Abstract
Recognition of pathogen-associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs) activates the innate immune response. The rice PRR, XA21, confers robust resistance at adult stages to most strains of the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo). Seedlings are still easily infected by Xoo, causing severe yield losses. Here we report that Xa21 is induced by Xoo infection and that ectopic expression of Xa21 confers resistance at three leaf stage (three-week-old), overcoming the developmental limitation of XA21-mediated resistance. Ectopic expression of Xa21 also up-regulates a larger set of defense-related genes as compared to Xa21 driven by the native promoter. These results indicate that altered regulation of Xa21 expression is useful for developing enhanced resistance to Xoo at multiple developmental stages.
Keywords: Oryza sativa, pathogen-associated molecular pattern, pattern recognition receptor, XA21, Xanthomonas oryzae pv. oryzae
1. Introduction
At least 10% of global food production is lost to plant disease [1]. Resistance to many of these diseases is often developmentally controlled such that only adult plants are resistant [2]. For example, development-controlled disease resistance is observed in wheat/Puccinia recondita [3], Maize/Puccinia sorghi [4], tomato/Cladosporium fulvum [5], and Arabidopsis/Pseudomonas syringae interactions [6]. Despite the economic importance of seedling resistance, little is known about the biochemical and molecular mechanisms involved in this regulation.
Plant innate immune systems rely on monitoring the presence of pathogen through PRRs (which recognize PAMPS) and nucleotide-binding site leucine-rich repeat (NB-LRR) type proteins (which recognize pathogen effectors) [7–9]. The rice PRR, XA21, recognizes the PAMP Ax21 (Activator of XA21-mediated immunity), present in all Xanthomonas and Xylella species [10–12]. XA21-mediated resistance has been shown to be under developmental control with incomplete resistance in early stages that gradually increases with development [13]. XA3/XA26-mediated resistance is also developmentally regulated [14]. The developmentally-controlled resistance is hypothesized to be due to transcript levels of Xa21 and Xa3/Xa26 [2, 14]. Their expressions are low at the two-leaf stage, and then gradually increase with development. The highest level of expression is reached at the maximum-tillering stage, consistent with an increased resistance at this stage. Based on the expression result of Xa21, we hypothesized that ectopic expression of Xa21 may overcome the developmental control of XA21-mediated resistance. Ectopic expression experiments are specifically designed to increase the abundance of the desired transcript through introduction of a transgene into the host. This strategy was applied to the study of several NB-LRR and PRR proteins. In the case of Arabidopsis RPS2, tomato Pto, and rice Xa3, overexpression leads to constitutive activation of downstream defense responses even in the absence of the corresponding effector or PAMPs [14–17].
Here we show that transgenic plants overexpressing Xa21 display significantly increased resistance at the seedling stage. These results support the hypothesis that Xa21 transcript levels are rate limiting in early stages of development.
2. Materials and methods
2.1 Plant Material and Growth Conditions
Rice (Oryza sativa L.) plants [cultivar Kitaake (Kit)] were maintained in the greenhouse. The growth chamber was set on a 16 h light and 8 h dark photoperiod, 28/26°C temperature cycle, and 85/90% humidity. Healthy and well-expanded leaves from three or six-week-old rice plants, Kit and transformed Kit were used for Xoo PXO99Az inoculation and nucleic acid or protein extraction.
2.2 Construction of the Ubi Myc-Xa21, Ubi Xa21-CFP, Nat Myc-Xa21 and Nat Xa21-CFP Plasmids for Expression in Rice
To construct the Ubi Myc-Xa21 plasmid, a 5´ fragment of Myc-Xa21 was PCR-amplified using primers, 5´-AAAGGATCCAACATCTCTCGCTGTCTT-3´ / 5´-GGCTGAGCTCCGGTGGTAT-3´ and template pC822-cMyc-Xa21 [18, 19]. This 420-bp 5´-fragment was cut with BamHI/SacI at the ends and cloned, together with a 4.2-kb SacI/SpeI Xa21 3´-fragment, into the pBluescript II SK- vector to create a promoterless full-length Myc-Xa21 gene. The 5´ end of this gene was confirmed by sequencing. This Myc-Xa21 gene was excised with BamHI/SpeI and subcloned into the Ubi-CAMBIA-1300 vector using the same enzyme sites to generate plasmid Ubi Myc-Xa21/C1300.
To fuse the XA21 protein to the cyan fluorescent protein (CFP), a 380-bp 3´ fragment of the Xa21 gene was PCR-amplified using primers, 5´-TGCATCAACGCATGGAGATA-3´ / 5´-AATTCCATGGGAAATTCAAGGCTCCCACCTT-3´. This fragment removed the stop codon and the EcoRI site located immediately in front of the stop codon. EcoRI was used to digest the 5’ region and NcoI to digest the 3’ region of Xa21 3´ PCR product. Meanwhile, the CFP gene was excised from the pECFP plasmid (Clontech) using NcoI/SpeI. These two fragments were joined at the NcoI site and cloned into pBluescript II SK-, predigested with EcoRI/SpeI, to create plasmid Xa21 3´-CFP/SK. The Xa21 portion and the junction were confirmed by sequencing. The Xa21 3´-CFP fragment was excised with EcoRI (the second EcoRI site, coming from the pECFP plasmid, is located downstream next to the end of the CFP gene) and used to replace the 380-bp EcoRI fragment in the original promoterless (as described above) Xa21 gene, creating Xa21-CFP. To create the Ubi XA21-CFP/C1300 construct, the Xa21-CFP fragment was excised with BamHI/SpeI and cloned into the Ubi-CAMBIA-1300 vector predigested with BamHI/SpeI as described above. The Nat XA21-CFP/C4300 construct was generated by ligating the SacI/SpeI Xa21-CFP 3’ fragment and the KpnI/SacI Xa21 5’ fragment jointly with CAMBIA-4300 pre-digested with KpnI/XbaI. The Nat Myc-XA21/C1300 construct was previously described [19].
2.3. Expression analysis
For reverse transcriptase-polymerase chain reaction (RT-PCR) analysis, total RNAs were extracted from leaves using TRIzol® reagent (Invitrogen) after each treatment. Then the RT reaction was performed following the manual for QuantumRNA 18S Internal Standards (Ambion). PCR analyses were performed with primers pairs, 5´-TCATCACTCTACTTGCTTATA-3´/5´-GAATTCAAGGCTCCCACCTTC-3´ (for Xa21_cds), 5´-GTTTTATTGCCACACTTCAGA-3´/5´-GGAAGCCCCTCCCCACCTCCCCTCATC-3´ (for Xa21_utr). After 28 cycles, the amplified products were resolved by gel electrophoresis.
For qRT-PCR, the total RNA was treated with RNase free DNase (Promega), purified using Macherey-Nagel Nucleospin RNA II kit and quantified using ND-1000 spectrophotometer (Nanodrop). cDNA was synthesized from 10 µg of total RNA in 40 µl volume using M-MLV RT (Invitrogen) followed by RNase treatment (NEB). The cDNA was cleaned using Zymo DNA clean and concentrator kit and eluted in 100 µl of 1mM Tris-HCl. The cDNA was diluted to 20 folds and 4 µl of it was used for each reaction. Gene-specific primers were designed using PRIMER EXPRESS version 2.0 (PE Applied Biosystems, USA) and checked for their specificity using Blast tool of National Center for Biotechnology Information (NCBI). Primer sequences have been given in supplementary table 1.
Each QPCR reaction was performed in 10 µl of volume using Bio-Rad CFX-96™ detection system (Bio-Rad) with Ssofast™ EvaGreen® supermix (Bio-Rad) using following PCR conditions: (95°C for 3 s and 60°C for 3 s) 40 cycles followed by melt curve analysis. The data was normalized using actin as endogenous control and analyzed to calculate relative expression values using ΔCt method. Three technical and three biological replicates were performed for each sample and standard error was calculated.
3. Results
3.1 Xa21 expression is induced by Xoo infection
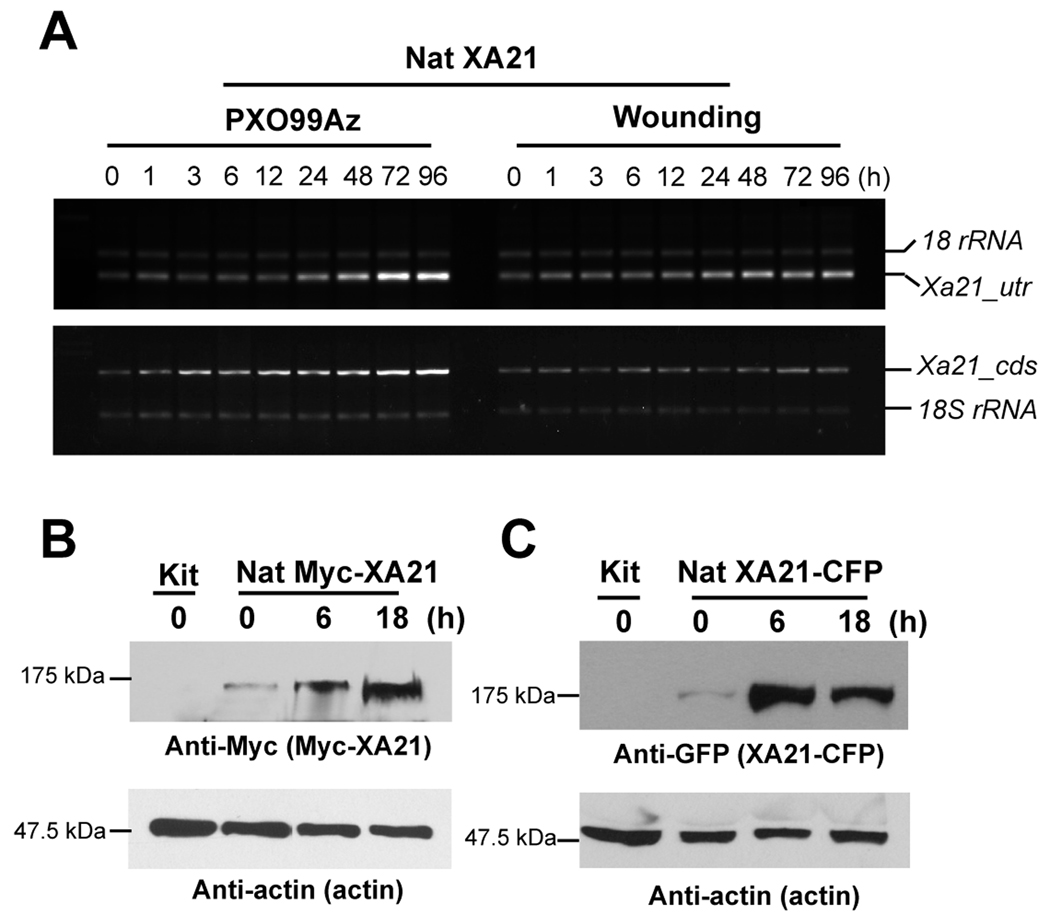
To determine if Xa21 expression is modulated by Xoo strain PXO99Az expressing Ax21, its expression pattern was monitored in six-week-old transgenic rice plants carrying Xa21 gene under the control of its native promoter (Nat XA21 23A-1-14, homozygous T2) (Fig. 1A). For reverse transcriptase (RT)-PCR analysis, specific primer sets in 3'-untranslated region (utr) and coding sequence (cds) of Xa21 were used. As expected, transcripts corresponding to Xa21 in wild type Kitaake (Kit) were not detectable at any time point after Xoo PXO99Az inoculation, indicating that primers for Xa21 are specific (data not shown). In contrast, Nat XA21 plant wounded by cutting the leaf tip with scissors induced Xa21 gene to moderate level. In Xoo-inoculated Nat XA21, Xa21 was induced in RT-PCR analysis, performed with both primer pairs (labeled as Xa21_utr and Xa21_cds) with maximum accumulation at 72 h after inoculation (HAI). This result suggests that Ax21 and XA21 interaction regulates XA21-mediated immune response as well as the transcription level of Xa21. 18S ribosomal RNA (18S rRNA) was used as an internal control.
Fig. 1. Xa21 is induced after Xoo strain PXO99Az inoculation.
(A) Total RNA was extracted from Kitaake, mock-treated Nat XA21 (Wounding), and Xoo PXO99Az-inoculated Nat XA21 (Xoo PXO99Az) at the indicated time points. RT-PCR was performed with the specific primers designed from Xa21 UTR (Xa21_utr) and Xa21 CDS (Xa21_cds) regions, respectively. Control RT-PCR reactions were carried out with 18S rRNA.
(B) Total protein extracts from Xoo PXO99Az-inoculated Nat Myc-XA21 (T3, line 20-4) at the indicated time points were used for protein gel blot analysis. Equal amounts of total protein from non-transgenic (Kit) and Nat Myc-XA21 were analyzed by SDS-PAGE, and immunoblotted with anti-Myc antibody. Myc-XA21 gives bands at about 140 kDa. Experiments were repeated over three times with similar results. Equal loading of total proteins was confirmed by anti-actin antibody.
(C) Total protein extracts from Xoo PXO99Az-inoculated Nat XA21-CFP (T2, line 15A-1-7) at the indicated time points were used for protein gel blot analysis using anti-GFP antibody. XA21-CFP gives bands at about 180 kDa. Experiments were repeated over three times with similar results. Equal loading of total proteins was confirmed by anti-actin antibody.
To investigate if accumulation of Xa21 transcripts correlates with the XA21 protein after Xoo PXO99Az inoculation, we generated transgenic Kit lines possessing an N-terminal Myc-epitope-tagged XA21 (Nat Myc-XA21) or a C-terminal CFP-tagged XA21 (Nat XA21-CFP), under the control of its native promoter. At the six-week-old adult stage (9 to 10 leaves), non-transgenic Kit plants are susceptible to Xoo PXO99Az. In contrast, the transgenic Nat Myc-XA21 (T3, 20-1) and Nat XA21-CFP (T1, 15A-1) plants were fully resistant to Xoo, indicating that the proteins are biologically equivalent to the native XA21 protein (Fig. S1A and B). We then investigated if XA21 is accumulated using anti-Myc or anti-GFP antibodies after Xoo PXO99Az inoculation (Fig. 1B and C). In the absence of Xoo (0 time point), Myc-XA21 protein of 140 kDa could be detected slightly, but not in Kit control plants (Fig. 1B). At 6 HAI, the protein started to accumulate and continuously increased till 18 HAI. We have previously reported that the XA21 protein accumulation is independent of mRNA levels after Xoo inoculation, because the stability of the XA21 protein is regulated through ER quality control mechanisms [20]. The anti-actin antibody has been used as an internal control to show equal loading in each lane. Anti-GFP antibody detected a 180-kDa polypeptide in transgenic plants carrying Nat XA21-CFP but not in the control line Kit (Fig. 1C). Similar accumulation pattern of XA21-CFP protein was observed after PXO99Az inoculation, indicating that XA21 protein synthesis and/or stability is increased during the defense response.
3.2 Generation of transgenic rice plants overexpressing Xa21
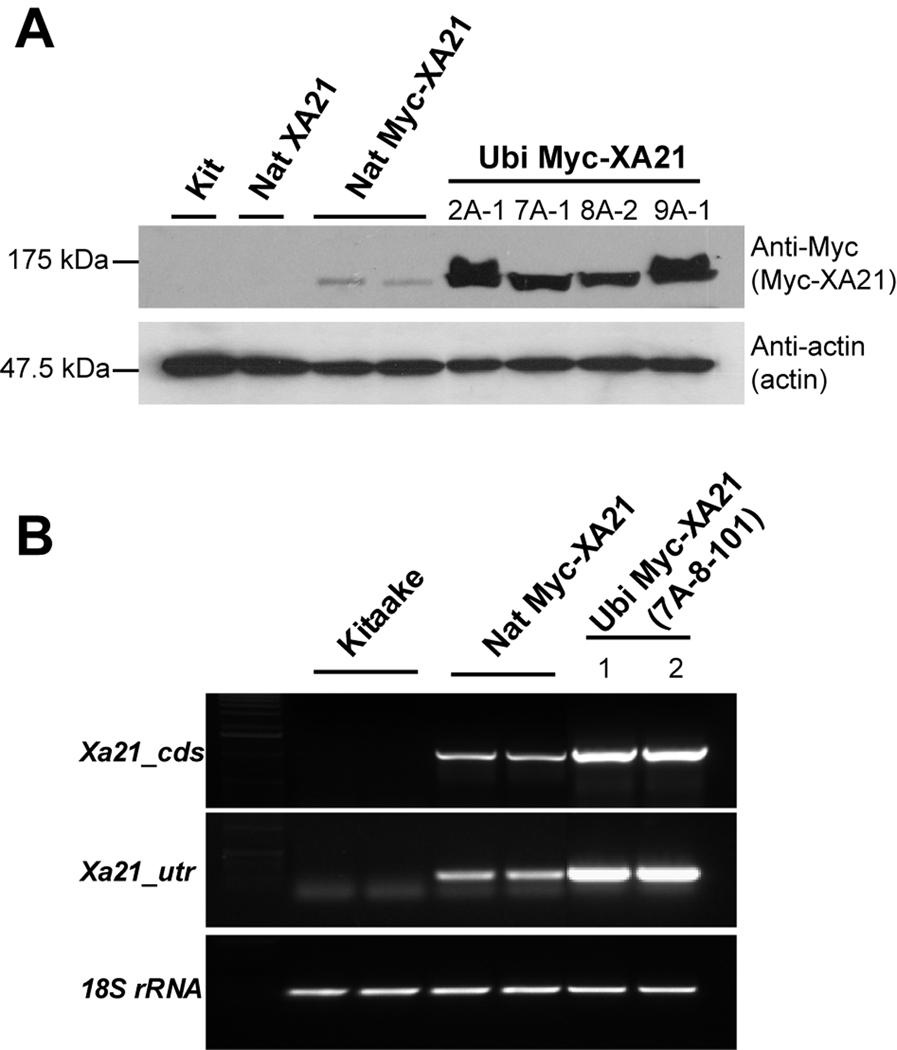
Based on the results of Xoo PXO99Az inoculation in Fig. 1, we hypothesized that accumulated Xa21 transcripts and XA21 proteins may affect the resistance level to Xoo. To evaluate the hypothesis, we transferred Xa21 driven by a strong constitutive promoter, maize ubiquitin gene promoter (Ubi) into Kitaake wild type and generated 21 independent transgenic lines carrying Myc-Xa21 under the control of the Ubi promoter (Ubi Myc-XA21). We then analyzed the transgenic lines (T0 generation) at six weeks of age for alterations in resistance to Xoo PXO99Az. Fourteen Ubi Myc-XA21 lines displayed similar level of enhanced resistance to Xoo. Seven lines showed no difference compared to Kitaake (data not shown). We generated T1 progeny from each of the T0 lines displaying enhanced resistance and analyzed the T1 progeny for Mendelian segregation of the transgene with the enhanced resistance phenotype. 2A, 7A, 8A, and 9A of lines were chosen for further analysis. The protein levels of Myc-XA21 in six-week-old transgenic lines (T1) displaying the enhanced resistance (2A-1, 7A-1, 8A-2, and 9A-1) were examined by western blot analysis using anti-Myc antibody (Fig 2A). The Ubi Myc-XA21 transgenic lines (T1) overexpress Myc-XA21 protein compared to the Nat Myc-XA21 lines. Anti-actin antibody was used as an internal loading control. Total RNAs from Kit, homozygous Nat Myc-XA21 (20-1), and homozygous Ubi Myc-XA21 (T3, 7A-8-101-1 and -2) were extracted. Then we performed RT-PCR with primers targeting specifically Xa21 (Fig. 2B). Internal control, 18S rRNA, showed constitutive expressions in all tested plant lines. Although the Xa21 was detected in Nat Myc-XA21 to moderate level, the levels of transcripts and protein were significantly increased in Ubi Myc-XA21, demonstrating that Xa21 under control of Ubi promoter was overexpressed constitutively.
Fig. 2. Rice plants overexpressing Myc-Xa21 under control of Ubi promoter (Ubi Myc-XA21) produce higher levels of XA21 protein.
(A) Total proteins were extracted from six-week-old plants (Kit, Nat XA21, Nat Myc-XA21, and Ubi Myc-XA21 (T1)) and protein gel blot analysis was performed with anti-Myc antibody. Equal loading of total proteins was confirmed by anti-actin antibody.
(B) Total RNA was extracted from six-week-old Kitaake wild type (Kit), Nat Myc-XA21, or Ubi Myc-XA21 (T3, 7A-8-101-1 and -2). RT-PCR was performed with specific primers for Xa21 UTR (Xa21_utr) and Xa21 CDS (Xa21_cds) regions, respectively. Control RT-PCR reactions were carried out with 18S rRNA.
3.3 Constitutive expression of Xa21 shows enhanced resistance to Xoo
We then examined if the increased XA21 protein caused by its constitutive expression results in the enhanced resistance to Xoo. After Xoo PXO99Az inoculation to six-week-old plants, the lesion lengths of two independent homozygous lines (Ubi Myc-XA21 T1, 7A-8 and 9A-12) were compared with homozygous Nat Myc-XA21 line (20-1) (Fig. S2A). At 12 days after inoculation (DAI), Kit wild type displayed susceptibility to Xoo PXO99Az with long lesions ranging in length over 15 cm, in contrast to the Nat Myc-XA21 line which showed 2 to 3 cm lesion lengths. Ubi Myc-XA21 displayed shorter lesion lengths (approximately 1 cm) compared to Nat Myc-XA21, indicating that overexpression of Xa21 confers enhanced resistance. To quantify the effect of XA21 overexpression, homozygous Ubi Myc-XA21 (T2, 7A-8-123), Nat Myc-XA21, and Kitaake were inoculated with Xoo PXO99Az and bacterial growth was monitored over time (Fig. S2B). At eight DAI, significant decrease in bacterial population was detected in the Ubi Myc-XA21 lines compared with the Nat Myc-XA21. At 12 DAI, Xoo strain PXO99Az populations in Ubi Myc-XA21 lines reached to 2.75×107 colony-forming units per leaf (cfu/leaf), which is approximately five-fold decrease compared to Nat-Myc-XA21 control (1.24×108 cfu/leaf).
This result was also confirmed with another Ubi XA21 lines which carry XA21 tagged with CFP (Ubi XA21-CFP). Six-week-old progenies (T1) from self-pollinated 7B, 10B, 11B, and 18B were inoculated with Xoo PXO99Az. Twelve DAI, we examined for co-segregation of genotype with phenotype by PCR analysis and enhanced resistance by measurement of the length of Xoo-induced lesions. All segregants carrying Ubi Xa21-CFP displayed enhanced resistance to Xoo PXO99Az compared to homozygous transgenic plants carrying XA21-CFP under control of native promoter (Nat XA21-CFP 15A-1) (Fig. S3). Segregants lacking Ubi Xa21-CFP showed susceptibility upon Xoo PXO99Az inoculation.
3.4. Overexpression of Xa21 overcomes developmentally-regulated resistance
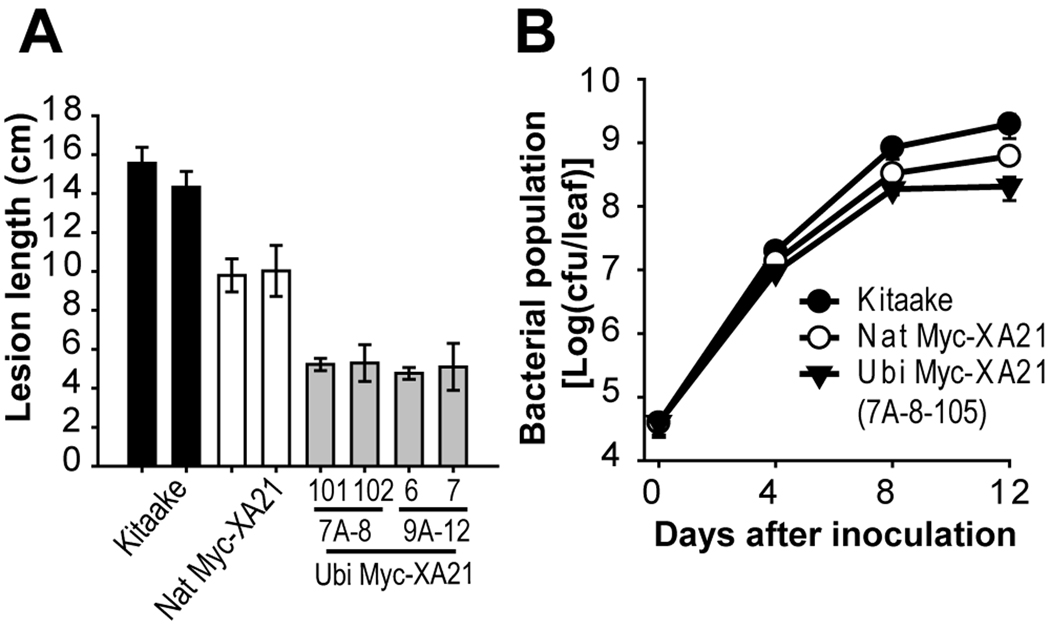
The resistance conferred by XA21 progressively increases from the susceptible juvenile two-leaf stage (approximately two-week-old) through later stages, with full resistance only at the adult stage [13]. To test if the enhanced resistance by the overexpression of Xa21 can overcome the juvenile stage susceptibility, we inoculate the three leaf stage plants (three-week-old). At this stage of development, XA21 rice plants are only partially resistant (approximately 40% of resistance to that of six-week-old plants) [13]. At 12 DAI, we measured the lesion length of three leaf stage plants inoculated with Xoo PXO99Az (Fig. 3A). The Kit control displayed susceptibility to Xoo PXO99Az with long lesions ranging in length from 14 to 15 cm. Homozygous Nat Myc-XA21 (20-1) developed relatively long lesion lengths (approximately 8 to 10 cm), confirming that juvenile stage XA21 plant does not possess full-resistance observed in adult-stage XA21 plants. However, significantly enhanced resistance was observed in two independent homozygous lines, Ubi Myc-XA21 (T2, 7A-8-101, 7A-8-102, 9A-12-6, and 9A-12-7) with only 4 to 5 cm lesion lengths.
Fig. 3. Rice plants overexpressing Myc-Xa21 (Ubi Myc-XA21, T2) overcome developmental control of XA21-mediated resistance to Xoo.
(A) Lesion length development of Xoo PXO99Az-inoculated three leaf stage plants (three-week-old), Kitaake control (Kit), Nat Myc-XA21 and Ubi Myc-XA21. Each data point represents the average and standard deviation of at least four samples.
(B) Plots of Xoo PXO99Az populations over 12 days in Kitaake control (Kit), Nat Myc-XA21, and Ubi My-XA21 (T2, 7A-8-105). For each time point, the bacterial populations were separately determined for three leaves. Capped, vertical bars represent standard deviation of values (cfu/leaf) from three samples. Experiments were repeated at least three times with similar results.
We quantified the effect of Xa21 over-expression on bacterial growth by monitoring bacterial populations on homozygous Ubi Myc-XA21 plants (T2, 7A-8-105) inoculated with Xoo PXO99Az (Fig. 3B). For all growth curves until four days after inoculation (DAI), there was no significant difference in bacterial populations in any of the lines. However, significant difference in bacterial growth was observed at 12 DAI. Susceptible Kit plants reached approximately 1.97×109 colony-forming units per leaf (cfu/leaf). The bacterial populations in Nat Myc-XA21 grew to 7.15×108 cfu/leaf, showing the partial resistance upon Xoo PXO99Az in three leaf stage. In Ubi Myc-XA21 plants (T2, 7A-8-105), the population leveled off at less than 2.05×108 cfu/leaf, indicating that developmental control of Xa21 can be overcome by its constitutive expression.
3.5 Expression of defense-related genes shows correlation with Xa21 expression levels
To elucidate the molecular mechanism to confer the enhanced resistance to juvenile and adult stages of Ubi XA21 transgenic lines, we examined if the constitutive expression of Xa21 activates defense signaling pathway. Total RNAs from Kit, homozygous Nat Myc-XA21 (20-1), and homozygous Ubi Myc-XA21 (T3, 7A-8-101-1 and -2) were extracted. We examined the expression of defense related genes in the transgenic plants carrying Xa21 under native or Ubi promoter using a real-time qRT-PCR analysis in the absence of Xoo treatment (Fig. 4). The list of genes is given in Table S1.
Fig. 4. Rice plants overexpressing Myc-Xa21 (Ubi Myc-XA21) show increased expression of defense-related genes.
Total RNA was extracted from six-week-old Kitaake wild type (Kit), Nat Myc-XA21, or Ubi Myc-XA21 (T3, 7A-8-101-1 and -2) plants. qRT-PCR was performed with specific primers for each genes. Gene expression level was normalized using actin as an internal reference.
Classical pathogen-induced genes, PR10b, PBZ1, WRKY09, peroxidase, hsp90, and SDF2, were induced in the both transgenic plants. COL1, involved in jasmonate signaling and defense response [21], OsMT2b, a negative regulator of oxidative burst [22], SSI2 implicated in cell death [23], and OsSGT1, associating with heat shock protein 90 for innate immune response, were up-regulated only in the Ubi Myc-XA21 transgenic plants. These results demonstrate that overexpression of XA21 up-regulates a new set of defense-related genes as compared to Nat Myc-XA21, thus enlarging the number of possible target genes that are induced.
3.6 Enhanced resistance mediated by overexpression of Xa21 is Ax21-dependent
To elucidate whether the enhanced resistance mediated by ectopic expression of Xa21 is Ax21- dependent, we inoculated Ubi Myc-XA21 with the Xoo PXO99ΔraxST, mutant strain defective in Ax21 biological activity [24]. At 12 DAI, Kit, Nat Myc-XA21, and Ubi Myc-XA21 (T3, 7A-8-117-1 and -2) displayed long lesions ranging in length around 12 to 14 cm, suggesting that Xa21 overexpression is not able to induce resistance to Xoo strains lacking Ax21 activity (Fig. S4).
We also investigated if overexpression of Xa21 can confer resistance to a normally virulent fungal pathogen (Fig. S5). For this experiment, we used Magnaporthe oryzae isolate R01-1, which is compatible with Kitaake. Ten DAI with M. oryzae, disease levels in Kit, Nat XA21, Nat Myc-XA21, and Ubi Myc-XA21 (T2, 7A-8-1 and 9A-12-1) were evaluated by measuring lesion lengths. No significant difference was observed in the tested rice lines. Taken together, these results indicate that enhanced resistance by overexpression of Xa21 still requires the presence of Ax21.
4. Discussion
4.1 Xa21 expression is regulated upon Xoo infection
It has been previously reported that the expression of PRR and NBS-LRR genes controlling plant immunity are transcriptionally regulated under various conditions, including Arabidopsis RPS2 and RPM1 [25] and rice Xa1, Xa27, and Xa3/Xa26 [14, 26, 27]. Although it was reported that expression of Arabidopsis PRR, FLS2, was not affected by flagellin treatment [16], a search in the gene expression database Genevestigator revealed that FLS2 is induced by bacterial LPS, fungal chitin, and the oomycete derived NPP1 [28, 29]. Those reports suggest that expression of the receptors may be regulated by their corresponding PAMPs and effector molecules.
Here we investigated if the transcript level of Xa21 changes during response to Xoo treatment. We found that Xa21 is induced in response to Xoo and wounding. Arabidopsis FLS2 is also activated by wounding [30]. Because wounds provide entry sites for potential pathogen, when wounded, plants respond quickly by protecting them from subsequent pathogen [30, 31]. Therefore, increased availability of PRRs such as XA21 and FLS2 in the plasma membrane may accelerate responding to subsequent bacterial infection through the wounding tissue.
4.2 XA21 up-regulates genes involved in pathogen-induced defense response
To explain the enhanced resistance to Xoo in transgenic plants overexpressing Xa21, we focused on the genes previously shown to be induced during the rice defense response [32–34] and analyzed their expressions in the transgenic lines carrying Xa21 under the control of its native or Ubi promoter. Our results indicate that PR and other defense-related genes are upregulated in Xa21-overexpressing lines compared with the Kitaake control.
Several previous reports have demonstrated that ectopic expressions of NB-LRR or PRR genes cause induction of downstream defense responses in the absence of the cognate effector or PAMP [14–17]. For example, overexpression of Pto displayed broad spectrum resistance to bacterial and fungal pathogens including Pseudomonas syringae pv tomato, Xanthomonas campestris pv vesicatoria, and Cladosporium fulvum [15]. Consistent with these previous studies, overexpression of Xa21 confers enhanced resistance and seedling stage and is accompanied by partial induction of defense-related genes. However, Xa21-overexpressing plants are still susceptible to an Xoo strain lacking Ax21 activity and to M. oryzae. We hypothesize that, in the absence of Ax21, the XA21-mediated resistance pathway in the Ubi Myc-XA21 plants is not fully activated.
In contrasts to the Pto-overexpressing tomato plants, which display spontaneous cell death [15], overexpression of Xa21 enhances resistance at the seedling stage with no observable detrimental effects on plant growth or development. Thus constitutive expression of Xa21 provides a useful strategy to engineer resistance to Xoo during multiple developmental stages.
Supplementary Material
Acknowledgments
We thank Dr. Kazunari Nozue in the Ronald laboratory for critical review of the manuscript. We also acknowledge Wei Bai, Seo Jung Yang and Kisung Jun for technical assistance.
Funding. This work was supported by grants from the National Institutes of Health (#GM055962), the U.S. Department of Agriculture (USDA) Rice Coordinated Agricultural Project (#2004-35317148), and World Class University program (R33-2008-000-10168-0) of the Korean Ministry of Education, Science and Technology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests. (The authors have declared that no competing interests exist.)
References
- 1.Cheatham MR, Rouse MN, Esker PD, Ignacio S, Pradel W, Raymundo R, Sparks AH, Forbes GA, Gordon TR, Garrett KA. Beyond yield: plant disease in the context of ecosystem services. Phytopathology. 2009;99:1228–1236. doi: 10.1094/PHYTO-99-11-1228. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J, Fu J, Li X, Xu C, Wang S. Dissection of the factors affecting development-controlled and race-specific disease resistance conferred by leucine-rich repeat receptor kinase-type R genes in rice, TAG. Theoretical and applied genetics. 2009;119:231–239. doi: 10.1007/s00122-009-1032-3. [DOI] [PubMed] [Google Scholar]
- 3.Pretorius ZA, Rijkenberg FHJ, Wilcoxson RD. Effects of growth stage, leaf position, and temperature on adult plant resistance of wheat infected by Puccinia recondita f.sp. tritici. Plant Pathol. 1988;37:36–44. [Google Scholar]
- 4.Abedon BG, Tracy WF. Corngrass1 of maize (Zea mays L) delays development of adult plant resistance to common rust (Puccinia sorghi Schw) and European corn borer (Ostrinia nubilalis hubner) J Hered. 1996;87:219–223. [Google Scholar]
- 5.Panter SN, Hammond-Kosack KE, Harrison K, Jones JD, Jones DA. Developmental control of promoter activity is not responsible for mature onset of Cf-9B-mediated resistance to leaf mold in tomato. Mol Plant Microbe Interact. 2002;15:1099–1107. doi: 10.1094/MPMI.2002.15.11.1099. [DOI] [PubMed] [Google Scholar]
- 6.Kus JV, Zaton K, Sarkar R, Cameron RK. Age-related resistance in Arabidopsis is a developmentally regulated defense response to Pseudomonas syringae. The Plant cell. 2002;14:479–490. doi: 10.1105/tpc.010481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ausubel FM. Are innate immune signaling pathways in plants and animals conserved? Nature immunology. 2005;6:973–979. doi: 10.1038/ni1253. [DOI] [PubMed] [Google Scholar]
- 8.Ingle RA, Carstens M, Denby KJ. PAMP recognition and the plant-pathogen arms race. Bioessays. 2006;28:880–889. doi: 10.1002/bies.20457. [DOI] [PubMed] [Google Scholar]
- 9.Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annual review of plant biology. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 10.Lee SW, Han SW, Bartley LE, Ronald PC. From the Academy: Colloquium review. Unique characteristics of Xanthomonas oryzae pv. oryzae AvrXa21 and implications for plant innate immunity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18395–18400. doi: 10.1073/pnas.0605508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science (New York, N.Y. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 12.Lee SW, Han SW, Sririyanum M, Park CJ, Seo YS, Ronald PC. A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science (New York, N.Y. 2009;326:850–853. doi: 10.1126/science.1173438. [DOI] [PubMed] [Google Scholar]
- 13.Century KS, Lagman RA, Adkisson M, Morlan J, Tobias R, Schwartz K, Smith A, Love J, Ronald PC, Whalen MC. Short communication: developmental control of Xa21-mediated disease resistance in rice. Plant J. 1999;20:231–236. doi: 10.1046/j.1365-313x.1999.00589.x. [DOI] [PubMed] [Google Scholar]
- 14.Cao Y, Ding X, Cai M, Zhao J, Lin Y, Li X, Xu C, Wang S. The expression pattern of a rice disease resistance gene xa3/xa26 is differentially regulated by the genetic backgrounds and developmental stages that influence its function. Genetics. 2007;177:523–533. doi: 10.1534/genetics.107.075176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang X, Xie M, Kim YJ, Zhou J, Klessig DF, Martin GB. Overexpression of Pto activates defense responses and confers broad resistance. The Plant cell. 1999;11:15–29. doi: 10.1105/tpc.11.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Molecular cell. 2000;5:1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 17.Tao Y, Yuan F, Leister RT, Ausubel FM, Katagiri F. Mutational analysis of the Arabidopsis nucleotide binding site-leucine-rich repeat resistance gene RPS2. The Plant cell. 2000;12:2541–2554. doi: 10.1105/tpc.12.12.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, Chern M, Canlas PE, Jiang C, Ruan D, Cao P, Ronald PC. A conserved threonine residue in the juxtamembrane domain of the XA21 pattern recognition receptor is critical for kinase autophosphorylation and XA21-mediated immunity. The Journal of biological chemistry. 2010 doi: 10.1074/jbc.M109.093427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang YS, Pi LY, Chen X, Chakrabarty PK, Jiang J, De Leon AL, Liu GZ, Li L, Benny U, Oard J, Ronald PC, Song WY. Rice XA21 binding protein 3 is a ubiquitin ligase required for full Xa21-mediated disease resistance. The Plant cell. 2006;18:3635–3646. doi: 10.1105/tpc.106.046730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park CJ, Bart R, Chern M, Canlas PE, Bai W, Ronald PC. Overexpression of the Endoplasmic Reticulum Chaperone BiP3 Regulates XA21-Mediated Innate Immunity in Rice. PLoS One. 2010;5:e9262. doi: 10.1371/journal.pone.0009262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feys B, Benedetti CE, Penfold CN, Turner JG. Arabidopsis Mutants Selected for Resistance to the Phytotoxin Coronatine Are Male Sterile, Insensitive to Methyl Jasmonate, and Resistant to a Bacterial Pathogen. Plant Cell. 1994;6:751–759. doi: 10.1105/tpc.6.5.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong HL, Sakamoto T, Kawasaki T, Umemura K, Shimamoto K. Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 2004;135:1447–1456. doi: 10.1104/pp.103.036384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kachroo P, Shanklin J, Shah J, Whittle EJ, Klessig DF. A fatty acid desaturase modulates the activation of defense signaling pathways in plants. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9448–9453. doi: 10.1073/pnas.151258398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.da Silva FG, Shen Y, Dardick C, Burdman S, Yadav RC, de Leon AL, Ronald PC. Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol Plant Microbe Interact. 2004;17:593–601. doi: 10.1094/MPMI.2004.17.6.593. [DOI] [PubMed] [Google Scholar]
- 25.Tan X, Meyers BC, Kozik A, West MA, Morgante M, St Clair DA, Bent AF, Michelmore RW. Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC plant biology. 2007;7:56. doi: 10.1186/1471-2229-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang ZX, Kono I, Kurata N, Yano M, Iwata N, Sasaki T. Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:1663–1668. doi: 10.1073/pnas.95.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu K, Yang B, Tian D, Wu L, Wang D, Sreekala C, Yang F, Chu Z, Wang GL, White FF, Yin Z. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- 28.Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 29.Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gomez-Gomez L, Boller T. Flagellin perception: a paradigm for innate immunity. Trends in plant science. 2002;7:251–256. doi: 10.1016/s1360-1385(02)02261-6. [DOI] [PubMed] [Google Scholar]
- 31.Takabatake R, Seo S, Ito N, Gotoh Y, Mitsuhara I, Ohashi Y. Involvement of wound-induced receptor-like protein kinase in wound signal transduction in tobacco plants. Plant J. 2006;47:249–257. doi: 10.1111/j.1365-313X.2006.02781.x. [DOI] [PubMed] [Google Scholar]
- 32.Vergne E, Ballini E, Marques S, Sidi Mammar B, Droc G, Gaillard S, Bourot S, DeRose R, Tharreau D, Notteghem JL, Lebrun MH, Morel JB. Early and specific gene expression triggered by rice resistance gene Pi33 in response to infection by ACE1 avirulent blast fungus. The New phytologist. 2007;174:159–171. doi: 10.1111/j.1469-8137.2007.01971.x. [DOI] [PubMed] [Google Scholar]
- 33.Shi BJ, Wang GL. Comparative study of genes expressed from rice fungus-resistant and susceptible lines during interactions with Magnaporthe oryzae. Gene. 2008;427:80–85. doi: 10.1016/j.gene.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Park CJ, Peng Y, Chen X, Dardick C, Ruan D, Bart R, Canlas PE, Ronald PC. Rice XB15, a Protein Phosphatase 2C, Negatively Regulates Cell Death and XA21-Mediated Innate Immunity. PLoS biology. 2008;6:e231. doi: 10.1371/journal.pbio.0060231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.