Abstract
The transmembrane protein Van gogh-like 2 (Vangl2) is a component of the non-canonical Wnt/Planar Cell Polarity (PCP) signaling pathway, and is required for tangential migration of facial branchiomotor neurons (FBMNs) from rhombomere 4 (r4) to r5–r7 in the vertebrate hindbrain. Since vangl2 is expressed throughout the zebrafish hindbrain, it might also regulate motor neuron migration in other rhombomeres. We tested this hypothesis by examining whether migration of motor neurons out of r2 following ectopic hoxb1b expression was affected in vangl2− (trilobite) mutants. Hoxb1b specifies r4 identity, and when ectopically expressed transforms r2 to an "r4-like" compartment. Using time-lapse imaging, we show that GFP-expressing motor neurons in the r2/r3 region of a hoxb1b-overexpressing wild-type embryo migrate along the anterior-posterior (AP) axis. Furthermore, these cells express prickle1b (pk1b), a Wnt/PCP gene that is specifically expressed in FBMNs and is essential for their migration. Importantly, GFP-expressing motor neurons in the r2/r3 region of hoxb1b-overexpressing trilobite mutants and pk1b morphants often migrate, even though FBMNs in r4 of the same embryos fail to migrate longitudinally (tangentially) into r6 and r7. These observations suggest that tangentially migrating motor neurons in the anterior hindbrain (r1–r3) can use mechanisms that are independent of vangl2 and pk1b functions. Interestingly, analysis of tri; val double mutants also suggests a role for vangl2-independent factors in neuronal migration, since the valentino mutation partially suppresses the trilobite mutant migration defect. Together, the hoxb1b and val experiments suggest that multiple mechanisms regulate motor neuron migration along the AP axis of the zebrafish hindbrain.
Keywords: motor neuron, neuronal migration, hoxb1, van gogh-like 2, prickle1
INTRODUCTION
Neuronal migration accompanies induction and specification of neurons in the developing nervous system. Precise migration is critical for nervous system function, and deficiencies in this process underlie many human genetic brain disorders (Copp and Harding, 1999). Newborn neurons migrate along distinct radial and tangential pathways to their final destinations in the developing neural tube. Radially migrating neurons are closely associated with radial glial fibers (Rakic, 1971), and the underlying mechanisms involving the secreted protein Reelin, its receptors ApoER2 and Vldlr, and the cytoplasmic protein Disabled have been studied intensively (see reviews by Hatten, 1999; Parnavelas, 2000; Kerjan and Gleeson, 2007; Kawauchi and Hoshino, 2008).
In addition to radial migration, a significant number of forebrain, cerebellar, and hindbrain neurons also migrate tangentially, independent of the radial glia (O’Rourke et al., 1992; Luskin, 1993; Lois et al., 1996; Pearlman et al., 1998; reviewed in Hatten, 1999, Marin and Rubenstein, 2003, and Chandrasekhar, 2004). However, given the diversity of cell types migrating tangentially, it is not surprising that a number of heterogeneous mechanisms have been implicated including growth factors, and axon guidance and cell adhesion molecules (Alcantara et al., 2000; Wong et al., 2001; Denaxa et al., 2001; Park et al., 2002; Flames et al., 2004; Pozas and Ibanez, 2005; Kawasaki et al., 2006). Indeed, our and other studies of facial branchiomotor neuron (FBMN) migration in the zebrafish and mouse hindbrain have defined novel roles for components of the non-canonical Wnt signaling pathway in tangential neuronal migration (Bingham et al., 2002; Jessen et al., 2002; Carreira-Barbosa et al., 2003; Wada et al., 2005, 2006; Rohrschneider et al., 2007; Song et al., 2006; Sittaramane et al., 2009; Vivancos et al., 2009; Derrick Glasco and A.C., unpublished data).
The FBMNs are a subset of cranial motor neurons found in the vertebrate brainstem (Lumsden and Keynes, 1989; Chandrasekhar, 2004). In mammals, the facial motor neurons compose the motor component of cranial nerve VII, and innervate muscles of facial expression, and of the middle ear and upper neck. During development, FBMNs in all vertebrates, except chick, undergo a stereotypic migration along the rostrocaudal axis (i.e., tangential to radial glia) from their birth place in rhombomere 4 (r4) into caudal rhombomeres (r5–r7) (Gilland and Baker, 1993; Goddard et al., 1996; Studer et al., 1996; Chandrasekhar et al., 1997; Higashijima et al., 2000; Bingham et al., 2002; for chick, see Studer, 2001). Inactivation of transcription factors like r4-expressed Hoxb1 and Pbx4 (Studer et al., 1996; McClintock et al., 2002; Cooper et al., 2003) or FBMN-expressed Ebf, Nkx6.1, Hdac1 and Tbx20 (Garel et al., 2000; Muller et al., 2003; Song et al., 2006; Nambiar and Henion, 2007; Pocock et al., 2008) causes defects in FBMN migration, and these factors appear to cell-autonomously regulate the ability of the motor neurons to respond to environmental cues. Conversely, inactivation of transcription factors like Krox20 and Kreisler/Valentino/MafB that results in the loss of rhombomere identity (r3 and r5 in Krox20 mutants (Schneider-Manoury et al., 1997), and r5 and r6 in kriesler/valentino mutants (Cordes et al., 1994; Moens et al., 1996)) leads to defective FBMN migration, likely a non-cell autonomous effect due to the potential loss of environmental cues caudal to r4 (Chandrasekhar et al., 1997; Garel et al., 2000; Studer, 2001). Interestingly, some proteins belonging to the non-canonical Wnt signaling pathway also appear to regulate FBMN migration in zebrafish through non cell-autonomous mechanisms (Bingham et al., 2002; Jessen et al., 2002; Carreira-Barbosa et al., 2003; Wada et al., 2005, 2006).
In mouse, targeted disruption of the r4-expressed Hoxb1 gene leads to the elimination of tangential migration of FBMNs (Studer et al., 1996). Similarly, hoxb1a, one of two zebrafish orthologs of mouse Hoxb1, is required for FBMN migration (McClintock et al., 2002). Furthermore, loss of function of the Wnt/PCP protein Strabismus (Stbm)/Van gogh-like 2 (Vangl2) in zebrafish trilobite (tri) mutants also eliminates tangential migration of FBMNs (Bingham et al., 2002; Jessen et al., 2002). In this study, we further investigated the mechanisms regulating neuronal migration by analyzing the effects of hoxb1a-induced changes on branchiomotor neuron development. Following ectopic expression of hoxb1a or hoxb1b in r2, trigeminal (nV) motor neurons are arranged in longitudinal columns in r2 and r3, reminiscent of migrating FBMNs in r4–r7 (McClintock et al., 2001, 2002). These nVII-like (nVII’) neurons express pk1b, an FBMN marker that is essential for migration (Rohrschneider et al., 2007). Using time-lapse imaging, we show that motor neurons in r2 and r3 of an ectopic hoxb1a-expressing embryo migrate along the anterior-posterior (AP) axis. Intriguingly, motor neurons in r2 and r3 can migrate along the AP axis in ectopic hoxb1a-expressing tri mutants and pk1b morphants, suggesting that tangential migration of BMNs can occur independently of Wnt/PCP signaling. Consistent with this, many FBMNs migrate out of r4 in trilobite; valentino double mutants, indicating that stbm/vangl2-deficient FBMNs can migrate out of r4 under specific conditions. Collectively, these data suggest that multiple mechanisms regulate tangential migration of motor neurons in the zebrafish hindbrain.
MATERIALS AND METHODS
Animals
Maintenance of zebrafish stocks, and collection and development of embryos in E3 embryo medium were carried out as described previously (Westerfield, 1995; Bingham et al., 2002). The tritc240a (Hammerschmidt et al., 1996) and valb337 (Moens et al., 1996) alleles were used in these studies. To facilitate analysis of branchiomotor neuron development, Tg(isl1:GFP) fish (Higashijima et al., 2000) were used for all experiments.
RNA and morpholino injections
Full-length synthetic mRNAs for hoxb1bWT and hoxb1bmut (McClintock et al., 2001) were synthesized using the mMessage mMachine kit (Ambion) and injected at a dose of 200 pg per embryo. Stbm/vangl2 and pk1b antisense morpholinos were injected at doses described previously (Jessen et al., 2002; Rohrschneider et al., 2007). Embryos at the one-four cell stage were injected with RNAs and/or morpholinos and examined at several ages from 18–36 hours post fertilization (hpf) for FBMN migration phenotypes.
Immunohistochemistry, in situ hybridization, and imaging
Whole-mount immunohistochemistry (zn5, 3A10, GFP antibodies) was performed as described previously (Vanderlaan et al., 2005; Sittaramane et al., 2009). Synthesis of the digoxygenin-labeled probes, and whole-mount in situ hybridization was carried out as described previously (Bingham et al., 2003). Embryos were deyolked, mounted in 70% glycerol, and examined with an Olympus BX60 microscope. For confocal imaging, fixed embryos were mounted in glycerol, and viewed under an Olympus IX70 microscope equipped with a BioRad Radiance 2000 confocal system. In all comparisons, at least ten embryos for each category were examined.
Time-lapse imaging and analysis
Embryos were mounted dorsally in 1% agarose, coverslipped, and bathed in E3 containing 0.002% tricaine to anesthetize the embryo and 10mM HEPES to buffer pH around the embryo. Images were acquired every five minutes using Cytos Imaging Software (ASI Inc., Eugene, OR) on an Olympus BX60 microscope equipped with shutters in the fluorescence and bright-field light paths. Recordings were carried out at 28.5°C for 5–6 hours per embryo. For generating cell tracks, movies were imported into imaging software (Dynamic Image Analysis System (DIAS), Soll Technologies Inc., Iowa City, IA), stabilized to correct for drift, cell outlines were manually outlined on individual frames, and cell paths were generated by joining centroids of cells in successive frames using DIAS.
RESULTS
Ectopic hoxb1b expression generates "r4-like" motor neurons in rhombomere 2
McClintock et al. (2001) demonstrated that ectopic expression of hoxb1b in the zebrafish anterior hindbrain led to the expression of an r4 marker, hoxb1a, in the r2 region. In addition, ectopic Mauthner cells, an r4-specific reticulospinal neuron, were found in the r2 region of hoxb1b-overexpressing embryos, suggesting strongly that this region had acquired r4 identity (McClintock et al., 2001). To further test this possibility, we examined the organization of branchiomotor neurons in hoxb1b-overexpressing embryos using the Tg(islet1:gfp) strain, which expresses GFP in cranial motor neurons (Higashijima et al., 2000). As shown previously, hoxb1a was often expressed ectopically in presumptive r2 in hoxb1b RNA-injected wild-type embryos (Fig. 1B; 37/84 embryos), indicative of changes in rhombomere identity. In most of these embryos (32/37), the hoxb1a expression domain spanned r2–r4, suggesting that the anterior hindbrain had been posteriorized extensively (Fig. 1B). Further, as shown previously, ectopic Mauthner cells were frequently found in the r2 region of hoxb1b RNA-injected embryos (Fig. 1D; 5/10). Consistent with these effects, GFP-expressing branchiomotor neurons in hoxb1b-expressing embryos also exhibited r4-like characteristics (Fig. 1D, F–H). In control embryos, facial branchiomotor neurons (FBMNs, nVII) were found predominantly in r5–r7 as a result of tangential (caudal) migration following induction in r4 (Fig. 1C, E; Table 1; Chandrasekhar et al., 1997; Higashijima et al., 2000). In addition, trigeminal motor neurons (nV) were found in characteristic mediolaterally organized clusters in r2 and r3 (Fig. 1C, E; Table 1). In contrast, in hoxb1b RNA-injected embryos, GFP-expressing cells in the r2–r3 region were frequently organized in longitudinal columns (Fig. 1D, F; Table 1). Since this arrangement is reminiscent of migratory FBMNs, we consider these cells to have FBMN-like (nVII') features. It is therefore possible that GFP-expressing putative motor neurons in the r2–r3 region migrate along the anterior-posterior (AP) axis following induction. Consistent with this interpretation, many GFP-expressing cells in the r2–r3 region of hoxb1b RNA-injected embryos extended axons from the anterior margin of the longitudinal column rather than laterally, as typical of trigeminal (nV) neurons (Fig. 1G, H). These axons subsequently turned laterally to exit the hindbrain, in a manner similar to FBMN axons located in r5–r7 (Fig. 1G, H). Importantly, injection of RNA encoding a non-functional Hoxb1b protein with a mutation in the homeodomain (Q257 to E257; McClintock et al., 2001) into wild-type embryos did not lead to ectopic hoxb1a expression in the r2 region (data not shown) or to the formation of FBMN-like (nVII') GFP-expressing cells in the r2–r3 region (Table 1). These results suggest strongly that ectopic hoxb1b expression in the anterior hindbrain alters the fate of resident motor neurons such that they resemble FBMNs found in r4–r7. In many hoxb1b RNA-injected embryos exhibiting the FBMN-like phenotype, there was a dramatic increase in the number of GFP-expressing motor neurons, particularly in the midbrain and r2–r3 (Fig. 1F, H; 9/21 embryos).
Figure 1.
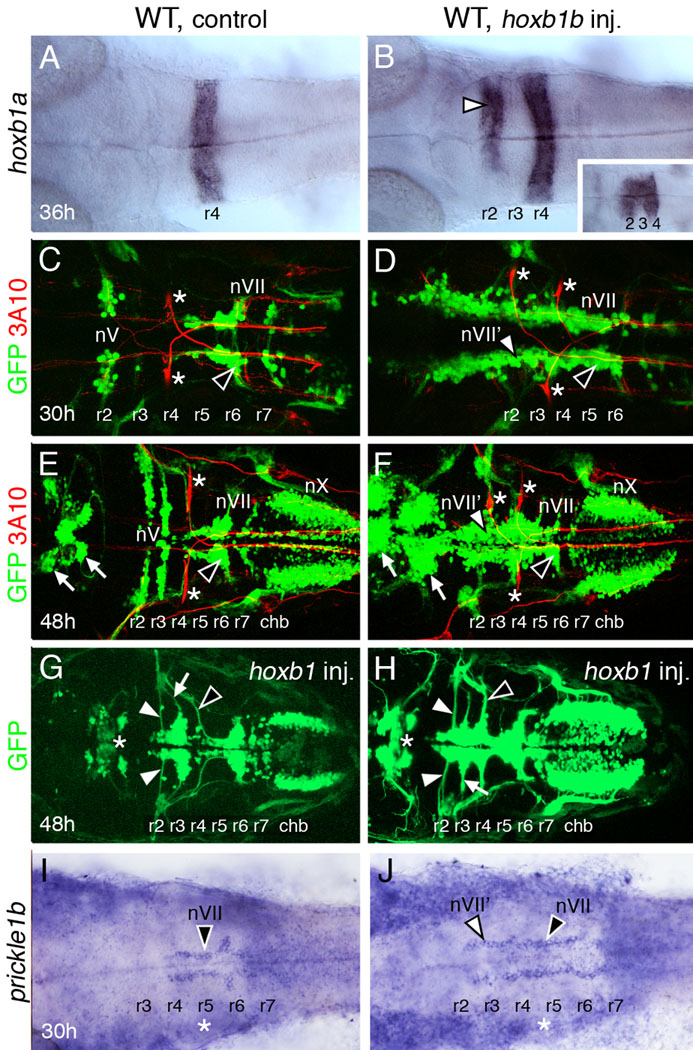
Ectopic expression of hoxb1b generates "r4-like" characteristics in rhombomere 2 (r2). All panels show dorsal views of the hindbrain with anterior to the left. Panels C–F show merged confocal images of GFP-expressing motor neurons (green) and 3A10 antibody-labeled reticulospinal and sensory neurons (red; Mauthner cells in r2 and r4 are indicated by asterisks). (A) In a 36 hpf control wild-type embryo, hoxb1a is expressed in r4. (B) In a wild-type embryo injected with hoxb1b RNA, hoxb1a is expressed in r4, but also ectopically in r2 (arrowhead). Hoxb1a expression frequently extends into r3, merging its normal and ectopic expression domains (inset). (C, E) In a 30 hpf control embryo (C), the trigeminal (nV) motor neurons are found in r2, and the facial branchiomotor neurons (nVII; FBMNs) have mostly migrated out of r4 into r6 (arrowhead) and r7. By 48 hpf (E), the nV motor neurons are found in characteristic mediolaterally elongated clusters in r2 and r3, and most of the FBMNs are located in r6 (arrowhead). The vagal (nX) motor neurons in the caudal hindbrain (chb) and midbrain motor neurons (arrows) are found in characteristic locations. (D, F) In a 30 hpf hoxb1b-injected wild-type embryo (D), GFP-expressing motor neurons are found in longitudinal columns extending the length of the hindbrain, with FBMNs (nVII; black arrowhead) in r4 through r6, and FBMN-like neurons (nVII'; white arrowhead) in the more rostral hindbrain including r2 and r3. In a 48 hpf embryo (F), the medially-positioned longitudinal columns are still evident, and are especially pronounced for the FBMN-like neurons (nVII'; white arrowhead) in r2 and r3. Many FBMNs (black arrowhead) have migrated into r6. The nX neurons in the caudal hindbrain are unaffected, but the midbrain motor neurons (arrows) are greatly increased in number. (G, H) In these 48 hpf hoxb1b-injected embryos, axon fascicles (white arrowheads) emerge from the anterior of the FBMN-like clusters in r2 and r3. The axons of unaffected nV neurons (white arrows) and of FBMNs (black arrowheads) are also indicated. The midbrain motor neurons (asterisks) are only slightly affected in these embryos. The apparent reduction in nX neurons in (G) results from embryo curvature, and is an artifact of the mounting process. (I, J) In a 30 hpf control embryo (I), pk1b is expressed in FBMNs (nVII, arrowhead) in r4–r6. In a hoxb1b-injected embryo, pk1b-expressing cells are found in longitudinal columns spanning r2–r6, and are likely composed of FBMN-like neurons (nVII’, white arrowhead) in r2 and r3, and FBMNs (black arrowhead) in r4–r6. Asterisks denote otic vesicle.
Table 1.
Formation of nVII’ neurons in r2/r3 appears to be independent of vangl2 and pk1b function
| Injected RNA* |
# of embryos& |
Organization of GFP-expressing motor neurons in r2 and r3 | |||||
|---|---|---|---|---|---|---|---|
| Wild-type embryos# | trilobite mutants# | pk1b morphants# | |||||
| Normal (nV)@ |
Abnormal (nVII')@ |
Normal (nV)@ |
Abnormal (nVII')@ |
Normal (nV)@ |
Abnormal (nVII')@ |
||
| none | 494 | 99% (419/422) |
1% (3/422) |
94% (28/30) |
6% (2/30) |
100% (42/42) |
0% (0/42) |
| hoxb1bmut | 275 | 98% (252/257) |
2% (5/257) |
100% (18/18) |
0% (0/18) |
-- | -- |
| hoxb1bWt | 1404 | 55% (653/1180) |
45% (527/1180) |
64% (128/199) |
36% (71/199) |
78% (19/25) |
24% (6/25) |
Approximately 200 pg of hoxb1b RNA (see Materials and Methods) was injected per embryo. Injection of the hoxb1bmut RNA (homeodomain mutant) had no effect, whereas injection of the hoxb1bWt RNA generated the ectopic hoxb1a expression phenotype in ~60% of injected wild-type embryos, as described previously (McClintock et al., 2001).
Embryos were scored at 30–48 hpf, and distinguished from wild-type embryos on the basis of body shape (tri mutants) and the loss of FBMN migration (tri mutants and pk1b morphants).
The characteristic (normal) organization of nV neurons, which is identical in wild-type and tri mutant embryos, has been described previously (Bingham et al., 2002), and is shown in Figures 1E and 3E. The abnormal pattern, indicative of partial nVII character (nVII’), is seen in the embryos shown in Figures 1 (G, H) and 3 (G, H).
Data from several experiments, some performed with only wild-type embryos, were pooled. Therefore, the ratio of wild-type to tri mutants is not 3:1.
To further examine motor neuron identity following hoxb1b overexpression, we monitored expression of prickle1b, a Wnt/PCP gene that is expressed in nVII but not nV motor neurons, and is required cell autonomously for FBMN migration (Rohrschneider et al., 2007). Hoxb1b RNA-injected embryos frequently contained ectopic pk1b-expressing cells in r2/r3 (3/5), whereas none (0/5) of the control embryos contained ectopic pk1b-expressing cells (Fig. 1I, J), suggesting strongly that the nVII’ neurons in the r2/r3 region of injected embryos have acquired FBMN identity.
FBMN-like neurons in the r2 region migrate along the AP axis
The longitudinal arrangement of FBMN-like (nVII’) neurons in the r2–r3 region of hoxb1b RNA-injected embryos may result either from the migration of the cells along the AP axis or from the de novo induction of GFP-expressing cells at these positions. To distinguish between these mechanisms, we examined the behaviors of GFP-expressing cells in the r2–r3 region by time-lapse fluorescence microscopy in control and hoxb1b RNA-injected embryos. Since the earliest GFP-expressing nV neurons in r3 of control embryos arise after 26 hpf (Higashijima et al., 2000), we compared behaviors of nV neurons in r2 of control embryos to nVII’ neurons in the broader r2/r3 region of hoxb1b-injected embryos. In 18–24 hpf control embryos, GFP-expressing trigeminal motor (nV) neurons in r2 changed shape frequently, but did not move along the AP axis (Fig. 2A–D; 7 cells, 3 embryos; Movies S1 and S2; Fig. S1). In contrast, in hoxb1b RNA-injected embryos, GFP-expressing cells in the r2–r3 region move along the AP axis (Fig. 2E–H; >8 cells, 3 embryos; Movies S3, S4 and S5; Fig. S1). This migratory behavior further supports the idea that GFP-expressing cells in the r2–r3 region of hoxb1b-overexpressing embryos have acquired an FBMN fate, as suggested previously by their axonal trajectories (Fig. 1G, H) and marker gene expression (Fig. 1J). Nevertheless, since hoxb1a expression often spans the r2–r4 region in hoxb1b RNA-injected embryos (Fig. 1B), it is possible that the longitudinal arrangement of FBMN-like neurons (Fig. 1D, F) is generated by a combination of migration and de novo induction at ectopic locations.
Figure 2.
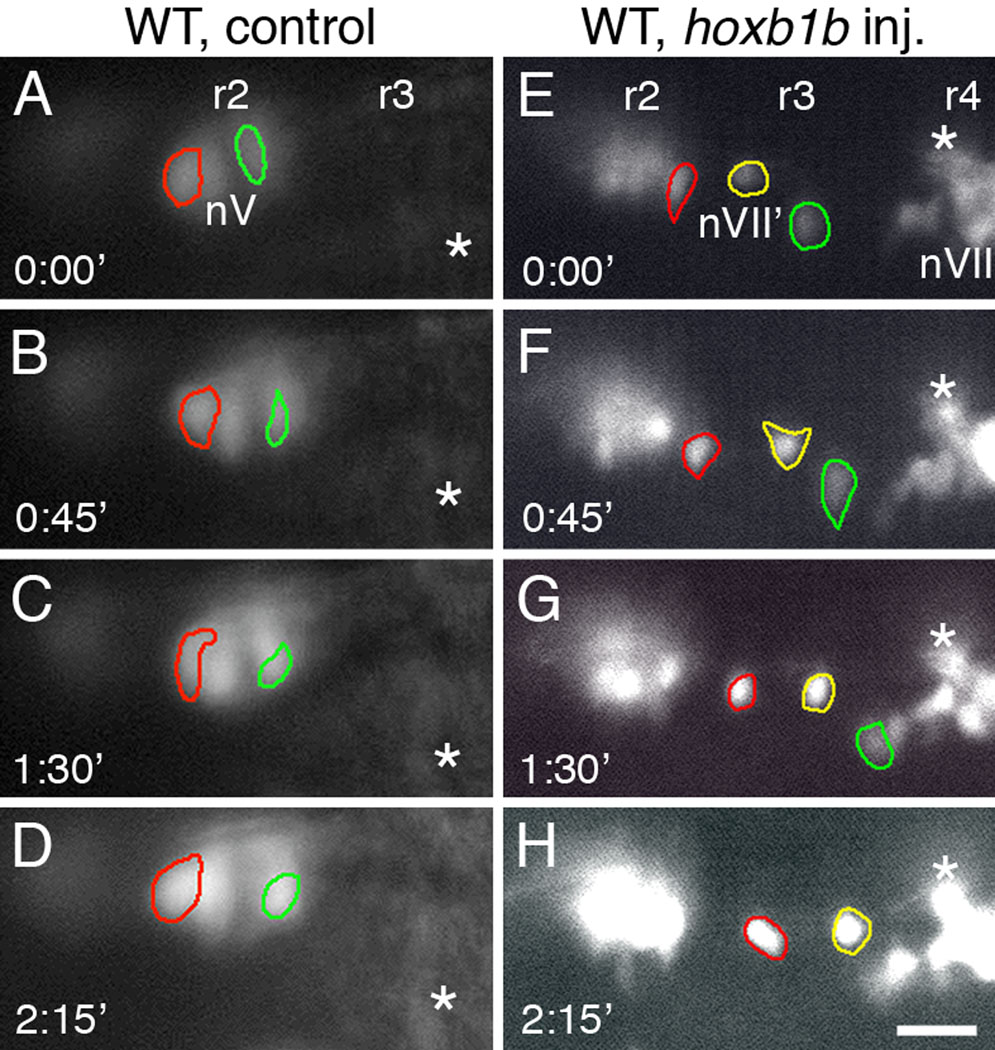
FBMN-like neurons in r2 and r3 move along the anterior-posterior (AP) axis of the hindbrain. All panels show dorsal views of the hindbrain, with anterior to the left. Panels A–D and E–H show a sequence of images, taken at 45 minute intervals, of GFP-expressing neurons in the r2/r3 region (on one side of the hindbrain) in a control wild-type embryo (A–D) and in a wild-type embryo injected with hoxb1b RNA (E–H). Since the earliest GFP-expressing nV neurons in r3 of control embryos arise after 26 hpf (Higashijima et al., 2000), we compared behaviors of nV neurons in r2 of control embryos to nVII’ neurons in the broader r2/r3 region of hoxb1b-injected embryos. Particular cells have been outlined in different colors to monitor their positions over the 135-minute observation period. Fiducial marks (asterisks) are indicated in successive frames. (A–D) In the control embryo, the two nV neurons (red and green outlines) in r2 change shape over time, but do not move along the AP axis. (E–H) In the hoxb1b-injected embryo, the FBMN-like cells (nVII'; red, green and yellow outlines) are located in r2 or r3, adjacent to a cluster of putative nV neurons in r2. The three cells change shape over time, but also move posteriorly, toward the cluster of FBMNs in r4 (nVII). Scale bar, 50 µm.
FBMN-like neurons in the r2–r3 region migrate in trilobite mutants and prickle1b morphants
Given that nVII’ neurons in the r2–r3 region migrated along the AP axis, we wondered whether this cell movement required the function of the transmembrane protein Strabismus/Van gogh-like 2/Trilobite (Jessen et al., 2002; Park and Moon, 2002), which is essential for FBMN migration from r4 into r5–r7 (Bingham et al., 2002). Therefore, we examined the distribution of GFP-expressing cells in the r2–r3 region of hoxb1b RNA-injected trilobite (vangl2−) mutant embryos. In control mutant embryos, trigeminal (nV) motor neurons were found in mediolaterally positioned clusters within r2 and r3 (Fig. 3C, E; Table 1). In hoxb1b RNA-injected trilobite (tri) mutant embryos, two effects were evident. First, the number of GFP-expressing cells in the anterior hindbrain in the presumptive r2–r3 region was greatly increased, as in hoxb1b-injected wild-type embryos (compare Fig. 3D, F to Fig. 1F). Second, many GFP-expressing cells in this region were positioned medially, in a longitudinal fashion (Fig. 3D, F–H; Table 1), which was never observed in control mutant embryos (Fig. 3C, E; Table 1). Although we were unable to obtain good quality time-lapse recordings of migrating GFP-expressing, nVII’ neurons in mutant embryos (data not shown), the similar arrangement of nVII’ neurons between hoxb1b RNA-injected tri mutants and wild-type embryos, where neuronal migration was confirmed (Fig. 2), indicates that the FBMN-like neurons migrate along the AP axis in tri mutant embryos. Importantly, FBMNs failed to migrate out of r4 in hoxb1b RNA-injected tri mutants (Fig. 3D, F–H; Table 1), as in control tri mutants (Fig. 3C,E; Bingham et al., 2002). These results suggest that FBMN-like motor neurons can undergo tangential migration in trilobite (vangl2−) mutants, apparently mediated by Stbm/Vangl2-independent mechanisms (Jessen et al., 2002).
Figure 3.
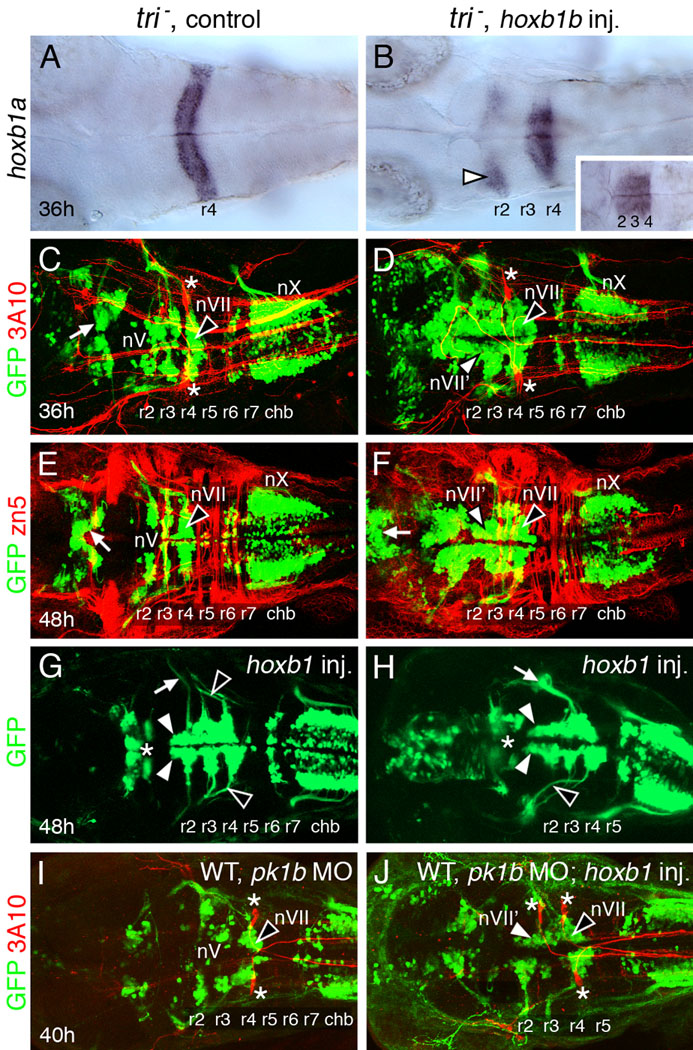
FBMN-like neurons can migrate normally in hoxb1b-injected trilobite mutants and pk1b morphants. All panels show dorsal views of the hindbrain with anterior to the left. Panels C–F, I, J show merged confocal images of GFP-expressing motor neurons (green) and 3A10 antibody-labeled (C, D, I, J) reticulospinal and sensory neurons (red; Mauthner cells in r4 are indicated by asterisks) or zn5 antibody-labeled (E, F) hindbrain commissural neurons and axons (red). (A) In a 36 hpf control tri mutant, hoxb1a is expressed in r4. (B) In a tri mutant injected with hoxb1b RNA, hoxb1a is expressed in r4, but also ectopically in r2 (arrowhead). Hoxb1a expression frequently extends into r3, merging its normal and ectopic expression domains (inset). (C, E) In a 36 hpf control mutant embryo (C), the trigeminal (nV) motor neurons are found in r2 and r3, and the facial branchiomotor neurons (nVII; FBMNs) fail to migrate caudally and remain in r4 (arrowhead). By 48 hpf (E), the nV motor neurons are found in characteristic mediolaterally elongated clusters in r2 and r3, and the FBMNs are still located in r4 (arrowhead). The vagal (nX) motor neurons in the caudal hindbrain (chb) and midbrain motor neurons (arrows) are found in characteristic locations. (D, F) In a 36 hpf hoxb1b-injected tri mutant embryo (D), GFP-expressing motor neurons are found in large numbers in the anterior hindbrain, including r2–r4. The cell bodies form longitudinal columns, suggestive of FBMN-like characteristics (nVII'; white arrowhead). The FBMNs (nVII; black arrowhead) remain in r4 and the vagal (nX) neurons are not affected. In a 48 hpf embryo (F), the medially-positioned longitudinal columns are still evident in r2 and r3 (nVII'; white arrowhead). FBMNs (black arrowhead) remain in r4, while the nX neurons in the caudal hindbrain and the midbrain motor neurons (arrow) are largely unaffected. (G, H) In these 48 hpf hoxb1b-injected tri mutants, motor neuron cell bodies in r2 form longitudinal columns (white arrowheads), suggestive of FBMN-like features. The axons of unaffected nV neurons (white arrow in G) and of FBMNs (black arrowheads) are indicated. For the embryo shown in (H), nV axon fascicles are missing. The nVII fascicle (white arrow) is unusually thick perhaps due to axons of nVII' neurons in r2 and r3 exiting out of r4 with nVII axons. The midbrain motor neurons (asterisks) are only slightly affected in these embryos. (I, J) In a 40 hpf WT embryo injected with pk1b MO (I), the trigeminal (nV) motor neurons are found in r2 and r3, and the facial branchiomotor neurons (nVII; FBMNs) fail to migrate caudally and remain in r4 (arrowhead). In a hoxb1b-injected pk1b morphant (J), GFP-expressing motor neurons in r2 and r3 on one side are located medially in a single elongated cluster that extends up to r4, suggestive of FBMN-like characteristics (nVII'; white arrowhead). The FBMNs (nVII; black arrowhead) remain in r4.
Since nVII’ neurons in the r2/r3 region expressed pk1b (Fig. 1J), which is also necessary for FBMN migration (Rohrschneider et al., 2007), we asked whether nVII’ neurons developed in Tg(isl1:gfp) embryos coinjected with pk1b MO and hoxb1b RNA. As expected, FBMNs failed to migrate out of r4 in control and hoxb1b RNA-injected pk1b morphants (Fig. 3I, J). Importantly, FBMN-like neurons formed in the r2/r3 region of hoxb1b-injected pk1b morphants (Fig. 3I, J; Fig. S2) albeit at a lower frequency (Table 1), suggesting that this event can, on occasion, occur independently of pk1b function. Taken together, the trilobite mutant and pk1b morphant data suggest that the tangential migration of nVII’ neurons in hoxb1b-overexpressing embryos is independent of Wnt/PCP signaling.
FBMNs migrate out of r4 in tri; val double mutants
Although mosaic analysis demonstrated that stbm/vangl2 functions non-cell autonomously for FBMN migration (Jessen et al., 2002), the identity of the cells in which gene function is required is not known because vangl2 is expressed ubiquitously during the period of FBMN migration (Jessen et al., 2002; Park and Moon, 2002). One possibility is that vangl2 functions in r5–r7 to promote migration out of r4, consistent with the observation that r5 and r6 can regulate FBMN migration in mouse (Studer, 2001). Furthermore, FBMNs migrate aberrantly in valentino (val)/kriesler/mafB mutants (Chandrasekhar et al., 1997), in which r5 and r6 (and adjacent boundaries) fail to develop normally, and are replaced by rX, a rhombomere with mixed identity (Cordes et al., 1994; Moens et al., 1996). Therefore, we examined FBMN migration in tri; val double mutants.
Embryos were obtained from incrosses of tritc240a +/−; valb337 +/−; Tg(islet1:gfp) fish, separated into the single mutant and double mutant categories on the basis of the somite (tri) and hindbrain (val) phenotypes at 18 hpf, and the distribution of GFP-expressing FBMNs in r4–r7 was examined at 35–40 hpf. To increase the number of double mutants analyzed, we performed some experiments by injecting stbm/vangl2 MO (Jessen et al., 2002) into embryos obtained from incrosses of val +/− fish (see below). In val mutants, FBMNs migrated aberrantly out of r4 into rX and r7, with the cells exhibiting a scattered distribution in the tissue (Fig. 4C, G; n=22). In contrast, by this stage, FBMNs were tightly clustered in both wild-type (r6 and r7, Fig. 4A, E; n=64) and tri mutant embryos (r4; Fig. 4B, F; n=106). Interestingly, FBMNs in tri; val double mutants exhibited a mixed phenotype (Fig. 4D, H). A majority of FBMNs remained tightly clustered in r4, as in tri single mutants. However, a significant number of GFP-expressing cells were found in rX and r7, suggestive of caudal migration (n=18). We also observed that FBMN axons exited aberrantly out of r6 rather than r4 in many double mutants (Fig. 4D, H; 7/18 embryos); by contrast, FBMN axons always exited from r4, and never from r6 in wild-type and single mutant embryos (0/192 embryos; all categories pooled). This axon pathfinding defect may reflect incorrect formation or specification of the cranial nerve exit points in r4 and r6, since the neural crest that originates in r6 normally expresses val/mafB and likely contributes to the r6 exit point (Moens et al., 1996).
Figure 4.
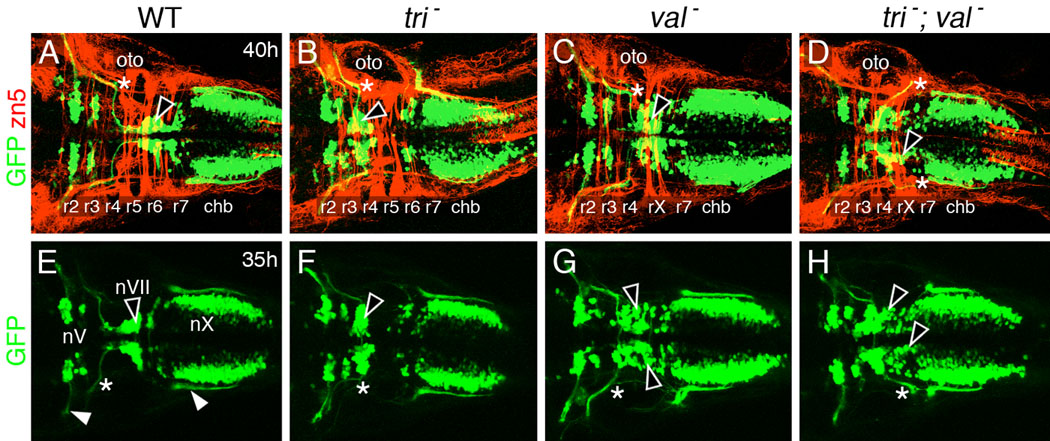
FBMNs appear to migrate caudally in tri mutants upon loss of val function. All panels show dorsal views of the hindbrain with anterior to the left. Panels A–D are merged confocal images of GFP-expressing motor neurons and zn5 antibody-labeled neurons and axons (red). (A, E) In wild-type siblings, FBMNs (nVII; black arrowheads) have mostly migrated into r6 and r7. The nV and nX neurons occupy characteristic positions in r2/r3 and the caudal hindbrain, respectively. The nV and nX axon fascicles (white arrowheads in E) exit the hindbrain from r2 and r7, respectively, and the nVII axon fascicles (asterisk) exit from r4, at the anterior margin of the otocyst (oto). (B, F) In trilobite mutants, FBMNs (arrowheads) remain in r4, and their axons (asterisks) exit the hindbrain from r4. (C, G) In valentino mutants, FBMNs (arrowheads) migrate out of r4 into the caudal rhombomeres rX and r7. However, the migration is disorderly, resulting in a scattered distribution of the motor neuron cell bodies. The nVII axons (asterisks) exit the hindbrain normally from r4. (D, H) In tri; val double mutants, a majority of FBMNs remains in r4 as in tri mutants, but many neurons (arrowheads) have migrated caudally into rX and r7, as in val mutants. In (D), both nVII axon fascicles (asterisks) exit aberrantly from r6, caudal to the otocyst, while in (H), one nVII fascicle exits correctly from r4, and the other aberrantly from rX/r7. The nV and nX axon fascicles exit from the correct rhombomeres in both single mutants and in the double mutant.
It is possible that FBMNs appear to migrate out of r4 in tri; val double mutants due to overcrowding within r4, pushing cells out of r4 at later time points as additional motor neurons differentiate and fail to migrate. Alternatively, the apparent migration defect may reflect misspecification of FBMNs in rX, since hoxb1b expression “leaks” into rX in val mutants (Prince et al., 1998). To distinguish between these causes, we examined the distribution of FBMNs in tri; val mutants (obtained by injecting vangl2 MO into val +/− incross embryos) between 18 and 24 hpf, when the number of FBMNs is much lower than at 36 hpf. Even at these younger ages, FBMNs were found in rX and r5/6 in val−; vangl2 MO and control embryos, respectively (data not shown), indicating that this phenotype is not a non-specific consequence of overcrowding in r4, but may reflect misspecification or caudal migration of neurons.
DISCUSSION
The mechanisms underlying facial branchiomotor neuron (FBMN) migration in the posterior hindbrain have been studied intensively. The roles of many components of the non-canonical Wnt/PCP signaling pathway such as Fzd3, Celsr1, Pk1, and Vangl2/Stbm are well established, although their modes of action remain to be clarified (Bingham et al., 2002; Carreira-Barbosa et al., 2003; Jessen et al., 2002; Rohrschneider et al., 2007; Song et al., 2006; Wada et al., 2006; Vivancos et al., 2009). Moreover, it is not known whether these molecules have the capacity to also regulate neuronal migration in the anterior hindbrain. We tested this possibility using a gain-of-function approach and show here that the Wnt/PCP components Vangl2 and Pk1b are likely not required for neuronal migration in the anterior hindbrain.
Identity of motor neurons in r2 and r3 of hoxb1b-overexpressing embryos
Ectopic expression of hoxb1b in zebrafish causes a transformation of r2 toward an r4 identity (McClintock et al., 2001), in a similar fashion to Hoxa1 overexpression in mice (Zhang et al., 1994). Rhombomere transformation is evident from the ectopic expression of the r4-specific marker hoxb1a in r2 and the ectopic induction of the r4-specific Mauthner reticulospinal neurons in r2 (Fig. 1; McClintock et al., 2001, 2002). In these embryos, the arrangement of islet1-expressing cells in r2 and r3 resembled the FBMN migratory pathway from r4 to r6/7, and suggested that these cells had acquired an FBMN-like (nVII’) identity. Our data strengthen this interpretation since we show that the nVII’ neurons express an nVII-specific marker (Fig. 1), the Wnt/PCP gene pk1b (Rohrschneider et al., 2007), and that nVII’ neurons migrate along the anterior-posterior axis in r2 and r3 (Fig. 2).
The branchiomotor neuron phenotypes of hoxb1b-overexpressing embryos (Fig. 1; McClintock et al., 2001) and lazarus mutants (pbx4−/−; Cooper et al., 2003) look similar in that motor neurons in the r2–r3 region are arranged in longitudinal columns at 30–36 hpf. But we believe that the similarities are merely coincidental for two reasons. First, in lazarus mutants, the trigeminal (nV) neurons do not migrate, and their axons aberrantly exit from r4. By contrast, in hoxb1b-overexpressing embryos, the transformed nVII’ neurons migrate and their axons exit from r2 (Figs. 1 and 2). Second, the lazarus nV axon guidance (and presumably the cell body) phenotype is cell non-autonomous, whereas the hoxb1b nVII’ phenotype is likely cell-autonomous (data not shown).
Motor neuron migration in the anterior hindbrain requires hoxb1a function but not Wnt/PCP signaling
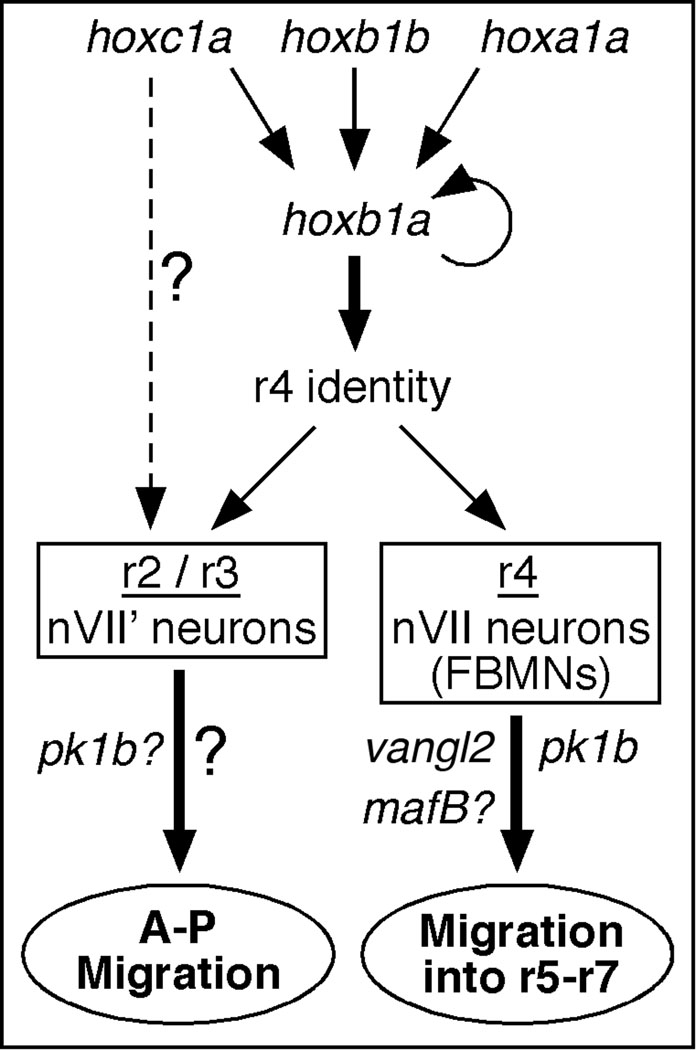
Since hoxb1a and its downstream target pk1b are both necessary for FBMN migration out of r4 (McClintock et al., 2002; Rohrschneider et al., 2007), we wondered whether their functions were also required for the migration of nVII’ neurons in the r2–r3 region. McClintock et al. (2002) showed that the hoxb1b-induced nVII’ phenotype was blocked in hoxb1a morphants. Other aspects of ectopic hoxb1b expression such as induction of ectopic Mauthner cells and hoxb1a expression in r2 were not affected in the hoxb1a morphants, indicating that hoxb1a (and its downstream-regulated genes) was necessary for motor neuron migration in r4–r6 (FBMNs) and in r2–r3 (nVII’). Surprisingly, however, the nVII’ neurons can migrate in hoxb1b-overexpressing embryos injected with vangl2 or pk1b MOs (Fig. 3), suggesting that these Wnt/PCP genes are not necessary for nVII’ neuron migration. These data present an interesting contrast: both hoxb1a and its downstream target pk1b are necessary for FBMN migration from r4, whereas only hoxb1a is required for nVII’ migration in r2. This suggests that hoxb1a regulates nVII’ migration through a pk1b-independent pathway (Fig. 5).
Figure 5.
Model depicting independent mechanisms for motor neuron migration in the anterior hindbrain. All hox paralog group 1 genes (hoxa1a, hoxb1a, hoxb1b, hoxc1a) can induce ectopic hoxb1a expression (and other aspects of r4 identity) in r2. Hoxc1a is less efficient than the other hox genes in inducing r4 identity, but is very efficient in inducing the nVII’ phenotype (McClintock et al., 2001), suggesting a parallel pathway that may be pk1b-dependent (broken arrow). The functions of val/mafB, and the Wnt/PCP genes vangl2 and pk1b are necessary for FBMN migration into r5–r7, but appear to be dispensable for anterior-posterior (A–P) migration of nVII’ neurons in the r2/r3 region.
Other observations are also consistent with the action of multiple mechanisms regulating tangential neuronal migration in the hindbrain. McClintock et al. (2001) found that ectopic expression of various hox paralog group 1 genes had different abilities to induce the r2 to r4 transformation and the nVII’ migration phenotype. Hoxc1a RNA-injected embryos showed ectopic hoxb1a expression only in narrow lateral domains within r2, and ectopic Mauthner neurons were induced at a lower frequency than in hoxb1b (or hoxb1a or hoxa1a)-injected embryos. In contrast, ectopic expression of hoxc1a induced the nVII’ phenotype at the same frequency as hoxa1a, hoxb1a, and hoxb1b misexpression, suggesting that hoxc1a can potentially induce nVII’ neuron migration through a hoxb1a-independent pathway (Fig. 5).
FBMN migration in stbm; mafB double mutants
Several genes have been shown to be necessary for FBMN migration in zebrafish. These include transcription factors (McClintock et al., 2001; Cooper et al., 2003, 2005) such as MafB (Moens et al., 1996), and components of signaling pathways affecting cell polarity and axon guidance, including Vangl2/Stbm (Bingham et al., 2002; Carreira-Barbosa et al., 2003; Wada et al., 2005, 2006; Paulus and Halloran, 2006; Nambiar et al., 2007; Rohrschneider et al., 2007; Sittaramane et al., 2009). In tri; val double mutants (vangl2−/−; mafB−/−), some FBMNs appear to migrate out of r4 into rX (Fig. 4). Since this putative migration occurs even in young embryos containing a small number of FBMNs, with the same time course as in wild-type embryos, it is not a consequence of overcrowding in r4 leading to “overflow” of neurons into rX. One possibility is that the presence of ectopic FBMNs in rX of double mutants reflects mis-specification rather than mis-migration, since hoxb1b is expressed ectopically in rX of val mutants (Prince et al., 1998). Interestingly, ectopic hoxb1b expression is not observed in ventral rX where the FBMNs would be located, leaving open a migratory basis for the double mutant phenotype. Based on these results, we propose that two independent mechanisms regulate FBMN migration: one is vangl2 (Wnt/PCP)-dependent, and promotes migration out of r4; the other is vangl2-independent and prevents migration out of r4 due to the action of repulsive cues (e.g., Eph-ephrin signaling) found in posterior rhombomeres and regulated by val/mafB. In wild-type embryos, the migration-promoting (positive) cue may suppress the response to the repulsive (negative) cue, resulting in FBMN migration out of r4. In tri (vangl2−) mutants, the positive cue is missing resulting in loss of migration. In val mutants, the negative cue is missing permitting migration. In double mutants, both the positive and negative cues are missing, enabling some FBMNs to migrate out of r4. These cues may act in concert with other guidance cues in adjacent rhombomeres (Studer, 2001; Cubedo et al., 2009; Vivancos et al., 2009), enabling effective migration of FBMNs.
In summary, we show that for FBMN-like neurons induced in the anterior hindbrain (r2/r3), migration appears to be independent of vangl2 and pk1b functions. For FBMNs migrating out of r4, a val/mafB-regulated cue may function independently of vangl2 to control migration. Collectively, these data suggest that multiple mechanisms regulate motor neuron migration along the anterior-posterior axis of the zebrafish hindbrain.
Supplementary Material
Acknowledgements
We thank Anagha Sawant for performing the image analysis shown in Fig. S1, and members of our labs for discussion and excellent fish care. The zn5 and 3A10 antibodies were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa.
Grant Support: NIH training grant fellowship (NIGMS T32 GM08396) to SMB, an NSF predoctoral fellowship to OM, a March of Dimes grant (FY07-410) to VEP, and NIH grant (NS040449) to AC.
REFERENCES
- Alcantara S, Ruiz M, De Castro F, Soriano E, Sotelo C. Netrin 1 acts as an attractive or as a repulsive cue for distinct migrating neurons during the development of the cerebellar system. Development. 2000;127:1359–1372. doi: 10.1242/dev.127.7.1359. [DOI] [PubMed] [Google Scholar]
- Bingham S, Chaudhari S, Vanderlaan G, Itoh M, Chitnis A, Chandrasekhar A. Neurogenic phenotype of mind bomb mutants leads to severe patterning defects in the zebrafish hindbrain. Dev. Dyn. 2003;228:451–463. doi: 10.1002/dvdy.10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham S, Higashijima S, Okamoto H, Chandrasekhar A. The Zebrafish trilobite gene is essential for tangential migration of branchiomotor neurons. Dev. Biol. 2002;242:149–160. doi: 10.1006/dbio.2001.0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira-Barbosa F, Concha ML, Takeuchi M, Ueno N, Wilson SW, Tada M. Prickle 1 regulates cell movements during gastrulation and neuronal migration in zebrafish. Development. 2003;130:4037–4046. doi: 10.1242/dev.00567. [DOI] [PubMed] [Google Scholar]
- Chandrasekhar A. Turning heads: development of vertebrate branchiomotor neurons. Dev. Dyn. 2004;229:143–161. doi: 10.1002/dvdy.10444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekhar A, Moens CB, Warren JT, Jr, Kimmel CB, Kuwada JY. Development of branchiomotor neurons in zebrafish. Development. 1997;124:2633–2644. doi: 10.1242/dev.124.13.2633. [DOI] [PubMed] [Google Scholar]
- Cooper KL, Armstrong J, Moens CB. Zebrafish foggy/spt5 is required for migration of facial branchiomotor neurons but not for their survival. Dev Dyn. 2005;234:651–658. doi: 10.1002/dvdy.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KL, Leisenring WM, Moens CB. Autonomous and nonautonomous functions for Hox/Pbx in branchiomotor neuron development. Dev. Biol. 2003;253:200–213. doi: 10.1016/s0012-1606(02)00018-0. [DOI] [PubMed] [Google Scholar]
- Copp AJ, Harding BN. Neuronal migration disorders in humans and in mouse models--an overview. Epilepsy Res. 1999;36:133–141. doi: 10.1016/s0920-1211(99)00047-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes SP, Barsh GS. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994;79:1025–1034. doi: 10.1016/0092-8674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- Cubedo N, Cerdan E, Sapede D, Rossel M. CXCR4 and CXCR7 cooperate during tangential migration of facial motoneurons. Mol Cell Neurosci. 2009;40:474–484. doi: 10.1016/j.mcn.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Denaxa M, Chan CH, Schachner M, Parnavelas JG, Karagogeos D. The adhesion molecule TAG-1 mediates the migration of cortical interneurons from the ganglionic eminence along the corticofugal fiber system. Development. 2001;128:4635–4644. doi: 10.1242/dev.128.22.4635. [DOI] [PubMed] [Google Scholar]
- Flames N, Long JE, Garratt AN, Fischer TM, Gassmann M, Birchmeier C, Lai C, Rubenstein JL, Marin O. Short- and long-range attraction of cortical GABAergic interneurons by neuregulin-1. Neuron. 2004;44:251–261. doi: 10.1016/j.neuron.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Garel S, Garcia-Dominguez M, Charnay P. Control of the migratory pathway of facial branchiomotor neurones. Development. 2000;127:5297–5307. doi: 10.1242/dev.127.24.5297. [DOI] [PubMed] [Google Scholar]
- Gilland E, Baker R. Conservation of neuroepithelial and mesodermal segments in the embryonic vertebrate head. Acta Anat. 1993;148:110–123. doi: 10.1159/000147530. [DOI] [PubMed] [Google Scholar]
- Goddard JM, Rossel M, Manley NR, Capecchi MR. Mice with targeted disruption of Hoxb-1 fail to form the motor nucleus of the VIIth nerve. Development. 1996;122:3217–3228. doi: 10.1242/dev.122.10.3217. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Pelegri F, Mullins MC, Kane DA, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Heisenberg C-P, Jiang YJ, Kelsh RN, Odenthal J, Warga RM, Nusslein-Volhard C. Mutations affecting morphogenesis during gastrulation and tail formation in the zebrafish, Danio rerio. Development. 1996;123:143–151. doi: 10.1242/dev.123.1.143. [DOI] [PubMed] [Google Scholar]
- Hatten ME. Central nervous system neuronal migration. Annu. Rev. Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. J Neurosci. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen JR, Topczewski J, Bingham S, Sepich DS, Marlow F, Chandrasekhar A, Solnica-Krezel L. Zebrafish trilobite identifies new roles for Strabismus in gastrulation and neuronal movements. Nat. Cell Biol. 2002;8:610–615. doi: 10.1038/ncb828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T, Ito K, Hirata T. Netrin 1 regulates ventral tangential migration of guidepost neurons in the lateral olfactory tract. Development. 2006;133:845–853. doi: 10.1242/dev.02257. [DOI] [PubMed] [Google Scholar]
- Kawauchi T, Hoshino M. Molecular pathways regulating cytoskeletal organization and morphological changes in migrating neurons. Dev Neurosci. 2008;30:36–46. doi: 10.1159/000109850. [DOI] [PubMed] [Google Scholar]
- Kerjan G, Gleeson JG. A missed exit: Reelin sets in motion Dab1 polyubiquitination to put the break on neuronal migration. Genes Dev. 2007;21:2850–2854. doi: 10.1101/gad.1622907. [DOI] [PubMed] [Google Scholar]
- Lois C, Garcia-Verdugo JM, Alvarez-Buylla A. Chain migration of neuronal precursors. Science. 1996;271:978–981. doi: 10.1126/science.271.5251.978. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Keynes R. Segmental patterns of neuronal development in the chick hindbrain. Nature. 1989;337:424–428. doi: 10.1038/337424a0. [DOI] [PubMed] [Google Scholar]
- Luskin MB. Restricted proliferation and migration of postnatally generated neurons derived from the forebrain subventricular zone. Neuron. 1993;11:173–189. doi: 10.1016/0896-6273(93)90281-u. [DOI] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Carlson R, Mann DM, Prince VE. Consequences of Hox gene duplication in the vertebrates: an investigation of the zebrafish Hox paralogue group 1 genes. Development. 2001;128:2471–2484. doi: 10.1242/dev.128.13.2471. [DOI] [PubMed] [Google Scholar]
- McClintock JM, Kheirbek MA, Prince VE. Knockdown of duplicated zebrafish hoxb1 genes reveals distinct roles in hindbrain patterning and a novel mechanism of duplicate gene retention. Development. 2002;129:2339–2354. doi: 10.1242/dev.129.10.2339. [DOI] [PubMed] [Google Scholar]
- Moens CB, Fritz A. Techniques in neural development. Methods Cell Biol. 1999;59:253–272. doi: 10.1016/s0091-679x(08)61829-4. [DOI] [PubMed] [Google Scholar]
- Moens CB, Yan YL, Appel B, Force AG, Kimmel CB. valentino: a zebrafish gene required for normal hindbrain segmentation. Development. 1996;122:3981–3990. doi: 10.1242/dev.122.12.3981. [DOI] [PubMed] [Google Scholar]
- Muller M, Jabs N, Lorke DE, Fritzsch B, Sander M. Nkx6.1 controls migration and axon pathfinding of cranial branchio-motoneurons. Development. 2003;130:5815–5826. doi: 10.1242/dev.00815. [DOI] [PubMed] [Google Scholar]
- Nambiar RM, Ignatius MS, Henion PD. Zebrafish colgate/hdac1 functions in the non-canonical Wnt pathway during axial extension and in Wnt-independent branchiomotor neuron migration. Mech. Dev. 2007;124:682–698. doi: 10.1016/j.mod.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke NA, Dailey ME, Smith SJ, McConnell SK. Diverse migratory pathways in the developing cerebral cortex. Science. 1992;258:299–302. doi: 10.1126/science.1411527. [DOI] [PubMed] [Google Scholar]
- Park HT, Wu J, Rao Y. Molecular control of neuronal migration. Bioessays. 2002;24:821–827. doi: 10.1002/bies.10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Moon RT. The planar cell-polarity gene stbm regulates cell behaviour and cell fate in vertebrate embryos. Nat. Cell Biol. 2002;4:20–25. doi: 10.1038/ncb716. [DOI] [PubMed] [Google Scholar]
- Parnavelas JG. The origin and migration of cortical neurones: new vistas. Trends Neurosci. 2000;23:126–131. doi: 10.1016/s0166-2236(00)01553-8. [DOI] [PubMed] [Google Scholar]
- Paulus JD, Halloran MC. Zebrafish bashful/laminin-alpha 1 mutants exhibit multiple axon guidance defects. Dev. Dyn. 2006;235:213–224. doi: 10.1002/dvdy.20604. [DOI] [PubMed] [Google Scholar]
- Pearlman AL, Faust PL, Hatten ME, Brunstrom JE. New directions for neuronal migration. Curr. Opin. Neurobiol. 1998;8:45–54. doi: 10.1016/s0959-4388(98)80007-x. [DOI] [PubMed] [Google Scholar]
- Pocock R, Mione M, Hussain S, Maxwell S, Pontecorvi M, Aslam S, Gerrelli D, Sowden JC, Woollard A. Neuronal function of Tbx20 conserved from nematodes to vertebrates. Dev Biol. 2008;317:671–685. doi: 10.1016/j.ydbio.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Pozas E, Ibanez CF. GDNF and GFRalpha1 promote differentiation and tangential migration of cortical GABAergic neurons. Neuron. 2005;45:701–713. doi: 10.1016/j.neuron.2005.01.043. [DOI] [PubMed] [Google Scholar]
- Prince VE, Moens CB, Kimmel CB, Ho RK. Zebrafish hox genes: expression in the hindbrain region of wild-type and mutants of the segmentation gene, valentino. Development. 1998;125:393–406. doi: 10.1242/dev.125.3.393. [DOI] [PubMed] [Google Scholar]
- Rakic P. Neuron-glia relationship during granule cell migration in developing cerebellar cortex. A Golgi and electronmicroscopic study in Macacus Rhesus. J. Comp. Neurol. 1971;141:283–312. doi: 10.1002/cne.901410303. [DOI] [PubMed] [Google Scholar]
- Rohrschneider MR, Elsen GE, Prince VE. Zebrafish Hoxb1a regulates multiple downstream genes including prickle1b. Dev Biol. 2007;309:358–372. doi: 10.1016/j.ydbio.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Schneider-Maunoury S, Seitanidou T, Charnay P, Lumsden A. Segmental and neuronal architecture of the hindbrain of Krox-20 mouse mutants. Development. 1997;124:1215–1226. doi: 10.1242/dev.124.6.1215. [DOI] [PubMed] [Google Scholar]
- Sittaramane V, Sawant A, Wolman MA, Maves L, Halloran MC, Chandrasekhar A. The cell adhesion molecule Tag1, transmembrane protein Stbm/Vangl2, and Lamininalpha1 exhibit genetic interactions during migration of facial branchiomotor neurons in zebrafish. Dev Biol. 2009;325:363–373. doi: 10.1016/j.ydbio.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MR, Shirasaki R, Cai CL, Ruiz EC, Evans SM, Lee SK, Pfaff SL. T-Box transcription factor Tbx20 regulates a genetic program for cranial motor neuron cell body migration. Development. 2006;133:4945–4955. doi: 10.1242/dev.02694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer M. Initiation of facial motoneurone migration is dependent on rhombomeres 5 and 6. Development. 2001;128:3707–3716. doi: 10.1242/dev.128.19.3707. [DOI] [PubMed] [Google Scholar]
- Studer M, Lumsden A, Ariza-McNaughton L, Bradley A, Krumlauf R. Altered segmental identity and abnormal migration of motor neurons in mice lacking Hoxb-1. Nature. 1996;384:630–634. doi: 10.1038/384630a0. [DOI] [PubMed] [Google Scholar]
- Vanderlaan G, Tyurina OV, Karlstrom RO, Chandrasekhar A. Gli function is essential for motor neuron induction in zebrafish. Dev. Biol. 2005;282:550–570. doi: 10.1016/j.ydbio.2005.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivancos V, Chen P, Spassky N, Qian D, Dabdoub A, Kelley M, Studer M, Guthrie S. Wnt activity guides facial branchiomotor neuron migration, and involves the PCP pathway and JNK and ROCK kinases. Neural Dev. 2009;4:7–22. doi: 10.1186/1749-8104-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Iwasaki M, Sato T, Masai I, Nishiwaki Y, Tanaka H, Sato A, Nojima Y, Okamoto H. Dual roles of zygotic and maternal Scribble1 in neural migration and convergent extension movements in zebrafish embryos. Development. 2005;132:2273–2285. doi: 10.1242/dev.01810. [DOI] [PubMed] [Google Scholar]
- Wada H, Tanaka H, Nakayama S, Iwasaki M, Okamoto H. Frizzled3a and Celsr2 function in the neuroepithelium to regulate migration of facial motor neurons in the developing zebrafish hindbrain. Development. 2006;133:4749–4759. doi: 10.1242/dev.02665. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book. Eugene, OR: University of Oregon; 1995. [Google Scholar]
- Wong K, Ren XR, Huang YZ, Xie Y, Liu G, Saito H, Tang H, Wen L, Brady-Kalnay SM, Mei L, Wu JY, Xiong WC, Rao Y. Signal transduction in neuronal migration: roles of GTPase activating proteins and the small GTPase Cdc42 in the Slit-Robo pathway. Cell. 2001;107:209–221. doi: 10.1016/s0092-8674(01)00530-x. [DOI] [PubMed] [Google Scholar]
- Zhang M, Kim HJ, Marshall H, Gendron-Maguire M, Lucas DA, Baron A, Gudas LJ, Gridley T, Krumlauf R, Grippo JF. Ectopic Hoxa-1 induces rhombomere transformation in mouse hindbrain. Development. 1994;120:2431–2442. doi: 10.1242/dev.120.9.2431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.