Abstract
Activation of the bHLH factor Math5 (Atoh7) is an initiating event for mammalian retinal neurogenesis, as it is critically required for retinal ganglion cell formation. However, the cis-regulatory elements and trans-acting factors that control Math5 expression are largely unknown. Using a combination of transgenic mice and bioinformatics, we identified a phylogenetically conserved regulatory element that is required to activate Math5 transcription during early retinal neurogenesis. This element drives retinal expression in vivo, in a cross-species transgenic assay. Previously, Pax6 was shown to be necessary for the initiation of Math5 mRNA expression. We extend this finding by showing that the Math5 retinal enhancer also requires Pax6 for its activation, via Pax6 binding to a highly conserved binding site. In addition, our data reveal that other retinal factors are required for accurate regulation of Math5 by Pax6.
Keywords: retina, neurogenesis, Math5, Pax6, transcriptional regulation
Introduction
Vertebrate eye development involves outgrowth, morphogenesis, specification and differentiation of multiple tissues such as the cornea, lens, iris, ciliary body, neuroretina and retinal pigmented epithelium. All of these tissues require the paired- and homeo-domain containing transcription factor Pax6 for their formation (reviewed in Hanson, 2003; Kozmik, 2005; Treisman, 2004). Both loss-of-function (Fujiwara et al., 1994; Glaser et al., 1992; Hill et al., 1991; Quiring et al., 1994) and gain-of-function studies (Chow et al., 1999; Halder et al., 1995) have demonstrated a critical role for Pax6 in metazoan eye formation. However, despite a wealth of studies on this transcription factor, the molecular mechanisms by which Pax6 controls the transcription of its downstream targets remain incompletely deciphered.
The vertebrate retina is an excellent system to investigate how multipotent cells give rise to neurons and glia, because it is comprised of only seven basic neuronal and glial cell types arranged in three cell layers (Pei and Rhodin, 1970). All retinal cells are derived from a common pool of retinal progenitor cells (RPCs) (Holt et al., 1988; Turner and Cepko, 1987; Turner et al., 1990; Wetts and Fraser, 1988). These RPCs express a cadre of transcription factors, including Pax6, Chx10, Rx/Rax, Six3, and Hes1 (reviewed in Levine and Green, 2004; Livesey and Cepko, 2001; Marquardt, 2003). RPCs give rise to specific cell types across time with retinal ganglion cells (RGCs) differentiating first, and Müller glia last (reviewed in Cayouette et al., 2006; Livesey and Cepko, 2001). The bHLH transcription factors Math5, Ngn2, Math3, NeuroD1, and Mash1 regulate neuronal specification, and they are activated in sequential order during mouse retinogenesis (reviewed in Vetter and Brown, 2001). Of these, Math5 (Atoh7) appears first and regulates RGC formation (Brown et al., 1998). Pax6 is genetically required for the transcriptional activation of Math5, Ngn2 and Mash1, and directly binds to Ngn2 regulatory DNA (Brown et al., 1998; Marquardt et al., 2001). But, Pax6 broadly activates these genes, meaning that other factors must regulate their precise spatiotemporal patterning. Importantly, Pax6 regulation of bHLH retinal factors appears highly conserved as in the Drosophila eye both eyeless (a Pax6 orthologue) and sine oculus (another retinal determination transcription factor) directly activate atonal expression (Zhang et al., 2006).
At the beginning of mouse retinal neurogenesis, one RPC population initiates Math5 expression as it becomes postmitotic (Le et al., 2006; Poggi et al., 2005). The Math5 retinal lineage contains all seven cell types (Brzezinski, 2005; Yang et al., 2003), but early this lineage predominantly produces RGCs (Brzezinski, 2005; Le et al., 2006). At later stages, nearby RPCs and differentiated neurons in non-Math5 lineages presumably direct Math5-expressing cells towards other retinal fates. Because RGCs differentiate first, their loss has a large effect on subsequent retinal organization and differentiation (Brown et al., 2001; Mu et al., 2005; Wang et al., 2001). Thus a key, initiating event in the mammalian eye is Math5 activation, but the regulation of this process has not been well characterized.
Here we show that Math5 is a direct transcriptional target of Pax6. Like Math5 mRNA, retinal expression in Math5-GFP transgenic reporter mice is sensitive to Pax6 gene dosage. We also demonstrate that Pax6 is required for Math5 expression beyond its initial activation at E11.5 and that this regulatory relationship is cell autonomous. Our in vivo transgenic analyses define a Math5 339bp distal regulatory element that drives retinal expression, wherein Pax6 specifically binds to one highly conserved binding site.
Methods
Transgenic Mice
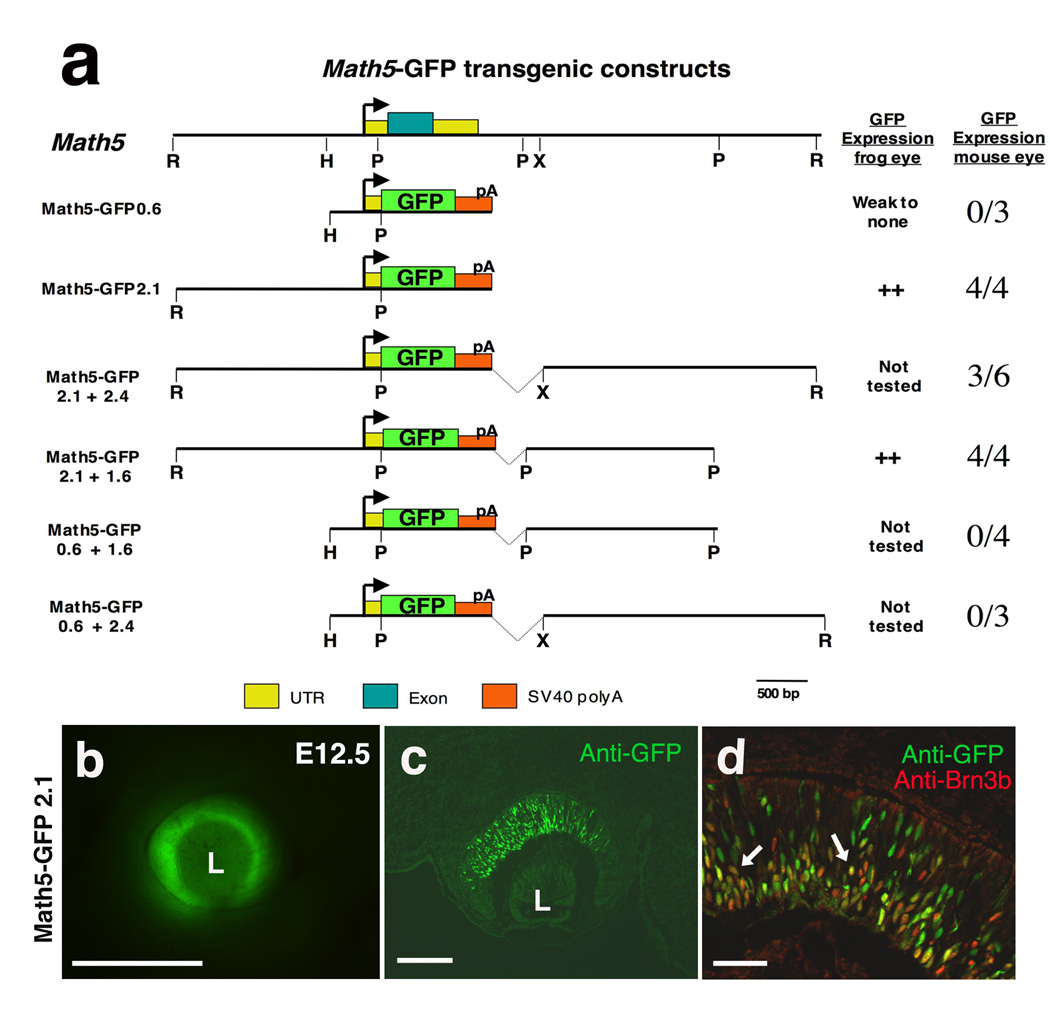
Six pG1-Math5-GFP reporters, with different combinations of 5’ and 3’ noncoding DNA plus the Math5 promoter, were generated (Figure 1). Reporter cassettes were released by SalI-NotI digests to generate transgenic mice via pronuclear injection at the CHRF Transgenic Core Facility. The Math5-GFP 2.1 transgene was previously reported as 2.3 KB in length (Hutcheson et al., 2005), but DNA sequencing indicates it is 2.1 KB. All transgenic lines were generated and maintained on a CD-1 background. The day vaginal plugs were observed was designated as E0.5. PCR genotyping for the GFP coding exon detected the presence of each transgene. Live GFP fluorescence was imaged using a Leica MZ12 dissecting microscope, and a Magnafire camera and image software. Xath5-GFP3.3 transgenic mice have been described (Hutcheson et al., 2005).
Figure 1. Retinal expression of Math5 transgenes.
A) Diagram of the Math5 locus, including the coding exon (blue box), and various transgenes containing different 5’ and 3’ Math5 noncoding fragments, driving GFP reporter expression. GFP expression was tested in the developing mouse or frog eye. An arrow denotes the Math5 TATA box. The right column shows number of independent mouse transgenic lines with GFP expression versus the number tested (n ≥ 3 litters scored per line). B) GFP expression in the optic cup of a living E12.5 Math5-GFP2.1 mouse embryo. C) Anti-GFP labeling of E12.5 retinal cryosection from same transgenic line as in B. D) Higher magnification images of anti-GFP (green) and anti-Brn3b (red) double labeling. Arrows point to coexpressing cells. Dorsal and scleral are up, rostral left in B–D; L = lens. Bar = 500 microns in B, 25 microns in C; 50 microns in D.
Pax6 mutant mice
Math5-GFP2.1, Math5-GFP2.1+2.4, Math5-GFP2.1+1.6 and Xath5-GFP3.3 transgenes were crossed into the Pax6Sey/+ background (Brown et al., 1998). Math5-GFP; Pax6Sey/+ or Xath5-GFP3.3; Pax6Sey/+ mice were then intercrossed to generate litters for analysis in Pax6 wild type, heterozygous and homozygous mutant embryos. Mice containing the Math5LacZ/+ allele (Brown et al., 2001) were crossed with Pax6 α-Cre; Pax6 CKO/CKO mice (maintained in a CD-1 background) to ascertain the retinal phenotypes of Math5LacZ/+; α-Cre; Pax6CKO/+ and Math5LacZ/+; α-Cre; Pax6CKO/CKO embryos. PCR genotyping assays have been described (Brown et al., 1998; Brown et al., 2001; Marquardt et al., 2001).
Transgenic frog embryos
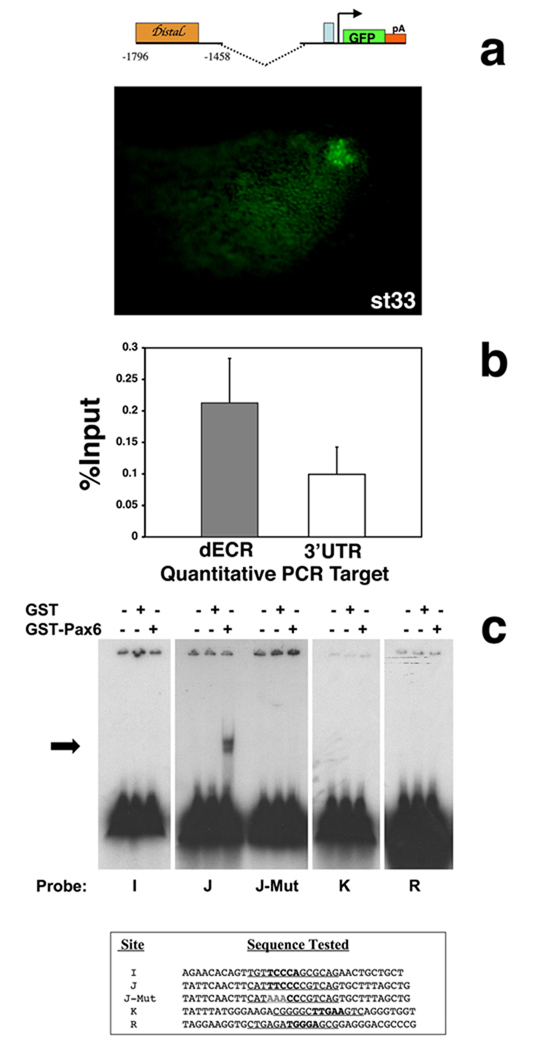
The generation of Xenopus transient transgenic embryos, containing Math5-GFP transgenes was reported (Hutcheson et al., 2005; Hutcheson and Vetter, 2002). pG1-M5-0.2dECR was made by separately PCR amplifying Math5 DNA fragments –1796 to −1458 and –503 to −339 and subcloning into the pG1 transgenic vector, verified by DNA sequencing. GFP fluorescence was scored in stage 33 embryos by live fluorescence and whole embryo anti-GFP labeling.
Immunohistochemistry and in situ hybridization
Mouse embryos were dissected in cold PBS, fixed one hour in cold 4% paraformaldehyde/PBS, washed into 15% sucrose/PBS and cryoembedded in OCT. Sections of embryonic or adult retinal tissue were antibody labeled as described (Le et al., 2006; Mastick and Andrews, 2001). Antibodies used were rabbit anti-GFP-Alexa488 (1:1000, Molecular Probes), goat anti-Brn3b (1:100, Santa Cruz), rat anti-βgal (1:1000, a gift from Tom Glaser), rabbit anti-Pax6 paired domain (1:5000, a gift from Simon Saulé) and rabbit anti-Pax6 C-terminus (1:1000, Covance). Directly conjugated or biotinylated secondary and streptavidin-conjugated tertiary fluorescent antibodies were obtained from Jackson Immunoresearch Laboratories or Molecular Probes. Whole mount in situ hybridization with alkaline phosphatase color development was used to visualize Math5 mRNA, followed by cryosectioning and Pax6 immunohistochemistry, using the Covance antibody and a streptavidin HRP tertiary antibody and DAB chromagen development (Brown et al., 1998). Imaging was performed on a Zeiss Axioplan microscope equipped with a Zeiss camera, Apotome deconvolution and Axiovision software or a Zeiss LS M510 confocal microscope and software.
Bioinformatics
Nucleotides −3000 to −1, directly upstream of the start codon, were compared for Math5 (Accession #AF418923), HATH5 (Accession #AF418922) and Xath5 (Accession #102561). A three species noncoding alignment was performed using Mulan (http://mulan.dcode.org/) (Loots and Ovcharenko, 2005; Ovcharenko et al., 2005), with a window of 25 bases and 70% identity. Pair-wise alignments among Math5, HATH5 and Xath5 3 Kb upstream DNA were generated with NCBI Blast 2 Sequences program (http://www.ncbi.nlm.nih.gov). A 339bp distal evolutionarily conserved sequence (ECR) was found in the 5’ genomic DNA of five species: mouse Math5 (Accession #AF418923), human HATH5 (Accession #AF418922), frog Xath5 (Accession #1025061), chick Cath5 (Accession #NW_001471715 Chr6, contig 30.299) and zebrafish Zath5 (Accession #AL627094). A Clustal W (v1.4) multiple sequence alignment of the distal ECR was executed using MacVector (v7.9) default parameters.
Potential Pax6 paired domain binding sites were identified using the Transfac MATCH program, version 10.3, (http://www.biobase-international.com) with matrices M00979 (V$PAX6_Q2)(Duncan et al., 1998; Duncan et al., 1996; Roth et al., 1991; Sander et al., 1997; Zhou et al., 2000) and M00097 (V$PAX6_01)(Epstein et al., 1994a). Nucleotides −3000 to −1 in Math5, HATH5, Xath5, Cath5 and Zath5 genes were tested, using 0.75 core similarity and 0.70 matrix similarity search parameter cutoffs. Twenty predicted binding sites within Math5 5’ regulatory DNA are listed in Supplementary Table 1.
EMSA
GST and GST-Pax6 paired domain proteins (Epstein et al., 1994a), were purified from BL21 bacterial lysates with glutathione agarose beads (Sigma) for 1 hour at 4°C, washed in PBS, eluted with 25 mM glutathione/0.1M Tris pH 8 and dialyzed into 10 mM Tris pH 7.5, 50 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 4% glycerol. Gel-shift reactions used a 5X binding buffer (50 mM Tris pH 7.5, 250 mM NaCl, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 0.25 mg/ml poly-dI-dC, and 20% glycerol). 20 µl reactions contained 4 µl of 5X binding buffer, 100 ng of recombinant protein and 75 fmol of γ32P end-labeled, annealed, double-stranded oligonucleotides (400,000 Cerenkov counts per reaction). After DNA probe addition, reactions were incubated for 20 minutes at room temperature, run on a 4% polyacrylamide gel in 0.5X Tris borate-EDTA buffer and dried gels exposed to x-ray film.
ChIP and Real-time PCR
ChIP was performed as described (Chen et al., 2004; Wells and Farnham, 2002) with minor modifications. Each ChIP assay used 100µg of fragmented chromatin (0.1–1 KB) from Ad12 HER10 human retinoblast cells (Grabham et al., 1988; Tucker et al., 2001), incubated with 1µg of either rabbit IgG (Jackson Immunoresearch) or anti-Pax6 antibody (Covance), along with mock (no antibody, no chromatin) and input (no IP) controls. Purified immunoprecipitated DNA was resuspended in 30 µl of 0.01M Tris/0.005M EDTA pH 8 and analyzed by real-time PCR, using a Biorad I Cycler, iQ SYBR Green PCR mixes (Biorad) and HATH5 dECR primers (227bp, FOR 5’ 5' CTGCTGTTCCCAACCAAGACTG 3’; REV 5' TAACCCCATTGTGACCGCCCTGAC 3') or HATH5 UTR negative control primers (159bp FOR 5' TTCGCATCATCAGACCTATGGACG 3'; REV 5' TGTTTTCCCTCAAAGTAGCCCAG 3'). A serial dilution series of 1% input chromatin generated a standard curve for calculating the percent input for each ChIP sample. Data plotted are the mean of eight independent assays, each performed in PCR duplicate, minus preimmune sera values.
Luciferase Assay
Various Math5 5’ noncoding DNA fragments, containing the Math5 TATA box (M5-0.2; −503 to −339) were PCR amplified, subcloned into pGL2 (Promega) and verified by DNA sequencing. Constructs with point mutations were made by site-directed mutagenesis using PCR amplification (Weiner et al., 1995). Pax6 site J was mutated from catTTCcccgtcag to catAAAcccgtcag and all constructs verified by DNA sequencing. A pCS2-Pax6 expression plasmid was created by BamHI-EcoRI double digestion and subcloning of the 1.6 Kb human Pax6 cDNA (Epstein et al., 1994b) into pCS2+ (Turner and Weintraub, 1994). HEK-293T human kidney epithelial cells or Ad12 HER10 cells were transfected using 10 µg of TransIT-LT1 (Mirus) and the manufacturer’s protocol. In these experiments 2 × 106 cells were plated into each well of a 6-well culture dish 48 hours prior to transfection. At 50% confluence, each well received an equivalent amount of pCS2+ expression plasmid (1 µg pCS2+; 25 ng pCS2-Pax6 plus 975 ng pCS2+; or 1 µg of pCS2-Pax6), 500 ng of each pGL2-Math5 construct and 250 ng of internal control pTK-Renilla plasmid (Promega). Cell extracts were harvested after 48 hours and assayed using the Dual-Luciferase Reporter Assay system (Promega) and a Veritas luminometer (Model #9100, Turner Biosystems, Inc). Firefly luciferase activity was normalized relative to Renilla luciferase activity. Three independent transfections were done in triplicate, and the Instat Statistics program (Graphpad Software, Inc, version 3.0b) used to perform ANOVA and a Bonferroni posthoc test to determine p values.
Results
Math5 5’ regulatory sequences control retinal expression
As vertebrate Ath5 is a critical regulator of retinal neurogenesis, it is essential to understand how this gene’s expression is regulated during eye development. For both the Xenopus Ath5 (Xath5) and chick Ath5 (Cath5) genes, a proximal activation element that contains conserved bHLH binding sites (E boxes) and a TATA box have been characterized (Hutcheson et al., 2005; Matter-Sadzinski et al., 2005; Skowronska-Krawczyk et al., 2004). An analogous mouse Ath5 (Math5/Atoh7) proximal transgene (M5-GFP0.6, Figure 1A) displayed weak GFP expression in the embryonic retina and retinal ciliary marginal zone of Xenopus embryos (Hutcheson et al., 2005). However, such expression only occurs in the context of the frog eye, as M5-GFP0.6 transgenic mice do not express GFP mRNA or protein in the E11.5–E15.5 embryonic retina (Figure 1A, n = 0/3 transgenic lines, n≥ 5 litters per line)(Hutcheson et al., 2005). We conclude that the M5-GFP0.6 transgene, containing the TATA-box, is insufficient for the activation of Math5 expression in the mouse eye.
To define the cis-regulatory sequences controlling Math5 expression more precisely, we created a series of Math5-GFP transgenes (Figure 1A). Previously, the upstream 2.1 Kb of Math5 regulatory DNA was shown to drive retinal GFP expression in both mouse and frog embryos (Hutcheson et al., 2005). However, only live GFP fluorescence was scored grossly in those E13.5 mouse embryos. Although 5’ regulatory DNA appears sufficient for retinal expression (Hutcheson et al., 2005), it was important to also test 3’ regulatory DNA, since both Drosophila atonal and mouse Math1 (a semi-orthologue of Math5) contain 3’ activation enhancers (Helms et al., 2000; Sun et al., 1998; Zhang et al., 2006). In particular, atonal has multiple retinal enhancers: one 3’ initiation enhancer and other 5’ and 3’ elements that control sequential refinement to the R8 photoreceptor cell (Sun et al., 1998; Zhang et al., 2006). By contrast, multiple Math5 transgenic lines with overlapping segments of 3’ regulatory DNA, plus 2.1 Kb of upstream DNA (M5-GFP2.1+2.4 and M5-GFP2.1+1.6), have identical reporter expression to M5-GFP2.1 (Figure 1A, n≥ 3 litters per transgene). Math5 5’ distal sequences are required for retinal expression, since transgenes lacking nucleotides −2100 to −600 exhibit no GFP mRNA or protein expression (Figure 1A, M5-GFP0.6+2.4, M5-GFP0.6+1.6; n ≥ 3 litters per transgenic line). This requirement for more distal Math5 regulatory sequences to achieve retinal expression is consistent with the expression of a zebrafish Ath5 transgene containing approximately 3.8 Kb of upstream regulatory DNA (Masai et al., 2005; Poggi et al., 2005).
Here, we further assayed transgenic embryos for live GFP fluorescence, GFP mRNA and protein expression from E11 to P0 (Figures 1B–D and data not shown). In M5-GFP2.1 (Figures 1B–D), M5-GFP2.1+2.4 and M5-GFP2.1+1.6 (not shown) embryos, GFP expression initiates at E11 and continues to at least birth. Reporter expression patterns are identical among constructs, and all reflect Math5 mRNA expression (compare Figure 1B,C to Figure 2F,I of Brown et al, 1998). An expression pattern comparison of M5-GFP2.1 and Math5LacZ has been reported (Hufnagel et al., 2007). In the E11.5 and E12.5 optic cup, all βgal+ cells are GFP+, with only a few cells expressing GFP alone.
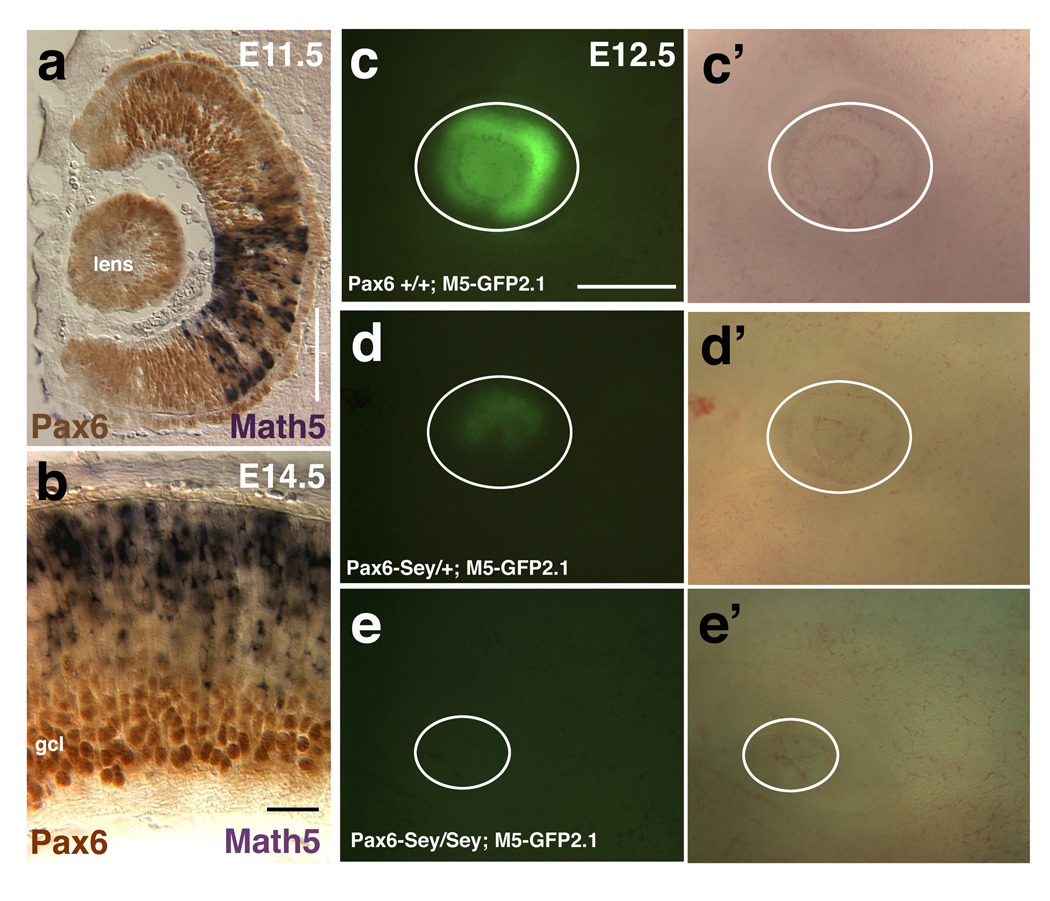
Figure 2. Math5 transgenes are sensitive to Pax6 gene dosage.
A) Math5 mRNA (purple) and Pax6 protein (brown) are completely coexpressed at E11.5. B) After the ganglion cell layer (gcl) forms, differentiated RGCs continue to express Pax6 strongly, but shut off Math5 mRNA. C–E) Math5-GFP2.1 expression in wild type (C,C’), Pax6Sey/+ (D,D’) and Pax6Sey/Sey (E,E’) E12.5 embryonic eyes. Panels C–E show GFP fluorescence and C’–E’ bright field images living embryonic eyes (white circles indicate eyes). In both E11.5 and E12.5 embryos, the GFP expression domain is smaller in Pax6Sey/+ eyes and completely lost in Pax6Sey/Sey eyes. Rostral is up in A, scleral is up in B, Dorsal is up in C–E. Bar in A, B = 100 microns; C–E = 500 microns.
We next compared GFP transgenic reporter expression to that of Brn3b, which is expressed in nascent RGCs and has been shown to be a downstream target of vertebrate Ath5 genes (Hutcheson and Vetter, 2001; Liu et al., 2001; Wang et al., 2001). Double antibody labeling for GFP and Brn3b shows Math5-GFP+ cells in the outer optic cup, positioned as post-mitotic, transitional cells (Dyer and Bremner, 2005; Le et al., 2006). As Math5-GFP cells migrate to the ganglion cell layer (GCL), they coexpress Brn3b (arrows in Figure 1D). At E12.5, Math5-GFP retinal cells were quantified with those expressing Brn3b. For the M5-GFP2.1 transgene there was an average (mean ± std error) of 277±0.5 GFP+ cells per section with 90% coexpressing Brn3b (n = 17 sections from 6 embryos representing 2 independent transgenic lines); and for the GFP2.1+1.6 transgene an average of 185±0.6 GFP+ cells per section, of which 82% coexpress Brn3b (n = 12 sections from 7 embryos representing 3 independent transgenic lines). Overall, we conclude that Math5-GFP transgenes are reliable reporters for the activation and expression of Math5 in the retina. Our data point to one set of regulatory sequences controlling Math5 activation in the optic cup situated between −2100 and −600.
Pax6 regulation of Math5 in vivo
Math5 mRNA expression is dosage-sensitive to the loss of Pax6. The number of E11.5 Math5+ RPCs is reduced in Pax6+/− eyes and completely absent in Pax6−/− eyes (Figures 3A–3F in Brown et al., 1998). However, it is unknown whether Pax6 directly regulates Math5 transcription. To begin to address this question, we compared the expression of Math5 and Pax6 during the initial period of retinal neurogenesis. In the E11.5 optic cup, those RPCs expressing Math5 mRNA (purple cells) completely coexpress Pax6 protein (brown nuclei) (Figure 2A). However, at E13.5 when the ganglion cell layer (GCL) laminates, Pax6 protein is downregulated in RPCs, but is maintained at high levels in differentiated GCL neurons (Figures 2B, 3H). Interestingly, from E13.5 onwards, cells expressing Math5 mRNA coexpress low Pax6 (Figures 2B, 3H and data not shown).
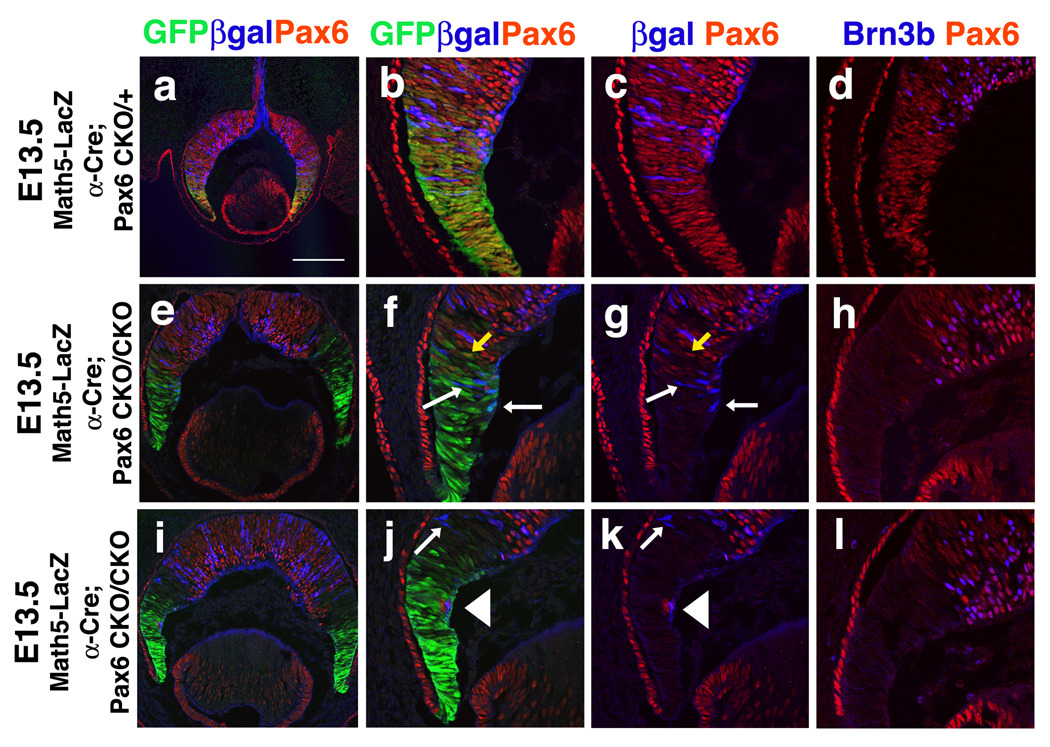
Figure 3. Pax6 activation of Math5 is cell autonomous.
Confocal images of E13.5 of triple labeled retinal sections. Comparison of Math5LacZ or Brn3b expression in E13.5 α-Cre;Pax6CKO/+ and α-Cre;Pax6CKO/CKO retinae. A–C) In α-Cre;Pax6CKO/+ eyes, Pax6 (red) and βgal (blue) proteins are coexpressed within the Cre expression domain (green). E–G, I–K) Where Pax6 is deleted, Math5LacZ expression is almost completely lost, but expressed at 1:1 correspondence with Pax6 protein in the central retina. Math5 dependence on Pax6 is highlighted in J and K where a single retinal cell lacking Cre-IRES-GFP autonomously retains both nuclear Pax6 and cytoplasmic βgal expression (arrowhead). White arrows in F,G,J,K point to Cre+; LacZ+; Pax6- cells. Yellow arrows in F,G point to Cre+;Pax6+ cells. See text for details. D,H,L) Two examples of E13.5 α-Cre;Pax6CKO/CKO peripheral retinal sections with complete loss of Brn3b (blue nuclei) in Pax6CKO/CKOcells (not red). Scleral is up; scale bar F = 400 microns.
To determine if Math5-GFP transgenes require Pax6 similarly to Math5 mRNA, we analyzed M5-GFP2.1 activity in Pax6Sey/Sey mutant mice. We compared live GFP expression among E11.5–E13.5 wild type, Pax6Sey/+ and Pax6Sey/Sey embryonic eyes (Figure 2C–E). The Pax6Sey mutation (Hill et al., 1991), causes smaller eyes in Pax6Sey/+ embryos and arrests eye development in Pax6Sey/Sey mutants. At all ages, the extent of the GFP expression domain was reduced in Pax6 heterozygote eyes (Figure 2D, 2D’) and missing from Pax6 mutant eyes (Figure 2E, 2E’; n = 8 litters with 19 mutants). The M5-GFP2.1+2.4 and M5-GFP2.1+1.6 transgenes were equally dependent on Pax6 gene dosage (not shown; for M5-GFP2.1+2.4 n = 2 litters with 5 mutants; for M5-GFP2.1+1.6 n = 3 litters with 3 mutants). To test whether Pax6 activation of Ath5 is conserved among vertebrates, we crossed mice carrying a Xath5-GFP transgene, containing 3.3 Kb of 5’ noncoding DNA (X5-GFP3.3) with Pax6Sey/+ mice (Hutcheson et al., 2005). Similar to M5-GFP2.1, X5-GFP3.3 the reporter expression domain was smaller in Pax6Sey/+ embryos and totally absent in Pax6Sey/Sey embryos (n = 4 litters with 9 mutant embryos, data not shown). We conclude that transgenes containing Math5 5’ distal noncoding DNA require Pax6 for their activation, and this relationship is conserved across a large phylogenetic distance (> 350 million years)(Roelants et al., 2007).
Although Math5-GFP and Xath5-GFP expression domains are clearly reduced in Pax6 heterozygotes, the loss of expression in Pax6 mutants could be attributed to nonspecific, early arrest of eye development. Therefore, we removed Pax6 function only in peripheral optic cup RPCs, using Cre/loxP conditional gene inactivation . Our goal was to understand if Math5 fails to activate at E11.5 in peripheral α-Cre;Pax6CKO/CKO RPCs. First we tested for complete deletion of Pax6 function at E11.5, since the α-Cre transgene initiates Cre recombinase and GFP expression only one day earlier (Marquardt et al., 2001). E11.5 optic cup sections from α-Cre;Pax6CKO/+ and α-Cre;Pax6CKO/CKO embryos were colabeled with anti-GFP and anti-Pax6 paired domain antibodies (Carriere et al., 1993). In Supplemental Figure 1B–E, GFP expression (green) indicates which RPCs have Cre activity, and the loss of anti-Pax6 paired-domain labeling (red) delineates RPCs without functional Pax6, since LoxP sites flank the paired-domain (Suppl. Fig 1A)(Marquardt et al., 2001). In E11.5 α-Cre; Pax6CKO/CKO peripheral RPCs, we observed many peripheral RPCs still coexpressing GFP and Pax6 (arrow in Suppl Fig 1E), indicating functional Pax6 has not yet been deleted (n = 5 α-Cre; Pax6CKO/CKO embryos).
Thus, our analysis was done at E13.5, when only occasional GFP/Pax6 double positive cells are found (yellow arrow in Figures 3F, 3G). Unfortunately, the α-Cre transgene IRES-GFP expression cassette prevents us from directly testing for a loss of Math5-GFP2.1 retinal expression. Instead, we created mice carrying the α-Cre transgene, Pax6CKO/+ or Pax6CKO/CKO alleles (Marquardt et al., 2001) and one allele of Math5LacZ (Brown et al., 2001), as a reporter of Math5 expression (Suppl. Fig 1A). In the peripheral retina of α-cre;Pax6CKO/CKO;Math5LacZ embryos, we observed dramatic loss of Math5LacZ (blue cells) (Figures 3F, 3G, 3J, 3K; n = 9 α-Cre; Pax6CKO/CKO embryos from 6 litters). Owing to mosaic expression of the α-Cre transgene (Yaron et al., 2006), we also observed isolated Pax6+;GFP- peripheral RPCs (arrowhead in Figures 3J,3K point to a red nucleus devoid of GFP labeling). These cells retain normal Pax6 function (no Cre expression), and autonomously express Math5LacZ (arrowheads in 3J, 3K point to cytoplasmic βgal surrounding nuclear Pax6), delineating the cell autonomous dependence of Math5 for Pax6. We also observe a third, rare class of GFP+, Pax6-, βgal+ RPCs in a subset of α-Cre;Pax6CKO/CKO;Math5LacZ eyes (white arrows in Figures 3F,3G,3J,3K). Because βgal protein has a long half-life, we presume that some Math5 transcription initiates prior to Pax6 deletion, followed by slow turnover of the βgal reporter. These rare GFP+, Pax6-, βgal+ RPCs do not express Brn3b, which acts immediately downstream of Math5 (Mu et al., 2005; Wang et al., 2001)(compare Figures 3D, 3I and 3L). Differentiated RGCs are also missing in the periphery of α-Cre;Pax6CKO/CKO mutant P21 retinas (Marquardt et al., 2001 and data not shown), meaning development was not simply delayed. We conclude that Math5 and Math5 transgenic expression requires Pax6.
Phylogenetic conservation and predicted Pax6 binding sites in Math5 5’ regulatory DNA
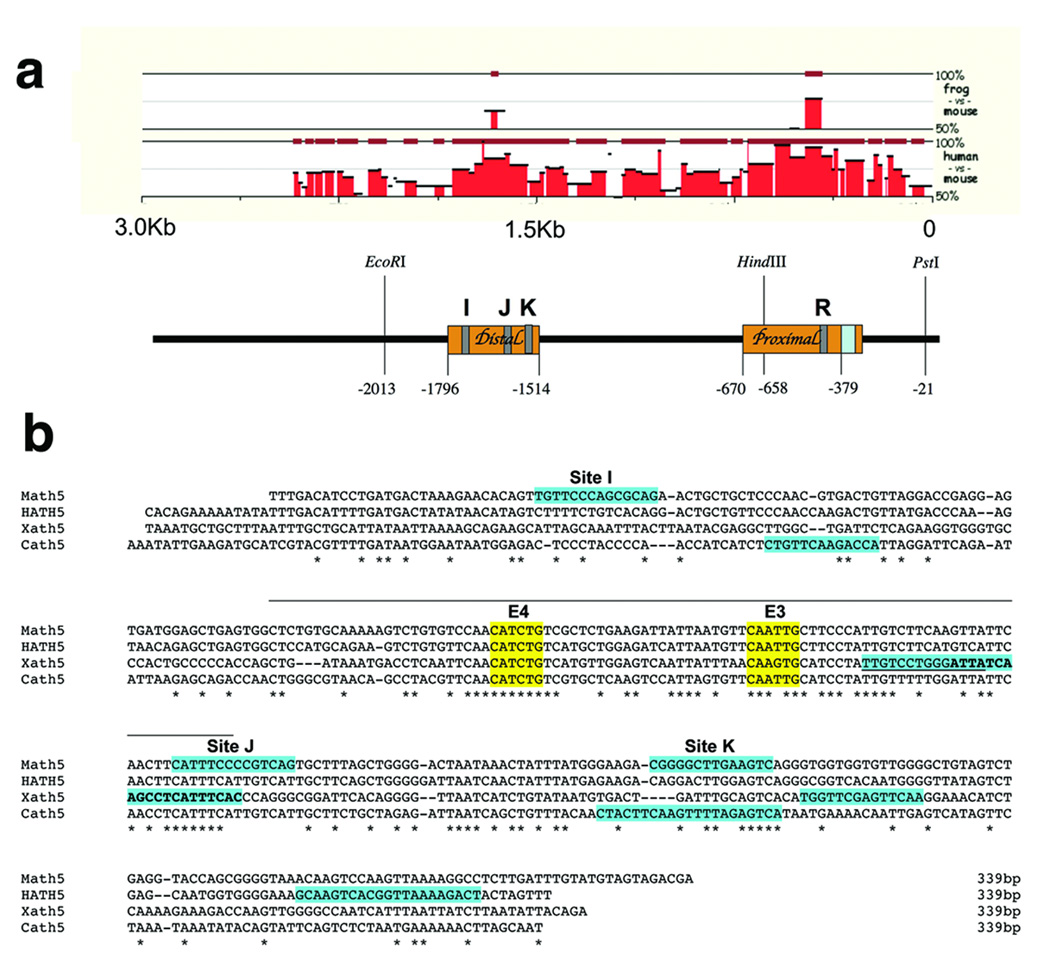
To understand if Pax6 directly regulates Math5, we wished to identify putative Pax6 binding sites in 5’ regulatory DNA. The first step was to determine the extent of nucleotide conservation among the Math5, Xath5 and human Ath5 (HATH5/ATOH7) 5’ DNA, since enhancers often reside in evolutionarily conserved regions. In Figure 4A a Mulan multiple-sequence algorithm (Loots and Ovcharenko, 2005; Ovcharenko et al., 2005) shows alignments between Xath5-Math5, and Math5-HATH5. The Xath5-HATH5 alignment was identical to that of Xath5-Math5 (not shown). Among all three species, only one distal and one proximal evolutionarily conserved region (ECR) are observed. Alignments with the NCBI BLAST 2 Sequences program identified the same two conserved regions, denoted by orange boxes in Figure 4A. The Math5 distal ECR is 282 bp (−1796 to −1514) and the proximal ECR is 291 bp (−670 to −379). Next, we searched Math5 5’ noncoding DNA for Pax6 paired domain sites, using the Transfac MATCH program. Multiple Pax6 paired domain matrices identified twenty putative binding sites in 3 Kb of upstream Math5 regulatory DNA (≥0.75 core and ≥0.7 matrix scores, Suppl. Table 1). However, only four predicted binding sites reside in the distal or proximal ECRs (I, J, K and R in Figure 4A).
Figure 4. Predicted Pax6 binding sites and phylogenetically conserved regions in mouse, human, frog and chick Ath5/Atoh7 5’ regulatory DNA.
A) Pictogram of Mulan alignments of upstream DNA (−3000 to –1), between Math5 vs. Xath5 and Math5 vs. HATH5. Underneath is a diagram of 3 Kb of upstream Math5 DNA (0 = A of ATG start codon). Orange boxes represent ≥70% nucleotide identity between Math5-HATH5, identified by NCBI BLAST. The blue box denotes the TATA box. Grey boxes indicate predicted Pax6 paired- domain binding sites determined by Transfac MATCH program (see Suppl. Table 1). EcoRI, HindIII and PstI restriction sites are the same as in Figure 1. B) Clustal W alignment of Ath5 distal ECRs, containing two completely conserved E boxes highlighted in yellow (E3 and E4, see Hutcheson et al., 2005). Transfac MATCH program predicted Pax6 binding sites are highlighted in teal. For Xath5, the two overlapping sites are distinguished by underlining versus bold faced type. Asterisks denote nucleotides completely conserved among the four species. A line, across the top, marks an 81 nucleotide stretch of high identity. Gaps (dashes) were inserted by the program for optimal alignment.
Our transgenic experiments demonstrated that a putative Math5 retinal expression element lies between −2100 to −600 bp, and the region of conserved nucleotide sequences within this interval suggests the distal ECR (dECR) to be the retinal element. The dECR contains two well-conserved E boxes, E3 and E4, (Hutcheson et al., 2005 and Figure 4B) and two putative Pax6 sites (I and J). However, we extended our working definition of the Math5 dECR to 339 bp (−1796 to −1458), to include another nearby putative Pax6 binding site K in subsequent experiments (Figures 4A,B). Figure 4B shows a Clustal W alignment of the dECR element from the human, mouse frog, and chick Ath5 genes. The zebrafish Zath5 dECR contained divergent nucleotide sequences, thus was omitted. The mouse, human, chick and frog dECRs all contain an 81 nucleotide stretch (marked by a line over the sequence in Figure 4B) with 57% identity among the four species, and 82% identity among any three species, including the E3 and E4 E-boxes and one putative Pax6 binding site (Figure 4B). Both the nucleotide sequence and relative spacing of these three sites are highly conserved. Although the Zath5 dECR is more divergent, it also contains two E-boxes and one putative Pax6 site, arranged differently (not shown).
For Math5, the most evolutionarily conserved Pax6 site, termed J, has the lowest MATCH score among all twenty predicted 5’ sites (Suppl. Table 1). The Xath5 dECR has three putative Pax6 sites. Of these, two are partially overlapping, with one aligning perfectly with the core nucleotides of Math5 site J (Figure 4B and Willardsen et al., in preparation). Paradoxically, the MATCH program failed to predict a site J in human or chick Ath5, despite conservation of 10/14 nucleotides, with the 7 critical core nucleotides completely conserved (Figures 4B). Based on the presence of well-conserved bHLH and Pax6 paired domain binding sites, the Math5 dECR should act as a retinal enhancer.
The Math5 339 bp distal fragment drives retinal expression in vivo
To test the idea that the Math5 distal ECR is a retinal enhancer, we asked if this regulatory DNA can direct retinal expression in vivo, by cloning these 339bp directly upstream of the Math5 TATA box in the promoterless pG1 vector (pG1-M50.2dECR), and making transient transgenic frog Xenopus embryos (Amaya and Kroll, 1999). Retinal GFP expression was scored at stage 33, when Ath5 is robustly expressed by retinal progenitors throughout much of the eye. Figure 5A shows that the Math5 dECR drives embryonic retinal GFP expression in vivo. Overall, 12.5% (20/160) of the embryos exhibited eye expression, of which nine had retinal-specific expression (Figure 5A), and the remainder displayed retinal plus other nervous system expression, including brain and spinal cord (not shown). These same two expression classes were previously seen for pG1-M5-2.1 (Hutcheson et al., 2005). Embryos with ectopic CNS expression could be explained by the absence of distal regulatory sequences that suppress a Math1-like expression pattern (Hufnagel et al., 2007). There was a lower percentage of GFP+ transgenic embryos for pG1-M50.2dECR (12.5%) than for pGL1-M52.1 (34%), or an analogous Xath5 distal 152bp retinal enhancer construct (35%), but this was higher than for pG1-M50.6 (6%) or pG1 vector alone (0%) (Hutcheson et al., 2005; Hutcheson and Vetter, 2002, Willardsen et al., in prep). Although pG1-M50.2dECR expressed the GFP reporter more weakly and/or at lower efficiency than other constructs, the mouse Math5 dECR directs retinal expression during frog eye development. Thus, we conclude it contains a retinal enhancer.
Figure 5. Pax6 interacts with the Ath5 distal ECR in vivo and in vitro.
A) GFP expression in pG1-M5-0.2 distal transient transgenic Xenopus embryos at stage 33. Embryos exhibited retinal expression alone (9/20) or retina plus nervous system expression (11/20) (not shown). pG1-Math5-2.1 also drives GFP expression in the same two expression pattern classes (Hutcheson et al., 2005). B) Real-time quantitative PCR using primers (targets) for the HATH5 dECR and a negative control sequence in HATH5 3’UTR. Results are expressed as a percentage of input chromatin isolated from Ad12 HER10 cells, showing averaged “enrichments” and the standard error of the mean. C) EMSAs of Pax6 paired domain-GST fusion protein with binding sites I,J,K and R in Math5 distal or proximal ECRs. For each binding site, the left lane contains free probe, the middle lane probe plus GST protein alone, and the right lane probe plus Pax6 paired domain-GST fusion protein. Only site J shifts in the presence of Pax6. This binding was completely lost by mutating 3 of 5 core nucleotides (J-Mut).
Pax6 binding to the Ath5 dECR in vivo and in vitro
To demonstrate Pax6 occupancy of the Ath5 dECR in vivo, we performed chromatin immunoprecipitation (ChIP) (Figure 5B). For this we used a specific antibody against Pax6 to immunoprecipitate cross-linked Ad12 HER10 cell chromatin. This cell line is a stably transformed, and clonally derived human embryonic retinoblast cell line (Grabham et al., 1988), which expresses PAX6, HATH5/ATOH7 and BRN3B mRNA by RT-PCR (Malgorzata Quinn and NLB, unpublished observations). Eight independent ChIP assays were quantified by real-time PCR, for the HATH5 dECR versus a negative control sequence in the HATH5 3’UTR (Figure 5B). These results show (Roelants et al., 2007) in vivo enrichment of Pax6 at the dECR.
Next, to identify which of the predicted sites Pax6 binds in the dECR, electrophoretic mobility assays were performed on sites I, J, K, along with site R in the proximal ECR since it is also evolutionarily conserved (Figure 4A). In these experiments, a Pax6 paired domain-GST fusion protein or GST protein alone (Epstein et al., 1994a) were incubated with 75 fmol of each radiolabeled, double stranded oligonucleotide (Figure 5C) and the electrophoretic mobility of potential protein-DNA complexes tested. Only the conserved site J was bound by 100 ng of paired domain protein (Figure 5B). A titration curve of Pax6 paired-GST protein, from 10 ng to 10 µg, demonstrated that 50 ng of protein weakly bound to site J, with more robust binding correlating with increasing protein concentrations (not shown). The specificity of Pax6 for site J was demonstrated by mutating 3 of 5 core nucleotides, changing TTCCC to AAACC. We observed a complete loss of paired domain binding (Figure 5C). None of the other three sites bound the Pax6 paired domain at any concentration tested. In a different study, we also tested Pax6 binding at sites H, S and T (Suppl. Table 1). Of these, Pax6 bound only to site T, but much more weakly than it does to site J (Hufnagel et al., 2007).
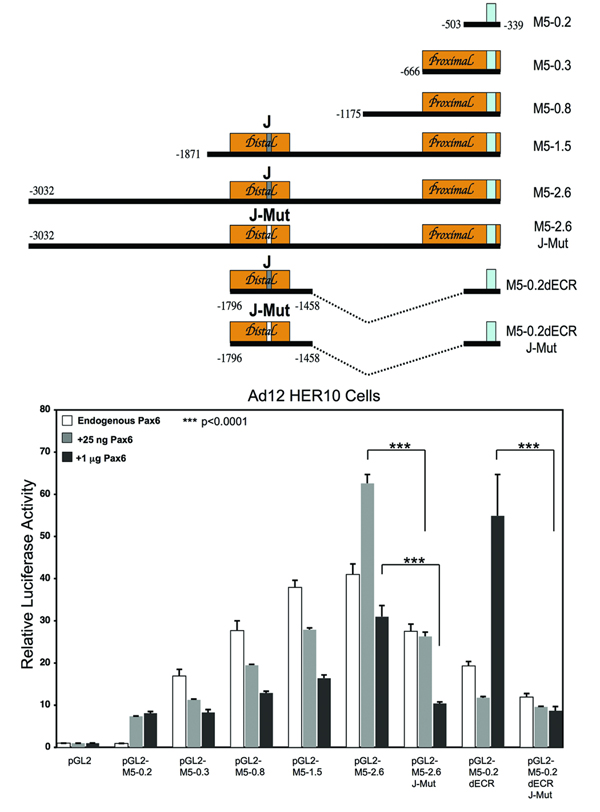
Finally, we also assessed the Pax6-dependent transcriptional activity of Math5 5’ DNA by performing luciferase assays in Ad12 HER10 cells, with constructs containing varying amounts of Math5 5’ DNA (Figure 6). In the mouse optic cup, RPCs coexpress Math5 mRNA with high levels of Pax6 protein at E11.5 (Figure 2A), but with low levels at E13.5 and beyond (Figure 2B). Therefore 25ng (low) and 1µg (high) of pCS2-Pax6 plasmid DNA were separately cotransfected with Math5 luciferase constructs, versus pCS2 alone as a control. These Pax6 DNA concentrations were chosen from a range known to regulate the transcriptional activity of other Pax6 target genes in analogous assays (Chauhan et al., 2004; Duncan et al., 1998). We first observed that the minimal TATA box and proximal ECR constructs (pGL2-M5-0.2 and pGL2-M5-0.3) exhibit a small stimulation in the presence of low and high Pax6. However, these DNA sequences contain only one putative binding site R, which did not bind the Pax6 paired domain (Figure 4A, Figure 5C). Thus, the low-level Pax6 activation appears indirect, but could also mean that Ad12 HER10 cells lack a retinal factor that acts in concert with Pax6 at this site in vivo.
Figure 6. Distinct Math5 5’ regulatory sequences are activated by different concentrations of Pax6 protein in vitro.
Relative promoter activities of Math5-luciferase reporter constructs co-transfected with pCS2, pCS2-Pax6 (25ng or 1µg) into Ad12 HER10 cells. Firefly luciferase values were normalized to pTK-Renilla luciferase values, and pGL2 empty vector activity set to 1.0. The mean values are shown, with error bars representing the standard error of the mean. n = 3 trials, each run in triplicate; *** = p<0.001.
By contrast, Pax6 induced high activation levels for constructs containing the dECR, pGL2-M5-2.6 and pGL2-M5-0.2dECR, although each displayed different concentration dependences. We found that high Pax6 activated pGL2-M5-0.2dECR 6.5-fold over the TATA box alone, while low Pax6 had no effect (Figure 6). This activation of the dECR is specific, since mutating the Pax6 J binding site (Figure 5C) reduced transcription levels back to that of the TATA box alone (Figure 6). Unexpectedly, Math5 5’ DNA sequences more distal to the dECR also respond to Pax6. pGL2-M5-2.6, which uniquely contains seven additional predicted Pax6 binding sites (A–G, Suppl. Table 1), had 8-fold higher activity than the TATA box alone in the presence of low Pax6, while high Pax6 only caused a 4-fold increase (Figure 7). Mutating site J in the context of this larger piece of Math5 upstream DNA, abolished the transcriptional activity induced by high Pax6, but only partially reduced that of low Pax6. Thus, we assume that low levels of Pax6 protein regulate Math5 regulatory sequences farther upstream.
The third construct containing the dECR, pGL2-M5-1.5, was repressed by additional Pax6 protein. Because pGL2-M5-0.3 and pGL2-M50.8 were similarly repressed, yet do not contain the dECR, Pax6 may bind to one or more putative binding sites between the proximal and distal ECRs (Suppl. Table 1). Finally, we observed that Pax6 regulation of Math5 5’ DNA requires a retinal-specific context. When the same dataset in Figure 6 was generated in HEK-293T human kidney epithelial cells, high Pax6 was unable to significantly stimulate the dECR (data not shown). Similarly, low Pax6 induced only a small increase in pGL2-M5-2.6 activity that was not statistically significant. Overall, we conclude that Pax6 directly binds to one well-conserved binding site to activate Math5 transcription. We further propose that Pax6 requires retinal-specific cofactor(s) for this regulatory activity, and these transcription factor complexes may further modulate Math5 levels via additional cis regulatory sequences.
Discussion
Math5 is a direct transcriptional target of Pax6
We initiated this study with two goals in mind. The first was to identify Math5 noncoding regions that control retinal expression. We also wished to understand if Pax6 directly activates Math5 expression. Several pieces of evidence demonstrate Pax6 as a direct transcriptional activator of Math5. Previously, initiation of Math5 mRNA retinal expression was shown to require Pax6 (Brown et al., 1998). This regulatory relationship was better defined by conditional deletion of Pax6 in the distal retina, which shows that Math5 activation requires Pax6 cell autonomously. We also mapped a Math5 retinal element to 339bp of 5’ distal regulatory DNA. Transgenes containing this distal DNA require Pax6 for their expression, and Pax6 protein interacts with the Math5 5’ distal element in vivo.
To define the Math5 retinal enhancer(s) more precisely, multi-species nucleotide alignments showed two evolutionarily conserved regions (ECRs), with the distal ECR (dECR) present in all transgenic lines exhibiting GFP reporter expression in the retina. The dECR nucleotides are well conserved among four vertebrate Ath5 genes. Interestingly, of the possible Pax6 sites within either the Math5 proximal or distal ECRs, only one binds Pax6 in vitro. Although this binding site is highly conserved, it has a low prediction score, and the prediction program failed to find the same site in the human or chick Ath5 dECR, despite complete conservation of 10 of 14 nucleotides. We attribute this to the limitations of computer programs in predicting germane transcription factor binding sites (Vavouri and Elgar, 2005) and the ability of the Pax6 paired domain to bind a variety of nucleotide sequences. Among 7 putative Pax6 sites tested for in vitro binding, the Pax6 paired domain binds well to site J in the dECR (this paper), and much more weakly to site T (Hufnagel et al., 2007). The biologic relevance of site T is unclear since it resides in proximal DNA that does not direct retinal expression in vivo. Instead, our studies demonstrate that in a retinal context differing amounts of Pax6 specifically stimulate two distinct Math5 5’ regulatory regions. These responses to Pax6 protein levels are meaningful, since in the mouse retina we observed obvious changes in the coexpression of Math5 mRNA and Pax6 protein across developmental time. When RPCs initiate Math5 expression at E11, they also express robust levels of Pax6. But, once differentiating RGCs accumulate in the GCL at E13.5, RPCs coexpress low Pax6 protein and Math5 mRNA, while RGCs maintain high Pax6 and no Math5 mRNA expression.
In principle, Pax6 may regulate Math5 in three different ways of which some, or all, are direct. First, Math5 requires high levels of Pax6 to initiate transcription in the first newly postmitotic cells within the dorsocentral optic cup. But, because Pax6 is expressed by all optic cup RPCs, other factors must direct the precise time and place of Math5 activation. Second, when the optic cup is transformed into a bilayered retina, Pax6 protein expression is lower in RPCs and higher in the GCL. Interestingly, only the RPCs express Math5 mRNA, suggesting that at older ages Math5 is activated or maintained by lower levels of Pax6. Moreover, in the GCL Pax6 might repress Math5 expression. An analogous dual role for Pax6 has been reported for the β-crystallin gene, where low levels of Pax6 is activating, yet high levels are inhibitory (Duncan et al., 1998). We propose that Pax6 acts within a protein complex to regulate Math5 via the dECR, since in HEK293T cells it was ineffective at stimulating transcriptional activity. Thus, other factor(s) that regulate Math5 have restricted dorsocentral expression, function to drive a subset of RPCs out of the cell cycle and/or simultaneously activate Math5. We propose that Pax6 is necessary for Math5 expression, but incapable of solely controlling crucial spatiotemporal and cell cycle phase expression. Overall, our data demonstrate that Math5 is a direct transcriptional target of Pax6.
Towards a comprehensive model of Math5 gene regulation
Multiple extrinsic signals act upstream of vertebrate Ath5 to instruct RGC formation. These include FGFs (Martinez-Morales et al., 2005), oep (Masai et al., 2000), Notch (Austin et al., 1995; Henrique et al., 1997; Lee et al., 2005; Schneider et al., 2001; Yaron et al., 2006), GDF11 (Kim et al., 2005), and shh (Masai et al., 2005; Stenkamp and Frey, 2003; Wang et al., 2005). In the zebrafish retina, FGF signaling activates Ath5 while oep (zebrafish), Notch (chick, frog, mouse), GDF11 (mouse), and shh (zebrafish, chick, mouse) pathways prohibit Ath5 expression. Signal integration instructs Ath5 spatiotemporal expression, although these signals cannot account for the entire expression pattern (reviewed in Mu and Klein, 2004; Vetter and Brown, 2001). Some transcription factors also genetically repress Math5, for example NeuroD, Math3 (Inoue et al., 2002) and Hes1 (Lee et al., 2005). Paradoxically, unlike most proneural bHLHs, mouse Math5 does not regulate its own expression (Brown et al., 2001; Hutcheson et al., 2005). This means that the initiation of mammalian retinogenesis is orchestrated by a combination of extracellular signals and transcription factors acting on Math5 cis-regulatory elements. But, it is unclear how many of these upstream, trans-acting factors are direct regulators of Math5.
Here we narrowed the cis-regulatory DNA needed to achieve Math5 retinal expression in vivo to a 339 bp fragment and identify a smaller 81 nucleotide segment with 57% nucleotide identity among Cath5, Xath5, Math5 and Hath5, and 82% identity among any three of these genes. Interestingly, in the Xath5 gene a 152 bp distal retinal enhancer, which also contains this 81 nucleotide segment, directs early retinal expression in the frog eye, although the 81 bp itself is insufficient to promote retinal expression (Willardsen, et al., in preparation). But, is this the only Math5 retinal enhancer? In the fruit fly eye, the atonal gene has at least three different eye enhancers, one located 5’ and the others 3’ (Sun et al., 1998; Zhang et al., 2006). Those elements controlling initial atonal activation in the eye disc reside downstream of the coding exon, while the 5’ atonal enhancer regulates refinement of a broad expression domain to a single cell, the R8 neuron (Sun et al., 1998). By analogy, additional 5’ or 3’ Math5 cis sequences may regulate other aspects of the Math5 retinal expression, for instance refinement of its precise spatiotemporal expression pattern. In particular, it will be important to identify Math5 repressor sequences that regulate its abrupt shutoff as postmitotic, transitional cells reach the GCL and differentiate as RGCs. Another important feature to define molecularly is how Math5 initiates in the dorsocentral optic cup and spreads outward to the periphery (Brown et al., 1998). Our in vitro transcription data strongly suggest additional 5’ regulatory sequences also contain Math5 regulatory information, and these will be tested in the future. The first region lies immediately 5’ to the distal end of the 2.1 KB transgene and is activated by low levels of Pax6 protein in vitro. The other lies between the proximal and distal ECRs and appears to have reduced activity in the presence of low and high Pax6. It will be interesting to understand how these other DNA segments require Pax6, search for additional factors that directly regulate aspects of Math5 expression via these regions of 5’ noncoding DNA, and discern how multiple regulatory enhancers may coordinately regulate Math5 transcription.
Evolutionary conservation of Ath5/atonal retinal elements
Conservation of Math5 5’ DNA, especially the distal ECR and Pax6 binding sites, are noteworthy in the context of metazoan eye development and evolution. For instance, Drosophila eyeless and mouse Pax6 retinal enhancers drive reporter expression with reasonable fidelity in cross-species transgenic experiments (Xu et al., 1999). Now, Pax6 direct regulation of atonal-Ath5 retinal expression is shown to be highly conserved (Zhang et al., 2006, Willardsen et al., in preparation and this paper). These are yet more examples, within a large body of evidence that cis-regulatory modules play important roles in phylogenetically conserved gene networks. We have identified a retinal element whose sequence is highly conserved among four vertebrate species that last shared a common ancestor more than 350 million years ago. This conservation probably goes even deeper since the zebrafish Zath5 distal ECR also contains a core of the two bHLH and one Pax6 binding site. Although the spacing and nucleotide sequences differ from the other vertebrate orthologues examined, these characteristics have been shown to be irrelevant for the conservation of cis regulation for the Drosophila eve stripe 2 enhancer (Ludwig et al., 2005). More recently, an extensive comparison of the direct regulation of four transcription factors among 4,000 target gene promoters, between mouse and human, demonstrated that 41–89% of the DNA binding events are species-specific (Odom et al., 2007). Therefore even when a transcription factor maintains its direct regulation of a target gene, the position of individual binding sites may have changed among species. Thus, the Zath5 distal enhancer would appear to represent coevolution of cis sequences with species-specific changes in the trans-acting factors, which nonetheless maintain conserved epistatic relationships among gene hierarchies.
It will be interesting to identify and test for other cis-regulatory binding sites in Ath5 genes for their phylogenetic conservation. Perhaps even more appealing than conserved aspects of retinal development however, are species-specific features such as the overall length of retinogenesis and cell-birth order. Interestingly, the mammalian retina develops more slowly and utilizes transcriptional regulation of bHLH factors more than posttranslational regulation, which is a significant feature of more rapid retinogenesis in frogs (Moore et al., 2002). Ultimately, the elucidation of genes and gene pathways with divergent regulation will allow us to focus in even greater depth on species-specific or potentially cell-type mechanisms of retinal neuron formation.
Supplementary Material
Acknowledgements
We thank Ruth Ashery-Padan, Peter Gruss and Derek van der Kooy for Pax6 α-cre and Pax6CKO/CKO mice; Simon Saulé for anti-paired domain Pax6 antibody; Tom Glaser for a Pax6 paired domain-GST fusion construct, Ad12 HER10 cells and anti-βgal antibody and Malgorzata Quinn for sharing RT-PCR data. We are indebted to Brian Gebelein and Tom Glaser for advice on EMSA, Jay Kormish and Shiming Chen for help with chromatin IP and real-time PCR, Michiya Sugimori for assistance with confocal microscopy; Emily Wroblewski and Ashley Riesenberg for technical support; and Dave Hutcheson, Richard Lang, Tiffany Cook, Brian Gebelein and Jim Lauderdale for helpful discussions and critical evaluation of this manuscript.
This work was supported by a graduate research fellowship to MIW, NIH grant EY12274 to MLV and NIH grant EY13612 to NLB
References
- Amaya E, Kroll KL. A method for generating transgenic frog embryos. Methods Mol Biol. 1999;97:393–414. doi: 10.1385/1-59259-270-8:393. [DOI] [PubMed] [Google Scholar]
- Austin CP, Feldman DE, Ida JA, Jr, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JA. Department of Human Genetics. Ann Arbor, MI: University of Michigan; 2005. The Role of Math5 in Retinal Development. [Google Scholar]
- Carriere C, Plaza S, Martin P, Quatannens B, Bailly M, Stehelin D, Saule S. Characterization of quail Pax-6 (Pax-QNR) proteins expressed in the neuroretina. Mol Cell Biol. 1993;13:7257–7266. doi: 10.1128/mcb.13.12.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayouette M, Poggi L, Harris WA. Lineage in the vertebrate retina. Trends Neurosci. 2006;29:563–570. doi: 10.1016/j.tins.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Chauhan BK, Yang Y, Cveklova K, Cvekl A. Functional properties of natural human PAX6 and PAX6(5a) mutants. Invest Ophthalmol Vis Sci. 2004;45:385–392. doi: 10.1167/iovs.03-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Peng GH, Wang X, Smith AC, Grote SK, Sopher BL, La Spada AR. Interference of Crx-dependent transcription by ataxin-7 involves interaction between the glutamine regions and requires the ataxin-7 carboxy-terminal region for nuclear localization. Hum Mol Genet. 2004;13:53–67. doi: 10.1093/hmg/ddh005. [DOI] [PubMed] [Google Scholar]
- Chow RL, Altmann CR, Lang RA, Hemmati-Brivanlou A. Pax6 induces ectopic eyes in a vertebrate. Development. 1999;126:4213–4222. doi: 10.1242/dev.126.19.4213. [DOI] [PubMed] [Google Scholar]
- Duncan MK, Haynes JI, 2nd, Cvekl A, Piatigorsky J. Dual roles for Pax-6: a transcriptional repressor of lens fiber cell-specific beta-crystallin genes. Mol Cell Biol. 1998;18:5579–5586. doi: 10.1128/mcb.18.9.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MK, Li X, Ogino H, Yasuda K, Piatigorsky J. Developmental regulation of the chicken beta B1-crystallin promoter in transgenic mice. Mech Dev. 1996;57:79–89. doi: 10.1016/0925-4773(96)00533-3. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Bremner R. The search for the retinoblastoma cell of origin. Nat Rev Cancer. 2005;5:91–101. doi: 10.1038/nrc1545. [DOI] [PubMed] [Google Scholar]
- Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994a;269:8355–8361. [PubMed] [Google Scholar]
- Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes Dev. 1994b;8:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Fujiwara M, Uchida T, Osumi-Yamashita N, Eto K. Uchida rat (rSey): a new mutant rat with craniofacial abnormalities resembling those of the mouse Sey mutant. Differentiation. 1994;57:31–38. doi: 10.1046/j.1432-0436.1994.5710031.x. [DOI] [PubMed] [Google Scholar]
- Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human Pax6 gene. Nature Genetics. 1992;2:232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- Grabham PW, Grand RJ, Byrd PJ, Gallimore PH. Differentiation of normal and adenovirus-12 E1 transformed human embryo retinal cells. Exp Eye Res. 1988;47:123–133. doi: 10.1016/0014-4835(88)90029-2. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hanson IM. PAX6 and congenital eye malformations. Pediatr Res. 2003;54:791–796. doi: 10.1203/01.PDR.0000096455.00657.98. [DOI] [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Henrique D, Hirsinger E, Adam J, Le Roux I, Pourquie O, Ish-Horowicz D, Lewis J. Maintenance of neuroepithelial progenitor cells by Delta-Notch signalling in the embryonic chick retina. Curr Biol. 1997;7:661–670. doi: 10.1016/s0960-9822(06)00293-4. [DOI] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BLM, Ton CCT, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse Small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Holt CE, Bertsch TW, Ellis HM, Harris WA. Cellular determination in the Xenopus retina is independent of lineage and birth date. Neuron. 1988;1:15–26. doi: 10.1016/0896-6273(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Hufnagel RB, Riesenberg AN, Saul SM, Brown NL. Conserved regulation of Math5 and Math1 revealed by Math5-GFP transgenes. Mol Cell Neurosci. 2007;36:435–448. doi: 10.1016/j.mcn.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson DA, Hanson MI, Moore KB, Le TT, Brown NL, Vetter ML. bHLH-dependent and -independent modes of Ath5 gene regulation during retinal development. Development. 2005;132:829–839. doi: 10.1242/dev.01653. [DOI] [PubMed] [Google Scholar]
- Hutcheson DA, Vetter ML. The bHLH factors Xath5 and XNeuroD can upregulate the expression of XBrn3d, a POU-homeodomain transcription factor. Dev Biol. 2001;232:327–338. doi: 10.1006/dbio.2001.0178. [DOI] [PubMed] [Google Scholar]
- Hutcheson DA, Vetter ML. Transgenic approaches to retinal development and function in Xenopus laevis. Methods. 2002;28:402–410. doi: 10.1016/s1046-2023(02)00259-1. [DOI] [PubMed] [Google Scholar]
- Inoue T, Hojo M, Bessho Y, Tano Y, Lee JE, Kageyama R. Math3 and NeuroD regulate amacrine cell fate specification in the retina. Development. 2002;129:831–842. doi: 10.1242/dev.129.4.831. [DOI] [PubMed] [Google Scholar]
- Kim J, Wu HH, Lander AD, Lyons KM, Matzuk MM, Calof AL. GDF11 controls the timing of progenitor cell competence in developing retina. Science. 2005;308:1927–1930. doi: 10.1126/science.1110175. [DOI] [PubMed] [Google Scholar]
- Kozmik Z. Pax genes in eye development and evolution. Curr Opin Genet Dev. 2005;15:430–438. doi: 10.1016/j.gde.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev Biol. 2006;295:764–778. doi: 10.1016/j.ydbio.2006.03.055. [DOI] [PubMed] [Google Scholar]
- Lee HY, Wroblewski E, Philips GT, Stair CN, Conley K, Reedy M, Mastick GS, Brown NL. Multiple requirements for Hes 1 during early eye formation. Dev Biol. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine EM, Green ES. Cell-intrinsic regulators of proliferation in vertebrate retinal progenitors. Semin Cell Dev Biol. 2004;15:63–74. doi: 10.1016/j.semcdb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Liu W, Mo Z, Xiang M. The Ath5 proneural genes function upstream of Brn3 POU domain transcription factor genes to promote retinal ganglion cell development. Proc Natl Acad Sci U S A. 2001;98:1649–1654. doi: 10.1073/pnas.98.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Loots GG, Ovcharenko I. Dcode.org anthology of comparative genomic tools. Nucleic Acids Res. 2005;33:W56–W64. doi: 10.1093/nar/gki355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MZ, Palsson A, Alekseeva E, Bergman CM, Nathan J, Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T. Transcriptional control of neuronal diversification in the retina. Prog Retin Eye Res. 2003;22:567–577. doi: 10.1016/s1350-9462(03)00036-3. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Martinez-Morales JR, Del Bene F, Nica G, Hammerschmidt M, Bovolenta P, Wittbrodt J. Differentiation of the vertebrate retina is coordinated by an FGF signaling center. Dev Cell. 2005;8:565–574. doi: 10.1016/j.devcel.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Masai I, Stemple DL, Okamoto H, Wilson SW. Midline signals regulate retinal neurogenesis in zebrafish. Neuron. 2000;27:251–263. doi: 10.1016/s0896-6273(00)00034-9. [DOI] [PubMed] [Google Scholar]
- Masai I, Yamaguchi M, Tonou-Fujimori N, Komori A, Okamoto H. The hedgehog-PKA pathway regulates two distinct steps of the differentiation of retinal ganglion cells: the cell-cycle exit of retinoblasts and their neuronal maturation. Development. 2005;132:1539–1553. doi: 10.1242/dev.01714. [DOI] [PubMed] [Google Scholar]
- Mastick GS, Andrews GL. Pax6 regulates the identity of embryonic diencephalic neurons. Mol Cell Neurosci. 2001;17:190–207. doi: 10.1006/mcne.2000.0924. [DOI] [PubMed] [Google Scholar]
- Matter-Sadzinski L, Puzianowska-Kuznicka M, Hernandez J, Ballivet M, Matter JM. A bHLH transcriptional network regulating the specification of retinal ganglion cells. Development. 2005;132:3907–3921. doi: 10.1242/dev.01960. [DOI] [PubMed] [Google Scholar]
- Moore KB, Schneider ML, Vetter ML. Posttranslational mechanisms control the timing of bHLH function and regulate retinal cell fate. Neuron. 2002;34:183–195. doi: 10.1016/s0896-6273(02)00666-9. [DOI] [PubMed] [Google Scholar]
- Mu X, Fu X, Sun H, Liang S, Maeda H, Frishman LJ, Klein WH. Ganglion cells are required for normal progenitor- cell proliferation but not cell-fate determination or patterning in the developing mouse retina. Curr Biol. 2005;15:525–530. doi: 10.1016/j.cub.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Mu X, Klein WH. A gene regulatory hierarchy for retinal ganglion cell specification and differentiation. Semin Cell Dev Biol. 2004;15:115–123. doi: 10.1016/j.semcdb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–732. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Giardine BM, Hou M, Ma J, Hardison RC, Stubbs L, Miller W. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei YF, Rhodin JA. The prenatal development of the mouse eye. Anat Rec. 1970;168:105–125. doi: 10.1002/ar.1091680109. [DOI] [PubMed] [Google Scholar]
- Poggi L, Vitorino M, Masai I, Harris WA. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J Cell Biol. 2005;171:991–999. doi: 10.1083/jcb.200509098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Roelants K, Gower DJ, Wilkinson M, Loader SP, Biju SD, Guillaume K, Moriau L, Bossuyt F. Global patterns of diversification in the history of modern amphibians. Proc Natl Acad Sci U S A. 2007;104:887–892. doi: 10.1073/pnas.0608378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth HJ, Das GC, Piatigorsky J. Chicken beta B1-crystallin gene expression: presence of conserved functional polyomavirus enhancer-like and octamer binding-like promoter elements found in non-lens genes. Mol Cell Biol. 1991;11:1488–1499. doi: 10.1128/mcb.11.3.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- Schneider ML, Turner DL, Vetter ML. Notch signaling can inhibit Xath5 function in the neural plate and developing retina. Mol Cell Neurosci. 2001;18:458–472. doi: 10.1006/mcne.2001.1040. [DOI] [PubMed] [Google Scholar]
- Skowronska-Krawczyk D, Ballivet M, Dynlacht BD, Matter JM. Highly specific interactions between bHLH transcription factors and chromatin during retina development. Development. 2004;131:4447–4454. doi: 10.1242/dev.01302. [DOI] [PubMed] [Google Scholar]
- Stenkamp DL, Frey RA. Extraretinal and retinal hedgehog signaling sequentially regulate retinal differentiation in zebrafish. Dev Biol. 2003;258:349–363. doi: 10.1016/s0012-1606(03)00121-0. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jan LY, Jan YN. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development. 1998;125:3731–3740. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- Treisman JE. How to make an eye. Development. 2004;131:3823–3827. doi: 10.1242/dev.01319. [DOI] [PubMed] [Google Scholar]
- Tucker P, Laemle L, Munson A, Kanekar S, Oliver ER, Brown N, Schlecht H, Vetter M, Glaser T. The eyeless mouse mutation (ey1) removes an alternative start codon from the Rx/rax homeobox gene. Genesis. 2001;31:43–53. doi: 10.1002/gene.10003. [DOI] [PubMed] [Google Scholar]
- Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- Turner DL, Snyder EY, Cepko CL. Lineage-independent determination of cell type in the embryonic mouse retina. Neuron. 1990;4:833–845. doi: 10.1016/0896-6273(90)90136-4. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes and Development. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Vavouri T, Elgar G. Prediction of cis-regulatory elements using binding site matrices--the successes, the failures and the reasons for both. Curr Opin Genet Dev. 2005;15:395–402. doi: 10.1016/j.gde.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Vetter ML, Brown NL. The role of basic helix-loop-helix genes in vertebrate retinogenesis. Semin Cell Dev Biol. 2001;12:491–498. doi: 10.1006/scdb.2001.0273. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Dakubo GD, Thurig S, Mazerolle CJ, Wallace VA. Retinal ganglion cell-derived sonic hedgehog locally controls proliferation and the timing of RGC development in the embryonic mouse retina. Development. 2005;132:5103–5113. doi: 10.1242/dev.02096. [DOI] [PubMed] [Google Scholar]
- Weiner H, Farres J, Rout UJ, Wang X, Zheng CF. Site directed mutagenesis to probe for active site components of liver mitochondrial aldehyde dehydrogenase. Adv Exp Med Biol. 1995;372:1–7. doi: 10.1007/978-1-4615-1965-2_1. [DOI] [PubMed] [Google Scholar]
- Wells J, Farnham PJ. Characterizing transcription factor binding sites using formaldehyde crosslinking and immunoprecipitation. Methods. 2002;26:48–56. doi: 10.1016/S1046-2023(02)00007-5. [DOI] [PubMed] [Google Scholar]
- Wetts R, Fraser SE. Multipotent precursors can give rise to all major cell types of the frog retina. Science. 1988;239:1142–1145. doi: 10.1126/science.2449732. [DOI] [PubMed] [Google Scholar]
- Xu PX, Zhang X, Heaney S, Yoon A, Michelson AM, Maas RL. Regulation of Pax6 expression is conserved between mice and flies. Development. 1999;126:383–395. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R. Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development. 2006;133:1367–1378. doi: 10.1242/dev.02311. [DOI] [PubMed] [Google Scholar]
- Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development. 2006;133:4881–4889. doi: 10.1242/dev.02669. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zheng JB, Gu X, Li W, Saunders GF. A novel Pax-6 binding site in rodent B1 repetitive elements: coevolution between developmental regulation and repeated elements? Gene. 2000;245:319–328. doi: 10.1016/s0378-1119(00)00019-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.