Summary
The circadian clock is the endogenous timer that coordinates physiological processes with daily and seasonal environmental changes. In Arabidopsis thaliana, establishment of the circadian period relies on targeted degradation of TIMING OF CAB EXPRESSION 1 (TOC1) by the 26S proteasome. ZEITLUPE (ZTL) is the F-box protein that associates with the SCF (Skp/Cullin/F-box) E3 ubiquitin ligase that is responsible for marking TOC1 for turnover. CULLIN1 (CUL1) is a core component of SCF complexes and is involved in multiple signaling pathways. To assess the contribution of CUL1-containing SCF complexes to signaling within the plant oscillator, circadian rhythms were examined in the recessive, temperature-sensitive CUL1 allele axr6-3. The activity of CUL1 in this mutant declines progressively with increasing ambient temperature, resulting in more severe defects in CUL1-dependent activities at elevated temperature. Examination of circadian rhythms in axr6-3 revealed circadian phenotypes comparable to those observed in ztl null mutants; namely, lengthened circadian period, altered expression of core oscillator genes, and limited degradation of TOC1. In addition, treatment of seedlings with exogenous auxin did not alter TOC1 stability. These results demonstrate that CUL1 is required for TOC1 degradation and further suggest that this protein is the functional cullin for the SCFZTL complex.
Keywords: circadian rhythms, CUL1, post-translational regulation, ubiquitin ligase, proteasome, Arabidopsis
Introduction
The circadian oscillator is the endogenous timekeeper responsible for generating and maintaining physiological rhythms with a period of approximately 24 h. Substantial progress has been made in understanding the molecular mechanism underlying the plant clock and the consequences of its rhythms for plant physiology (Harmer et al., 2005; Harmon et al., 2005). Beyond providing a simple list of components for the oscillator, circadian research in Arabidopsis thaliana has established that the clock serves to enhance plant fitness by influencing myriad physiological processes (Hotta et al., 2007). The circadian clock is perhaps best known for its contribution to the measurement of day length, which triggers flowering during inductive photoperiods (Imaizumi and Kay, 2006). The clock also participates in responses to additional external stimuli, including 568 neighbor shading, low temperature, and other abiotic stresses (Fowler et al., 2005; Kreps et al., 2002; Salter et al., 2003). In addition, circadian regulation has been observed for many aspects of metabolism, including starch production/utilization, transpiration, and nitrate assimilation (Blasing et al., 2005; Kreps and Kay, 1997). The plant growth rate is also closely tied to circadian clock function (Dodd et al., 2005; Green et al., 2002).
The ubiquitin/26S proteasome pathway is a key means of controlling protein activity in plants, fungi, and animals (Hershko and Ciechanover, 1998; Smalle and Vierstra, 2004). The central regulatory mechanism of this pathway is selective degradation of proteins by ubiquitinylation, which involves covalent attachment of the small protein ubiquitin to a specific target protein. Ubiquitin serves as a signal for target protein degradation, mediated by the 26S proteasome (Pickart, 2001). Plants have significantly expanded the use of this pathway to control the activity and expression of proteins: genes encoding components of the ubiquitin/26S proteasome comprise approximately 5% of the Arabidopsis genome (Bachmair et al., 2001; Gagne et al., 2002).
Ubiquitinylation takes place at the end of a multi-step process and is mediated by specific E3 ubiquitin ligases that are responsible for identifying cognate target proteins and for final transfer of ubiquitin to the target protein (Pickart, 2001). Four categories of E3 ubiquitin ligases have been confirmed in plants, the most abundant being the SCF (for SKP/CULLIN/F-box) complex (Vierstra, 2003). As its name indicates, the SCF complex is a multi-protein assembly composed of individual CULLIN (CUL), SKP (ASK), and F-box subunits, in addition to one member of a class of RING finger proteins known as RBX. The function of the F-box protein is to identify the target protein; therefore, the F-box protein provides target specificity to the E3 ligase. Among the SCF subunits in plants, F-box proteins exhibit the greatest diversity, as over 700 of these proteins are encoded in the Arabidopsis genome (Gagne et al., 2002). CULs are the core subunits that act as scaffolds for assembly of the complex (Zheng et al., 2002). Of the eleven CUL genes predicted in Arabidopsis, only five (CUL1, CUL2, CUL3A, CUL3B, and CUL4) encode canonical CUL proteins (Risseeuw et al., 2003). The activity of CUL1 is required for embryogenesis, senescence, floral development and responses to phytohormones and light (Durfee et al., 2003; Gray et al., 2002; Guo and Ecker, 2003; Hellmann et al., 2003; Potuschak et al., 2003; Shen et al., 2002; Woo et al., 2001). Homozygous null alleles of CUL1 confer an embryolethal phenotype, suggesting an essential role for SCF ubiquitin ligases early in plant development (Shen et al., 2002). Although CUL1 was thought to participate in the circadian clock (Han et al., 2004), there is no functional evidence confirming its role in the oscillator.
axr6-3 is a temperature-sensitive CUL1 mutant that is recessive and fully viable (Quint et al., 2005). This mutant has defects in flower development and in responses to the phytohormones auxin and jasmonate. The severity of the axr6-3 phenotype is progressively stronger with increasing ambient temperature; for example, the concentrations of auxin required to inhibit root growth by 50% in axr6-3 at 24 and 28°C are 10- and 100-fold greater, respectively, than that needed at 20°C (Quint et al., 2005). axr6-3 harbors a missense mutation in CUL1, substituting a glutamic acid at position 159 for a lysine, which renders the protein unstable at elevated temperatures and limits the formation of active SCF complexes harboring CUL1 (Quint et al., 2005). The temperature-sensitive and viable nature of axr6-3 makes it a unique tool to investigate the contribution of CUL1 to plant signaling pathways.
The ubiquitin/26S proteasome pathway plays an important role in the establishment of period length in the Arabidopsis circadian system (Kim et al., 2007; Mas et al., 2003b). ZEITLUPE (ZTL) is an F-box/LOV/Kelch protein (Han et al., 2004; Somers et al., 2000) that acts as a circadian clock-specific blue-light photoreceptor (Kim et al., 2007). TIMING OF CAB EXPRESSION 1 (TOC1) is a pseudo-response regulator protein that is presumed to function as a transcription factor (Strayer et al., 2000). TOC1 lengthens the circadian period (Makino et al., 2002; Mas et al., 2003a), and ZTL acts antagonistically to shorten period (Somers et al., 2004). Strong constitutive overexpression of TOC1 drives the clock to arrhythmicity (Makino et al., 2002; Mas et al., 2003a). ztl loss-of-function mutants exhibit long periods (Jarillo et al., 2001; Somers et al., 2000), whereas toc1 mutants have short periods (Mas et al., 2003a; Somers et al., 1998; Strayer et al., 2000). The ztl toc1 double mutant also has a short period (Mas et al., 2003b), indicating that the ztl phenotype requires TOC1 function. TOC1 and ZTL physically interact in vivo, and TOC1 accumulation is constitutively high in a ztl background (Mas et al., 2003b); hence, the F-box protein ZTL targets TOC1 for proteasome-mediated degradation. ZTL is critical for controlling TOC1 activity, which, in turn, is important for modulation of the length of the circadian period.
Although ZTL is associated with CUL1 in vivo (Han et al., 2004), CUL1 has not been directly demonstrated to be functionally important for SCFZTL activity. Furthermore, it remains unclear whether SCF complexes harboring CUL1 or other CULs contribute at any point to the generation and maintenance of circadian rhythms. To determine the role that SCF complexes reliant on CUL1 play in the Arabidopsis molecular clock, we examined circadian rhythms in axr6-3. We found that axr6-3 plants exhibited strong circadian phenotypes, which correlated with the loss of SCFZTL activity associated with ztl null alleles, including a substantial phase delay for circadian gene expression, altered expression levels for core clock genes, and a long circadian period. Circadian rhythms in the CUL1 mutant were also less precise; the period varied substantially within a group of seedlings, such that a larger proportion of the individuals were scored as arrhythmic. Furthermore, inactivation of CUL1 by elevated temperature blocked turnover of TOC1. These findings indicate that CUL1 activity is required for TOC1 degradation, consistent with CUL1 acting as the primary CUL protein in SCFZTL.
Results
Leaf movement rhythms in axr6-3 are temperature-dependent
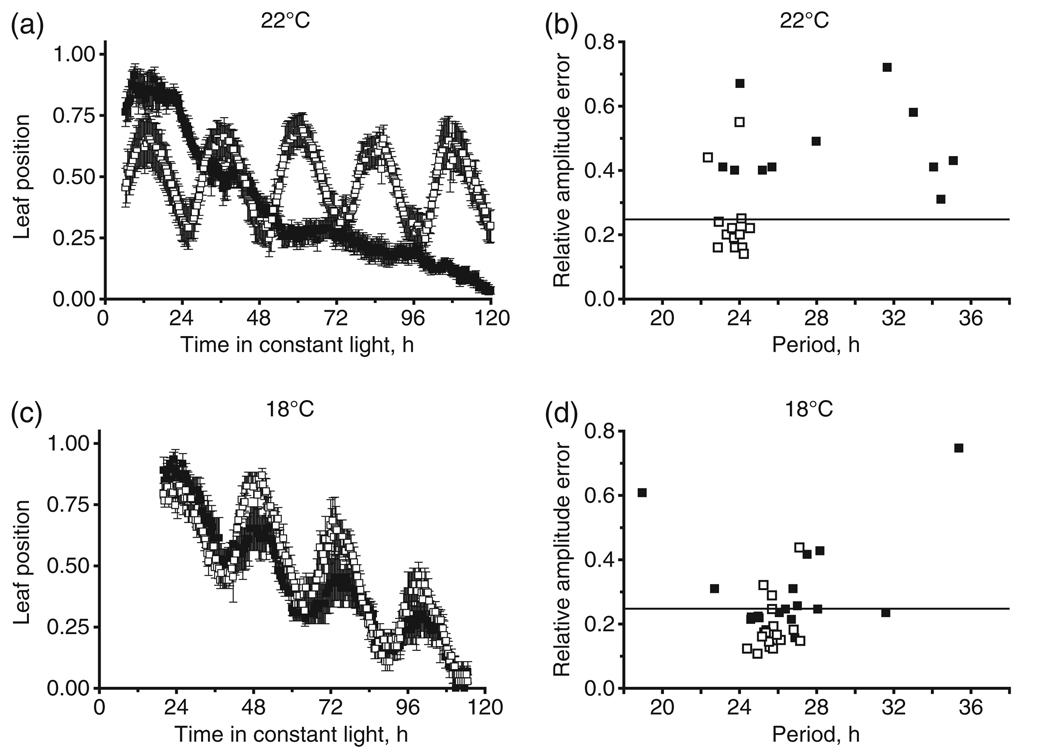
To assess the impact of diminished CUL1 activity on circadian rhythms, axr6-3 and wild-type (WT) seedlings were assayed for leaf movement rhythms at 22°C, which is a restrictive temperature for axr6-3. Seedlings were grown in 12 h light:12 h dark cycles for 5 days, and then transferred at Zeitgeiber Time 0 (ZT0; ZT is expressed in hours, and ZT0 represents 08:00 h) to constant light conditions. Seedlings were assayed for rhythmic leaf movements for 5 days under these free-running conditions. axr6-3 seedlings were arrhythmic for leaf movement when assayed at 22°C (Figure 1a). The average leaf movement trace for mutant seedlings clearly shows that these seedlings could not sustain rhythms after the first day in constant light. Conversely, WT seedlings exhibited obvious leaf movement rhythms that persisted over the entire 5 days of the experiment (Figure 1a). axr6-3 appeared capable of entrainment, however, as a peak of the same approximate phase as in WT was apparent during the first day in constant light.
Figure 1. Rhythmic leaf movement in axr6-3 is temperature-dependent.
(a, c) Traces of leaf position for seedlings of axr6-3 (filled squares), and WT (open squares) in constant light at 22°C (a) and 18°C (c). Mean values (n = 6) are shown for each time point, and error bars indicate SEM. Data in (a) and (c) are from one of two independent experiments. The leaf position under each condition was normalized to the highest value in that experiment.
(b, d) Relative amplitude error versus estimated free-running period for axr6-3 (filled squares) and WT (open squares) seedlings as determined by FFT/NLLS analysis at 22°C (b) and 18°C (d). Relative amplitude error values ≤0.25 were considered rhythmic, and this cut-off is indicated by the horizontal line. Data in (b) and (d) are from two independent experiments.
Curve-fitting analysis of the leaf movement traces confirmed that rhythmic leaf movement for all axr6-3 seedlings was compromised under these conditions (Table 1 and Figure 1b). The estimated period length for axr6-3 individuals was widely distributed across a broad spectrum of estimates, and a greater proportion of the seedlings were defined as arrhythmic, consistent with a lack of robust circadian behavior in axr6-3 (Figure 1b). On the other hand, all the WT seedlings, with the exception of two, were rhythmic under the same conditions, and the period estimates tightly clustered around a value of 23.8 ± 0.5 h (Table 1 and Figure 1b). These results suggest that axr6-3 lacks circadian leaf movements at this restrictive temperature, and therefore that CUL1 activity is important for maintenance of circadian rhythms in constant light.
Table 1.
Circadian leaf movement in WT and axr6-3 seedlings at 22°C and 18°C
| Genotype | 22°Ca |
18°Ca |
||
|---|---|---|---|---|
| Period ± SDb (h) |
Rhythmic/ total |
Period ± SDb (h) |
Rhythmic/ total |
|
| WT | 23.8±0.5 | 11/13 | 25.7±0.7 | 14/17 |
| axr6-3 | ARRc | 0/12 | 26.0±1.1 | 11/18 |
Plants were entrained for 5 days under 12 h light:12 h dark cycles at the indicated temperature, then held in continuous white light (50 µmol m−2 sec−1) for 5 days.
Data from two independent replicates.
The variance-weighted mean period was determined as described previously (Millar et al., 1995b; Plautz et al., 1997). Only those period estimates with relative amplitude error values ≤0.25 and periods between 20 and 30 h were included in this analysis. SD, standard deviation.
ARR, arrhythmic (indicates the absence of period estimates with relative amplitude error values ≤0.25 for any of the seedlings).
To verify that the arrhythmic phenotype in axr6-3 was due to temperature-dependent inactivation of CUL1 activity, rhythms in cotyledon movement were examined in seedlings exposed to 18°C (Figure 1c,d). Previous work demonstrated that growth of axr6-3 at 20°C enhances the auxin response in mutant seedlings, but does not restore full CUL1 activity (Quint et al., 2005). We found that, at 18°C, cotyledon movement in axr6-3 was more rhythmic than at 22°C, and the rhythms were superficially comparable to those of WT (Figure 1c). Curve-fit analysis demonstrated that the average period of the oscillator in rhythmic axr6-3 seedlings was 26.0 ± 1.1 h, which is similar to the 25.7 ± 0.7 h period observed for WT (Table 1). However, axr6-3 individuals exhibited a larger range of rhythmic period estimates than did WT (Figure 1d), and more axr6-3 seedlings were arrhythmic than WT (Table 1); thus, this less restrictive temperature only partially suppressed the axr6-3 circadian defect in leaf movement. Clearly, robust leaf movement rhythms require full CUL1 activity, which suggests an important role in the circadian oscillator for those SCF E3 ubiquitin ligases that require CUL1 activity.
axr6-3 affects circadian period and maintenance of rhythms
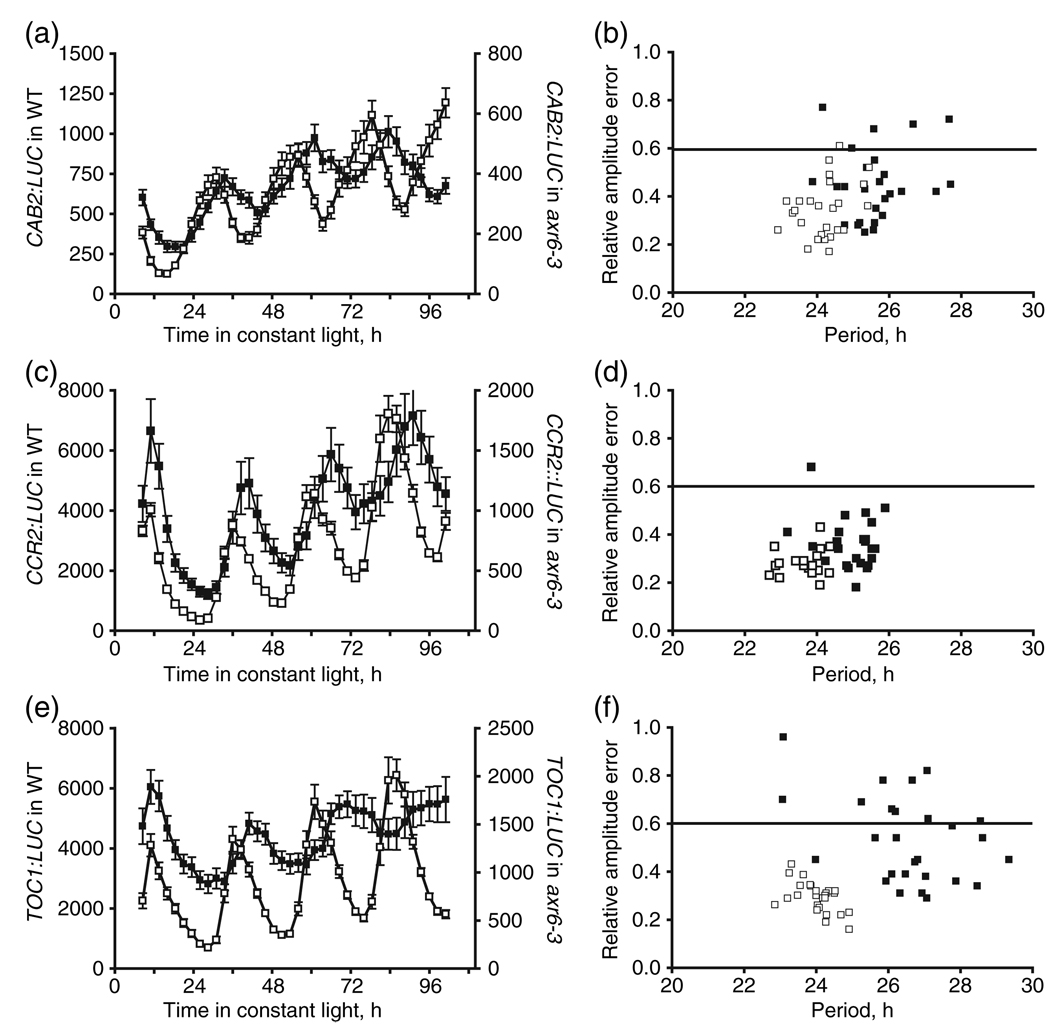
Leaf movement is an indirect measure of oscillator function, and the physiological changes driving leaf movement could have relied on CUL1-dependent processes separate from the circadian clock. Therefore, the leaf movement phenotype in axr6-3 may have represented an amalgam of phenotypes to which the oscillator was one of several contributors. To more directly assess the state of the circadian clock in axr6-3, circadian gene expression was examined in the WT and mutant backgrounds by well-characterized circadian promoter:luciferase (LUC) reporter constructs. The axr6-3 allele was crossed into plants harboring DNA constructs of CHLOROPHYLL a/b-BINDING PROTEIN 2 (CAB2):LUC, COLD CIRCADIAN RHYTHM–RNA BINDING 2 (CCR2):LUC, and TOC1:LUC, and these lines were examined for rhythmic gene expression under constant light conditions after entrainment under 12 h light:12 h dark cycles at 22°C for 7 days. The bioluminescence signal from axr6-3 seedlings was consistently lower than from WT for each of the reporter constructs (Figure 2a,c,e). The lower bioluminescence signal from axr6-3 seedlings appeared to be due to the small size of the axr6-3 seedlings rather than changes in the amplitude of gene expression, as the area of the cotyledons from mutant seedlings was consistently smaller than in WT seedlings (data not shown), and transcript levels for CAB2, CCR2, and TOC1 in axr6-3 were comparable to WT levels when examined by real-time PCR (Figure 3c and Figure S1a,b).
Figure 2. Circadian gene expression in axr6-3 is long-period and rhythms lack the precision of those in WT.
(a, c, e) Bioluminescence rhythms for CAB2:LUC (a), CCR2:LUC (c), and TOC1:LUC (e) in axr6-3 (filled squares) and WT (open squares) seedlings at 22°C. The left y axis shows the bioluminescence signal for WT and the right y-axis shows that for axr6-3. Data are mean values (n = 15 for axr6-3 and n = 10 for WT), and error bars indicate SEM. Representative data in (a), (c), and (e) are from one of two independent experiments.
(b, d, f) Relative amplitude error versus estimated free-running period for CAB2:LUC (b), CCR2:LUC (d), and TOC1:LUC (f) for axr6-3 (filled squares) and WT (open squares) seedlings as determined by FFT/NLLS analysis. Relative amplitude error values ≤0.6 were considered rhythmic, and this cut-off is indicated by the horizontal line. Data in (b), (d), and (f) are from two independent experiments.
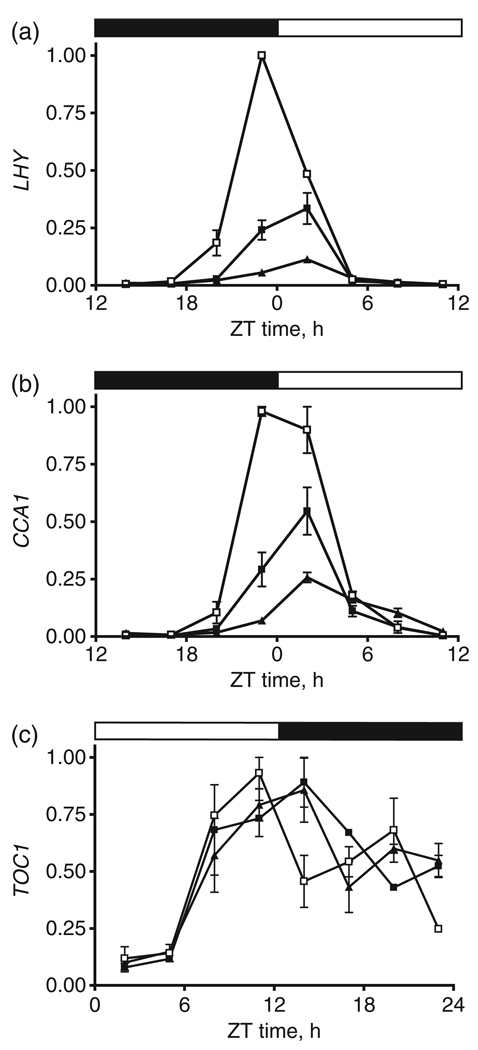
Figure 3. Clock gene expression in axr6-3 is comparable to that of ztl-4.
LHY(a), CCA1(b), and TOC1(c) transcript abundance in 1-week-old seedlings of WT (open squares), axr6-3 (filled squares), and ztl-4 (filled triangles), each grown under diurnal conditions. The black and white bars above the graphs indicate periods of dark and light, respectively. Data are the means of two independent replicates and error bars indicate range. Gene expression was determined by real-time PCR, and expression of IPP2 (see Experimental procedures) was used as a normalization control. The expression level for each experimental gene was calculated using the formula 2[CT(IPP2)–CT(experimental)] where CT is the average threshold cycle for three technical replicates. The expression value for each gene was normalized to the highest value for each experiment.
Clock-driven gene expression in axr6-3 clearly deviated from that in WT with respect to both period length and precision of the rhythms, regardless of the reporter construct assayed (Figure 2). Visual inspection of the curves revealed a consistent phase delay in the traces from axr6-3 seedlings, which ranged from 2.5 to 5 h, depending upon the reporter (Figure 2a,c,e). TOC1:LUC expression also exhibited an obvious lengthening of period, such that, under constant light conditions, expression in axr6-3 was anti-phase to WT by the fourth day (Figure 2e). A comparable period change was not apparent in the curves for either CAB2:LUC or CCR2:LUC (Figure 2a,c). The estimated period of expression for each reporter was determined, and these results indicated that the pace of the clock in those axr6-3 seedlings exhibiting detectable rhythms was slower than in WT (Table 2). In axr6-3, the average period for expression of the two reporters representing the circadian output genes CAB2:LUC, and CCR2:LUC was longer by about 1.5 h (Table 2). As inferred from visual inspection, the strongest period phenotype was observed for TOC1:LUC expression, where the average period for rhythmic axr6-3 seedlings was 3 h longer than WT and 1.5 h longer than expression of the other two reporters (Table 2). The strong effect of axr6-3 on TOC1 expression suggests that CUL1 may participate directly in the regulation of this gene, whereas the changes observed for CCR2 and CAB2 expression may reflect contributions of CUL1 to overall clock function.
Table 2.
Estimates of free-running periods for CAB2:LUC, CCR2:LUC, and TOC1:LUC expression, and the proportion of seedlings exhibiting detectable rhythms in WT and axr6-3 backgrounds
| Genotype |
CAB2:LUCa |
CCR2:LUCa |
TOC1:LUCa |
|||
|---|---|---|---|---|---|---|
| Period ± SDb (h) | Rhythmic/total | Period ± SDb (h) | Rhythmic/total | Period ± SDb (h) | Rhythmic/total | |
| WT | 24.2 ± 0.6 | 25/26 | 23.6 ±0.6 | 18/18 | 24.0 ± 0.5 | 26/26 |
| axr6-3 | 25.6± 0.8 | 23/27 | 25.0 ± 0.7 | 24/25 | 27.2±1.4 | 19/29 |
Plants were entrained for 6 days under 12 h light:12 h dark cycles at 22°C, then held in continuous white light (50 µmol m−2 sec−1) at 22°C for 5 days.
Data are from two independent replicates.
The variance-weighted mean period was determined as described previously (Millar et al., 1995b; Plautz et al., 1997). Only those period estimates with relative amplitude error values ≤0.6 and period estimates between 20 and 30 h were included in this analysis. SD, standard deviation.
The impact of axr6-3 on circadian gene expression was more apparent when the rhythms for individual seedlings were compared within each genotype. In general, period length in axr6-3 seedlings was less uniform than in WT, and a greater proportion of seedlings exhibited poor rhythms, with relative amplitude error values ≤0.6 (Figure 2b,d,g and Table 2). For CAB2:LUC, the period length was spread over a broad range of values that overlapped with those for WT, and more mutant individuals were scored as arrhythmic (20% in axr6-3 versus 5% in WT; Figure 2b and Table 2). In contrast, the period of CCR2:LUC expression in most of the axr6-3 seedlings clustered around a common period value that was 1.5 h longer than in WT (Figure 2f). Furthermore, only one axr6-3 seedling was considered arrhythmic for CCR2:LUC expression (Table 2). The period of TOC1:LUC expression was consistently 3 h longer than WT (Table 2), but the spread of these longer period values was much greater than in WT (Figure 2d). In addition, 30% of axr6-3 individuals showed arrhythmic TOC1 expression, whereas all WT seedlings were scored as rhythmic. These findings clearly demonstrate that CUL1 activity is important for establishment of the clock period and phase, as well as for maintenance of circadian rhythms under constant light.
Expression of core circadian clock genes in axr6-3 mirrors that in ztl mutants
ZEITLUPE is a component of an SCF-type E3 ubiquitin ligase that plays a critical role in setting the circadian period (Mas et al., 2003b; Somers et al., 2004). Purification of the SCFZTL complex from plants yielded CUL1, which is consistent with these proteins participating in a common SCF complex (Han et al., 2004). As full CUL1 activity is needed for correct setting of the circadian period and maintenance of accurate rhythms, the phenotype in axr6-3 is potentially due to a reduction in ZTL activity, arising from a difficulty in assembling the SCFZTL complex. This possibility is supported by the observation that axr6-3 seedlings capable of maintaining rhythms exhibit a long circadian period, similar to that seen in ztl mutants (Jarillo et al., 2001; Somers et al., 2000, 2004). Therefore, we assessed the phenotypic parallels between axr6-3 and the loss-of-function mutant ztl-4 by examining the levels of expression for the circadian clock genes CCA1, LHY, and TOC1 in both mutant backgrounds.
ZEITLUPE is required in diurnal conditions for upregulation of the morning expression of CCA1 and LHY, each encoding myb-like transcription factors that function at the core of the plant oscillator (Green and Tobin, 1999; Schaffer et al., 1998; Somers et al., 2004; Wang and Tobin, 1998). A phenotypic consequence of ZTL inactivation is a strong reduction in CCA1 and LHY transcript levels. As expected, we found that CCA1 and LHY levels in ztl-4 were significantly reduced in seedlings grown under 12 h light:12 h dark cycles at 22°C. The concentrations of CCA1 and LHY mRNA were 4- and 10-fold lower, respectively, in ztl-4 than in WT (Figure 3a,b). A comparable reduction in mRNA levels was observed in axr6-3: LHY and CCA1 transcripts were down-regulated by approximately threefold in the CUL1 mutant, relative to their levels in WT (Figure 3a,b). The diminished accumulation of LHY and CCA1 transcripts in both mutant backgrounds was specific to these clock genes, as peak expression levels of TOC1 in axr6-3 and ztl-4 remained close to those in WT (Figure 3c), as did expression levels of CCR2 and CAB2 (Figure S1a,b). Furthermore, the strength of the reduction in CCA1 and LHY expression correlated well with the severity of the mutation in each background, as axr6-3 showed a milder phenotype than the loss-of-function mutant ztl-4. In addition, both ztl-4 and axr6-3 showed a pronounced 4 h phase delay in the expression of CCA1, LHY, and TOC1 (Figure 3a–c). The magnitude of this phase delay was similar to that observed in axr6-3 for expression from promoter:LUC reporters under constant light (Figure 3). Thus, clock gene expression in axr6-3 largely matched that observed in ztl-4, which suggests that the circadian phenotype of axr6-3 resulted in large part from a reduction in ZTL activity.
Inhibition of CUL1 function in axr6-3 limits TOC1 degradation
In the late night, SCFZTL targets TOC1 for degradation by the ubiquitin/26S proteasome pathway (Mas et al., 2003b). Circadian accumulation of TOC1 peaks at approximately 4 h after dusk, and the activity of ZTL is needed to impose this circadian rhythm on TOC1 levels (Mas et al., 2003b). In the absence of ZTL, TOC1 levels are not rhythmic and the protein accumulates to high levels during the night.
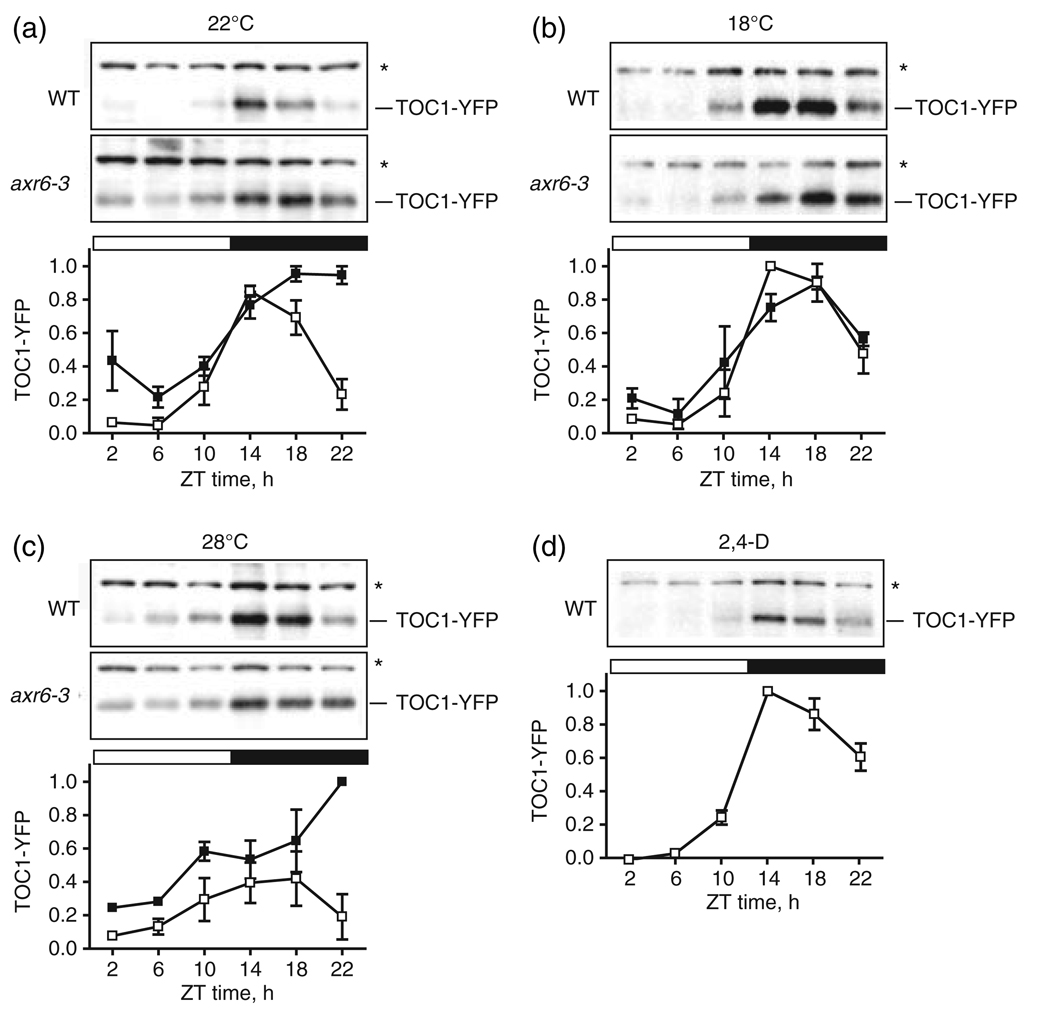
We examined TOC1 stability in axr6-3 to determine whether the circadian phenotype in axr6-3 correlated with changes in TOC1 turnover, which would suggest an inhibition of SCFZTL activity in this mutant. The axr6-3 allele was introduced into the well-studied TOC1:TOC1–YFP mini-gene (TMG) line, in which expression of a translational fusion of TOC1 and YFP is driven by the native TOC1 promoter (Mas et al., 2003b). This line was used to assess the TOC1 expression waveform. In WT, TOC1–YFP exhibited the expected rhythmic expression profile at 22°C in 12 h light:12 h dark cycles; peak protein levels occurred in the evening at ZT14 and then declined with the approaching dawn (Figure 4a). Under the same conditions, TOC1 attained noticeably higher levels in axr6-3 and exhibited an altered expression waveform (Figure 4a). The greatest TOC1 concentration differences between axr6-3 and WT occurred between ZT22 and ZT2. In axr6-3, TOC1 did not exhibit the typical decline in concentration between ZT18 and ZT22 that marks the activity of SCFZTL; instead TOC1 levels fell only after the lights came on at ZT0. A trivial explanation for the modified TOC1 waveform in axr6-3 TMG was that this new protein profile resulted from underlying changes in the expression pattern for the TOC1–YFP mRNA; however, TOC1 mRNA was expressed in axr6-3 TMG seedlings with a diurnal pattern comparable to that in the WT TMG background (Figure S2). In fact, TOC1 expression levels were somewhat lower in the mutant than in WT. Thus, the altered TOC1 waveform in axr6-3 was due to post-transcriptional changes in TOC1 accumulation and not to alterations in the abundance of TOC1 mRNA. These data are consistent with the hypothesis that a defect in SCFZTL activity in axr6-3 blocks TOC1 degradation during the night.
Figure 4. TOC1 turnover is altered in axr6-3, consistent with changes in SCFZTL activity.
(a–c) Normalized TOC1–YFP levels in WT and axr6-3 TOC1:TOC1–YFP mini-gene seedlings grown under 12 h light:12 h dark conditions at 22°C (a), 18°C (b) or shifted from 22 to 28°C (c). (d) WT TMG seedlings grown at 22°C were sprayed at ZT0 with 0.3 µm 2,4-D. Samples were taken at the indicated ZT times.
Upper panels, representative Western blots probed with anti-GFP. Lower panel, normalized average levels of TOC1-YFP in axr6-3 (filled squares) and WT (open squares) from two independent replicates. Error bars represent the range. Asterisks indicate the nonspecific band used as a loading control and for normalization of TOC1-YFP levels. TOC1-YFP levels represent the ratio of the TOC1-YFP band intensity to that of the loading control. The TOC1-YFP level was then normalized to the highest value for each experiment.
To confirm that the axr6-3 allele affects the activity of the SCFZTL complex, the waveform of TOC1 accumulation under 12 h light:12 h dark cycles was determined at 18 and 28°C. These two conditions are expected to have opposite effects on TOC1 accumulation if CUL1 is critical for SCFZTL activity. The TOC1 profile at 18°C was anticipated to more closely match that of WT, as this condition improves axr6-3 leaf movement rhythms (Figure 1b). Conversely, we expected that TOC1 degradation in axr6-3 would be reduced at 28°C, as this condition diminishes root growth inhibition by auxin and abolishes CUL1 association with the SCFTIR1 complex (Quint et al., 2005).
The TOC1–YFP waveform in axr6-3 TMG seedlings grown continuously at 18°C was comparable to that observed for WT under the same conditions (Figure 4b). In contrast to the stable levels of TOC1 exhibited in axr6-3 during the late night at 22°C, a clear reduction in TOC1–YFP levels was apparent at the ZT22 time point in the mutant background, which is the time at which SCFZTL directs TOC1 to the 26S proteasome. Interestingly, peak TOC1 accumulation in axr6-3 occurred 4 h later than in WT, which correlated well with the phase delay observed for TOC1 expression in the mutant background (Figure S2). These results confirm that conditions permitting greater CUL1 activity in axr6-3 resulted in a more substantial loss of TOC1–YFP at the time of night when SCFZTL normally promotes TOC1 turnover.
To test the effect of a temperature that considerably reduced CUL1 activity in axr6-3, mutant and WT TMG seedlings were grown under 12 h light:12 h dark cycles at 22°C and transferred to 28°C at ZT0 on the day of sample collection. The waveform of TOC1 accumulation in WT at this elevated temperature generally followed the same profile as observed at 22°C: maximal amounts of protein were present at ZT18 and then rapidly declined to trough levels by ZT2 (Figure 4c). In contrast, the expression profile of TOC1 in axr6-3 was comparable to that observed in a ztl mutant, in which TOC1 degradation is inhibited (Mas et al., 2003b). TOC1x02013;YFP accumulated throughout the day to levels significantly higher than in WT. In addition, the concentration of the protein did not wane at ZT22, as it did in WT; instead, the amount of TOC1 continued to climb throughout the night. Clearly, temperature inactivation of CUL1 in the axr6-3 background stabilizes TOC1 at the time when SCFZTL usually acts to remove this protein. Thus, ZTL appears unable to direct TOC1 for degradation in the absence of functional CUL1, consistent with CUL1 acting as the cullin protein in the assembly and activity of SCFZTL.
Exposure of seedlings to exogenous natural or artificial auxin moderately lengthens the free-running period and reduces the accuracy of rhythms (Covington and Harmer, 2007; Hanano et al., 2006). For example, the presence of the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D) in growth medium at a concentration of either 0.3 or 5 µM slows the pace of the clock by approximately 1 h and noticeably reduces the amplitude of rhythms (Covington and Harmer, 2007). Another synthetic auxin, 1-napthalene-acetic acid (NAA), has similar effects on rhythms (Hanano et al., 2006). Therefore, it is possible that the circadian phenotype in axr6-3 might be explained by an auxin signaling defect instead of direct inhibition of SCFZTL activity. To examine this possibility, we determined the effect of 2,4-D application on the waveform of TOC1 accumulation. 2,4-D is a potent herbicide and its toxic effects might have indirectly changed TOC1 levels; therefore, we treated seedlings with 0.3 µM 2,4-D, which is the lowest concentration shown to effectively lengthen the circadian period (Covington and Harmer, 2007). WT TMG seedlings sprayed with 0.3 µM 2,4-D at ZT0 exhibited a TOC1 accumulation profile in the 24 h following application of the hormone that matched the behavior of the protein in untreated seedlings (compare Figure 4a,d). As this acute 2,4-D treatment did not elicit a change in TOC1 degradation comparable to the effect in axr6-3 at restrictive temperatures, defective auxin signaling appears unlikely to be a major contributor to the inhibition of TOC1 turnover that is evident in axr6-3.
Discussion
The temperature-sensitive axr6-3 allele of CUL1 was used to directly investigate the functional contribution of CUL1 to the plant circadian oscillator. We show that rhythmic leaf movements in axr6-3 exhibit temperature-dependent arrhythmia. In addition, the period of circadian gene expression in axr6-3 under restrictive temperature conditions is 1–3 h longer than in WT; furthermore, individual seedlings display substantial variability in period length and the robustness of their rhythms. Many aspects of the axr6-3 phenotype correlated with those of a ztl mutant, including changes in the expression of core clock genes and a reduction in TOC1 degradation. Together, our results demonstrate that CUL1 is needed to set and maintain the circadian period, and that this is accomplished, at least in part, through its role in TOC1 turnover, which likely involves participation in the SCFZTL complex.
Circadian leaf movements in axr6-3 were profoundly compromised, but less dramatic changes in circadian rhythms were apparent for CCR2, CAB2, and TOC1 gene expression. A potential explanation for the difference between these two phenotypes is the fact that circadian leaf movement relies on multiple signaling pathways that each use CUL1. For example, petioles are the organs that support cotyledons in young Arabidopsis seedlings. Rhythmic leaf movements are produced by antiphase expansion of the abaxial (bottom) and adaxial (top) cells in the petioles (Engelmann and Johnsson, 1998). Auxin signaling contributes to regulation of cell expansion, and the auxin response is substantially blunted in axr6-3 (Quint et al., 2005). It follows that the auxin signaling defect in axr6-3 is likely to limit the extent of petiole cell expansion, which, in turn, would render circadian rhythms difficult to detect because of weak overall leaf movements. Our data clearly demonstrate that circadian leaf movement in axr6-3 is strongly compromised. For example, the circadian period for leaf movement varied substantially at the permissive temperature (18°C), and a greater proportion of seedlings were arrhythmic. These phenotypes were comparable to those seen for gene expression, indicating that leaf movement reflects important aspects of the axr6-3 circadian phenotype.
The pace of the clock was slowed by axr6-3, and this change in period was observed for three circadian promoter:LUC reporter constructs; however, the severity of the phenotype varied by reporter. The strongest gene expression phenotype was observed for TOC1:LUC. The period length for this reporter was approximately 3 h longer than for WT, which was two times greater than the change observed for CAB2:LUC and CCR2:LUC, which shared a 1.5 h slowing of the pace of the oscillator. In addition, the period length indicated by TOC1:LUC expression in the axr6-3 population was distributed over a greater range than in WT, and a higher proportion of the seedlings exhibited poor rhythms. This same trend was seen when TOC1:LUC expression was compared to that of the other two bioluminescence reporters. Known repressors of TOC1 expression are the two myb-like transcription factors, CCA1 and LHY (Alabadi et al., 2002; Matsushika et al., 2002). In axr6-3, CCA1/LHY transcript levels were significantly reduced, but this did not result in an increase in TOC1 expression. In fact, TOC1 mRNA levels in the mutant were essentially the same as in WT. Therefore, the marked effect of axr6-3 on the TOC1:LUC period cannot be explained by loss of LHY/CCA1-dependent control of TOC1 expression.
Given that TOC1 is a core clock component, it seems counter-intuitive that the strong effects observed for TOC1 expression were not directly translated into comparable changes in the rhythms exhibited by the bioluminescence reporters of the output genes CAB2 and CCR2. A potential explanation for this situation is that CUL1 has separable contributions to the control mechanisms responsible for sculpting the TOC1 expression profile. The TOC1 waveform that ultimately contributes to clock parameters such as period length is shaped by transcriptional regulation at the TOC1 promoter (Alabadi et al., 2001; Matsushika et al., 2002; Perales and Mas, 2007) and post-transcriptional control of TOC1 stability (Kim et al., 2007; Mas et al., 2003b). CUL1 appears to contribute to both these aspects of TOC1 regulation; the sensitivity of TOC1 expression to the axr6-3 allele suggests CUL1 activity may directly contribute to transcriptional control of TOC1 expression, and functional CUL1 is clearly important for TOC1 degradation. As TOC1 protein is the active species needed to set the pace of output rhythms, the TOC1 degradation phenotype caused by axr6-3 may mask the underlying transcriptional changes in TOC1 expression. We propose that the expression of CCR2 and CAB2 reflect the ultimate effect of axr6-3 on TOC1 accumulation, and that TOC1:LUC indicates the effects of diminished CUL1 activity on transcription of TOC1.
CUL1 activity appeared to be required to correctly set the circadian period and maintain robust rhythms under constant light conditions because axr6-3 seedlings exhibited arrhythmic or long-period circadian gene expression. The molecular basis for this phenotype was most consistent with the limited TOC1 degradation observed in axr6-3. In the mutant, TOC1 accumulated to high levels at restrictive temperatures, and the severity of this phenotype was temperature-dependent. TOC1 turnover is known to be mediated by the F-box protein ZTL, which suggests that the major effect of CUL1 inactivation on the clock was to impede SCFZTL activity. The RING protein RBX1 is a core component of SCF complexes, and is associated with SCFZTL in vivo (Han et al., 2004). Transient silencing of RBX1 by RNA interference (RNAi) in Arabidopsis seedlings lengthens the circadian period for CCA1 accumulation (Han et al., 2004). The RBX1 RNAi phenotype is analogous to the effect of axr6-3 observed here for gene expression rhythms; thus, inhibition of SCFZTL, either by inactivation of RBX1 or CUL1, produces a long-period phenotype, similar to that of a ztl mutant. Our findings are consistent with the hypothesis that CUL1 contributes to TOC1 degradation by acting as the cullin constituent of SCFZTL, in agreement with previous work demonstrating that ZTL is physically associated with CUL1 in vivo (Han et al., 2004).
Circadian rhythms in Arabidopsis are influenced by several phytohormones, including auxin (Covington and Harmer, 2007; Hanano et al., 2006). Exposure of seedlings to exogenous natural auxin indoleacetic acid (IAA) or the artificial auxins NAA, and 2,4-D lengthens the free-running period by an average of 1 h for bioluminescence rhythms exhibited by six promoter:LUC reporters constructs, including CCR2:LUC, CAB2:LUC, and TOC1:LUC (Covington and Harmer, 2007; Hanano et al., 2006). Also, Arabidopsis seedlings grown on medium containing 2,4-D exhibit a marked decrease in the amplitude of bioluminescence rhythms (Covington and Harmer, 2007), which notably includes the expression of CCA1 and TOC1. NAA has similar effects on the amplitude of expression of CCA1, CCR2, and CAB2, and this auxin also causes reduced clock precision (Hanano et al., 2006).
The pharmacological effects of auxin application on circadian rhythms are reminiscent of the clock phenotypes observed here for axr6-3. Given that axr6-3 is an auxin signaling mutant, the circadian phenotype in this background could be potentially explained by auxin signaling defects that feed back to influence the circadian oscillator. We do not favor this possibility because of the molecular phenotype exhibited by axr6-3. We found that acute treatment of WT seedlings with a concentration of 2,4-D known to slow the pace of the oscillator did not produce an inhibition of TOC1 degradation comparable to that observed in axr6-3 at either of the restrictive temperatures tested here. Furthermore, the fact that axr6-3 exhibits a marked phase delay for both gene expression and TOC1–YFP accumulation is inconsistent with an auxin signaling defect, as this phytohormone does not contribute to setting of the clock phase or promote phase shifting of circadian rhythms (Covington and Harmer, 2007). Although a contribution of auxin signaling to the axr6-3 phenotype remains a formal possibility, the data presented here are most consistent with a reduction in TOC1 degradation arising from a temperature-dependent inhibition of CUL1 that affects ZTL activity.
If the major circadian defect in axr6-3 stems from reduced turnover of TOC1, then is it possible that this is the only regulatory point where CUL1 participates in circadian signaling? We think that is unlikely. Three explanations can be advanced to account for the predominance of the ztl-like phenotype in axr6-3. First, sufficient CUL1 activity may exist in axr6-3 such that other clock-associated SCF complexes can still assemble and bind to their cognate targets, either because the targets for these SCF complexes are expressed at relatively low levels, or because the associated F-box protein is highly expressed. On the other hand, effective TOC1 turnover may require nearly full SCFZTL activity, because TOC1 and SCFZTL exist at stoichiometric levels in the cell. If this were the case, then TOC1 levels would be tightly coupled to the absolute amount of SCFZTL activity, and, as a result, TOC1 accumulation would respond proportionally to changes in activity of the E3 ligase. As circadian period is responsive to the balance between the activities of TOC1 and ZTL, this system would act as a molecular rheostat that finely controls clock period in response to changes in either ZTL activity or TOC1 expression.
Second, one or more of the alternative CUL proteins in Arabidopsis may substitute for CUL1 in other clock-associated SCF complexes. In support of this possibility, ethylene and GA responses are not dramatically affected in axr6-3, even though EBF1/2 and SLY1 associate with CUL1 in vitro (Potuschak et al., 2003; Xu et al., 2002). Possibly, the SCFEBF1/2 and SCFSLY1 complexes are capable of incorporating a cullin other than CUL1, and thereby maintain a proper response to these hormones in axr6-3. Finally, the phenotype produced by inhibition of TOC1 degradation may be sufficiently strong to mask any other phenotypic effects resulting from CUL1 inactivation.
In conclusion, our results demonstrate that CUL1 is required for TOC1 degradation, most likely by affecting SCFZTL activity, and therefore CUL1 functions as part of the system responsible for defining clock period and ensuring precise rhythms. These findings show the importance of selective protein turnover mediated by the ubiquitin/26S proteasome pathway in the molecular mechanism of the plant circadian clock.
Experimental procedures
Plant materials and growth conditions
All plants were in the Columbia background. ztl-4 corresponds to the T-DNA line SALK_35701 (Alonso et al., 2003); the insert is present at base pair 2216, which interrupts the portion of the gene encoding the second kelch repeat (Figure S3a). ZTL transcript expression was diminished fourfold in this background (Figure S3b). The period for CAB2:LUC expression in ztl-4 was 27.45 ± 0.38 h compared with 24.19 ± 0.45 h in WT (Figure S3c). Surface-sterilized seeds were plated on 1× MS medium (Murashige and Skoog, 1962; Caisson Laboratories, http://www.caissonlabs.com) with 0.8% agar and 3% sucrose, stratified in the dark at 4°C for 3–5 days, and then transferred to the indicated conditions. Light was provided by cool white fluorescent bulbs, at an intensity of approximately 50 µmol m−1 sec−1.
Leaf movement
Seedlings for experiments determining leaf movement were grown under 12 h light:12 h dark cycles for 5 days at the indicated temperature. On day 6, seedlings were transferred to continuous white light (approximately 50 µmol m−2 sec−1) for 5 days. Video-recorded images of seedlings under constant light were acquired every 20 min. Curve-fit analysis was used to determine estimated period lengths from the cotyledon position information (Plautz et al., 1997). The data for each seedling were analyzed by the Fast Fourier Transform/Nonlinear Least Squares method (FFT/NLLS), which fits the data to a modified cosine curve, to obtain estimated period length and relative amplitude error. Relative amplitude error is a statistical measure of how well the data conform to an ideal cosine curve, where a perfect match corresponds to a value of 0 and no correlation is indicated by a value of 1 (Millar et al., 1995a). A cut-off of ≤0.25 for the relative amplitude error was used to identify robust rhythms in this data.
Bioluminescence assay
axr6-3 was introduced into CCR2:LUC (Alabadi etal., 2001), CA-B2:LUC (Millar etal., 1992), and TOC1:LUC (Alabadi et al., 2001) reporter lines by crossing. Seedlings harboring the reporter construct were grown for 6 days under 12 h light:12 h dark cycles at the indicated temperature. On the morning of the seventh day, seedlings were transferred to continuous light conditions. Imaging of bioluminescence rhythms was performed as described previously (Millar etal., 1992;Somers etal., 1998). Estimated period length was determined by FFT/NLLS analysis. A cut-off of ≤0.6 for the relative amplitude error was used to identify robust rhythms in this data.
Whole-cell extracts and Western blots
Whole 10–14-day-old seedlings grown under the indicated conditions were harvested at the indicated times and flash frozen in liquid nitrogen. For experiments at 28°C, seedlings were transferred to 28°C at ZT0 (08:00 h) on the day of sample collection. Auxin treatments were applied atZT0 and consisted of spraying approximately 100 seedlings on a 90 cm2 square Petri plate with 1.5 ml of 0.3 µm 2,4-D (Sigma, http://www.sigmaaldrich.com/) diluted in 0.1% Triton X-100. The 2,4-D solution used was prepared from a 20 µm stock in ethanol. Tissue was disrupted by grinding in a mortar and pestle cooled with liquid nitrogen. Whole-cell extracts were prepared by vortexing ground tissue with an equal volume of extraction buffer (50 mm HEPES/NaOH, pH 7.5, 100 mm NaCl, 10% glycerol, 5 mm EDTA, 5 mm DTT, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 5 mm benzamidine, EDTA-free protease inhibitor (Roche, http://www.roche.com, 1 mm PMSF (phenylmethanesulfonyl fluoride), 50 µm MG132 (Peptides International, http://www.pepnet.com) and 1% polyvinylpolypyrrolidone). The lysate was cleared by centrifugation at 17 000 g for 30 min at 4°C. Total protein concentration was determined using the reducing agent-compatible BCA protein assay (Pierce, http://www.piercenet.com). Equal amounts of total protein (40–80 µg) were separated on 6% SDS–PAGE gels as described previously (Sambrook and Russell, 2001). Proteins were transferred to a 0.2-nm nitrocellulose membrane, and the TOC1–YFP fusion was detected using rabbit anti-GFP horseradish peroxidase (HRP) conjugate (Abcam, http://www.abcam.com). HRP activity was determined using SuperSignal West Pico chemiluminescent substrate (Pierce) and BioMax film (Kodak, http://www.kodak.com). TOC1–YFP levels were determined as the ratio of the integrated density of the TOC1–YFP band to that of a nonspecific band serving as the loading control (indicated by an asterisk in Figure 4). The integrated density of the protein bands was measured using imagej software (Abramoff et al., 2004) from scanned tiff images of the films from each blot.
Expression analysis by real-time PCR
Seedlings were grown, harvested and ground as described above for immunoblot analyses. Total RNA was isolated using the Plant RNeasy Mini Kit (Qiagen, http://www1.quagen.com). First-strand cDNA was generated from 1 µg of total RNA using the SuperScript II first-strand synthesis for RT-PCR kit (Invitrogen, http://www.invitrogen.com). This cDNA was diluted 1:4 to produce a working stock, and 2 µl of the working stock was used as template for PCR amplification using a iCycler iQ Real-Time PCR Detection System (Bio-Rad, http://www.bio-rad.com), to determine the expression of TOC1, LHY, and CCA1 using the conditions, primers and probes described previously (Hazen et al., 2005). Expression of IPP2 (isopentenyl pyrophosphate:dimethylallyl pyrophosphate isomerase, At3g02780) was used as a normalization control, and its expression was determined using the primers and probes described previously (Hazen et al., 2005). The expression level for each gene was calculated using the formula 2[CT(IPP2)–CT(experimental)], where CT is the average threshold cycle for three technical replicates. ZTL expression was determined using 5′-TCAGCGTCTCAGCTTTATCTAC-3′ as the forward primer and 5′-GAGGCGGTCTTCCTGGAATG-3′ as the reverse primer. For this primer set and the corresponding IPP2 controls, PCR conditions were 95°C for 1 min, followed by 40 cycles of 10 sec at 95°C and 30 sec at 60°C in a buffer consisting of 1 × Ex Taq buffer (Takara Bio USA, http://www.takarabiousa.com), 1 × EvaGreen (Biotium, http://www.biotium.com), 0.1% v/v Tween-20, 5% v/v DMSO, 50 µg ml−1 BSA (New England Biolabs, http://www.neb.com), 0.2 mM dTNPs, 0.3 µm primers and 0.5 U Taq polymerase (New England Biolabs).
Supplementary Material
Acknowledgements
We thank Steve A. Kay (University of California, San Diego) for early support of this work provided by NIH grants GM56006 and GM67837. We also thank Sheila McCormick and Sarah Hake for critical reading of the manuscript. This work was supported by USDA grant CRIS 5335-21000-025-00D (to F.G.H.) and NIH grant GM067203 (to W.M.G.). As a postdoctoral fellow, F.G.H was a DOE-Energy Biosciences fellow of the Life Sciences Research Foundation. T.I. is supported by NIH grant GM079712 and W.M.G. is supported by NIH grant GM067203.
Footnotes
Supporting Information
Additional supporting information may be found in the online version of this article.
Figure S1. Expression of CAB2 and CCR2 under light:dark cycles.
Figure S2. TOC1 expression in axr6-3 and WT TMG lines.
Figure S3. Characterization of ztl-4.
Please note: Blackwell publishing are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics Int. 2004;11:36–42. [Google Scholar]
- Alabadi D, Oyama T, Yanovsky MJ, Harmon FG, Mas P, Kay SA. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- Alabadi D, Yanovsky MJ, Mas P, Harmer SL, Kay SA. Critical role for CCA1 and LHY in maintaining circadian rhythmicity in Arabidopsis. Curr. Biol. 2002;12:757–761. doi: 10.1016/s0960-9822(02)00815-1. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Bachmair A, Novatchkova M, Potuschak T, Eisenhaber F. Ubiquitylation in plants: a post-genomic look at a post-translational modification. Trends Plant Sci. 2001;6:463–470. doi: 10.1016/s1360-1385(01)02080-5. [DOI] [PubMed] [Google Scholar]
- Blasing OE, Gibon Y, Gunther M, Hohne M, Morcuende R, Osuna D, Thimm O, Usadel B, Scheible WR, Stitt M. Sugars and circadian regulation make major contributions to the global regulation of diurnal gene expression in Arabidopsis. Plant Cell. 2005;17:3257–3281. doi: 10.1105/tpc.105.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Harmer SL. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol. 2007;5:e222. doi: 10.1371/journal.pbio.0050222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kevei E, Toth R, Nagy F, Hibberd JM, Millar AJ, Webb AA. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Durfee T, Roe JL, Sessions RA, Inouye C, Serikawa K, Feldmann KA, Weigel D, Zambryski PC. The F-box-containing protein UFO and AGAMOUS participate in antagonistic pathways governing early petal development in Arabidopsis. Proc. Natl Acad. Sci. USA. 2003;100:8571–8576. doi: 10.1073/pnas.1033043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann W, Johnsson A. Rhythms in organ movement. In: Lumsden PJ, Millar AJ, editors. Biological Rhythms and Photoperiodism in Plants. Oxford: Bios Scientific Publishers; 1998. pp. 35–50. [Google Scholar]
- Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137:961–968. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne JM, Downes BP, Shiu SH, Durski AM, Vierstra RD. The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl Acad. Sci. USA. 2002;99:11519–11524. doi: 10.1073/pnas.162339999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Hellmann H, Dharmasiri S, Estelle M. Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell. 2002;14:2137–2144. doi: 10.1105/tpc.003178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tobin EM. Loss of the circadian clock-associated protein I in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl Acad. Sci. USA. 1999;96:4176–4179. doi: 10.1073/pnas.96.7.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Tingay S, Wang ZY, Tobin EM. Circadian rhythms confer a higher level of fitness to Arabidopsis plants. Plant Physiol. 2002;129:576–584. doi: 10.1104/pp.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- Han L, Mason M, Risseeuw EP, Crosby WL, Somers DE. Formation of an SCF(ZTL) complex is required for proper regulation of circadian timing. Plant J. 2004;40:291–301. doi: 10.1111/j.1365-313X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- Hanano S, Domagalska MA, Nagy F, Davis SJ. Multiple phytohormones influence distinct parameters of the plant circadian clock. Genes Cells. 2006;11:1381–1392. doi: 10.1111/j.1365-2443.2006.01026.x. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Covington MF, Blasing OE, Stitt M. Circadian regulation of global gene expression and metabolism. In: Hall A, McWatters HG, editors. Endogenous Plant Rhythms. Oxford: Blackwell Publishing Ltd; 2005. pp. 133–166. [Google Scholar]
- Harmon FG, Imaizumi T, Kay SA. The plant circadian clock: review of a clockwork Arabidopsis. In: Hall AJW, McWatters HG, editors. Endogenous Plant Rhythms. Oxford: Black-well Publishing Ltd; 2005. pp. 1–23. [Google Scholar]
- Hazen SP, Schultz TF, Pruneda-Paz JL, Borevitz JO, Ecker JR, Kay SA. LUX ARRHYTHMO encodes a Myb domain protein essential for circadian rhythms. Proc. Natl Acad. Sci. USA. 2005;102:10387–10392. doi: 10.1073/pnas.0503029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H, Hobbie L, Chapman A, Dharmasiri S, Dharmasiri N, del Pozo C, Reinhardt D, Estelle M. Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J. 2003;22:3314–3325. doi: 10.1093/emboj/cdg335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AA. Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ. 2007;30:333–349. doi: 10.1111/j.1365-3040.2006.01627.x. [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 2006;11:550–558. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR. An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature. 2001;410:487–490. doi: 10.1038/35068589. [DOI] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE. ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature. 2007;449:356–360. doi: 10.1038/nature06132. [DOI] [PubMed] [Google Scholar]
- Kreps JA, Kay SA. Coordination of plant metabolism and development by the circadian clock. Plant Cell. 1997;9:1235–1244. doi: 10.1105/tpc.9.7.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Wu Y, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol. 2002;43:58–69. doi: 10.1093/pcp/pcf005. [DOI] [PubMed] [Google Scholar]
- Mas P, Alabadi D, Yanovsky MJ, Oyama T, Kay SA. Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell. 2003a;15:223–236. doi: 10.1105/tpc.006734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003b;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Yamashino T, Mizuno T. The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: II. Characterization with CCA1-overexpressing plants. Plant Cell Physiol. 2002;43:118–122. doi: 10.1093/pcp/pcf006. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua NH, Kay SA. A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell. 1992;4:1075–1087. doi: 10.1105/tpc.4.9.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AJ, Carre IA, Strayer CA, Chua NH, Kay SA. Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science. 1995a;267:1161–1163. doi: 10.1126/science.7855595. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua NH, Kay SA. The regulation of circadian period by phototransduction pathways in Arabidopsis. Science. 1995b;267:1163–1166. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–497. [Google Scholar]
- Perales M, Mas P. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. Plant Cell. 2007;19:2111–2123. doi: 10.1105/tpc.107.050807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA. Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms. 1997;12:204–217. doi: 10.1177/074873049701200302. [DOI] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- Quint M, Ito H, Zhang W, Gray WM. Characterization of a novel temperature-sensitive allele of the CUL1/AXR6 subunit of SCF ubiquitin-ligases. Plant J. 2005;43:371–383. doi: 10.1111/j.1365-313X.2005.02449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risseeuw EP, Daskalchuk TE, Banks TW, Liu E, Cotelesage J, Hellmann H, Estelle M, Somers DE, Crosby WL. Protein interaction analysis of SCF ubiquitin E3 ligase subunits from Arabidopsis. Plant J. 2003;34:753–767. doi: 10.1046/j.1365-313x.2003.01768.x. [DOI] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature. 2003;426:680–683. doi: 10.1038/nature02174. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schaffer R, Ramsay N, Samach A, Corden S, Putterill J, Carre IA, Coupland G. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photo-periodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/s0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- Shen WH, Parmentier Y, Hellmann H, Lechner E, Dong A, Masson J, Granier F, Lepiniec L, Estelle M, Genschik P. Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol. Biol. Cell. 2002;13:1916–1928. doi: 10.1091/mbc.E02-02-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- Somers DE, Webb AA, Pearson M, Kay SA. The short-period mutant, toc1-1, alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development. 1998;125:485–494. doi: 10.1242/dev.125.3.485. [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R. The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell. 2004;16:769–782. doi: 10.1105/tpc.016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. doi: 10.1126/science.289.5480.768. [DOI] [PubMed] [Google Scholar]
- Vierstra RD. The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci. 2003;8:135–142. doi: 10.1016/S1360-1385(03)00014-1. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/s0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell. 2001;13:1779–1790. doi: 10.1105/TPC.010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, et al. Structure of the Cul1-Rbx1-Skp1-F box(Skp2)SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.