the following concentrations: kanamycin, 100 μg/ml; nalidixic acid, 20 μg/ml; chloramphenicol, 12.5 μg/ml.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U169 deoP recA1 endA1 hsdR17 (rK− mK−) | Invitrogen |
| S17-1λpir | Tpr StrrrecA thi pro hsdR hsdM+ RP4::2-Tc::Mu::Km Tn7 λ pir lysogen | 33 |
| Y. enterocolitica | ||
| JB580v | 8081v (r− m+ Nalr) serotype O:8 | 24 |
| YVM925 | JB580v ysaE-lacZYA | 46 |
| YVM1320 | JB580v ΔysrS ysaE-lacZYA | This work |
| YVM1323 | JB580v ΔysrT ysaE-lacZYA | This work |
| YVM1250 | JB580v ΔysrR ysaE-lacZYA | This work |
| YVM1239 | JB580v ΔrcsB ysaE-lacZYA | This work |
| YVM1378 | JB580v ysrT-H38A ysaE-lacZYA | This work |
| YVM1419 | JB580v ysrR-D75A ysaE-lacZYA | This work |
| 2440-87 | Y. enterocolitica biovar 1B serotype O:8 | 32 |
| 9286-78 | Y. enterocolitica biovar 1B serotype O:20 | 32 |
| 9287-78 | Y. enterocolitica biovar 1B serotype O:20 | 32 |
| 634-83 | Y. enterocolitica biovar 1B serotype O:4,32 | 32 |
| 655-83 | Y. enterocolitica biovar 1B serotype O:18 | 32 |
| 657-83 | Y. enterocolitica biovar 1B serotype O:20 | 32 |
| 658-83 | Y. enterocolitica biovar 1B serotype O:21 | 32 |
| Y295 | Y. enterocolitica biovar 1B serotype O:8 | 32 |
| Plasmids | ||
| pSR47S | Kanr MobRP4 oriR6K cloning vector | 29 |
| pWKS130 | Kanr low-copy-number cloning vector | 48 |
| pKW62 | Kanr; ysrT in-frame deletion (codons 8-78 deleted) in pSR47S | This work |
| pKW77 | KanrysrS in-frame deletion (codons 28-768 deleted) in pSR47S | This work |
| pKW31/pYsrSWT | KanrysrS coding sequence in pWKS130 | 46 |
| pKW79/pYsrSH320A | Kanr pKW31 with H320A substitution | This work |
| pKW80/pYsrSD714A | Kanr pKW31 with D714A substitution | This work |
| pKW63/pYsrTWT | KanrysrT coding sequence in pWKS130 | This work |
| pKW76/pYsrTH38A | Kanr pKW63 with H38A substitution | This work |
| pKW24/pYsrRWT | KanrysrR coding sequence in pWKS130 | 46 |
| pKW72/pYsrRD75A | Kanr pKW24 with D75A substitution | This work |
| pKW78 | KanrysrT-H38A in pSR47S | This work |
| pKW75 | KanrysrR-D75A in pSR47S | This work |
Plasmid and strain construction.
The plasmids and strains used in this study are listed in Table 1, and the primers used are listed in Table 2. All of the plasmids made were confirmed by restriction digest patterns and sequencing.
TABLE 2.
Primers used in this work
| Name | Sequencea (5′→3′) |
|---|---|
| ysrT-delA | ACGCGTCGACGCCCTATTACTCTGCCGCTG |
| ysrT-delB | CGGGATCCCTGAAGGTGGCATCAGTCATGTTC |
| ysrT-delC | CGGGATCCCTAGATAACAGCTAAATAAATAGCCCCGGC |
| ysrT-delD | ATAAGAATGCGGCCGCCATAAAAACCGGAAATAGTGGCCC |
| ysrS-delA2 | GCGTCGACCTCGTAGCGGGTGAAACAGATGAG |
| ysrS-delB2 | CGGGATCCCTCGGCAGCATAAACAGCCGTCG |
| ysrS-delC2 | CGGGATCCCCGGTTGCACGCGAACAATGGCAG |
| ysrS-delD2 | ATAAGAATGCGGCCGCCTAAATTGATCTCGAACCTGTAGG |
| KW197 | GCGTCGACCAATGGCAGCAATTGATACG |
| KW198 | CGGGATCCTTAGCTGTTATCTAGCAAGGC |
| KW199 | GAAGCTCGGCGCTAAAATGCTGGG |
| KW209 | GCGTCGACGGCGTCGGGTTAATCAAGCGC |
| KW210 | ATAAGAATGCGGCCGCCTCTGGCAATTGTTAATGCAGAG |
| D75A-R | CGCTACGAGGGAATATGCCAGTAATAGTACTTCAATCTCAGGGTTGC |
| D75A-F | GTACTATTACTGGCATATTCCCTCGTAGCGGGTGAAACAGATGAGAACGAG |
| H320A-R | GCGCAATTCGGCCGCAATAACCGACAGCTCCAGATTGTTTTTTGAACCC |
| H320A-F | GTTATTGCGGCCGAATTGCGCACGCCGCTGATTGGTATTTTAACC |
| D714A-R | AAGCCGACAGGCGGTCACAATAATAGTATGGTGGTGTTGTAACCAGC |
| D714A-F | ATTATTGTGACCGCCTGTCGGCTTGATGAGAGCGACGGCTTTGAAC |
| YsrS OE-F1b | GCGTCGACGGGCTTACTTCAAACACTGATTTC |
| YsrS RP3b | ATAAGAATGCGGCCGCTCAGTCATGTTCTTTTTCCTTAG |
| YsrR delAb | ACGCGTCGACGCAGGATAATCCGATGAAATCTCG |
| YsrR delDb | ATAAGAATGCGGCCGCGCTTGGTAAACCACTCAATCAGCG |
Restriction sites are underlined (six- to eight-base sequences). In primers used to introduce alanine substitutions, the mutated codons are italicized and the bases that were substituted to code for alanine are bolded.
Published elsewhere (46) and included here for completeness.
In-frame deletions were constructed as described previously (46). Briefly, for ysrS, fragments of approximately 500 bp upstream and downstream were independently amplified using primers ysrS-delA2/delB2 (upstream) or ysrS-delC2/delD2 (downstream). These fragments were digested with SalI and BamHI (upstream) or BamHI and NotI (downstream), ligated into pSR47S cut with SalI and NotI, and transformed into S17-1 λ pir. The resulting plasmid, pKW77, was introduced into the desired Y. enterocolitica strains by conjugation. Following counterselection, confirmation of the deleted gene was determined by diagnostic PCR using at least one primer outside the region cloned in pKW77. For ysrT, the same procedure was followed; the primer pairs used were ysrT-delA/delB and ysrT-delC/D, and the resulting plasmid was pKW62.
Complementing clone for ysrT.
The complementing clone for ysrT was constructed by amplifying the gene with some additional upstream sequence to include the native ribosome binding site using primers KW197 and KW198. The resulting 300-bp product was digested with SalI and BamHI and ligated into those sites of pWKS130 to generate pYsrTWT (pKW63).
Point mutations made by overlap extension for ysrS.
Point mutations were introduced into the ysrS gene using overlap extension PCR (18). Overlapping forward and reverse primers were designed with the desired mutation. These primers were used in two separate PCRs with appropriate cognate primers. The two products were gel purified and used as template DNA for PCR with the flanking forward and reverse primers to amplify the intact full-length product. The product was digested with SalI and NotI and cloned into those sites of pWKS130 to yield pYsrSH320A (pKW79) and pYsrSD714A (pKW80). These plasmids are identical to pYsrSWT, except that they contain the alanine substitutions. The primer pairs used were as follows. For H320A, the primers used to introduce the point mutation were H320A-R and H320A-F, used with cognate primers ysrS-OE F1 and ysrS-RP3, respectively. For D714A, the primers used to introduce the point mutation were D714A-R and D714A-F, used with cognate primers ysrS-OE F1 and ysrS-RP3, respectively.
Point mutations by megaprimer PCR for ysrT and ysrR.
To generate an alanine substitution for His38 in ysrT, primer KW199 (containing the mutation) was used with KW198 to amplify an ∼150-bp product that was gel purified and used as a reverse megaprimer in a second PCR with forward primer KW197. The resulting 300-bp fragment was digested with SalI and BamHI and ligated into those sites of pWKS130 to generate pYsrTH38A (pKW76). A second construct was made for generation of the chromosomal copy of ysrT-H38A in a similar fashion. Primer KW199 was used with ysrT-delD to make a 370-bp megaprimer that was subsequently used in a second PCR with ysrT-delA. This product was digested with SalI and NotI and ligated into those sites of pSR47S to generate pKW78. The D75A mutation in ysrR was similarly constructed. Primers M13R and D75A-F were used with pYsrRWT as template DNA to generate a megaprimer that was subsequently used with ysrR-delA to amplify the full coding sequence. This product was digested with SalI and KpnI and ligated into those same sites of pWKS130 to generate pYsrRD75A (pKW72). To generate a chromosomal copy of ysrR-D75A, the megaprimer was synthesized using genomic DNA for the template and primers ysrR-delA and D75A-R. The full-length product was amplified using the megaprimer and ysrR-delD. This product was digested with SalI and NotI and cloned into those same sites of pSR47S to generate pKW75.
Introduction of chromosomal point mutations for ysrR and ysrT.
Chromosomal copies of genes encoding point mutations were constructed using the same methods employed to delete genes. Plasmids pKW78 (ysrT) and pKW75 (ysrR) were conjugated into the Y. enterocolitica strain bearing a deletion of the targeted gene. Following counterselection, individual colonies were screened by PCR for the presence of the full-length gene. The regions surrounding the engineered point mutations were amplified and sequenced to verify that the desired strain had been constructed without errors. The resulting strains are referred to as ysrT-H38A (YVM1378) and ysrR-D75A (YVM1419).
β-Galactosidase assays.
Saturated cultures grown overnight in L broth were diluted into fresh L broth or LB-290 to an initial optical density at 600 nm (OD600) of 0.2 and grown for 2 h at 26°C with aeration. Antibiotics were added as necessary to retain plasmids and chromosomal integrations. Assays were performed as described previously (31). Individual assays were conducted with at least three independent cultures for each strain tested, and the assays were repeated at least three times to ensure reproducibility. Representative assays are shown.
Preparation of secreted proteins.
Proteins secreted into culture supernatants were collected essentially as described previously (46), except that the base growth medium was BHI or BHI with 490 mM NaCl. Briefly, overnight cultures grown in LB at 26°C were subcultured to an OD of 0.2 into BHI or BHI-490 mM NaCl and grown for 5 to 6 h at 26°C on a roller drum. The cells were pelleted and discarded. Supernatants were filtered and precipitated with 10% trichloroacetic acid at 4°C overnight. Protein pellets were resuspended in sample buffer at a concentration of 1 OD equivalent per 10 μl. Approximately 2.5 OD equivalents was subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Coomassie blue.
DNA sequencing and analysis.
All DNA sequencing was performed by Eton Bioscience, Inc., at their North Carolina facility. For sequencing of the ysrS-ysrT region of the biovar 1B isolates (listed in Table 1), genomic DNA was prepared and used as templates for PCR using primers ysrS-delC and ysrT-delD. The PCR products were gel purified and sequenced with primer ysrS-delC2. The sequences for Y. mollaretii (ATCC 43969, accession no. AALD00000000) (6), Y. aldovae (ATCC 35236, accession no. ACCB00000000) (6), JB580v (Y. enterocolitica 8081, accession no. NC008800) (41), and Y. enterocolitica WA-C (WA-314, accession no. AJ344214) (21) were obtained from the NCBI repository. Alignments were conducted using the CLUSTAL W algorithm (40) in the Geneious Pro version 5 software package (BioMatters, Ltd., New Zealand).
Secondary structural analysis and modeling.
Analysis of the secondary structure prediction for YsrT was performed using the Phyre server (http://www.sbg.bio.ic.ac.uk/phyre) (23). The sequences for the designated HPt-containing proteins in Y. enterocolitica were obtained using the following accession numbers: ArcB (YE3733), YP_001007887.1; BarA (YE0742), YP_001005086.1; RcsD (YE1398), YP_001005711.1; CheA (YE2577), YP_001006780.1; BvgS (YE2663), YP_001008383.1. The residues that make up the HPt domain were obtained from the MiST database (43), and the alignment was generated using CLUSTAL W as described above. The YsrS receiver domain was modeled using SWISS-MODEL (16) and generated using PyMOL (The PyMOL Molecular Graphics System, version 1.2; Schrödinger, LLC). Domain sequences from YsrR, YsrS, and YsrT were identified with the simple modular architecture research tool (SMART) and used to perform a BLASTp search (1). A subset of homologous proteins whose domains had been defined either genetically or biochemically were aligned using the CLUSTAL W algorithm (40) in the Geneious Pro version 5 software package (BioMatters, Ltd., New Zealand).
RESULTS
Identification of YsrT.
We and others previously demonstrated that YsrS and YsrR are required to activate expression of the ysaE promoter (30, 45, 46). While these proteins were predicted to be a hybrid TCS, we noticed that YsrS lacks an HPt domain. Hybrid TCS have four sites of phosphorylation in a His(H1)→Asp(D1)→His(H2)→Asp(D2) phosphorelay. Typically, H1, D1, and H2 are contained on the sensor and D2 is found on the RR but examples exist where this is not the case (49). Initial attempts to identify the protein containing the HPt domain for the Ysr phosphorelay were unsuccessful. All of the genes that could be identified at the time as encoding HPt proteins in Y. enterocolitica (YE2577 [cheA], YE0724 [barA], YE3733 [arcB], and YE1398 [rcsD]) were inactivated by plasmid insertion within the gene and the resulting mutants were tested for the ability to secrete Ysps into culture supernatants. None of these proteins appeared to be the intermediate between YsrS and YsrR, as Ysp secretion was unaffected by these mutations (data not shown). Upon release of the annotated Y. enterocolitica genome, we noticed that a small open reading frame (ORF), designated YE3561a, was located adjacent to ysrS (Fig. 1A). BLASTp results revealed that this hypothetical protein had homology to HPt domains, and this is supported by aligning YE3561a with all of the known HPt domains found in Y. enterocolitica (Fig. 1B). YE3561a encodes an 82-amino-acid protein that contains a single histidine residue (H38) located within a short stretch of somewhat conserved residues found in HPt domains (13). Protein structure prediction using the Phyre server (23) predicted a high helical content for YE3561a, with the conserved H located on an exposed region of an alpha helix. These findings are consistent with the notion that YE3561a serves as an HPt protein.
To investigate if YE3561a encodes a protein important for the function of the Ysa T3SS, we constructed an in-frame deletion of this gene and tested the resulting strain for the presence of Ysps in culture supernatants. We found that no secreted proteins could be detected from cultures grown under inducing conditions when YE3561a was deleted (Fig. 1C). This result is the same as that observed in strains lacking ysrR and ysrS (45-47). We further tested this mutant in a strain carrying a chromosomal ysaE-lacZ reporter and found that ysaE expression was not activated when the bacteria were grown under inducing conditions. These data are described in detail in the following sections and shown in Fig. 2. Because of the critical role in ysa expression and its putative function as part of a phosphorelay, we have renamed YE3561a ysrT and refer to it as such here.
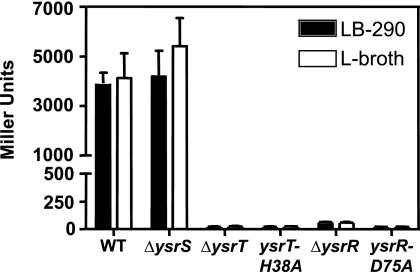
FIG. 2.
Genetic analysis of the YsrS-YsrT-YsrR phosphorelay. Assays of β-galactosidase activity in strains harboring a ysaE-lacZ reporter were used to evaluate the role of each protein. Saturated cultures grown in L broth were subcultured into fresh L broth (noninducing) or LB-290 (inducing) and grown for 2 h at 26°C on a roller drum. Each protein is represented in cartoon form, with a different shape for each domain type. Solid, gene is single copy (chromosomal); hatched, gene is carried on a multicopy plasmid. Point mutations are indicated above the respective domains. The relevant genotype is given on the left, and the relative promoter activity is given on the right in Miller units with the standard deviation in parentheses. The strains and plasmids used to evaluate (A) YsrS, (B) YsrT, and (C) YsrR are listed in Table 1. WT, wild type.
YsrS, YsrT, and YsrR are phosphorelay proteins.
With all of the components of a hybrid two-component system identified and shown to be requisite factors in transcriptional activation of the ysaE promoter, we wanted to determine if YsrSTR were indeed functioning as phosphorelay proteins. To this end, alanine substitutions were made at the histidine and aspartate residues predicted to participate in the phosphorelay. These residues were chosen based on alignments with other phosphorelay proteins in which the key residues had been defined (Fig. 1B; see also Fig. 3 and 4) (9). Strains carrying either plasmid or chromosomal copies of the mutated genes were used to evaluate their ability to activate ysaE expression.
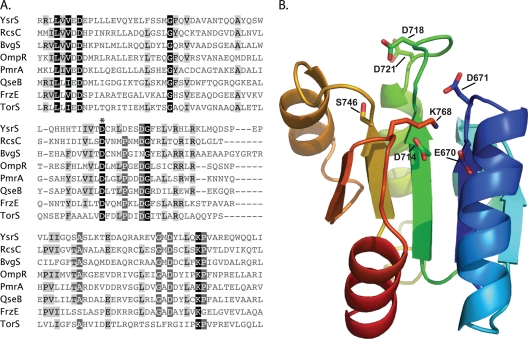
FIG. 3.
Analysis of domains and critical residues in the YsrS receiver domain. (A) Sequences of selected REC domains are aligned with the predicted REC domain of YsrS. Conserved residues are shaded such that the darker boxes are more highly conserved. The phosphorylation site (D1) is designated with an asterisk (D714 for YsrS). The accession numbers for the sequences used are as follows: BvgS of Bordetella bronchiseptica, P26762; FrzE of Myxococcus xanthus, P18769; OmpR of E. coli, P0AA16; PmrA of Pectobacterium carotovorum, Q70FH0; QseB of E. coli, Q8XBS3; RcsC of E. coli, P14276; TorS of E. coli, P39453; YsrS, A1JQC5. (B) Model of the YsrS REC domain generated using SWISS-MODEL and edited using PyMOL. Highly conserved residues important for phosphorylation, as well as unusual residue D718, are shown as sticks.
FIG. 4.
Analysis of the YsrR receiver domain. Sequences of selected REC domains are aligned with the predicted REC domain of YsrR. Conserved residues are shaded such that the darker boxes are more highly conserved. The phosphorylation site (D2) is designated with an asterisk (D75 for YsrR). The accession numbers for the sequences used are as follows: BvgA of Bordetella pertussis, P0A4H2; CheY of Thermotoga maritima, Q56312; DegU of Brevibacillus brevis, P54662; FixJ of Bradyrhizobium japonicum, P23221; GacA of Pseudomonas aeruginosa, Q51373; NtrC of Salmonella typhimurium, P41789; RegR of Rhodobacter capsulatus, P42508; Spo0F of Bacillus subtilis, P06628; UhpA of Salmonella typhimurium, P27667; YsrR, A1JQC4.
Analysis of YsrS.
In the wild-type strain (YVM925), ysaE-lacZ expression levels are very low in the absence of NaCl (L broth), resulting in about 25 Miller units (MU). In the presence of 290 mM NaCl (LB-290), ysaE-lacZ expression levels reach about 1,085 MU, a 43-fold activation (Fig. 2A). Consistent with previous observations with different ysrS mutant strains, no activation of ysaE-lacZ is observed in LB-290 in the ΔysrS mutant strain (YVM1320), indicating that YsrS is required for the salt-dependent activation of ysaE expression (45-47). This phenotype can be complemented by expressing ysrS on a low-copy-number plasmid (pYsrSWT), resulting in 420 MU in L broth and 2,614 MU in LB-290 (Fig. 2A). The elevated levels are likely the consequence of the multicopy plasmid. Plasmid pYsrSH320A carries ysrS with an alanine substitution at H320, the predicted site of autophosphorylation (H1). When pYsrSH320A is transformed into YVM1320 (ΔysrS), ysaE expression is the same as in a strain with no plasmid or the vector only, with about 20 MU in L broth or LB-290 (Fig. 2A). This indicates that H320 is a critical residue for the function of YsrS in its role in activating ysaE expression in response to NaCl. A similar result was expected with pYsrSD714A; D714 is the predicted site of phosphorylation in the receiver domain of YsrS (D1) (Fig. 3 A). However, a very different result was observed. This mutation caused levels of ysaE to be very high both in LB-290 and in L broth, with 4,253 and 5,472 MU, respectively (Fig. 2A). These results indicate that both H320 and D714 are required for the normal function of YsrS, but only H320A interferes with the ability of YsrS to activate ysaE expression.
The “constitutive on” phenotype from pYsrSD714A is seemingly contradictory to the assumption that YsrS is a hybrid sensor kinase, as one would have predicted a loss of function. We had noticed that there are two neighboring aspartate residues: D718 and D721 (Fig. 3). Curiously, the residue located four amino acids from the phosphorylated D is typically a proline; in the case of YsrS, this is D718. One hypothesis to explain the unusual phenotype associated with D714A is that one of these other sites could be the actual site of phosphorylation or could be phosphorylated if D714 were unavailable. To test this notion, a plasmid was generated that contained alanine substitutions at all three aspartate residues. When transformed into the ΔysrS mutant strain, this plasmid had the same impact on ysaE expression as pYsrSD714A: very high expression, independent of the NaCl concentration (data not shown). In addition, the YsrS receiver domain was modeled (Fig. 3B). D714 is located at the end of predicted β-strand 3, as is common for D1 phosphorylation sites, whereas D718 and D721 are located in the loop between β-strand 3 and α-helix 3. Other highly conserved residues are in the proper positions, such as magnesium ion coordinating residues E670 and D671, as well as S746 and K768, which are important for signal transduction (4). Taking all of these data together, we conclude that YsrS is indeed a hybrid sensor kinase, that D714 is the D1 phosphorylation site, and that neither of the neighboring aspartate residues can substitute for D714 as a site for phosphorylation.
Analysis of YsrT.
A similar analysis was conducted with YsrT. As mentioned above, deletion of ysrT prevented activation of ysaE expression, yielding 24 MU in L broth and 29 MU in LB-290 (Fig. 2B). When wild-type ysrT was provided in trans on pYsrTWT, ysaE levels were slightly activated in LB-290, with 73 MU. However, with pYsrTH38A, which expresses ysrT with an alanine substitution at H38, no activation of ysaE expression was observed (21 MU in L broth and LB-290). While the activation with the wild-type plasmid is subtle, the absence of this activation with H38A supports the hypothesis that YsrT functions as an HPt.
The lack of full complementation by pYsrTWT could be the result of the HPt domain playing a role in dephosphorylation of YsrR, which is a known function of these domains (14, 35). To test this notion, pYsrTWT was transformed into YVM925 (wild type). Expression of ysaE was markedly decreased, yielding only 70 MU in LB-290 (Fig. 2B). Transformation of pYsrTH38A into YVM925 had no negative impact on ysaE levels, which measured 29 and 988 MU in L broth and LB-290, respectively. Taken together, these data imply that YsrT possesses both kinase and phosphatase activities and that H38 is critical for both activities.
Because of the concerns of plasmid copy number, we constructed a strain with the ysrT-H38A mutation on the chromosome, designated ysrT-H38A (YVM1378). β-Galactosidase activity from the ysaE reporter in this strain is similarly low as with the ΔysrT mutant strain, further supporting the idea that H38 is required for YsrT to function properly (Fig. 2B). As was also observed with the ΔysrT mutant strain, addition of pYsrTWT to ysrT-H38A results in about a 2-fold increase in ysaE expression (not shown).
Analysis of YsrR.
YsrR has two key domains, a C-terminal DNA-binding domain of the LuxR family of helix-turn-helix proteins and an N-terminal receiver domain. Compared to other RRs, YsrR appears to be somewhat unusual in that it has two stretches of sequence not found in other RR proteins (Fig. 4). Secondary structural analysis predicts that these regions of 15 and 12 amino acids are α-helical and indicates that they form additional helices. It is possible, however, that they may form extensions of conserved helix 1 and helix 3, respectively. The receiver domain contains the probable site of phosphorylation (D2), which was predicted to be D75 in YsrR by alignment with other RRs (Fig. 4). In the ΔysrR mutant strain (YVM1250), the activity of ysaE-lacZ is very low, with 64 and 72 MU in L broth and LB-290, respectively (Fig. 2C). The ΔysrR deletion can be complemented by expressing ysrR in trans (pYsrRWT), resulting in 149 and 3,053 MU in L broth and LB-290, respectively. When the ΔysrR mutant strain is transformed with pYsrRD75A, the activation of ysaE in LB-290 is markedly less than with pYsrRWT, producing 733 MU in LB-290 (data not shown). This suggests that D75 is important for activation of ysaE expression.
As with ysrT, there were concerns of pleiotropic effects resulting from overexpression of transcriptional regulators. We therefore introduced ysrR with D75A into the chromosome of YVM1250 to generate a single copy of the mutant gene, designated ysrR-D75A (YVM1419). The levels of ysaE expression in this strain were measured at 32 and 33 MU in L broth and LB-290, respectively. Transformation of this strain with pYsrRWT restores ysaE promoter activity, yielding 315 and 2,866 MU in L broth and LB-290, respectively. This level of activity is similar to that seen when the ΔysrR mutant strain was complemented. The inability of the ysrR-D75A mutant strain to show activation of ysaE under inducing conditions supports the hypotheses that this protein is an RR and that phosphorylation of D75 is critical to its function as a transcriptional regulator.
Effect of pYsrSD714A requires all other phosphorelay proteins.
The curious result of high ysaE-lacZ levels in the strain overexpressing pYsrSD714A (Fig. 2A) begs for further analysis. There are reports in the literature of a hybrid two-component system that has a phosphorylation event from H1 to D2, bypassing the D1 and H2 residues (15, 42). Although this was subsequently found to occur only when the respective proteins were overexpressed (25), we wanted to address if YsrSD714A acts independently of YsrT. The ΔysrT mutant strain was transformed with pYsrSD714A, and ysaE-lacZ levels in this strain were the same as with the vector, indicating that YsrT is required for the constitutive on phenotype of pYsrSD714A (Fig. 5). To determine if YsrR is also required and if phosphorylation of the conserved residues is an important part of this phenotype, we transformed pYsrSD714A into the ysrT-H38A, ΔysrR, and ysrR-D75A mutant strains (Fig. 5). We observed that YsrT and YsrR must be present and phosphorylatable. If either gene is deleted or if the phosphorylation sites are mutated, ysaE levels are the same as the background (∼25 to 60 MU). These results indicate that the peculiar phenotype observed with pYsrSD714A requires wild-type copies of YsrT and YsrR, indicating that no step in the phosphorelay is bypassed.
FIG. 5.
The peculiar phenotype produced by pYsrSD714A does not bypass any step of the phosphorelay. pYsrSD714A was transformed into the specified strain and evaluated for the ability to activate ysaE expression, as measured by β-galactosidase activity. WT, wild type.
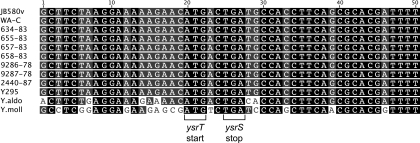
The genetic structure of YsrRST is conserved in yersiniae.
In JB580v, the ysrT coding region begins within the ysrS coding region such that there is a 7-bp overlap. One theory for YsrT being a separate protein from YsrS is that a frameshift mutation was acquired at some point in our laboratory strain. To address if this is a likely explanation, we examined the genetic arrangement of this region in several Yersinia strains. With the knowledge that several Y. enterocolitica biotype 1B isolates contain genes encoding the Ysa T3SS apparatus and effector genes (19, 41), we sequenced the region surrounding ysrT to determine if it is a separate gene in these strains. In every strain examined, ysrT was a separate gene (Fig. 6). A recent bioinformatic study of some of the nonpathogenic Yersinia strains has provided whole genome sequences for Yersinia mollaretii, Yersinia aldovae, and Yersinia ruckerii, among others (6). In these three species, genes with homology to TCS are located adjacent to remnants of what is likely the ysa locus. Closer examination revealed that Y. aldovae and Y. mollaretii have genes with reasonable similarity to ysrR, ysrS, and ysrT, and in these Yersinia strains, ysrT is encoded as a separate gene (Fig. 6). These results indicate that ysrT has been separate from ysrS since before the evolutionary divergence of these strains from the progenitor, indicating that there may be a functional purpose to YsrT being an isolated HPt protein.
FIG. 6.
The genetic structure of the ysrS-ysrT region is conserved in multiple Yersinia strains. Sequences for Y. mollaretii, Y. aldovae, and Y. enterocolitica strains JB580v and WA-C were obtained from NCBI (accession numbers are given in Materials and Methods), and the others were sequenced from PCR products generated in our laboratory as described in Materials and Methods. Alignments were generated using the CLUSTAL W algorithm in the Geneious Pro software package. The start codon for YsrT and the stop codon for YsrS are indicated by brackets.
DISCUSSION
In this report, we present genetic evidence that the predicted two-component proteins YsrR and YsrS do indeed conduct a phosphorelay and that YsrT encodes a small protein providing the previously unidentified HPt domain that is necessary to shuttle the phosphoryl group from the sensor to the RR. This phosphorelay leads to the activation of genes encoding the Ysa type III secretion apparatus and its effectors. These, however, are not the only genes playing a regulatory role in ysa expression, but we believe they are at or near the top of the regulatory cascade. In addition to YsrRST, RcsB is also required for activation of the ysaE promoter (45, 47). RcsB is the RR of the RcsCDB phosphorelay system (discussed below), and the mechanism by which it acts at the ysaE promoter is the current focus of research in our laboratory. YsaE itself is a transcriptional regulator that acts in conjunction with the chaperone/regulator SycB to stimulate the expression of six of the nine ysp genes (46, 47). Thus, YsrRST and RcsB are requisite factors in the transcription of the ysa and ysp genes.
While YsrR and YsrS have been predicted to conduct a phosphorelay, we sought to demonstrate this phenomenon and to understand how the phosphate was transferred from YsrS to YsrR. The identification of YsrT solved this mystery, establishing that YsrRST contain all of the predicted domains known to be required for phosphorelay. To determine if YsrRST are indeed phosphorelay proteins, we mutated all four predicted phosphorylation sites. Each mutation ablated the normal function of the proteins, and all but YsrSD714A (D2) produced the expected phenotype, with an inability to activate ysaE expression. While the phenotype produced by the YsrR D56A mutation was exactly as predicted, there are two structural deviations that indicate that the mechanism of how YsrR responds to phosphorylation may be atypical. First, there are two insertions of 12 and 15 amino acids, respectively, and second, a highly conserved proline residue located four resides from the phosphorylation site is absent. Both features may contribute to an altered conformation that could confer a unique function(s).
The constitutive on phenotype of the YsrSD714A mutant is not easily understood but is very intriguing. It has been proposed that the evolutionary advantage of hybrid sensors is the increased number of regulatory checkpoints to minimize phosphorylation of the REC under noninducing conditions (13, 20, 34). In addition to kinase activity, some HKs also appear to possess phosphatase activities (7, 14). One plausible explanation is that this mutation may have impaired the phosphatase function of YsrS without impairing the kinase activity. However, one would have to assume that a neighboring residue could substitute for D714 for the kinase activity but not for the phosphatase activity. YsrS has D resides at positions 718 and 721, but these are unlikely to become phosphorylated due to their locations just outside the active site. A curious feature of this region of YsrS is the absence of a highly conserved proline four residues from D714. It is tempting to speculate that the architecture of this loop may be altered and confer an atypical function. We are continuing experimentation with this mutant in an effort to understand this unusual phenotype. However, biochemical analyses often used to complement the genetic experiments and more definitively show transfer of phosphoryl groups have proven to be challenging for YsrS.
Two-component and phosphorelay systems are common mechanisms by which prokaryotes and fungi can rapidly adapt to their environments, and many bacterial strains contain 20 to 30 such systems (MiST2 database; 43). The majority of these systems are composed of two proteins, a sensor kinase and an RR, but in a few systems, multiple proteins are involved in a phosphorelay cascade. The first phosphorelay system described consisted of the sporulation regulators KinABC, Spo0F, Spo0B, and Spo0A of Bacillus subtilis, which have each domain encoded in a separate protein (5). More common among phosphorelay systems are the hybrid two-component systems which are composed of two proteins. However, many examples exist where the phosphorelay requires three proteins: a sensor kinase containing the HK (H1) and REC (D1) domains, an HPt (H2) protein, and an RR (D2). One such system is the Rcs system. In this system, RcsC is the sensor, RcsD has the HPt domain, and RcsB is the DNA-binding RR (reviewed in reference 26). In this case, the HPt protein is a large membrane-bound protein that has features suggesting that at one time it may have been a complete hybrid sensor (39). In the Vibrio harveyi Lux system, the HPt-containing protein, LuxU, is encoded as a small cytosolic protein (11). Intriguingly, LuxU serves as the HPt for two sensors (LuxN and LuxQ), allowing cross communication and versatility in these important quorum-sensing regulators. While these two examples are cases of independent HPt domains, this is a relatively rare situation. Examination of the MiST2 database (43) shows that very few organisms have proteins that contain only an HPt domain. Pseudomonas aeruginosa PAO1 is a rare example of an organism that has several such free HPt proteins, with four. Y. enterocolitica has an average number of HPt-containing proteins (six) compared to other bacteria, and all are part of hybrid HKs. This database does not identify YsrT as an HPt protein (search last performed on 24/6/2010), perhaps because it is such a small ORF. This suggests that there may indeed be more such proteins, but since the similarity between the known HPt domains is weak, they may be difficult to identify in silico.
The location of ysrT led us to question if a frameshift mutation may have occurred in our strain, resulting in YsrT as a protein independent of YsrS. Sequence analysis indicates that this genetic organization is conserved not only in Y. enterocolitica 1B isolates but in more distantly related nonpathogenic Yersinia strains. Whole-genome sequencing has recently been performed for several nonpathogenic Yersinia strains, and Y. ruckerii and Y. mollaretii have reasonably well-conserved ysrR and ysrS sequences and Y. mollaretii has a gene that is likely ysrT (6). The region downstream of ysrS is more divergent in Y. ruckerii, and the protein sequence for the ORF downstream of ysrS is quite different from YsrT but still has homology to HPt domains (not shown). Y. aldovae appears to contain this locus, but the sequence suggests that a number of frameshift mutations have been acquired; the DNA sequence is well conserved, however, and indicates that ysrT would have encoded a separate protein. Thus, the conservation of the genetic structure of the ysrRST genes among not only the pathogenic 1B isolates but also environmental isolates implies that there is some functional importance to YsrT being produced as a separate protein. However, one should be careful about assuming that the presence of a ysrRST locus, as well as the ysa and ysp genes, will result in secretion of Ysps. Although we have shown here that biotype 1B isolates of Y. enterocolitica have ysrT and Howard et al. previously demonstrated by microarray analysis that these biotype 1B isolates have the ysa and ysp genes (19), we have found significant differences in the abilities of these same strains to secrete Ysps under laboratory conditions (K. A. Walker, S. E. Witowski, and V. L. Miller, unpublished results).
This body of research serves to define genetically that the YsrRST system comprises a phosphorelay system and that phosphorylation is a requisite element in the normal function of these proteins in their role as activators of the ysa type III secretion genes. Several elements of this system are unusual in that (i) the hybrid sensor lacks an HPt domain, (ii) the HPt domain is provided on the small cytosolic protein YsrT, (iii) ysrT is encoded immediately downstream of ysrS, and (iv) YsrT is unique to Y. enterocolitica and nonpathogenic Yersinia species. In addition, YsrR contains stretches of amino acids that appear to be insertions not found in similar RRs, indicating that it may have some unique properties/functions as well. Like many Gram-negative bacteria, Y. enterocolitica has about 30 TCS, with 24 HKs, 6 of which are hybrid HKs, and 31 RRs. Of the six hybrid HKs, two are unique to yersiniae: YsrRST and YE3578-YE3579. Curiously, YE3578 and YE3579 are nearly identical to YsrR and YsrS, respectively, on both the amino acid and DNA levels. Adding to the intrigue of this second system is that there is no HPt-containing protein associated with YE3578-YE3579. In analogy to the LuxNUO and LusQUO systems, it is tempting to speculate that YsrT could function as the HPt for both YsrS and YE3579, thus explaining the significance of ysrT encoding a protein detached from its cognate sensor. Efforts to elucidate whether YE3578-YE3579 utilizes YsrT for regulating ysaE or other promoters are ongoing in our laboratory.
Acknowledgments
We thank Ruth E. Silversmith for critical reading of the manuscript and Ruth E. Silversmith, Bob Bourret, Joshua Hall, and Matthew Lawrenz for stimulating discussions. We are indebted to Brittany Fogarty and R. Patrick Summers for technical assistance.
This research was supported by National Institutes of Health grant AI063299 awarded to V. L. Miller.
Footnotes
Published ahead of print on 24 September 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Appleby, J. L., J. S. Parkinson, and R. B. Bourret. 1996. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell 86:845-848. [DOI] [PubMed] [Google Scholar]
- 3.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourret, R. B. 2010. Receiver domain structure and function in response regulator proteins. Curr. Opin. Microbiol. 13:142-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burbulys, D., K. A. Trach, and J. A. Hoch. 1991. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell 64:545-552. [DOI] [PubMed] [Google Scholar]
- 6.Chen, P. E., C. Cook, A. C. Stewart, N. Nagarajan, D. D. Sommer, M. Pop, B. Thomason, M. P. Thomason, S. Lentz, N. Nolan, S. Sozhamannan, A. Sulakvelidze, A. Mateczun, L. Du, M. E. Zwick, and T. D. Read. 2010. Genomic characterization of the Yersinia genus. Genome Biol. 11:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clarke, D. J., S. A. Joyce, C. M. Toutain, A. Jacq, and I. B. Holland. 2002. Genetic analysis of the RcsC sensor kinase from Escherichia coli K-12. J. Bacteriol. 184:1204-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cover, T. L., and R. C. Aber. 1989. Yersinia enterocolitica. N. Engl. J. Med. 321:16-24. [DOI] [PubMed] [Google Scholar]
- 9.Foussard, M., S. Cabantous, J. Pedelacq, V. Guillet, S. Tranier, L. Mourey, C. Birck, and J. Samama. 2001. The molecular puzzle of two-component signaling cascades. Microbes Infect. 3:417-424. [DOI] [PubMed] [Google Scholar]
- 10.Fredriksson-Ahomaa, M., M. Bucher, C. Hank, A. Stolle, and H. Korkeala. 2001. High prevalence of Yersinia enterocolitica 4:O3 on pig offal in southern Germany: a slaughtering technique problem. Syst. Appl. Microbiol. 24:457-463. [DOI] [PubMed] [Google Scholar]
- 11.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 12.Freeman, J. A., and B. L. Bassler. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao, R., and A. M. Stock. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgellis, D., O. Kwon, P. De Wulf, and E. C. Lin. 1998. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J. Biol. Chem. 273:32864-32869. [DOI] [PubMed] [Google Scholar]
- 15.Georgellis, D., A. S. Lynch, and E. C. Lin. 1997. In vitro phosphorylation study of the arc two-component signal transduction system of Escherichia coli. J. Bacteriol. 179:5429-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 17.Haller, J. C., S. Carlson, K. J. Pederson, and D. E. Pierson. 2000. A chromosomally encoded type III secretion pathway in Yersinia enterocolitica is important in virulence. Mol. Microbiol. 36:1436-1446. [DOI] [PubMed] [Google Scholar]
- 18.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 19.Howard, S. L., M. W. Gaunt, J. Hinds, A. A. Witney, R. Stabler, and B. W. Wren. 2006. Application of comparative phylogenomics to study the evolution of Yersinia enterocolitica and to identify genetic differences relating to pathogenicity. J. Bacteriol. 188:3645-3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inclán, Y. F., S. Laurent, and D. R. Zusman. 2008. The receiver domain of FrzE, a CheA-CheY fusion protein, regulates the CheA histidine kinase activity and downstream signalling to the A- and S-motility systems of Myxococcus xanthus. Mol. Microbiol. 68:1328-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwobi, A., A. Rakin, E. Garcia, and J. Heesemann. 2002. Representational difference analysis uncovers a novel IS10-like insertion element unique to pathogenic strains of Yersinia enterocolitica. FEMS Microbiol. Lett. 210:251-255. [DOI] [PubMed] [Google Scholar]
- 22.Jayarao, B. M., and D. R. Henning. 2001. Prevalence of foodborne pathogens in bulk tank milk. J. Dairy Sci. 84:2157-2162. [DOI] [PubMed] [Google Scholar]
- 23.Kelley, L. A., and M. J. Sternberg. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363-371. [DOI] [PubMed] [Google Scholar]
- 24.Kinder, S. A., J. L. Badger, G. O. Bryant, J. C. Pepe, and V. L. Miller. 1993. Cloning of the YenI restriction endonuclease and methyltransferase from Yersinia enterocolitica serotype O8 and construction of a transformable R-M+ mutant. Gene 136:271-275. [DOI] [PubMed] [Google Scholar]
- 25.Kwon, O., D. Georgellis, and E. C. Lin. 2000. Phosphorelay as the sole physiological route of signal transmission by the arc two-component system of Escherichia coli. J. Bacteriol. 182:3858-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379-405. [DOI] [PubMed] [Google Scholar]
- 27.Matsumoto, H., and G. M. Young. 2006. Proteomic and functional analysis of the suite of Ysp proteins exported by the Ysa type III secretion system of Yersinia enterocolitica Biovar 1B. Mol. Microbiol. 59:689-706. [DOI] [PubMed] [Google Scholar]
- 28.McNally, A., T. Cheasty, C. Fearnley, R. W. Dalziel, G. A. Paiba, G. Manning, and D. G. Newell. 2004. Comparison of the biotypes of Yersinia enterocolitica isolated from pigs, cattle and sheep at slaughter and from humans with yersiniosis in Great Britain during 1999-2000. Lett. Appl. Microbiol. 39:103-108. [DOI] [PubMed] [Google Scholar]
- 29.Merriam, J. J., R. Mathur, R. Maxfield-Boumil, and R. R. Isberg. 1997. Analysis of the Legionella pneumophila fliI gene: intracellular growth of a defined mutant defective for flagellum biosynthesis. Infect. Immun. 65:2497-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mildiner-Earley, S., K. A. Walker, and V. L. Miller. 2007. Environmental stimuli affecting expression of the Ysa type three secretion locus. Adv. Exp. Med. Biol. 603:211-216. [DOI] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Miller, V. L., J. J. Farmer III, W. E. Hill, and S. Falkow. 1989. The ail locus is found uniquely in Yersinia enterocolitica serotypes commonly associated with disease. Infect. Immun. 57:121-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson, J. S., and E. C. Kofoid. 1992. Communication modules in bacterial signaling proteins. Annu. Rev. Genet. 26:71-112. [DOI] [PubMed] [Google Scholar]
- 35.Peña-Sandoval, G. R., O. Kwon, and D. Georgellis. 2005. Requirement of the receiver and phosphotransfer domains of ArcB for efficient dephosphorylation of phosphorylated ArcA in vivo. J. Bacteriol. 187:3267-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Posas, F., S. M. Wurgler-Murphy, T. Maeda, E. A. Witten, T. C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86:865-875. [DOI] [PubMed] [Google Scholar]
- 37.Robins-Browne, R. M., and J. K. Prpic. 1985. Effects of iron and desferrioxamine on infections with Yersinia enterocolitica. Infect. Immun. 47:774-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, R. E., A. M. Carey, J. M. Damare, F. M. Hetrick, R. W. Johnston, and W. H. Lee. 1981. Evaluation of iron dextran and mucin for enhancement of the virulence of Yersinia enterocolitica serotype O:3 in mice. Infect. Immun. 34:550-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeda, S., Y. Fujisawa, M. Matsubara, H. Aiba, and T. Mizuno. 2001. A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC→YojN→RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol. Microbiol. 40:440-450. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomson, N. R., S. Howard, B. W. Wren, M. T. Holden, L. Crossman, G. L. Challis, C. Churcher, K. Mungall, K. Brooks, T. Chillingworth, T. Feltwell, Z. Abdellah, H. Hauser, K. Jagels, M. Maddison, S. Moule, M. Sanders, S. Whitehead, M. A. Quail, G. Dougan, J. Parkhill, and M. B. Prentice. 2006. The complete genome sequence and comparative genome analysis of the high pathogenicity Yersinia enterocolitica strain 8081. PLoS Genet. 2:e206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuzuki, M., K. Ishige, and T. Mizuno. 1995. Phosphotransfer circuitry of the putative multi-signal transducer, ArcB, of Escherichia coli: in vitro studies with mutants. Mol. Microbiol. 18:953-962. [DOI] [PubMed] [Google Scholar]
- 43.Ulrich, L. E., and I. B. Zhulin. 2010. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic Acids Res. 38:D401-D407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vantrappen, G., E. Ponette, K. Geboes, and P. Bertrand. 1977. Yersinia enteritis and enterocolitis: gastroenterological aspects. Gastroenterology 72:220-227. [PubMed] [Google Scholar]
- 45.Venecia, K., and G. M. Young. 2005. Environmental regulation and virulence attributes of the Ysa type III secretion system of Yersinia enterocolitica biovar 1B. Infect. Immun. 73:5961-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker, K. A., and V. L. Miller. 2004. Regulation of the Ysa type III secretion system of Yersinia enterocolitica by YsaE/SycB and YsrS/YsrR. J. Bacteriol. 186:4056-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker, K. A., and V. L. Miller. 2009. Synchronous gene expression of the Yersinia enterocolitica Ysa type III secretion system and its effectors. J. Bacteriol. 191:1816-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 49.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 50.Young, B. M., and G. M. Young. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184:1324-1334. [DOI] [PMC free article] [PubMed] [Google Scholar]