Abstract
The region-specific homeotic gene spalt (sal) acts in two separate domains in the head and tail region of the Drosophila melanogaster embryo. Based on comparative morphology, sal is likely to be involved in the establishment of the head during the evolution of invertebrates and thus, it should be conserved. We have analyzed the conservation of the segmentation genes Krüppel (Kr) and even-skipped (eve) in parallel with sal coding sequences in several Drosophila species that are evolutionarily separated by up to 60 million years. To our surprise, sal sequences appear to be conserved in the Sophophora subgenus of the Drosophila genus but not in the Drosophila subgenus. On the other hand, the segmentation and other homeotic genes are conserved in the Drosophila subgroup as well. Our data suggest that sal encodes an accessory function that evolved relatively late during Drosophila speciation rather than playing a fundamental evolutionary role similar to that of other homeotic genes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987 Sep;101(1):1–22. [PubMed] [Google Scholar]
- Blackman R. K., Meselson M. Interspecific nucleotide sequence comparisons used to identify regulatory and structural features of the Drosophila hsp82 gene. J Mol Biol. 1986 Apr 20;188(4):499–515. doi: 10.1016/s0022-2836(86)80001-8. [DOI] [PubMed] [Google Scholar]
- Bodmer M., Ashburner M. Conservation and change in the DNA sequences coding for alcohol dehydrogenase in sibling species of Drosophila. 1984 May 31-Jun 6Nature. 309(5967):425–430. doi: 10.1038/309425a0. [DOI] [PubMed] [Google Scholar]
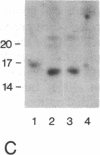
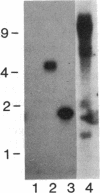
- Frei E., Schuh R., Baumgartner S., Burri M., Noll M., Jürgens G., Seifert E., Nauber U., Jäckle H. Molecular characterization of spalt, a homeotic gene required for head and tail development in the Drosophila embryo. EMBO J. 1988 Jan;7(1):197–204. doi: 10.1002/j.1460-2075.1988.tb02800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaul U., Jäckle H. Pole region-dependent repression of the Drosophila gap gene Krüppel by maternal gene products. Cell. 1987 Nov 20;51(4):549–555. doi: 10.1016/0092-8674(87)90124-3. [DOI] [PubMed] [Google Scholar]
- Hayashida H., Miyata T. Unusual evolutionary conservation and frequent DNA segment exchange in class I genes of the major histocompatibility complex. Proc Natl Acad Sci U S A. 1983 May;80(9):2671–2675. doi: 10.1073/pnas.80.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham P. W. The molecular genetics of embryonic pattern formation in Drosophila. Nature. 1988 Sep 1;335(6185):25–34. doi: 10.1038/335025a0. [DOI] [PubMed] [Google Scholar]
- Jürgens G. Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. EMBO J. 1988 Jan;7(1):189–196. doi: 10.1002/j.1460-2075.1988.tb02799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis J. A., Poole S. J., Wright D. K., O'Farrell P. H. Sequence conservation in the protein coding and intron regions of the engrailed transcription unit. EMBO J. 1986 Dec 20;5(13):3583–3589. doi: 10.1002/j.1460-2075.1986.tb04686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman T. C., Lewis R., Wakimoto B. Cytogenetic Analysis of Chromosome 3 in DROSOPHILA MELANOGASTER: The Homoeotic Gene Complex in Polytene Chromosome Interval 84a-B. Genetics. 1980 Jan;94(1):115–133. doi: 10.1093/genetics/94.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Hoey T. Homeobox proteins as sequence-specific transcription factors. Cell. 1988 Nov 18;55(4):537–540. doi: 10.1016/0092-8674(88)90209-7. [DOI] [PubMed] [Google Scholar]
- Lewis E. B. A gene complex controlling segmentation in Drosophila. Nature. 1978 Dec 7;276(5688):565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Ouweneel W. J. Developmental genetics of homoeosis. Adv Genet. 1976;18:179–248. doi: 10.1016/s0065-2660(08)60439-3. [DOI] [PubMed] [Google Scholar]
- Schuh R., Aicher W., Gaul U., Côté S., Preiss A., Maier D., Seifert E., Nauber U., Schröder C., Kemler R. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Krüppel, a Drosophila segmentation gene. Cell. 1986 Dec 26;47(6):1025–1032. doi: 10.1016/0092-8674(86)90817-2. [DOI] [PubMed] [Google Scholar]
- Treier M., Pfeifle C., Tautz D. Comparison of the gap segmentation gene hunchback between Drosophila melanogaster and Drosophila virilis reveals novel modes of evolutionary change. EMBO J. 1989 May;8(5):1517–1525. doi: 10.1002/j.1460-2075.1989.tb03536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde C. D., Akam M. Conserved sequence elements in the 5' region of the Ultrabithorax transcription unit. EMBO J. 1987 May;6(5):1393–1401. doi: 10.1002/j.1460-2075.1987.tb02380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]