Abstract
In this paper we give the details of the numerical solution of a three-dimensional multispecies diffuse interface model of tumor growth, which was derived in (Wise et al., J. Theor. Biol. 253 (2008)) and used to study the development of glioma in (Frieboes et al., NeuroImage 37 (2007) and tumor invasion in (Bearer et al., Cancer Research, 69 (2009)) and (Frieboes et al., J. Theor. Biol. 264 (2010)). The model has a thermodynamic basis, is related to recently developed mixture models, and is capable of providing a detailed description of tumor progression. It utilizes a diffuse interface approach, whereby sharp tumor boundaries are replaced by narrow transition layers that arise due to differential adhesive forces among the cell-species. The model consists of fourth-order nonlinear advection-reaction-diffusion equations (of Cahn-Hilliard-type) for the cell-species coupled with reaction-diffusion equations for the substrate components. Numerical solution of the model is challenging because the equations are coupled, highly nonlinear, and numerically stiff. In this paper we describe a fully adaptive, nonlinear multigrid/finite difference method for efficiently solving the equations. We demonstrate the convergence of the algorithm and we present simulations of tumor growth in 2D and 3D that demonstrate the capabilities of the algorithm in accurately and efficiently simulating the progression of tumors with complex morphologies.
Keywords: cancer, necrosis, computer simulation, diffuse interface method, mixture model, nonlinear multigrid method, three dimensional model
1 Introduction
The morphological evolution of a growing solid tumor is the result of the dynamics of a complex system that includes many nonlinearly interacting factors, including cell-cell and cell-matrix adhesion, mechanical stress, cell motility, clonal instability, angiogenesis and the degree of heterogeneity of cell proliferation just to name a few [3]. Mathematical modeling may yield important insight on tumor progression, help to explain experimental and clinical observations and to help assess optimal treatment strategies. Numerous mathematical models have been developed to study various aspects of tumor progression and this has been an area of intense research interest. See the recent reviews by Ribba et al. [73]; Quaranta et al. [69]; Hatzikirou et al. [51]; Nagy [67]; Byrne et al. [18]; Fasano et al. [35]; van Leeuwen et al. [80]; Roose et al. [74]; Graziano and Preziosi [48]; Harpold et al. [50]; Drasdo and Höhme [34]; Friedman et al. [42]; Sanga et al. [75]; Deisboeck et al. [31]; Anderson and Quaranta [7]; Bellomo et al. [14]; Cristini et al. [28]; and Lowengrub et al. [61]. Because of the complexity of tumor progession and the associated mathematical models, numerical simulation has become an indispensible tool in studying solid tumor growth. Further, recent experimental and theoretical evidence suggests that understanding the morphological stability of a cancerous tumor may be important for controlling its spread to surrounding tissue. See for example the references [8,13,27,30,40,44,62].
Recently, we introduced a continuum diffuse interface model of multispecies tumor growth and tumor-induced angiogenesis in two and three dimensions for investigating morphological evolution [83]. Three dimensional simulations of nonlinear tumor growth using this model were presented in [83] and in [13,40,39] where neovascularization was coupled with growth. However, the full details of the numerical algorithm used for the diffuse-interface model in [83] have not been presented. This is done here.
In the diffuse approach, sharp interfaces are replaced by narrow transition layers that arise due to differential adhesive forces among the cell-species. In [83] we introduced a continuum model of adhesion using the Cahn-Hilliard framework [24], which originated from the theory of phase transitions, and is used extensively in materials science and multiphase fluid flow. In related work, Khain and Sander [56] recently presented a Cahn-Hilliard type model for a single tumor species. Armstrong et al. [10] and later Gerisch and Chaplain [45] recently developed nonlocal models of cell-adhesion.
A diffuse interface formulation (i) eliminates the need to enforce complicated boundary conditions across the tumor/host-tissue and other species/species interfaces that would have to be satisfied if the interfaces were assumed sharp, and (ii) eliminates the need to explicitly track the position of interfaces, as is required in the sharp interface framework. The diffuse interface model in [83] is thermodynamically consistent and capable of providing a detailed description of tumor progression, including variable cell-cell and cell-extracellular matrix interactions. The model is related to recently developed multicomponent mixture models, e.g., [4,5,9,15,16,22,23,26,29,36–38,78] that were introduced to study the dynamics of multispecies tumor growth. In the mixture approach, mass, momentum and energy balances are introduced for each species and, as such, provide the potential for significantly greater modeling detail than in single phase tumor models like those considered earlier in [15,16,19,20,22,25,30,43,49,52,62,64,65,85] to list a few. Other references may be found in the review articles listed above.
There are a variety of methods now available for performing nonlinear simulations of solid tumor growth. These include cellular automaton and agent based models (e.g., see [1,2,6,8,11,12,21,32,33,46,47,55,59,70–72]), single-phase continuum tumor growth models (e.g., see [30,52,60,62–65,76,85]) and continuum multiphase mixture models (e.g., see [29,68] and the mixture model references listed above). While discrete models may translate biological processes into model rules more easily than continuum approaches, discrete models can be difficult to parametrize with biophysical data and study analytically. Further, the associated computational cost rapidly increases with the number of cells modeled, which makes it difficult to simulate millimeter or greater sized tumors. In larger-scale systems (millimeter to centimeter scale), continuum methods provide a good modeling alternative and mixture models provide the capability of simulating in detail the interactions among multiple cell species and extracellular matrix. Hybrid continuum-discrete models that combine both discrete and continuum descriptions of cells (e.g., [13,58,77]) are very promising and have the potential to exploit the best features of both approaches, although more work is necessary to make these models competitive with the continuum approach at large scales.
Because the diffuse interface model involves high-order equations and the solutions are characterized by having narrow transition layers, it is challenging to develop accurate and efficient numerical methods. For example, explicit methods would result in severe time step restrictions, while the presence of transition layers requires the resolution multiple spatial scales. To solve the diffuse interface system numerically, we develop a new and very efficient, fully adaptive, nonlinear multigrid/finite difference method. The algorithm is semi-implicit in time and the nonlinear equations at the implicit time level are solved using a nonlinear multigrid finite difference method with nearly optimal complexity, meaning that the computational cost of the algorithm is nearly proportional to the total number of mesh points. To efficiently resolve the multiple spatial scales, an adaptive block-structured Cartesian mesh is used. In particular, an adaptive mesh can enable one to resolve the small-scale transition layers of the cell density functions, as well as the large-scale morphological structure of the tumor using as few spatial mesh points as possible. Locally refined block-structured Cartesian meshes strike a balance between grid structure and efficiency and are natural to use in conjunction with multilevel, multigrid methods. Here, we follow the methodology we recently developed in [82].
We present numerical simulations of tumor growth in 2D and 3D that demonstrate the capabilities of the algorithm in accurately and efficiently simulating the progression of tumors with complex morphologies. We show that the algorithm using an adaptively refined mesh is significantly more efficient than that using a uniform mesh, and we demonstrate numerically the convergence of the algorithm in space and time.
The outline of the remainder of the paper is as follows. In Sec. 2, we present the nondimensional model from [83]. In Sec. 3, the adaptive nonlinear multigrid method for solving the model is fully described and in Sec. 4 some numerical results are presented. In Sec. 5 the paper is summarized and future work is discussed.
2 The mathematical model
In this section we recall the modeling framework from [39,83] for describing multispecies tumor growth. Consider a bounded, open tissue domain Ω ⊂ R3 in which a tumor is evolving. We describe the tumoral and healthy tissues using continuous volume fractions that are defined over the whole of Ω. Because adhesive forces bind the tumor cells together, the tumoral and healthy tissues tend to phase separate, and a boundary layer of finite thickness forms between the spatially localized healthy and tumoral tissue domains. See Fig. B.1. This is referred to as a diffuse interface framework and is one that is common in materials science and multiphase fluids. To keep the presentation to a reasonable length, we will use the simplest formulation of the multispecies model from [83] that involves densities for viable and dead tumor tissue, healthy tissue, and extracellular fluid. In [39] we generalized this model by adding more than one viable tumor species, so that, for example, one could simulate mutation from one viable tumor species to another. In [39,40] we also included angiogenesis, chemotaxis, haptotaxis, and other phenomena involved in realistic in vivo tumor growth that we will neglect in the discussion here. The model below should therefore be viewed within a much more general context. However, the numerical algorithm presented here is essentially independent of the specific modeling choices within our more general framework.
B.1.
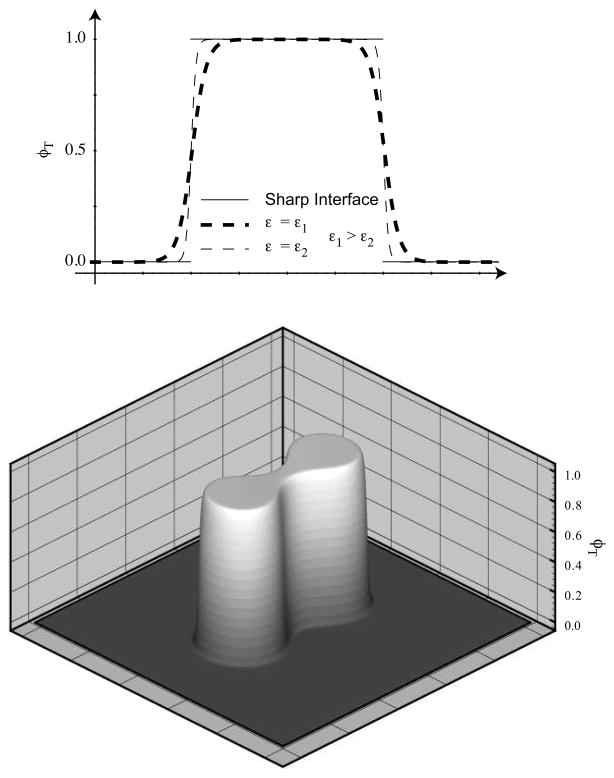
Top: Representation of a one dimensional tumor by the continuous function φT. As ε is reduced (from ε1 to ε2), the thickness of the diffuse interfacial region is reduced. For comparison we plot the sharp interface representation of the tumor as well. Bottom: Representation of a two dimensional tumor by the function φT. Apart from the region through which it has a large gradient, the range of φT is essentially {0, 1}. φT = 1 in the tumor region, and φT = 0 in the host tissue region.
Following [83], the dimensionless dependent variables defined on Ω are
φW, the volume fraction of extracellular fluid,
φV, the volume fraction of viable tumor tissue,
φD, the volume fraction of dead tumor tissue,
φH, the volume fraction of healthy tissue,
uW, the velocity of extracellular fluid,
uS, the tissue velocity,
q, the extra-cellular fluid pressure,
p, the cell-to-cell (solid) pressure,
n, the concentration of nutrient.
We assume that the densities of the components are matched (set to 1). In [83], we assumed that the extracellular fluid volume fraction is everywhere constant:
| (1) |
This limits the coupling between the solid fraction (cells) and water. We further assume that cells close-pack, i.e.,
| (2) |
where 1 is the normalized close-packing volume fraction. Hence the sum of all volume fractions is a constant. The model simplifies if we utilize the total tumor volume fraction
| (3) |
We refer the reader to [83] for further explanation.
The evolution equation for φT is the following Cahn-Hilliard-type advection-reaction-diffusion equation:
| (4) |
| (5) |
where M > 0 is the mobility constant related to phase separation between tumoral and healthy tissue; ST is the net source of tumor cells specified later in section 4; f(φ) = φ2(1 − φ)2/4 is the quartic double-well potential; ε > 0 is the parameter specifying the thickness of the interface between healthy and tumoral tissue (Fig B.1).
Rather than solving for φV, a dynamical equation for the volume fraction of dead tissue φD is used instead:
| (6) |
where SD is the net source of dead cells, which is also specified later in section 4. Knowing φT and φD, the viable tumor tissue volume fraction is calculated as φV = φT − φD, following Eq. (3).
The tissue velocity is
| (7) |
where κ > 0 is the tissue motility function and γ ≥ 0 is a measure of the excess adhesion force at the diffuse tumor/host-tissue interface. Assuming no proliferation or death of the host tissue, the velocity is constrained to satisfy
| (8) |
Together, Eqs. (7) and (8) constitute a Poisson equation for the solid pressure p:
| (9) |
Knowing the pressure p we back calculate uS using Eq. (7).
The extracellular fluid velocity satisfies
| (10) |
where κW > 0 is the fluid motility; the velocity is constrained such that
| (11) |
Thus the extracellular fluid flows contrary to the flow of tissue cells. Moreover, the exchange of mass between water and cells precisely accounts for tissue growth (both positive and negative). Equations (10) and (11) constitute a Poisson equation for the fluid pressure:
| (12) |
We note that the fluid velocity and pressure may be calculated as a postprocessing step because the solid and fluid problems are decoupled by assumption.
The nondimensional quasi-steady nutrient equation is
| (13) |
where we have assumed (i) that nutrient diffusion occurs on a much faster time scale (e.g., minutes) than the cell-proliferation time scale (e.g., day or more) and (ii) that nutrient uptake by healthy or dead tumor tissue is negligible compared with the uptake by viable tumor tissue. The diffusion coefficient and nutrient capillary source term are, respectively,
| (14) |
| (15) |
where DH is the nutrient diffusivity in the healthy tissue (relative to the tumoral tissue); and are constants specifying the degree of pre-existing uniform vascularization; and nC ≥ 0 is the nutrient level in the capillaries. Q(φ) is an interpolation function defined as
| (16) |
In either case Q(1/2) = 1/2, although one choice is smoother than the other.
The model equations are valid throughout Ω, and there are no internal boundary conditions required for φT, φD, or any other variables. For outer-boundary conditions, we choose
| (17) |
where ζ is the outward-pointing unit normal on the outer boundary ∂Ω. The conditions μ = p = q = 0 allow for the free flow of cells and water across the outer boundary in order to accommodate growth. As discussed in [83], classical single-phase tumor models (e.g., [30,60,63]) may be recovered as sharp-interface limits of this model as ε → 0.
3 Details of the numerical algorithm
Here we describe the discretization of governing equations and the adaptive multigrid method that solves the resulting coupled nonlinear system of equations. Without loss of generality, since the adaptivity is based on block-structured refinement, which uses a collection of rectangular, uniform grids, we may restrict some of the description of the method to a uniform grid. The essential difference between the classical uniform multigrid and block-structured, locally-refined multigrid may be gleaned from Fig. B.2. The general procedure for extending uniform multigrid to the locally-refined, block-structured Cartesian mesh setting is described fully in [82]. We will only give a brief description of this extension later.
B.2.
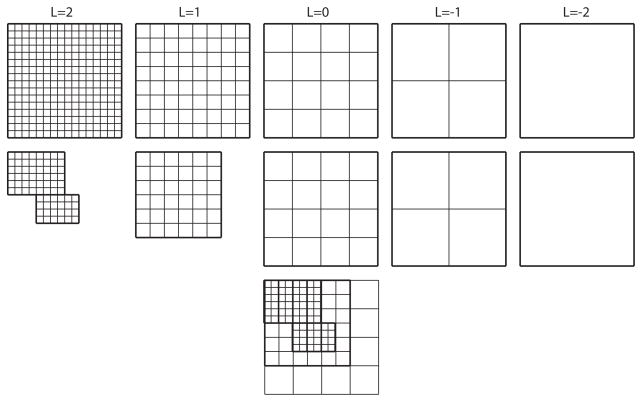
A hierarchical representation of grid data in the uniform multigrid setting (top), and in the block-structured, locally-refined multigrid setting (middle). The composite mesh related to the locally-refined case is shown at the bottom. Level L = 0 is called the root level. Levels L = 1 and L = 2 are called refinement levels. At the root level and below the grids of the uniform and locally refined multigrid hierarchy are the same.
3.1 Uniform FAS multigrid
3.1.1 Finite difference discretization
To shorten the exposition, we assume some familiarity on the part of the reader with the principle components of multigrid, as can be obtained from [17,79,82]. Suppose that the computational tissue domain Ω is rectangular, i.e., Ω = (0, Nxh) × (0, Nyh), where Nx and Ny are positive integers, and h > 0. h is called the grid spacing. Define
| (18) |
where r is an integer or half integer. We specify three sets of uniform grid points: (i) east-west edge points, Eew, (ii) north-south edge points, Ens, (iii) cell-centered points, C, defined via
| (19) |
| (20) |
| (21) |
Real-valued grid functions whose domains equal Eew are called east-west edge-centered functions and are identified via ; those whose domains equal Ens are called north-south edge-centered functions and are identified via ; and those whose domains equal C are called cell-centered functions and are identified via φi,j := φ(xi, yj). The velocities uS and uW are approximated as edge-centered functions. For example, writing is approximated as an east-west edge-centered function, and is approximated as a north-south edge-centered function. All of the other dependent variables are approximated as cell-centered functions.
To complete the spatial discretization, we replace spatial derivatives by difference operators. The Laplacian operator is approximated to second order by
| (22) |
where φ is cell-centered. The Laplacian with non-constant diffusivity/mobility is approximated to second order via
| (23) |
where both φ and m are assumed to be cell-centered, and Ax and Ay are the averaging operators defined component-wise as
| (24) |
To calculate the advection term ∇ · (uSφ), where φ is cell-centered and uS is the edge-centered cell velocity, we use the third-order WENO reconstruction method [54], with the simple upwind flux. In particular, we approximate ∇ · (uSφ) by
| (25) |
The field f = (few, fns) is the numerical upwind flux and is determined by
| (26) |
| (27) |
where is the upwind WENO reconstruction of φ to the east-west cell edges, and is the upwind WENO reconstruction of φ to the north-south cell edges.
For notational simplicity, we define
| (28) |
where the * is a place-holder for any superscripted indices. To denote the array of cell centered values of a particular field we drop the subscripts. For example, ψk is the 5 · Nx · Ny vector of cell-centered values of the fields at time step k. Note that we drop uS from the solution vector since it is a derived quantity of the pressure and other fields. We also do not consider the water problem herein, since it decouples from the problem for the solid volume factions. Water pressure, q, and velocity, uW, can be calculated as a post-processing step using techniques similar to those described below.
Since the equations are fourth-order in space (i.e., of Cahn-Hilliard-type), they are numerically “stiff” and explicit time step methods require the time step restriction s < Ch4 for stability. We therefore discretize the equations in time using an implicit Crank-Nicholson-type approximation [57,66,82]. Denoting the fixed time step-size s, and indexing the time iterations by a superscript k, the discrete equations corresponding to (4), (5), and (6) are
| (29) |
| (30) |
| (31) |
Equations (9) and (13) are discretized in a fashion similar to that above:
| (32) |
| (33) |
where
| (34) |
is a nutrient source term due to a uniform, preexisting capillary network. Note that our discretization is second-order in both time and space, apart from the explicit treatment of the excess surface adhesion term in Eq. (32). The latter reduces the local truncation error to only first-order in time. Extensive numerical tests indicated very poor multigrid convergence when the term ∇d · (κ (φT, φD) μ∇d φT)i,j was treated implicitly in Eq. (32). The convergence test reported on later in Sec. 4.2 (see also Tab. A.1) suggests that the global convergence rate is first-order. As we explain later, even an improvement of the treatment of the excess surface adhesion term does not improve the global convergence rate.
Table A.1.
Nondimensional parameters used in the two-dimensional numerical simulation shown in Fig. B.3 and the three-dimensional simulations shown in Figs. B.5 and B.5.
|
|
0.5 |
|
0.0 | ||
| nC | 1.0 | κ | 1.0 | ||
| DH | 1.0 | nN | 0.4 | ||
| λA | 0.0 | λN | 3.0 | ||
| λL | 1.0 | λ | −0.1 (Fig. B.3) 0.0 (Figs. B.5a and B.6) 0.1 (Fig. B.5b) |
||
| M | 10.0 | ε | 0.1 |
We finally mention a modification that is incorporated in order to avoid negative values of the mobility functions at the numerical level. This is especially needed in the smoothing, detailed later, since this operation does not guarantee positivity of iterates. Denote MT (φT) = M × φT and, likewise, MD (φD) = M × φD. These functions are replaced by
| (35) |
| (36) |
respectively. We suppress writing these replacements in the sequel to preserve the clarity of the exposition.
3.1.2 Separation into source and operator terms
As is standard in multigrid methods, the discrete equations (29)–(33) are split into source terms F̃(ν) and operator terms, Ñ(ν), where Ñ(ν) = F̃(ν), for ν = 1, … 5. The F̃(ν) are defined as
| (37) |
| (38) |
| (39) |
| (40) |
| (41) |
Note that there are terms that depend on the solution at the implicit time step k. These terms will be lagged in the multigrid V-cycle iteration as described below.
The operator terms Ñ(ν) are defined as
| (42) |
| (43) |
| (44) |
| (45) |
| (46) |
Defining Ñ = (Ñ(1), …, Ñ(5)), and F̃ = (F̃(1), …, F̃(5)), it is clear that the discrete system (29)–(33) is equivalent to Ñ = F̃, a 5 × Nx × Ny nonlinear vector equation. Note that we have neglected writing the discrete boundary conditions for brevity. These may be handled in a straightforward way.
3.1.3 V-cycle algorithm
We shall assume that, for some reasonable time step-size s, a unique solution ψk exists for the equation Ñ = F̃. We aim to construct a sequence that converges to the solution as m → ∞. To construct this sequence we redefine the source and operator terms as,
| (47) |
| (48) |
| (49) |
| (50) |
| (51) |
and
| (52) |
| (53) |
| (54) |
| (55) |
| (56) |
To construct the next iterate ψk,m from the previous iterate ψk,m−1 we solve the equation
| (57) |
for ψk,m. This is written in operator form as
| (58) |
It should be clear that a fixed point solution of Eq. (57) is a solution to Ñ = F̃. The following pseudo-code gives the essence of the algorithm for generating an approximate solution to Ñ = F̃ by this fixed point method:
Fixed Point Algorithm
| set φk,0 = ψk−1 |
| Solve loop: for m = 1 until mmax |
| ψk,m = Solve (N(·, ψk,m−1) = F(ψk,m−1, ψk−1) |
| if ||F(ψk,m, ψk−1) − N(ψk,m, ψk,m)|| < tol exit Solve loop |
| end for Solve loop |
| set ψk = ψk,m |
In the algorithm above we have abused notation by setting ψk = ψk,m, because the object on the right-hand-side of the equal sign is only an approximate solution to the original discrete system (29)–(33). Also we are neglecting the theoretical justification that the sequence generated by this algorithm does indeed converge to a unique fixed point solution of the system (29)–(33). In practice, we find that solutions exist, with reasonable constraints on the time step size s, and that our algorithm converges rapidly to a solution.
It is not practical to expect to be able to solve Eq. (57) exactly. In other words, the operator in (58) must be replaced with an approximate solver. Here, as in [82], we use a single FAS (Full Approximation Storage) multigrid V-cycle iteration as an approximate solver. In particular, we define
| (59) |
to mean obtain an approximate solution ψ★ of Eq. (57) using a single FAS V-cycle iteration with ψk,m−1 as the initial guess.
The FAS V-cycle framework in multigrid is a way to accelerate a classical local iterative method (such as the Jacobi, Gauss-Seidel, and SOR method) by applying it on hierarchy of representations of the grid data. See Fig B.2 (top). This local iterative method is called the smoother in the multigrid setting. In FAS multigrid, nonlinearity is handled by performing local Newton linearizations inside the smoother [57,79], rather than performing global Newton linearizations. Herein, we use Gauss-Seidel as our smoother. In the V-cycle, the smoother is performed first on the finest grid, and then on successively coarser grids in the mesh, until a coarsest grid is reached. (See again Fig. B.2 (top), and note that coarser grids are viewed as being “below” the finer grids.) Then the smoother is performed on successively finer grids, where the fine grid solutions are corrected by the coarser grid results, until the finest grid is reached.
The upshot is that, by applying the smoother to a hierarchy of representations of the data, communication among grid points is effectively accelerated compared to that of the classical iterative methods, and hence convergence may be accelerated.
The solution algorithm becomes
FAS Algorithm
| set ψk,0 = ψk−1 |
| Vcycle loop: for m = 1 until mmax |
| ψ★ = Vcycle (ψk,m−1, N(·, ψk,m−1) = F(ψk,m−1, ψk−1) |
| set ψk,m = ψ★ |
| if ||F(ψk,m, ψk−1) − N(ψk,m, ψk,m)|| < tol exit Vcycle loop |
| end for Vcycle loop |
| set ψk = ψk,m |
We comment that it has not been shown rigorously that the sequence generated by this algorithm converges to the solution of Ñ = F̃, however, convergence is seen in practice with reasonable constraints on s. The norm used in the test for convergence is a scaled discrete L2 norm.
3.1.4 Nonlinear smoothing
To complete the description of the FAS V-cycle scheme, we finally detail the nonlinear Gauss-Seidel smoother used in the method. Here we describe the Gauss-Seidel scheme using the lexicographic ordering, but in practice we use the Red-Black ordering. The latter is harder to describe, but can speed up the convergence significantly with respect to the former [79].
Let ℓ be the index of the smoothing sweeps. One full sweep will consist of visiting every point of the grid one time in lexicographic order. The description simplifies if we introduce the following center-to-edge averages:
and, similarly,
The center-to-edge averages for κ (·, ·) and D(·) are defined similarly and are suppressed for brevity.
One sweep of the smoothing operator is defined iteratively in the following way: stepping lexicographically from (i, j) = (1, 1) to (i, j) = (Nx, Ny), at each cell center (i, j) find the vector solution of
| (60) |
| (61) |
| (62) |
| (63) |
| (64) |
Note that the five unknowns , and appear on the left-hand-sides of the five equations; everything on the right-hand-sides is known. The last two equations, (63) and (64), are completely decoupled from the others, and are solved by simple division. The first two equations, (60) and (61), are weakly coupled with the third, (62). We invert (60) and (61) using 2×2 Cramer’s rule [57], solving for and , then subsequently solve (62) for . We represent ℓ = λ full sweeps of the smoothing operator via
| (65) |
where ψk,m,0 is the initialization (ℓ = 0) of the smoother.
3.2 Block-structured, locally-refined FAS multigrid
With all of the components of the uniform FAS V-cycle defined, we refer the reader to [82, Sec. 4.2] for the precise details of how to put these pieces into the block-structured, adaptive mesh framework. Here we touch briefly on some of the aspects of the adaptive mesh. The basic rationale for using the adaptive block-structured framework is (i) adaptivity allows one to accurately resolve phenomena on various spatial scales, and (ii) it is straightforward to extend uniform FAS multigrid to this setting. In particular, the algorithm from [82] allows for a very fine resolution of diffuse interfaces without expending computational resources where fine resolution may be unnecessary. The numerical degrees of freedom can be very nearly minimized, and since the solver is based on the multigrid algorithm, the resulting discrete equations can be solved with nearly optimal complexity [79,82]. Here, we very briefly describe the algorithm for a two-dimensional computational domain. The extension to three dimensions is straightforward.
Consider the simplified block-structured, locally-refined Cartesian mesh in Fig. B.2 (middle and bottom). The composite mesh (bottom of Fig. B.2) consists of the grids from levels L = 0, L = 1, and L = 2, where finer resolution data replaces coarser data underneath. Level L = 0 is called the root level. The root-level grid consists of a single rectangular, uniform 4 × 4 grid of mesh spacing h0. The next level, the level-1 mesh, consists of a union of rectangular grid patches. In this case, there is only one 6×6 grid in the level-1 mesh. The uniform spacing of cells in the level-1 mesh, h1, is always half of h0. The next level of refinement is called level 2, and again consists of a union of uniform rectangular grids, with mesh spacing h2 = h1/2. In Fig. B.2, there are two grids in the level-2 mesh. Each new “child” level mesh lives over top of the coarser “parent” level mesh [82] with the mesh-spacings always decreasing by with each increasing level of refinement. Here, the level L= 1 grid is the parent of the two level L = 2 child grids. We say that the composite mesh in Fig B.2 has an effective resolution of 16 × 16, meaning that to obtain the equivalent resolution using a uniform grid, the grid would have size 16 × 16 (Fig. B.2 (top)). It is clear that the composite mesh can have significantly fewer cells than than an equivalent uniform mesh. The grids that go below the root level, e.g., the level L = −1, and L = −2 grids in Fig. B.2, are not seen in the composite mesh, but are necessary for the multigrid algorithm, as explained in [79,82].
Where to add child level mesh in the parent depends upon certain refinement criteria. Herein we use a single, simple refine criterion. We refine the mesh in areas where the volume fractions have a steep gradient. In particular, the volume fraction of tumor cells has a steep gradient at the tumor/healthy tissue interface. The refinement in Fig. B.3 is such that there are approximately 8 mesh cells in the finest level between the level curves φT = 0.1 and φT = 0.9. We also refine the mesh so as to have sufficient resolution of the necrotic interface, i.e., the region between the level curves φD = 0.1 and φD ≈ 0.9.
B.3.
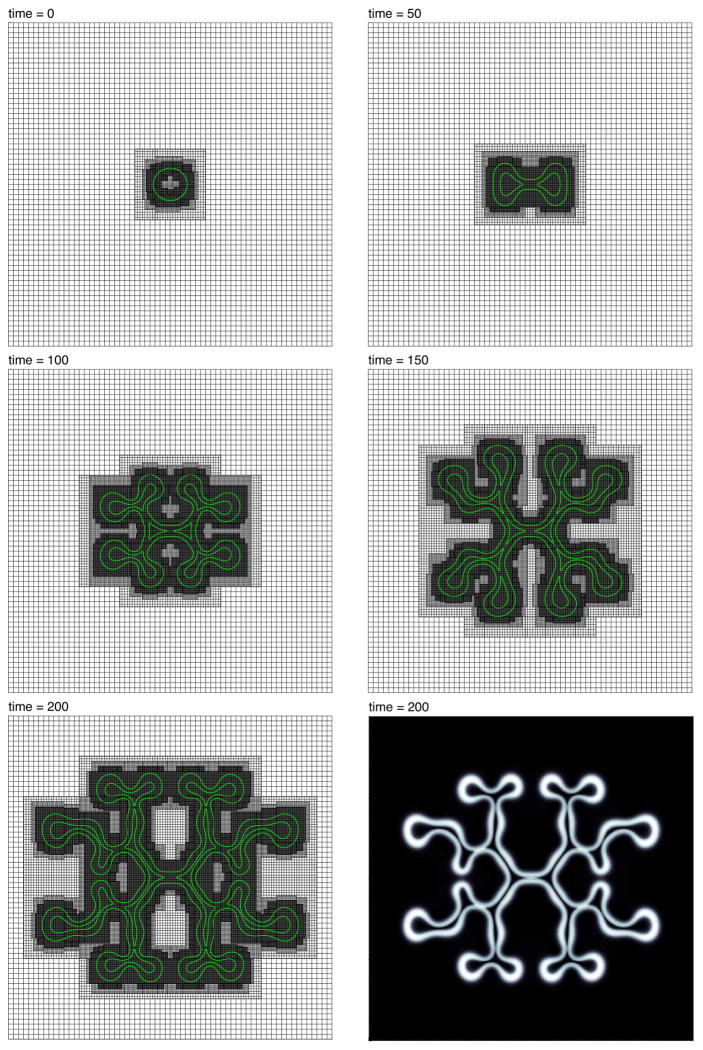
(color figure) The evolution of the contours φV = 0.5 (green), together with the adaptive mesh, during growth. The viable cells are primarily contained between the inner and outer contours. The biophysical parameters are given in Tab. A.1. There are three levels of refinement; each level of refinement has half the grid spacing of the one on the level below. The coarsest level, level 0, is 642. The finest level, level 3, has the equivalent resolution of a 5122 mesh. The step sizes are s = 1 × 10−2 and h = 40/512. The bottom right image shows the full viable cell region. White corresponds to φV ≈ 1, black, φV ≈ 0.
Note that the refinement criterion may be modified when an angiogenesis algorithm, that describes tumor induced neovascularization, is coupled to the tumor solver, as was done in [40,13]. With neovascularization, refinement may be initiated around the growing blood vessels in addition to the interfaces in the system. A similar refinement scheme was used in [85] for coupled growth and neovascularization in 2D.
Once a new, locally-refined, block-structured Cartesian mesh is constructed data are copied and/or interpolated into the new composite mesh, the governing equations are discretized on the composite mesh, and the resulting nonlinear system of equations is solved according to the nonlinear FAS multi-grid method. The basic methodology of refining, interpolating, and solving is detailed in [82].
4 Numerical results
By h we denote the smallest mesh-spacing in the composite mesh. In other words, h = hLmax, where Lmax is the number of levels or refinement. By s we denote the fixed time-step size for a simulation. We will assume for simplicity that the cell motility is κ (·, t) ≡ 1. Following [83], we assume that the net source of tumors cells ST is due to cell proliferation (with proliferation rate normalized to 1) and the lysing of dead cells to form water with rate λ L ≥ 0:
| (66) |
where G(x) is a continuous cut-off function,
| (67) |
which suppresses the growth of small clusters of viable tumor cells outside the tumor environment. Such a cutoff is not needed when ε is sufficiently small. Equation (4) drives the phase separation of healthy and tumoral tissue and also accounts for the growth of the tumor. The net source of dead cells is assumed to arise from apoptosis and necrosis with respective rates λA ≥ 0 and λN ≥ 0:
| (68) |
where 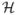 is the Heaviside function; and nN ≥ 0 is the necrotic limit, below which tumor tissue dies for lack of nutrient.
is the Heaviside function; and nN ≥ 0 is the necrotic limit, below which tumor tissue dies for lack of nutrient.
4.1 2D simulations
We first consider a two-dimensional simulation of a tumor growing in a nutrient-rich environment. The simulation is depicted in Fig. B.3, and the parameters are given in Tab. A.1. Note that γ = −0.1, the significance of which we explain below. The grid parameters are the following: the domain is Ω = (0, 40) × (0, 40) and a root-level grid of size 642 covers all of Ω. Three levels of refinement are used, so that the mesh has an equivalent resolution of a uniform grid of size 5122. The step sizes are s = 1.0×10−2 and h = 40/512. As pointed out earlier, refinement is sought where the gradients of the cell densities are steep.
By setting we model a constant source of nutrient coming from a uniform, pre-existing vasculature outside the growing tumor. The coefficient indicates that no nutrient source exists within the tumor itself. Dead tumor cells begin to accumulate due to necrosis, at the rate indicated in Eq. (68), since λN > 0. This is the only non-negligible source of dead cells, since the apoptosis rate is zero (λA = 0).
By setting γ = −0.1, we have introduced negative excess surface adhesion, which manifests itself mathematically as negative diffusion. This however is offset to some degree by the positive diffusion naturally inherent in the diffuse interface framework. It is well-known that there is a positive surface-tension-like force due to the presence of the diffuse interface. This force diminishes to zero, in the present scaling, as ε → 0, but is certainly non-zero for ε ≠ 0. The effect of negative γ is to make perturbations of the tumor/healthy-tissue interface more unstable than for γ ≥ 0, as may be seen comparing Figs. B.3 and B.5. The net surface adhesion “term” (or contribution) for the present parameters is still positive diffusive and thus well-posed. However, for γ sufficiently negative, the net term may become negative diffusive, and lead to ill-posedness.
B.5.
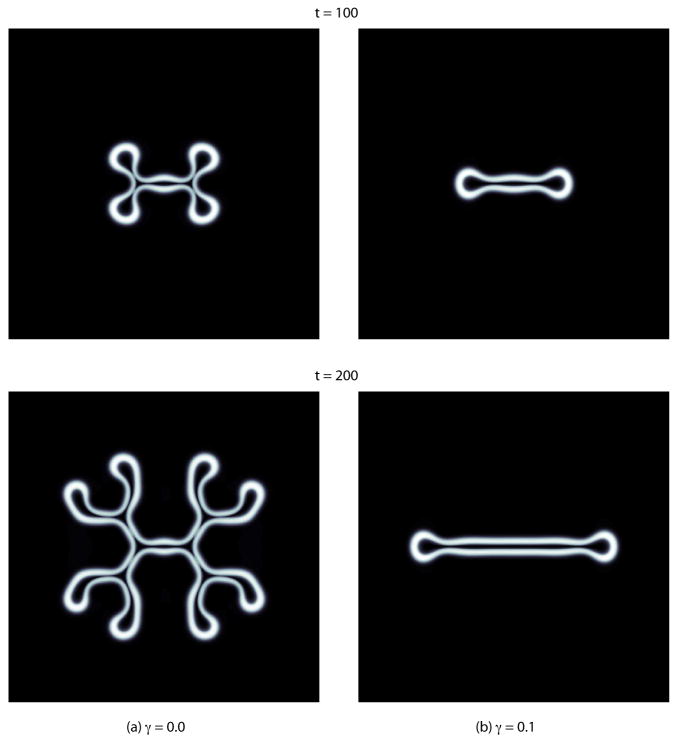
The evolution of the φV fields for (a) γ = 0.0, and (b) γ = 0.1, where all other parameters (biophysical and mesh related) are the same as for the simulation depicted in Fig. B.3. See Tab. A.1 for the biophysical parameters. White corresponds to φV ≈ 1, black, φV ≈ 0. The enclosed black regions indicate necrotic (dead) tumoral tissue.
The initial tumor shape for the simulation in Fig. B.3 is a slightly elliptical mass with a diffuse interface. The φT = 0.5 level curve is given by {(x, y)|(x − 20)2/1.1+(y − 20)2 = 22}; thus, the shape bulges slightly along the x-direction. There are initially no dead cells and, hence, φD(·, t) ≡ 0. Dead cells do accumulate as the mass grows due to necrosis. The dark region in the interior of the tumor has a high concentration of dead cells (see grayscale images in Fig. B.3, t = 200, and Fig. B.5). Due to the limitation of nutrient, and the resulting mass loss in the interior of shape, the tumor grows unstably as predicted in the context of sharp-interface models by Cristini et al. [30] and Li et al. [60]. In particular, the tumor increases its boundary length to interior area ratio in order to flux as much nutrient across the tumor/healthy tissue interface as possible. The effect is explained in more detail in [30,60,63,83].
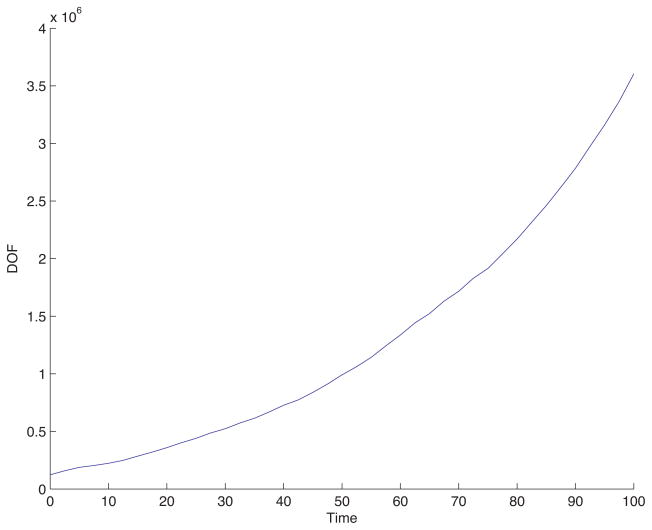
The adaptive mesh, follows the growth and morphological instability of the tumor, as it seeks to adequately resolve the gradients of the cell densities. Since the mass is small initially, the degrees of freedom (DOF) of the mesh are similarly small. Figure B.4 shows a plot of the DOF of the mesh versus time for the simulation in Fig. B.3. The composite mesh initially has 9,445 DOF, and ends with 108,508 DOF, more than a 10 fold increase. Near t = 0 the solver completed 250 time steps in under one minute, whereas, near t = 200, 250 time steps were completed in about 13 minutes. The total computation time was 11 hours, 38 minutes, where the simulation was executed as a serial process on a 3.0GHz Dual Quad-Core Intel Xeon processor, with 8GB of memory. The same computation using a uniform mesh of equivalent resolution, 5122, would require about 41 hours of computation time, a conservative estimate, which represents nearly a 4 fold increase with respect to the adaptive simulation. The computational savings in 3D may be much more dramatic as seen below.
B.4.
The degrees of freedom (number of cell-centered mesh points) in the spatially adaptive simulation from Fig. B.3 as a function of time.
Figure B.5 demonstrates that increasing the excess surface adhesion γ decreases the degree of instability in the morphological growth of the tumor. The biophysical parameters for the simulations in Fig. B.5 are given in Tab. A.1, and are the same as those for the simulation shown in Fig. B.3, except that γ = 0.0 in Fig. B.5a and γ = 0.1 in Fig. B.5b. The initial condition for both simulations is the same as that depicted in Fig. B.3, a slightly elliptical initial tumor. The mesh parameters and step sizes are likewise the same. The domain is Ω = (0, 40) × (0, 40).
The dynamics of the simulations depicted in Figs. B.3 and B.5a are comparable, though the rate of growth of the instability is markedly faster in Fig. B.3. Note that the frame at t = 150 in Fig. B.3 compares well with the frame at t = 200 in Fig. B.5a. On the other hand, for γ = 0.1, the simulation for which is depicted in Fig. B.5b, the tumor mass is only unstable along the x-direction, and a long, finger-like shape emerges. For γ sufficiently large, the initial shape will become circular, develop an inner core filled with dead cells (a necreotic core), and will thereafter remain circular. These results are consistent with earlier predictions of sharp-interface tumor models (e.g. [30,60,63]).
4.2 A 2D convergence test
Here, we report a numerical convergence test to determine empirically whether on decreasing h (the smallest grid spacing in the adaptive mesh) and decreasing s (the temporal time step size) the error in the approximation decreases, and at what rate the error is decreasing. The test is the same as described in [82, Sec. 5.1]; we test only the convergence of the field φT. To compare the values of φT on coarse and fine meshes, averaging is used because φT is defined on cell-centers. Further, since the composite meshes are typically different for the coarse and fine meshes, we compare results by interpolating the data on the composite mesh to a uniform mesh with the grid size corresponding to the finest mesh hLmax. The biophysical parameters used in the convergence test are the same as those for the simulation in Fig. B.5b, and are given in Tab. A.1. The initial shape of the tumor is elliptical, exactly the same as for the simulations in Fig. B.3 and B.5, such that the φT = 0.5 level curve is given by {(x, y)|(x − 20)2/1.1 + (y − 20)2 = 22}. The domain is Ω = (0, 40) × (0, 40).
Three computations are performed for the test, where the root-level grid has sizes 322, 642, and 1282, respectively. In each calculation there are three levels of refinement. The step sizes are, respectively, h = 40/256 and s = 4 × 10−3 (root level grid 322); h = 40/512 and s = 2 × 10−3 (root level grid 642), and h = 40/1024 and s = 1 × 10−3 (root level grid 1282). In other words, we have taken the linear refinement path s = 0.0256 h. The results are given in Tab. A.2. A first-order convergence rate is observed for the test.
Table A.2.
Errors and convergence rate for an adaptive simulation of a two-dimensional growing tumor. Parameters are given in Tab. A.1. In particular, λ = 0.1, as in Fig. B.5b. The final time is tf = 20, and three levels of refinement are used. The step sizes are, respectively, h = 40/256 and s = 4 × 10−3 (root level grid 322); h = 40/512 and s = 2 × 10−3 (root level grid 642), and h = 40/1024 and s = 1 × 10−3 (root level grid 1282). The initial data are described in the text. First-order convergence is observed.
| root-level grid sizes | 322 − 642 | 642 − 1282 |
| error | 5.34807×10−3 | 2.67776 × 10−3 |
| rate | 0.99807 | |
The first-order result should be expected because of the treatment of the excess surface adhesion term. (See the discussion following Eq. (32)). However, even when this term is omitted (by setting γ = 0), first-order convergence is still observed, when one might have expected second-oreder convergence. There are several possible explanations for why this might be observed. First, our refinement path for our step sizes (s, h) might still be outside of the region where asymptotic convergence is observed. Thus we could take a different path using smaller time and space steps and retest the convergence. Second, because we use non-smooth functions in our model, one would not expect high-order convergence for any refinement path. This second reason is by far the more plausible. Indeed, in our definitions of G (67), Q (16), (35), and (36) we use non-smooth functions. While these definitions are reasonable in the sense of formal asymptotics (as ε → 0), they are very likely reducing the effective order of convergence. In principle, one could use smooth functions in the model, especially in the cases of G and Q. Finding the root cause of the reduced convergence will be the subject of future work.
4.3 A 3D simulation
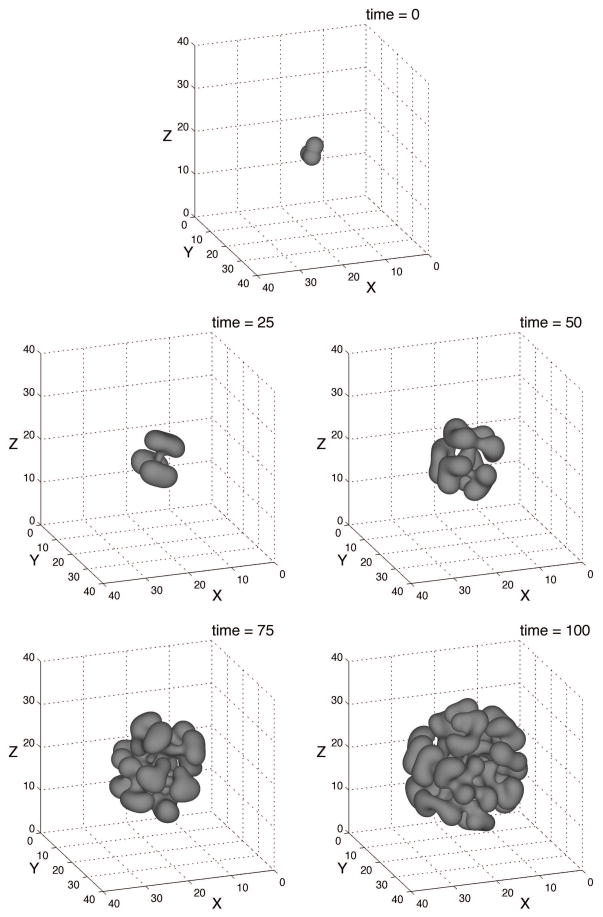
Finally we present a three-dimensional simulation of tumor growth in Figs. B.6, B.7, and B.8. The biophysical parameters are given in Tab. A.1, and are, for the most part, the same as those used in the previous two-dimensional simulation. Note that γ = 0.0 is used, as in the simulation depicted in Fig. B.5a. The initial shape of the φT = 0.5 isosurface is displayed in Fig. B.6 and the domain is Ω = (0, 40)3. The mesh parameters are the following: the root-level grid is 323, and there are three levels of refinement, as shown in Fig. B.7. Hence, h = 40/256 and the mesh has resolution equivalent to that of a uniform grid of size 2563. The time step size is s = 1 × 10−2.
B.6.
The evolution of the φT = 0.5 isosurface during the growth of an asymmetrical 3D tumor. The biophysical parameters are given in Tab. A.1 There are three levels of refinement. The root level, level 0, is 323, hence the composite mesh has the equivalent resolution of a uniform 2563 grid. The time step is s = 1 × 10−2.
B.7.
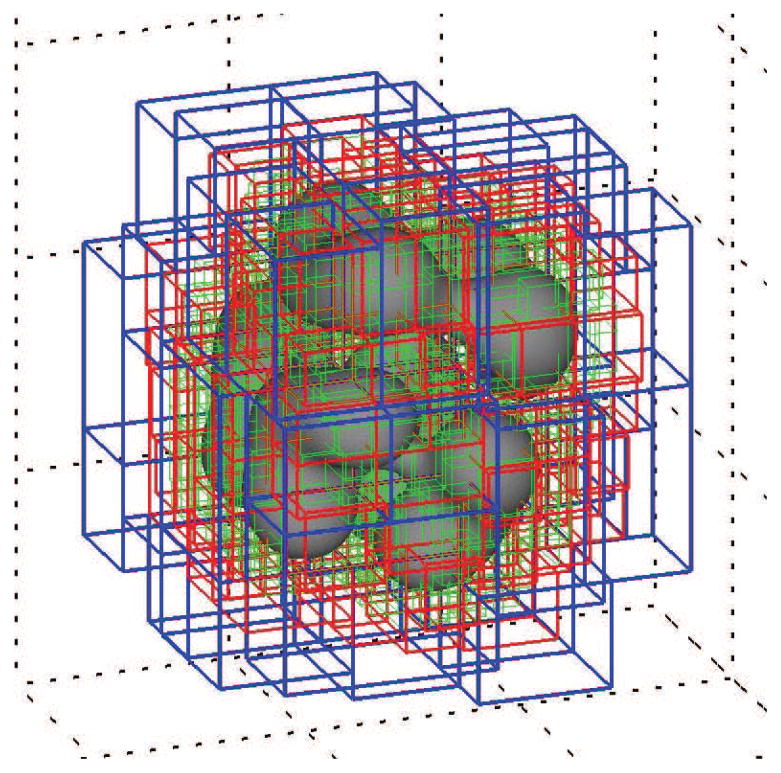
(color figure) The φT = 0.5 isosurface at time t = 50 from Fig. B.5 together with the bounding boxes of the block-structured mesh. The mesh spacings of the respective levels are h1 = 40/64 (blue), h2 = 40/128 (red), and h = h3 = 40/256 (green). There are precisely 991,201 degrees of freedom (number of cell-centered mesh points) in this composite mesh. See Fig. B.8.
B.8.
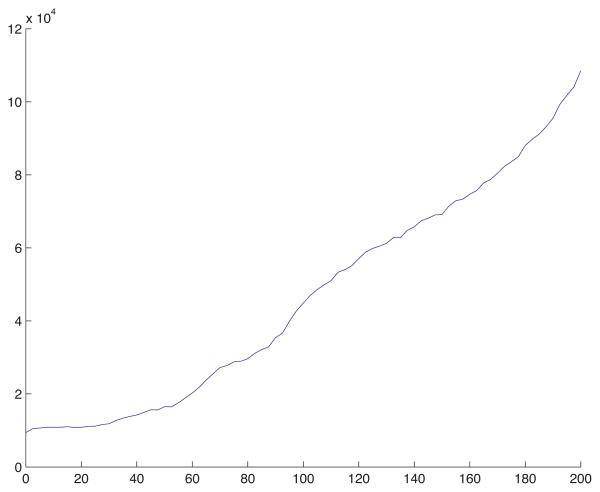
The degrees of freedom (number of cell-centered mesh points) in the 3D spatially adaptive simulation from Fig. B.6 as a function of time. This plot indicates a 30 fold increase in the DOF during the simulation.
The evolution of the φT = 0.5 isosurface is shown in Fig. B.6. As in the two dimensional case the tumor boundary is unstable due to weak adhesion forces, the limited nutrient supply and the resulting mass loss in the interior of the tumor. But, as expected from the analysis of a similar sharp-interface model [30,60], the three-dimensional tumor is even more unstable than its two-dimensional counterpart in Fig. B.5a, and a highly complex tumor morphology develops. The tumor repeatedly forms buds (similar to the sub-spheroid growth reported in experiments on glioblastoma tumor spheroids [41]) to increase its surface to volume ratio and increase access of interior cells to nutrients.
Figures B.7 and B.8 show some statistics of the adaptive mesh for the simulation presented in Fig. B.6. Figure B.7 shows the bounding boxes of the refinement patches in the mesh at t = 50. The blue are level-1 patches with step size h1 = 40/64; the red are level-2 patches with step size h2 = 40/128; and the green are level-3 patches with step size h = h3 = 40/256. There are 991,201 DOF in the composite mesh shown in Fig. B.7. Figure B.8 displays the degrees of freedom (DOF) as a function of time for the simulation in Fig. B.6. Initially the mesh has 122,872 DOF; at t = 50, as indicated in the caption of Fig. B.7, there are 991,201 DOF; and at the final time t = 100 there are 3,604,140 DOF. This represents a start-to-finish increase by a factor nearly 30.
Near t = 0 the solver completed 250 time steps in about 30 minutes, whereas, near t = 100, 250 time steps were completed in about 16 hours, 49 minutes. The total computation time was 211 hours, 35 minutes (about 9 days), where the simulation was executed as a serial process on a 3.0GHz Dual Quad-Core Intel Xeon processor, with 8GB of memory. The same computation using a uniform mesh of equivalent resolution, 2563, would require more than 3100 hours (about 130 days or just over 4 months) of computation time, a conservative estimate, which represents about a 14 fold increase with respect to the adaptive simulation.
5 Summary and Future Work
In this work we have described in detail the adaptive finite-difference, nonlinear multigrid solution of the diffuse interface tumor model, which was presented in [83] and was utilized in [39,40] and elsewhere. To overcome the severe time-step restrictions of explicit methods, we employed a semi-implicit time integration scheme. To resolve the small-scale transition layers in the cell densities, and, at the same time solve the discrete equations efficiently, we employed an adaptive multigrid scheme using a block structured Cartesian mesh. We previously used this mesh to efficiently solve a modified Cahn-Hilliard equation in [82].
We presented numerical simulations that suggest the convergence of the algorithm in space and time. The results indicated that the method is at least first-order convergent along a refinement path s ~ h. We also presented simulations of tumor growth in 2D and 3D that demonstrate the capabilities of the algorithm in accurately and efficiently simulating the progression of tumors with complex morphologies. We further showed that the algorithm using an adaptively refined mesh is significantly more efficient than that using a uniform mesh. In a 2D calculation we showed about a factor of 4 true computational savings was achieved using the adaptive implementation while and in a 3D calculation, about a factor of 14 savings was achieved.
In the future, we plan to extend this algorithm to incorporate adaptive time step control. We also plan to incorporate more biophysical effects such as multiple tumor cell and host species and to build upon recent work [13,58] to develop novel algorithms that simultaneously combine both continuum and discrete representations of cells. Also, we will work to make the solver fully second-order in time and space. We plan to redesign some elements to make use of the “energy stable” algorithms that we developed in our work [53,84] for the phase field crystal (PFC) equation and in our work [81] for the Cahn-Hilliard-Hele-Shaw (CHHS) system of equations. The numerical algorithms in [53,81,84] are known as convex-splitting schemes and apply to a general class of equations that includes the Cahn-Hilliard equation, which is a foundation of our diffuse interface model. The convex-splitting schemes have desirable stability, solvability, and accuracy properties that would benefit our tumor simulator greatly, with the potential to make the simulation algorithm cleaner and eliminate (or at least to mitigate) the issue with convergence.
The CHHS system that we studied in [81] is intimately related to the tumor model herein; but it models only a single tumor species and has no volumetric growth term:
| (69) |
| (70) |
Still, it embodies much of the essential structure of the tumor model. We were able to design a stable first-order accurate (in time) convex-splitting scheme for the CHHS system, and we demonstrated that the resulting discrete equations could be solved efficiently using a nonlinear multigrid scheme, like that described here. In [53] we developed a second-order convex-splitting scheme and a practical, efficient multigrid solver for PFC equations. Using the ideas from [53,81,84] it seems that a stable, fully second-order accurate scheme for our tumor model should be available. This is the subject of ongoing work.
Acknowledgments
SW and JL acknowledge funding from the National Science Foundation-Division of Mathematical Sciences (NSF-DMS). JL also acknowledges support from the National Institutes of Health (NIH) through grant P50GM76516 for a Center of Excellence in Systems Biology at the University of California, Irvine. VC acknowledges funding from NSF-DMS, and from the NIH–National Cancer Institute. The authors thank Hermann Frieboes, Jun-Seok Kim, Xiangrong Li, John Sinek and Xiaoming Zheng for many useful discussions. They also thank the anonymous reviewer whose suggestions improved the paper significantly.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Abbott RG, Forrest S, Pienta KJ. Simulating the hallmarks of cancer. Artif Life. 2006;12:617–634. doi: 10.1162/artl.2006.12.4.617. [DOI] [PubMed] [Google Scholar]
- 2.Alarcón T, Byrne HM, Maini PK. A cellular automaton model for tumour growth in inhomogeneous environment. J Theor Biol. 2003;225:257–274. doi: 10.1016/s0022-5193(03)00244-3. [DOI] [PubMed] [Google Scholar]
- 3.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4. Garland Science; New York: 2002. [Google Scholar]
- 4.Ambrosi D, Preziosi L. On the closure of mass balance models for tumor growth. Math Models Meth Appl Sci. 2002;12:737–754. [Google Scholar]
- 5.Ambrosi D, Preziosi L. Cell adhesion mechanisms and stress relaxation in the mechanics of tumours. Biomech Model Mechanobiol. 2009;8:397–413. doi: 10.1007/s10237-008-0145-y. [DOI] [PubMed] [Google Scholar]
- 6.Anderson ARA. A hybrid mathematical model of solid tumour invasion: The importance of cell adhesion. Math Med Biol. 2005;22:163–186. doi: 10.1093/imammb/dqi005. [DOI] [PubMed] [Google Scholar]
- 7.Anderson ARA, Quaranta V. Integrative mathematical oncology. Nature Reviews Cancer. 2008;8:227–244. doi: 10.1038/nrc2329. [DOI] [PubMed] [Google Scholar]
- 8.Anderson ARA, Weaver AM, Commmings PT, Quaranta V. Tumor morphology and phenotypic evolution driven by selective presure from the microenvironment. Cell. 2006;127:905–915. doi: 10.1016/j.cell.2006.09.042. [DOI] [PubMed] [Google Scholar]
- 9.Araujo RP, McElwain DLS. A mixture theory for the genesis of residual stresses in growing tissues I: A general formulation. SIAM J Appl Math. 2005;65:1261–1284. [Google Scholar]
- 10.Armstrong NJ, Painter KJ, Sherratt JA. A continuum approach to modeling cell-cell adhesion. J Theor Biol. 2006;243:98–113. doi: 10.1016/j.jtbi.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartha K, Rieger H. Vascular network remodeling via vessel cooption, regression and growth in tumors. J Theor Biol. 2006;241:903–918. doi: 10.1016/j.jtbi.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 12.Bauer AL, Jackson TL, Jiang Y. A cell-based model exhibiting branching and anastomosis during tumor-induced angiogenesis. Biophys J. 2007;92:3105–3121. doi: 10.1529/biophysj.106.101501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bearer EL, Lowengrub JS, Chuang YL, Frieboes HB, Jin F, Wise SM, Ferrari M, Agus DB, Cristini V. Multiparameter computational modeling of tumor invasion. Cancer Res. 2009;69:4493–4501. doi: 10.1158/0008-5472.CAN-08-3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bellomo N, Li NK, Maini PK. On the foundations of cancer modeling: Selected topics, speculations, and perspectives. Math Models Meth Appl Sci. 2008;4:593–646. [Google Scholar]
- 15.Breward CJW, Byrne HM, Lewis CE. The role of cell-cell interactions in a two-phase model for avascular tumour growth. J Math Biol. 2002;45:125–152. doi: 10.1007/s002850200149. [DOI] [PubMed] [Google Scholar]
- 16.Breward CJW, Byrne HM, Lewis CE. A multiphase model describing vascular tumour growth. Bull Math Biol. 2003;65:609–640. doi: 10.1016/S0092-8240(03)00027-2. [DOI] [PubMed] [Google Scholar]
- 17.Briggs WL, Henson VE, McCormick SF. A Multigrid Tutorial. 2. SIAM; Philadelphia: 2000. [Google Scholar]
- 18.Byrne HM, Alarcón T, Owen MR, Webb SW, Maini PK. Modeling aspects of cancer dynamics: A review. Phi Trans R Soc A. 2006;364:1563–1578. doi: 10.1098/rsta.2006.1786. [DOI] [PubMed] [Google Scholar]
- 19.Byrne HM, Chaplain MAJ. Growth of nonnecrotic tumors in the presence and absence of inhibitors. Math Biosci. 1995;130:151–181. doi: 10.1016/0025-5564(94)00117-3. [DOI] [PubMed] [Google Scholar]
- 20.Byrne HM, Chaplain MAJ. Growth of necrotic tumors in the presence and absence of inhibitors. Math Biosci. 1996;135:187–216. doi: 10.1016/0025-5564(96)00023-5. [DOI] [PubMed] [Google Scholar]
- 21.Byrne HM, Drasdo D. Individual-based and continuum models of growing cell populations: A comparison. J Math Biol. 2009;58:657–687. doi: 10.1007/s00285-008-0212-0. [DOI] [PubMed] [Google Scholar]
- 22.Byrne HM, King JR, McElwain DLS, Preziosi L. A two-phase model of solid tumour growth. Appl Math Lett. 2003;16:567–573. [Google Scholar]
- 23.Byrne HM, Preziosi L. Modelling solid tumour growth using the theory of mixtures. Math Med Biol. 2003;20:341–366. doi: 10.1093/imammb/20.4.341. [DOI] [PubMed] [Google Scholar]
- 24.Cahn JW, Hilliard JE. Free energy of a nonuniform system I. Interfacial free energy. J Chem Phys. 1958;28:258. [Google Scholar]
- 25.Chaplain MAJ. Avascular growth, angiogenesis and vascular growth in solid tumours: The mathematical modelling of the stages of tumour development. Math Comput Model. 1996;23:47–87. [Google Scholar]
- 26.Chaplain MAJ, Graziano L, Preziosi L. Mathematical modelling of the loss of tissue compression responsiveness and its role in solid tumor development. Math Med Biol. 2006;23:192–229. doi: 10.1093/imammb/dql009. [DOI] [PubMed] [Google Scholar]
- 27.Cristini V, Frieboes HB, Gatenby R, Caserta S, Ferrari M, Sinek J. Morphologic instability and cancer invasion. Clin Cancer Res. 2005;11:6772–6779. doi: 10.1158/1078-0432.CCR-05-0852. [DOI] [PubMed] [Google Scholar]
- 28.Cristini V, Frieboes HB, Li X, Lowengrub JS, Macklin P, Sanga S, Wise SM, Zheng X. Nonlinear modeling and simulation of tumor growth. In: Bellomo N, Chaplain M, de Angelis E, editors. Selected Topics in Cancer Modeling: Genesis, Evolution, Immune Competition, and Therapy, Modeling and Simulation in Science, Engineering and Technology. Birkhäuser; Boston: 2008. [Google Scholar]
- 29.Cristini V, Li X, Lowengrub JS, Wise SM. Nonlinear simulations of solid tumor growth using a mixture model: Invasion and branching. J Math Biol. 2009;58:723–763. doi: 10.1007/s00285-008-0215-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cristini V, Lowengrub JS, Nie Q. Nonlinear simulation of tumor growth. J Math Biol. 2003;46:191–224. doi: 10.1007/s00285-002-0174-6. [DOI] [PubMed] [Google Scholar]
- 31.Deisboeck TS, Zhang L, Yoon J, Costa J. In silico cancer modeling: Is it ready for prime time? Nature Clin Pract Oncol. 2009;6:34–42. doi: 10.1038/ncponc1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drasdo D, Höhme S. Individual-based approaches to birth and death in avascular tumors. Math Comput Model. 2003;37:1163–1175. [Google Scholar]
- 33.Drasdo D, Höhme S. A single-scale-based model of tumor growth in vitro: Monolayers and spheroids. Phys Biol. 2005;2:133–147. doi: 10.1088/1478-3975/2/3/001. [DOI] [PubMed] [Google Scholar]
- 34.Drasdo D, Höhme S. On the role of physics in the growth and pattern formation of multicellular systems: What we learn from individual-cell based models? J Stat Phys. 2007;128:287–345. [Google Scholar]
- 35.Fasano A, Bertuzzi A, Gandolfi A. Complex Systems in Biomedicine. Biomedical and Life Science; Springer; Milan: 2006. Mathematical modelling of tumour growth and treatment; pp. 71–108. [Google Scholar]
- 36.Franks SJ, Byrne HM, King JR, Underwood JCE, Lewis CE. Modelling the early growth of ductal carcinoma in situ of the breast. J Math Biol. 2003;47:424–452. doi: 10.1007/s00285-003-0214-x. [DOI] [PubMed] [Google Scholar]
- 37.Franks SJ, Byrne HM, Mudhar HS, Underwood JCE, Lewis CE. Mathematical modelling of comedo ductal carcinoma in situ of the breast. Math Med Biol. 2003;20:277–308. doi: 10.1093/imammb/20.3.277. [DOI] [PubMed] [Google Scholar]
- 38.Franks SJ, King JR. Interactions between a uniformly proliferating tumor and its surrounding, uniform material properties. Math Med Biol. 2003;20:47–89. doi: 10.1093/imammb/20.1.47. [DOI] [PubMed] [Google Scholar]
- 39.Frieboes HB, Jin F, Chuang YL, Wise SM, Lowengrub JS, Cristini V. Three-dimensional multispecies nonlinear tumor growth–II: Tissue invasion and angiogenesis. J Theor Biol. 2010;264:1254–1278. doi: 10.1016/j.jtbi.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frieboes HB, Lowengrub JS, Wise SM, Zheng X, Macklin P, Bearer EL, Cristini V. Computer simulation of glioma growth and morphology. NeuroImage. 2007;37:S59–S70. doi: 10.1016/j.neuroimage.2007.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frieboes HB, Zheng X, Sun C-H, Tromberg BJ, Gatenby R, Cristini V. An integrated computational/experimental model of tumor invasion. Cancer Res. 2006;66:1597–1604. doi: 10.1158/0008-5472.CAN-05-3166. [DOI] [PubMed] [Google Scholar]
- 42.Friedman A, Bellomo N, Maini PK. Mathematical analysis and challenges arising from models of tumor growth. Math Models Meth Appl Sci. 2007;17:1751–1772. [Google Scholar]
- 43.Friedman A, Reitich F. Analysis of a mathematical model for the growth of tumors. J Math Biol. 1999;38:262–284. doi: 10.1007/s002850050149. [DOI] [PubMed] [Google Scholar]
- 44.Gatenby RA, Smallbone K, Maini PK, Rose F, Averill J, Nagle RB, Worrall L, Gillies RJ. Cellular adaptations to hypoxia and acidosis during somatic evolution of breast cancer. Br J Cancer. 2007;97:646–653. doi: 10.1038/sj.bjc.6603922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerisch A, Chaplain MA. Mathematical modelling of cancer cell invasion of tissue: Local and non-local models and the effect of adhesion. J Theor Biol. 2008;250:684–704. doi: 10.1016/j.jtbi.2007.10.026. [DOI] [PubMed] [Google Scholar]
- 46.Gerlee P, Anderson ARA. An evolutionary hybrid cellular automaton model of solid tumor growth. J Theor Biol. 2007;246:583–603. doi: 10.1016/j.jtbi.2007.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gerlee P, Anderson ARA. A hybrid cellular automaton model of clonal evolution in cancer: The emergence of the glycolytic phenotype. J Theor Biol. 2008;250:705–722. doi: 10.1016/j.jtbi.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graziano L, Preziosi L. Mechanics in tumor growth. In: Mollica F, Preziosi L, Rajagopal KR, editors. Modeling of Biological Materials, Modeling and Simulation in Science, Engineering and Technology. Birkhäuser; Boston: 2007. pp. 263–321. [Google Scholar]
- 49.Greenspan HP. On the growth and stability of cell cultures and solid tumors. J Theor Biol. 1976;56:229–242. doi: 10.1016/s0022-5193(76)80054-9. [DOI] [PubMed] [Google Scholar]
- 50.Harpold HL, Alvord EC, Jr, Swanson KR. The evolution of mathematical modeling of glioma proliferation and invasion. J Neuropath Exp Neurol. 2007;66:1–9. doi: 10.1097/nen.0b013e31802d9000. [DOI] [PubMed] [Google Scholar]
- 51.Hatzikirou H, Deutsch A, Schaller C, Simon M, Swanson K. Mathematical modeling of glioblastoma tumour development: A review. Math Models Meth Appl Sci. 2005;15:1779–1794. [Google Scholar]
- 52.Hogea CS, Murray BT, Sethian JA. Simulating complex tumor dynamics from avascular to vascular growth using a general level-set method. J Math Biol. 2006;53:86–134. doi: 10.1007/s00285-006-0378-2. [DOI] [PubMed] [Google Scholar]
- 53.Hu Z, Wise SM, Wang C, Lowengrub JS. Stable and efficient finite-difference nonlinear-multigrid schemes for the phase-field crystal equation. J Comput Phys. 2009;228:5323–5339. [Google Scholar]
- 54.Jiang G-S, Shu C-W. Efficient implementation of weighted ENO schemes. J Comput Phys. 1996;126:202–228. [Google Scholar]
- 55.Jiang Y, Pjesivac-Grbovic J, Cantrell C, Freyer JP. A multiscale model for avascular tumor growth. Biophys J. 2005;89:3884–3894. doi: 10.1529/biophysj.105.060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khain E, Sander LM. Generalized cahn-hilliard equation for biological applications. Phys Rev E. 2008;77:051129. doi: 10.1103/PhysRevE.77.051129. [DOI] [PubMed] [Google Scholar]
- 57.Kim JS, Kang K, Lowengrub JS. Conservative multigrid methods for ternary Cahn-Hilliard systems. Comm Math Sci. 2004;2:53–77. [Google Scholar]
- 58.Kim Y, Stolarska MA, Othmer HG. A hybrid model for tumor spheroid growth in vitro I: Theoretical development and early results. Math Models Meth Appl Sci. 2007;17:1773–1798. [Google Scholar]
- 59.Lee DS, Rieger H. Flow correlated percolation during vascular remodeling in growing tumors. Phys Rev Lett. 2006;96:058104. doi: 10.1103/PhysRevLett.96.058104. [DOI] [PubMed] [Google Scholar]
- 60.Li X, Cristini V, Nie Q, Lowengrub JS. Nonlinear three-dimensional simulation of solid tumor growth. Disc Dyn Contin Dyn Syst B. 2007;7:581–604. [Google Scholar]
- 61.Lowengrub JS, Frieboes HB, Jin F, Chuang Y-L, Li X, Macklin P, Wise SM, Cristini V. Nonlinear modeling of cancer: Bridging the gap between cells and tumors. Nonlinearity. 2010;23:R1–R91. doi: 10.1088/0951-7715/23/1/r01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Macklin P, Lowengrub JS. An improved geometry-aware curvature discretization for level set methods: Application to tumor growth. J Comput Phys. 2006;215:392–401. [Google Scholar]
- 63.Macklin P, Lowengrub JS. Nonlinear simulation of the effect of microenvironment on tumor growth. J Theor Biol. 2007;245:677–704. doi: 10.1016/j.jtbi.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 64.Macklin P, Lowengrub JS. A new ghost cell/level set method for moving boundary problems: Application to tumor growth. J Sci Comp. 2008;35:266–299. doi: 10.1007/s10915-008-9190-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Macklin P, McDougall S, Anderson ARA, Chaplain MAJ, Cristini V, Lowengrub J. Multiscale modelling and simulation of vascular tumour growth. J Math Biol. 2008;58:765–763. doi: 10.1007/s00285-008-0216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Morton KW, Mayers DF. Numerical Solution of Partial Differential Equations. 2. Cambridge University Press; Cambridge: 2005. [Google Scholar]
- 67.Nagy JD. The ecology and evolutionary biology of cancer: A review of mathematical models of necrosis and tumor cell diversity. Math Biosci Eng. 2005;2:381–418. doi: 10.3934/mbe.2005.2.381. [DOI] [PubMed] [Google Scholar]
- 68.Preziosi L, Tosin A. Multiphase modeling of tumor growth and extracellular matrix interaction: Mathematical tools and applications. J Math Biol. 2009;58:625–656. doi: 10.1007/s00285-008-0218-7. [DOI] [PubMed] [Google Scholar]
- 69.Quaranta V, Weaver AM, Cummings PT, Anderson ARA. Mathematical modeling fo cancer: The future of prognosis and treatment. Clinica Chimica Acta. 2005;357:173–179. doi: 10.1016/j.cccn.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 70.Ramis-Conde I, Drasdo D, Anderson ARA, Chaplain MAJ. Modeling the influence of the E-cadherin-beta-catenin pathway in cancer cell invasion: A multiscale approach. Biophys J. 2008;95:155–165. doi: 10.1529/biophysj.107.114678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rejniak K. A single-cell approach in modeling the dynamics of tumor microregions. Math Biosci Eng. 2005;2:643–655. doi: 10.3934/mbe.2005.2.643. [DOI] [PubMed] [Google Scholar]
- 72.Rejniak K. An immersed boundary framework for modeling the growth of individual cells: An application to the early tumour development. J Theor Biol. 2007;247:186–204. doi: 10.1016/j.jtbi.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 73.Ribba B, Alarcón T, Marron K, Maini PK, Agur Z. The use of hybrid cellular automaton models for improving cancer therapy. In: Chopard B, Sloot PMA, Hoekstra AG, editors. Cellular Automata, Lecture Notes in Computer Science. Springer; Berlin: 2004. pp. 444–453. [Google Scholar]
- 74.Roose T, Chapman SJ, Maini PK. Mathematical models of avascular tumor growth. SIAM Review. 2007;49:179–208. [Google Scholar]
- 75.Sanga S, Frieboes HB, Zheng X, Gatenby R, Bearer EL, Cristini V. Predictive oncology: A review of multidisciplinary, multiscale in silico modeling linking phenotype, morphology and growth. NeuroImage. 2007;37:S120–S134. doi: 10.1016/j.neuroimage.2007.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sinek J, Frieboes HB, Zheng X, Cristini V. Two-dimensional chemotherapy simulations demonstrate fundamental transport and tumor response limitations involving nanoparticles. Biomed Microdevices. 2004;6:297–309. doi: 10.1023/B:BMMD.0000048562.29657.64. [DOI] [PubMed] [Google Scholar]
- 77.Stolarska MA, Kim Y, Othmer HG. Multi-scale models of cell and tissue dynamics. Phil Trans R Soc A. 2009;367:3525–3553. doi: 10.1098/rsta.2009.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tosin A. Multiphase modeling and qualitative analysis of the growth of tumor cords. Networks Heterogen Media. 2008;3:43–84. [Google Scholar]
- 79.Trottenberg U, Oosterlee CW, Schüller A. Multigrid. Academic Press; New York: 2001. [Google Scholar]
- 80.van Leeuwen IMM, Edwards CM, Ilyas M, Byrne HM. Towards a multiscale model of colorectal cancer. World J Gastroenterol. 2007;13:1399–1407. doi: 10.3748/wjg.v13.i9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wise SM. Unconditionally stable finite difference, nonlinear multigrid simulation of the Cahn-Hilliard-Hele-Shaw system of equations. J Sci Comput. 2010;44:38–68. [Google Scholar]
- 82.Wise SM, Kim JS, Lowengrub JS. Solving the regularized, strongly anisotropic Cahn-Hilliard equation by an adaptive nonlinear multigrid method. J Comput Phys. 2007;226:414–446. [Google Scholar]
- 83.Wise SM, Lowengrub JS, Frieboes HB, Cristini V. Three-dimensional multispecies nonlinear tumor growth–I: Model and numerical method. J Theor Biol. 2008;253:524–543. doi: 10.1016/j.jtbi.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wise SM, Wang C, Lowengrub JS. An energy stable and convergent finite-difference scheme for the phase field crystal equation. SIAM J Numer Anal. 2009;47:2269–2288. [Google Scholar]
- 85.Zheng X, Wise SM, Cristini V. Nonlinear simulation of tumor necrosis, neo-vascularization and tissue invasion via an adaptive finite-element/level-set method. Bull Math Biol. 2005;67:211–259. doi: 10.1016/j.bulm.2004.08.001. [DOI] [PubMed] [Google Scholar]