Abstract
Protein biomarkers are essential in assessing pathogenic processes. The impetus for finding new biomarkers has been accelerated by the arrival of the “omics” technologies. However, equally important is the re-discovery of existing biomarkers with these new approaches as novel variants can be discovered that can improve their utility. Presented here is a mass spectrometric immunoassay method for quantitative determination of beta-2-microglobulin - an established biomarker used in the diagnosis of active rheumatoid arthritis and kidney disease, and its structural variant - cleaved at and deficient in lysine-58 (ΔK58-b2m). Beta lactoglobulin was incorporated into the assay as an internal reference standard, serving as normalization point for beta-2-microglobulin quantification. The precision, linearity, and recovery characteristics of the assay were established. The new assay was also benchmarked against existing beta-2-microglobulin ELISA. The assay was utilized to determine the individual concentration of beta-2-microglobulin and its variant across a larger cohort of samples, demonstrating the ability to simultaneously quantify both proteins.
Keywords: biomarker, beta-2-microglobulin, immunoassay, mass spectrometry, posttranslational modification
Introduction
Protein biomarkers are critical for early detection of disease, prognosis and therapy monitoring. However, the number of new FDA approved biomarkers has greatly decreased in recent years 1, which has been attributed to the high false-discovery rate of the “omics” methods, and the lack of robust methods for biomarker verification 2. While studies are being conducted to identify and standardize approaches that will circumvent these issues 3–5, it is equally important to use these new approaches for re-evaluation of existing biomarkers. Mass spectrometric methods of detection can shed light on the existence of structural variants that might serve as biomarkers with better sensitivity and specificity than their wild-type proteins. As an example, structural isoforms have recently been discovered for some well-established biomarkers such as cardiac troponin I 6 and B-type natriuretic peptide 7. More importantly, a structural variant of hemoglobin (glycated hemoglobin - HbA1c) is currently being used to assess glycemic control in people with diabetes, and has been proposed as a useful method of screening for and diagnosing diabetes 8.
Beta 2-microglobulin (b2m) is an FDA approved biomarker used in the diagnosis of active rheumatoid arthritis and kidney disease 9, 10. B2m is the light chain of the major histocompatibility complex class I (MHC-I) molecules 11. It is expressed by all nucleated cells, and circulates in blood as a monomeric 99 amino acids protein (MW=11,731.2). B2m also forms amyloids in articular structures in kidney failure patients that undergo chronic hemodyalisis treatment 12, 13, and has recently been indicated as a potential therapeutic target for prostate cancer and renal cell carcinoma 14. A structural variant of b2m is generated in vivo by the action of complement component C1s which cleaves b2m at position 58, after which the C-terminal Lys58 is removed by a carboxypeptidase B-like activity 15. The variant, termed ΔK58-b2m 16, has been associated with autoimmune disease and small-cell lung cancer 17, 18, and shown to be both less stable and more amyloidogenic than native b2m 19, 20. The variant also exhibits decreased affinity for the heavy chain of the MHC-I molecules 21. To further study the role of this variant (and possibly others) it is important to have means for its detection and quantification across large sample cohorts.
The detection of post-translationally modified proteins and their distinction from their wild-type pro-forms is not a straightforward proposition. These isoforms are only slightly chemically different from their wild-type proteins. Normally, separation approaches are employed to fractionate the proteins isoforms. Alternatively, the same result can be achieved using detection methods that allow for simultaneous observation of all protein isoforms. MALDI-TOF mass spectrometry is one such method of detection. When coupled to immunoaffinity separations that utilize antibodies toward invariable epitopes in the protein sequence, it yields a high-throughput approach for simultaneous detection of protein isoforms. Presented here is a quantitative mass spectrometric immunoassay for beta-2-microglobulin and its isoforms. We have in the past reported on the detection of the ΔK58-b2m isoform using a qualitative beta-2-microglobulin assay 22, 23. In this work we develop a fully quantitative beta-2-microglobulin mass spectrometric immunoassay and apply it to determine the human plasma concentration of b2m and its ΔK58-b2m isoform.
Materials and Methods
Reagents
Rabbit anti-human polyclonal antibody to beta-2-microglobulin (b2m) was obtained from DAKO (Carpinteria, CA, USA, Catalog No. A0072, 5.7 g/L), and rabbit anti-human polyclonal antibody to beta-lactoglobulin (BL) was obtained from GeneTex (Irvine, CA, Cat. No. GTX77272, 1 mg/mL). Recombinant human beta-2-microglobulin was purchased from Cell Scieces (Canton, MA, Cat. No. CSI19620). Beta-lactoglobulin from bovine milk (Cat. No. L8005), 1,1’ Carbonyldiimidazole (115533), TWEEN 20 (P7949) and α-cyano -4-hydroxycinnamic acid (476870) were all obtained from Sigma-Aldrich (St. Lous, MO). Phosphate buffered saline was obtained from Thermo Scientific (Rockford, IL, Cat. No. 28374). Sterile water (Cat. No. AB02120), acetone (AB00636), MES (AB01235), acetonitrile (AB00120) and trifluoracetic acid (AB02010) were purchased from American Bionalytical (Natick, MA). Methyl-1 pyrrolidone-2 was obtained from EMD Chemicals (Gibbstown, NJ, Cat. No. MX1932-5). N-octylglucoside was obtained from Roche Applied Science (Indianapolis, IN, Cat. No. 10634425001). Affinity pipettes fitted with porous microcolumns were obtained from Intrinsic Bioprobes (Tempe, AZ, Cat No. IBI-CMD-R96). Beta-2-microglobulin ELISA kit was obtained from ALPCO (Salem, NH, Cat. No. 25-BMGHU-E01).
Instrumentation
Immobilization of antibodies on the affinity pipettes and high-throughput mass spectrometric immunoassays were performed on a Multimek 96 automated 96-channel pipettor (Beckman Coulter, Brea, CA). Manual mass spectrometric immunoassays were performed with an 8-channel Finnpipette Novus multichannel pipetter (Thermo Fisher Scientific, Waltham, MA). Mass spectrometry was performed on Autoflex II MALDI-TOF mass spectrometer (Bruker, Billerica, MA). ELISA readouts were obtained on Cary 50 sprectophotometer equiped with a microplate reader accesory (Varian Instruments, Walnut Creek, CA).
Human plasma and serum samples
Forty-four Na-heparin human plasma samples and twenty human serum samples were obtained from ProMedDX (Norton, MA, USA). The samples were collected at certified blood donor and medical centers and designated as normal based on their non-reactivity for common blood infectious agents and the donor information itself. The samples were labeled only with a barcode and supplied with an accompanying specification sheet containing information only about the gender and age, ensuring proper privacy protection.
Preparation of affinity pipettes
Ninety-six affinity pipettes were mounted on the head of the Multimek 96 pipettor and initially rinsed with 200 mM HCl (20 cycles, each cycle consisting of an aspiration and dispense of a 150 µL volume), followed by a water rinse (5 cycles) and acetone rinse (5 cycles). To activate the surface of the microcolumns contained within, the pipettes were immersed into a tray containing 100 mg/mL 1,1’ Carbonyldiimidazole (in methyl-1 pyrrolidone-2), and 500 cycles of 100 µL aspirations and dispenses through each affinity pipette were performed. Two rinses with methyl-1 pyrrolidone-2 (10 cycles each, 150 µL volumes) and a final rinse with acetone (10 cycles, 150 µL) followed. The affinity pipettes were then immediately immersed into a microwell plate containing the antibodies solutions (0.057 mg/mL beta-2-microglobulin antibody, and 0.01 mg/mL beta-lactoglobulin antibody, in 10 mM MES) and 800 cycles of aspirations and dispenses of 50 µL volumes were performed to bind the antibodies to the activated microcolumns surfaces. Two rinses with 60 mM HCl followed (30 cycles each, 100 µL), ending with two final rinses with assay buffer (PBS w/0.1% TWEEN, 30 cycles each, 100 µL). The antibody-derivatized pipettes were stored at 4°C until used.
Preparation of standards and analytical samples
For the generation of the standard curve, a solution containing 1.0 mg/L recombinant beta-2-microglobulin was prepared (Standard #1) and serially diluted with assay buffer to 0.5 mg/L (Standard #2), 0.25 mg/L (Standard #3), 0.125 mg/L (Standard #4), 0.0625 mg/L (Standard #5), and 0.0312 mg/L (Standard #6). Five microliters of each of these standards were added to microtubes containing 140 µL assay buffer and 5 µL of 10 mg/L beta-lactoglobulin. For the analytical samples, the standard solutions were substituted with human plasma or serum sample (diluted 10-fold).
Mass spectrometric immunoassay
During the method development, an 8-channel electronic pipettor was used, while for the high-throughput screening the Multimek 96 was used. Regardless of the instrument, identical assay steps were always followed. The antibody-derivatized affinity pipettes were mounted onto the head of the Multimek pipettor and initially rinsed with assay buffer (10 aspirations and dispense cycles, 100 µL volumes each). Next, the pipettes were immersed into a microplate containing the samples and 100 aspirations and dispense cycles were performed (100 µL volumes each) allowing for affinity capture of beta-2-microglobulin and beta lactoglobulin. The pipettes were then rinsed with assay buffer (100 cycles), and twice with water (10 cycles each). In preparation of elution, the affinity pipettes containing the retrieved protein were rinsed with 1 mM N-octylglucoside (single cycle with a 150 µL aliquot) in order to homogenize the subsequent MALDI matrix draw and elution by completely wetting the porous microcolumns inside the pipettes. For elution of the captured proteins, 6 µL aliquots of MALDI matrix (25 g/L α-cyano-4-hydroxycinnamic acid in aqueous solution containing 33% (v/v) acetonitrile, and 0.4 % (v/v) trifluoroacetic acid) were aspired into the affinity pipettes, and after a 10 second delay (to allow for the dissociation of the protein from the capturing antibody), the eluates from all 96 affinity pipettes containing the targeted proteins were dispensed directly onto a 96-well formatted MALDI target. Following air-drying and visual inspection of the sample spots, linear mass spectra were acquired with a delayed extraction mode using a 1.7 kV draw out pulse, 200 ns delay, and a full accelerating potential of 20 kV. Five mass spectra were acquired from each sample spot, each consisting of three-hundred laser shots. The mass spectra were processed (baseline subtracted and smoothed) with Flex Analysis software (Bruker Daltonics). The peak heights for the beta-2-microglobulin and beta-lactoglobulin signals were measured and entered into an Excel spreadsheet. The ratios of the b2m/BL peak heights were calculated, and the average ratio for each sample determined. A standard curve was generated by plotting the b2m/BL ratios against the concentration of the human b2m standards, and the data was fitted with a linear trendline using Sigma Plot (Systat Software, San Jose, CA). This standard curve was then utilized to determine the absolute concentration of beta-2-microglobulin and its isoforms in the analytical samples.
RESULTS AND DISCUSSION
Internal Reference Standard
The first step in the development of a quantitative assay is the selection of internal reference standard (IRS). We have used homologous proteins from other animal species as IRS in the past for several protein assays, including b2m 24, 25. These animal homologs are recognized by the anti-human protein antibody and are spiked into the analytical sample. The single antibody then retrieves both the target protein and the IRS, and they register in the same region of the mass spectrum, but at a slightly different m/z value. The shortcomings of this approach is that the concentration of the IRS can vary in various lots of animal sera (which are often used because purified proteins are unavailable), and that for different protein assays different IRS is required, which can increase the assay development time and cost. One way of alleviating these issues is to use universal and exogenous IRS. A single protein may serve as an IRS if an antibody toward that protein is co-immobilized with the antibody toward the targeted human protein, and the analytical samples are spiked with constant amounts of that IRS. An important prerequisite for the universal IRS is that it can not be endogenous to human plasma or serum - its spiked concentration in the analytical samples shall always be constant and not influenced by the human serum components. The IRS should also produce signals in the mass spectra that are in vicinity of the targeted protein signal, so that the same MS acquisition parameters can be used for both proteins. Based on these criteria, and having in mind the molecular weight of b2m (MW=11,731.2), we have selected beta-lactoglobulin as an IRS for this assay (and possibly others as well). Beta-lactoglobulin (MW=18,281) is major whey protein of cow's milk, present in many other mammalian species, but not in humans. It can be obtained in large quantities, and is relatively affordable. To assess the ionization efficiencies and confirm the eligibility of BL as an IRS, equimolar mixtures of b2m and BL were prepared and directly analyzed via MALDI-TOF MS, producing signals at relatively equal intensities (not shown).
Assay optimization
In the first step of the assay optimization we determined the ratios of the immobilized antibodies in the affinity pipettes. Generally, the affinity pipettes should have more affinity for the targeted protein than the IRS because the targeted protein varies in concentration across the samples, while the IRS can be spiked and kept constant at levels that saturate the anti-IRS antibody and produce constant signal in the mass spectra. After several empirical iterations, the optimal ratio of the two antibodies in the affinity pipettes was determined to be 5.7:1 (b2m:BL), and the optimal volume and concentration of BL spiked in the samples was determined to be 5 µL, and 10 mg/L, respectively. Next, we experimented with the volumes and dilutions of human serum or plasma to determine the optimal sample volume into the analytical samples. It is very important not to saturate the anti-b2m antibody so that accurate quantification can be achieved. It was determined that 5 µL of 10-fold diluted serum or plasma provided the best results. At normal concentrations of b2m in plasma (~2 mg/L), these volume and concentration translate into less than 100 fmoles of beta-2-microglobulin in the analytical samples.
Standard Curve
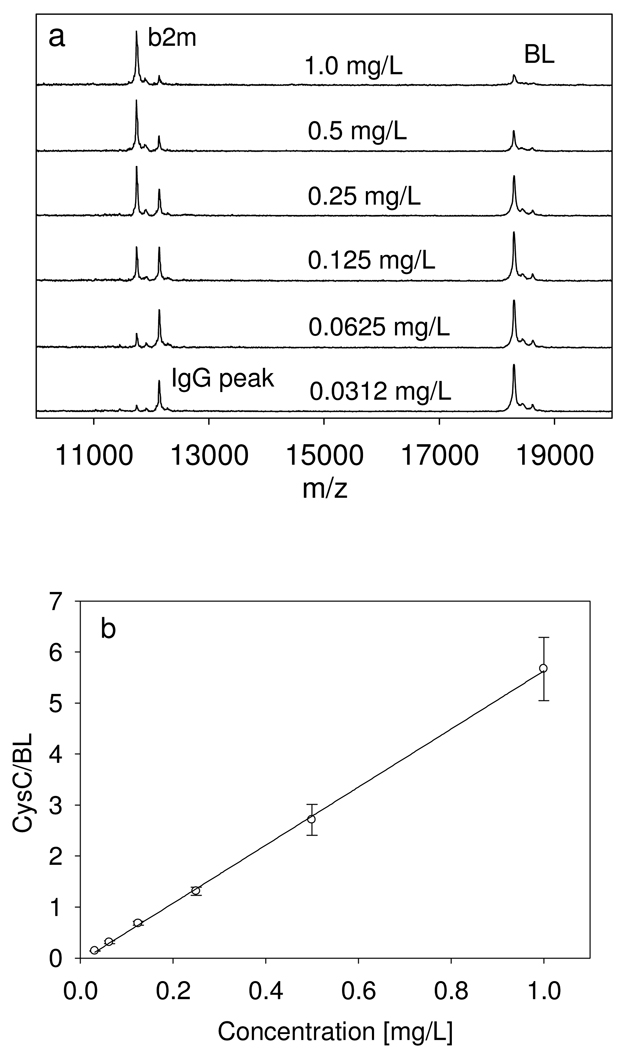
A 6-point standard curve was used for the assay, spanning the concentration range from 0.0312 to 1.0 mg/L. The range of the standard curve was sufficient to determine the concentration of beta-2-microglobulin in all of the examined human plasma and serum samples (diluted appropriately). An example of the standard curve, along with representative mass spectra, is shown in Fig. 1. The response is linear across the entire range, with a coefficient of determination (R2=0.999) and standard error of estimate (SEE=0.0534). A standard curve was run with each analytical sample mass spectrometric immunoassay.
Fig. 1.
(a) Representative beta-2-microglobulin standards mass spectra, and (b) Standard curve generated with the beta-2-microglobulin mass spectrometric immunoassay.
Assay parameters
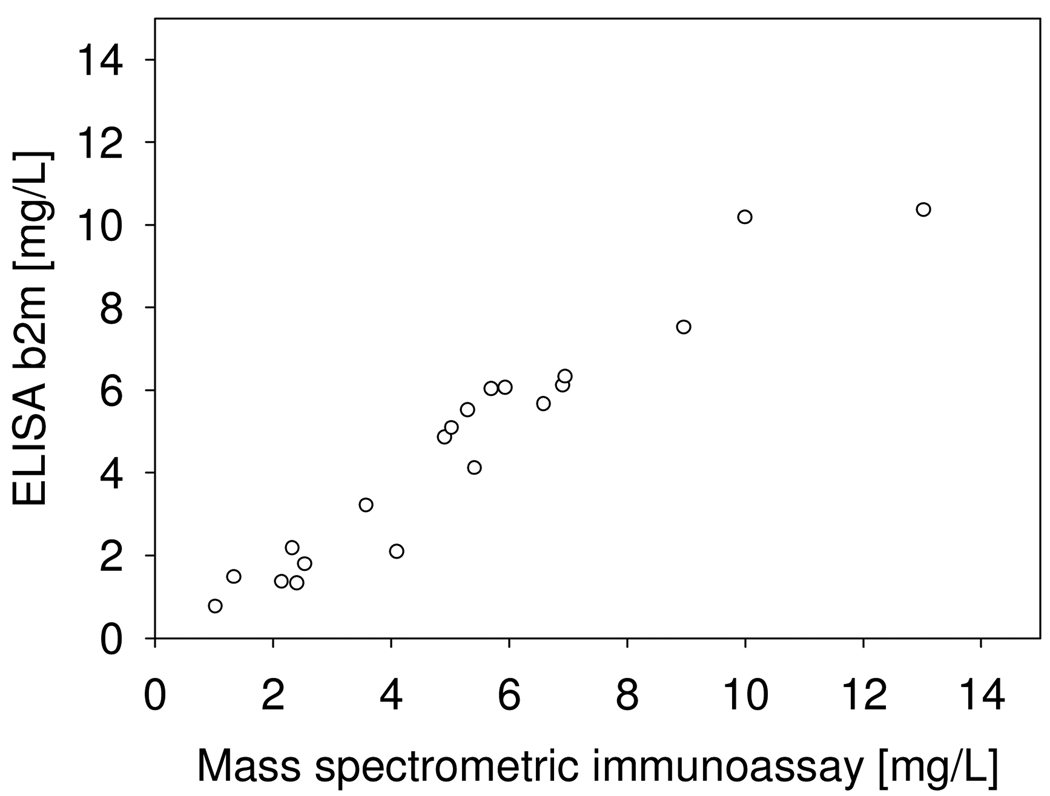
The intra-assay precision (within an assay) was determined by analyzing three plasma samples, in triplicates, each with a single standard curve. The inter-assay precision (between assays) was determined by analyzing one plasma sample three times, on different days, with separate standard curves each time. The results are shown in Table 1, and indicate CVs of less than 10%. To determine the linearity of the assay, serum samples with known beta-2-microglobulin concentration were serially diluted, analyzed with the mass spectrometric immunoassay to determine the b2m concentrations, and the results compared to those expected (Table 2). Spiking recovery experiments were also performed by spiking serum samples with different amounts of recombinant human b2m, followed by analysis with the assay to determine the total b2m concentration, and comparison of the results with those expected (Table 3). In a final test of the assay, twenty human serum samples were analyzed both by the mass spectrometric immunoassay and a commercially available ELISA, and the beta-2-microglobulin concentrations determined with the two methods were compared. The graph shown in Fig. 2 indicates good correlation between the two set of numbers (Passing & Bablok fit of 0.49+1.01×, and Cusum linearity p-value of >0.1), validating the results obtained with the new beta-2-microglobulin assay.
Table 1.
Intra-and inter-assay precision.
| Intra-assay CVs | |||
|---|---|---|---|
| Sample | 1 | 2 | 3 |
| STDV: | 0.074 | 0.031 | 0.078 |
| MEAN: | 1.39 | 1.84 | 1.09 |
| CV: | 5.29 | 1.68 | 7.15 |
| Inter-assay CV | |
|---|---|
| STDV | 0.15 |
| MEAN | 1.69 |
| CV | 8.71 |
Table 2.
Assay linearity.
| Sample | Dilution | Observed mg/L |
Expected mg/L |
Recovery O/E % |
|---|---|---|---|---|
| 1 | 1.50 | |||
| 2× | 0.706 | 0.751 | 94.0 | |
| 4× | 0.356 | 0.376 | 94.7 | |
| 8× | 0.158 | 0.188 | 84.0 | |
| 2 | 1.75 | |||
| 2× | 0.782 | 0.874 | 89.5 | |
| 4× | 0.387 | 0.437 | 88.6 | |
| 8× | 0.253 | 0.218 | 116 | |
Table 3.
Spiking recovery.
| Sample | Observed mg/L |
Expected mg/L |
Recovery O/E% |
|---|---|---|---|
| 1 | 1.19 | ||
| 1.70 | 1.49 | 114 | |
| 1.89 | 1.79 | 105 | |
| 2.65 | 2.39 | 111 | |
| 2 | 1.87 | ||
| 2.19 | 2.17 | 101 | |
| 2.56 | 2.47 | 104 | |
| 3.06 | 3.07 | 99.8 | |
Fig. 2.
Beta-2-microglobulin mass spectrometric immunoassay and standard ELISA method comparison.
Human plasma samples screening
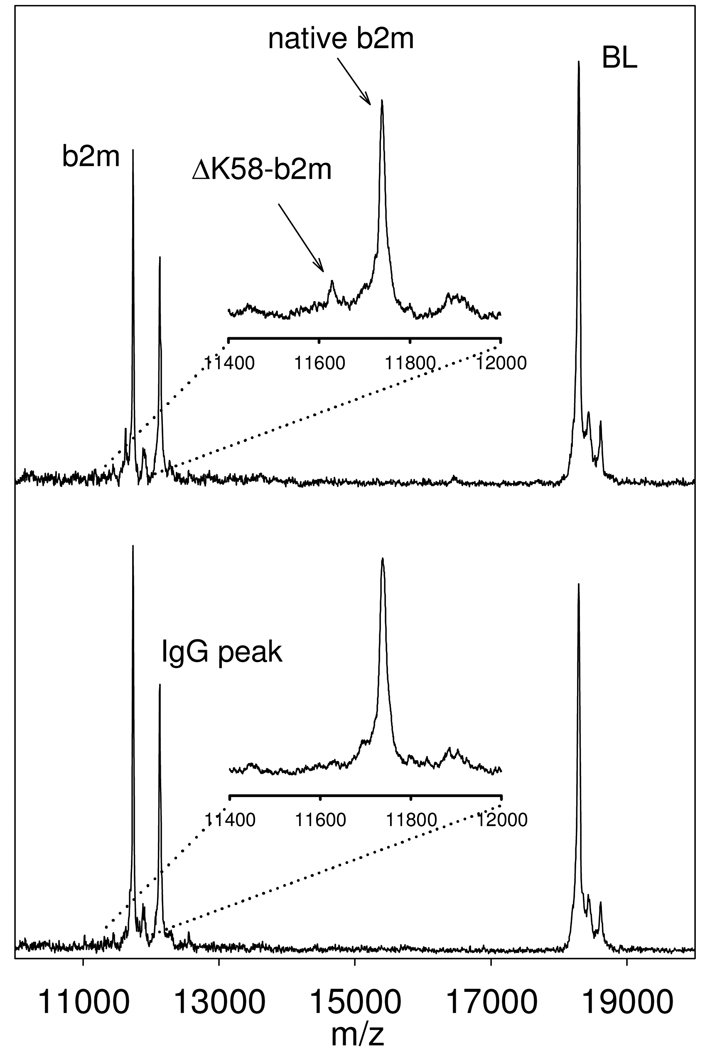
The fully-developed, characterized, and validated beta-2-microglobulin mass spectrometric immunoassay was used to analyze a set of human plasma samples in a high throughput mode. Forty-four sodium heparin plasma samples were utilized to prepare 88 analytical samples (each sample was prepared in duplicate, at different dilutions) and placed in columns 2–12 of a 96-well microplate. The standard curve samples were placed in column 1, along with a control sample with a known beta-2-microglobulin concentration. The 96 beta-2-microglobulin assays were executed in parallel using the Multimek 96 channel pipettor. Following mass spectrometry analysis and spectra processing, a standard curve was constructed from the data in column 1, and the performance of the assay verified with the control sample. The concentrations of beta-2-microglobulin were determined from the b2m/BL ratios in the analytical samples mass spectra, and are presented in Table 4. The average concentration of beta-2-microglobulin in the samples (2.19 mg/L) correlates well with a previously established value (~2.0 mg/L) obtained using standard ELISA approaches 10. The ΔK58-b2m isoform was detected in only one sample (Fig 3), which is also in line with our previous qualitative data that showed its frequency to be 2–6% in the normal population. The concentration of the ΔK58-b2m isoform in the single sample was 0.28 mg/L, and represented ~15% of the total b2m in that sample.
Table 4.
B2m concentrations in normal human plasma samples.
| Sample | native b2m [mg/L] |
ΔK58-b2m [mg/L] |
|---|---|---|
| 1 | 1.91 | |
| 2 | 1.85 | |
| 3 | 2.00 | |
| 4 | 2.05 | |
| 5 | 2.72 | |
| 6 | 2.42 | |
| 7 | 1.93 | |
| 8 | 2.66 | |
| 9 | 3.61 | |
| 10 | 1.89 | |
| 11 | 2.42 | |
| 12 | 2.58 | |
| 13 | 1.80 | |
| 14 | 1.67 | |
| 15 | 2.53 | |
| 16 | 1.50 | |
| 17 | 2.17 | |
| 18 | 2.32 | |
| 19 | 2.25 | |
| 20 | 2.59 | |
| 21 | 2.35 | |
| 22 | 2.80 | |
| 23 | 1.96 | |
| 24 | 1.84 | |
| 25 | 1.52 | 0.28 |
| 26 | 1.15 | |
| 27 | 2.11 | |
| 28 | 2.50 | |
| 29 | 2.48 | |
| 30 | 1.37 | |
| 31 | 2.98 | |
| 32 | 2.52 | |
| 33 | 2.17 | |
| 34 | 2.88 | |
| 35 | 1.94 | |
| 36 | 1.49 | |
| 37 | 2.43 | |
| 38 | 2.20 | |
| 39 | 3.32 | |
| 40 | 1.75 | |
| 41 | 1.74 | |
| 42 | 1.86 | |
| 43 | 1.36 | |
| 44 | 2.32 |
Fig. 3.
Mass spectrum showing the detection of the ΔK58-b2m isoform (upper trace). For comparison, also shown is a mass spectrum containing signals only from native b2m (lower trace).
CONCLUSIONS
The work presented here demonstrates the ability to fully quantify individual forms of post-translationally modified proteins using MALDI-TOF mass spectrometry. Quantitative MALDI 26 offers a unique advantage of detecting and quantifying multiple proteins and protein variants in a single spectrum, with the help of an exogenous internal reference standard that is introduced into the analytical process. The MALDI format is also amenable to high-throughput analysis - more so than LC-MS. The affinity pipettes are processed in parallel (96 at a time) with the help of multi-channel pipetting workstations, and the protein-containing eluates stamped directly onto a 96-well formatted MALDI target. The resulting mass spectrometric immunoassays are fully automated and fast - the 96 samples incubation and deposition onto the MALDI target was executed in 30 min, while the mass spectra acquisition/data processing was performed in 3 hours.
The quantitative mass spectrometric immunoassay for beta-2-microglobulin described in this work can simultaneously detect and quantify both the native protein and the ΔK58-b2m variant. The assay can be used to screen specifically for the ΔK58-b2m variant across larger disease cohorts to assess its importance and diagnostic value. Moreover, these types of assays are ideally suited for detection and quantification of novel variants, not known a priori. The mass spectrometric immunoassays are opening the realms of population proteomics wherein protein diversity is examined in the context of the individuals or a group, either as part of a wide-ranging protein variants cataloguing effort, or a disease-specific biomarker variants (re)discovery endeavor.
ACKNOWLEDGEMENT
The project described was supported by Grant Number 1 R43 RR025701 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
REFERENCES
- 1.Anderson NL. Clin Chem. 2010;56:177–185. doi: 10.1373/clinchem.2009.126706. [DOI] [PubMed] [Google Scholar]
- 2.Carr SA, Anderson L. Clin Chem. 2008;54:1749–1752. doi: 10.1373/clinchem.2008.114686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Addona TA, Abbatiello SE, Schilling B, Skates SJ, Mani DR, Bunk DM, Spiegelman CH, Zimmerman LJ, Ham AJ, Keshishian H, Hall SC, Allen S, Blackman RK, Borchers CH, Buck C, Cardasis HL, Cusack MP, Dodder NG, Gibson BW, Held JM, Hiltke T, Jackson A, Johansen EB, Kinsinger CR, Li J, Mesri M, Neubert TA, Niles RK, Pulsipher TC, Ransohoff D, Rodriguez H, Rudnick PA, Smith D, Tabb DL, Tegeler TJ, Variyath AM, Vega-Montoto LJ, Wahlander A, Waldemarson S, Wang M, Whiteaker JR, Zhao L, Anderson NL, Fisher SJ, Liebler DC, Paulovich AG, Regnier FE, Tempst P, Carr SA. Nat Biotechnol. 2009;27:633–641. doi: 10.1038/nbt.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aebersold R. Nat Methods. 2009;6:411–412. doi: 10.1038/nmeth.f.255. [DOI] [PubMed] [Google Scholar]
- 5.Bell AW, Deutsch EW, Au CE, Kearney RE, Beavis R, Sechi S, Nilsson T, Bergeron JJ. Nat Methods. 2009;6:423–430. [Google Scholar]
- 6.Jaffe AS, Van Eyk JE. In: Cardiovascular biomarkers - pathophysiology and disease management. Morrow DA, editor. Totowa: Humana Press; 2006. pp. 161–174. [Google Scholar]
- 7.Niederkofler EE, Kiernan UA, O'Rear J, Menon S, Saghir S, Protter AA, Nelson RW, Schellenberger U. Circulation: Heart Failure November. 2008;1:258–264. doi: 10.1161/CIRCHEARTFAILURE.108.790774. [DOI] [PubMed] [Google Scholar]
- 8.Saudek CD, Brick JC. J Diabetes Sci Technol. 2009;3:629–634. doi: 10.1177/193229680900300402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manicourt D, Brauman H, Orloff S. Ann Rheum Dis. 1978;37:328–332. doi: 10.1136/ard.37.4.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schardijn GH, Statius van Eps LW. Kidney Int. 1987;32:635–641. doi: 10.1038/ki.1987.255. [DOI] [PubMed] [Google Scholar]
- 11.Grey HM, Kubo RT, Colon SM, Poulik MD, Cresswell P, Springer T, Turner M, Strominger JL. J Exp Med. 1973;138:1608–1612. doi: 10.1084/jem.138.6.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drueke TB. Nephrol Dial Transplant. 2000;15 Suppl 1:17–24. [Google Scholar]
- 13.Heegaard NH. Amyloid. 2009;16:151–173. doi: 10.1080/13506120903151775. [DOI] [PubMed] [Google Scholar]
- 14.Freeman MR. J Urol. 2007;178:10–11. doi: 10.1016/j.juro.2007.03.203. [DOI] [PubMed] [Google Scholar]
- 15.Nissen MH, Roepstorff P, Thim L, Dunbar B, Fothergill JE. Eur J Biochem. 1990;189:423–429. doi: 10.1111/j.1432-1033.1990.tb15505.x. [DOI] [PubMed] [Google Scholar]
- 16.Nissen MH, Thim L, Christensen M. Eur J Biochem. 1987;163:21–28. doi: 10.1111/j.1432-1033.1987.tb10731.x. [DOI] [PubMed] [Google Scholar]
- 17.Nissen MH, Plesner T, Rorth M. Clin Chim Acta. 1984;141:41–50. doi: 10.1016/0009-8981(84)90165-7. [DOI] [PubMed] [Google Scholar]
- 18.Plesner T, Wiik A. Scand J Immunol. 1979;9:247–254. doi: 10.1111/j.1365-3083.1979.tb02728.x. [DOI] [PubMed] [Google Scholar]
- 19.Heegaard NH, Jorgensen TJ, Rozlosnik N, Corlin DB, Pedersen JS, Tempesta AG, Roepstorff P, Bauer R, Nissen MH. Biochemistry. 2005;44:4397–4407. doi: 10.1021/bi047594t. [DOI] [PubMed] [Google Scholar]
- 20.Mimmi MC, Jorgensen TJ, Pettirossi F, Corazza A, Viglino P, Esposito G, De Lorenzi E, Giorgetti S, Pries M, Corlin DB, Nissen MH, Heegaard NH. Febs J. 2006;273:2461–2474. doi: 10.1111/j.1742-4658.2006.05254.x. [DOI] [PubMed] [Google Scholar]
- 21.Wang M, Harhaji L, Lamberth K, Harndahl M, Buus S, Heegaard NH, Claesson MH, Nissen MH. Scand J Immunol. 2009;69:203–212. doi: 10.1111/j.1365-3083.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- 22.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Proc Natl Acad Sci U S A. 2005;102:10852–10857. doi: 10.1073/pnas.0500426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nedelkov D, Phillips DA, Tubbs KA, Nelson RW. Mol Cell Proteomics. 2007;6:1183–1187. doi: 10.1074/mcp.M700023-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Niederkofler EE, Tubbs KA, Gruber K, Nedelkov D, Kiernan UA, Williams P, Nelson RW. Anal. Chem. 2001;73:3294–3299. doi: 10.1021/ac010143j. [DOI] [PubMed] [Google Scholar]
- 25.Tubbs KA, Nedelkov D, Nelson RW. Anal Biochem. 2001;289:26–35. doi: 10.1006/abio.2000.4921. [DOI] [PubMed] [Google Scholar]
- 26.Duncan MW, Roder H, Hunsucker SW. Brief Funct Genomic Proteomic. 2008;7:355–370. doi: 10.1093/bfgp/eln041. [DOI] [PMC free article] [PubMed] [Google Scholar]