Introduction
Hypersensitivity to foods is now recognized as a world-wide problem in the industrialized world and it’s prevalence may be increasing [1, 2]. Reactions to foods are thought to be the most common cause of anaphylaxis when it occurs outside of the hospital [3].
Elevated allergen-specific IgE is the hallmark of allergic sensitization. The fine specificity of IgE binding is important as IgE from those peanut-allergic patients with the most severe clinical reactions recognizes a more diverse array of linear peptides within the family of peanut proteins [4]. Over the last 3 decades there have been multiple reports that allergen-specific IgG (and, when measured, IgG1 and IgG4) tends to be higher in atopic individuals than in non-atopics [5]. This has been reported for grass (Rye 1) [6], ragweed (AgE) [7], birch (Bet v 1) [8], and peanut (crude peanut extract (CPE)) [9, 10]. Lesser differences were reported for cat (Fel d 1) although the allergic subjects again had higher levels [11] and no differences were reported with IgG to ovalbumin [10]. Jones et al. have demonstrated increasing peanut-specific IgG in patients undergoing oral desensitization but did not compare these findings with subjects who are naturally tolerant to peanuts [12].
HLA (human leukocyte antigen) proteins were originally discovered due to their importance in rejection of transplants and eventually understood as the major histocompatiblilty complex (MHC) for their importance in presenting antigen to T cells [13]. Presentation of exogenous antigens to CD4+ T cells occurs via MHC class II on the surface of antigen-presenting cells (APCs) [13, 14]. Peanut proteins are clearly exogenous antigens and are presumably presented via MHC class II [14]. MHC class II molecules have four distinct pockets that serve to anchor peptides and interact with T cell receptors [15, 16]. Shared epitopes (small groups of amino acids having similar surface properties) within these pockets may lead to shared recognition of important peptides by otherwise disparate alleles [15, 17-21].
Two previous studies have examined the possible relationship between HLA class II alleles in peanut-allergic individuals compared with peanut tolerant siblings using low-resolution HLA typing [22, 23]. Howell et al. studied 50 subjects with peanut allergy, 34 non-peanut allergic siblings, and 77 non-peanut allergic parents and compared their findings with those from 293 unrelated controls [22]. After Bonferroni corrections, there were no significant differences between the peanut allergic patients and their non-peanut allergic siblings at any allele. However, they found significant differences between the peanut-allergic subjects and the unrelated controls for the presence of DRB1*08 [22]. Shreffler et al. performed a similar study, enrolling 73 peanut-allergic subjects and 75 peanut-tolerant siblings and found no significant differences between the peanut-allergic and peanut-tolerant siblings [23]. They did not compare their subjects to unrelated controls. These two studies were limited because analyses were performed at a serologic level and this can miss the important details within the peptide-binding groove.
Given reports that peanut-specific IgG is higher in allergic than in non-allergic subjects we reasoned that differences in peanut-specific IgG production between peanut-allergic subjects and those who are tolerant to peanuts may be related to differences in the ability to present allergens via HLA class II. To test this hypothesis, we determined anti-peanut IgG, peanut-specific IgE, and HLA-Class II expression at high resolution in a population of well-characterized peanut-allergic subjects and their peanut-tolerant siblings including 14 sibling pairs who are identical at HLA-Class II. We compared the HLA findings to those from a large public database [24]. Our primary finding is that differences in anti-peanut IgG and IgE are unequivocally independent of HLA class II. A secondary finding is that HLA DRB1*08 appears to be increased in the peanut allergic subjects and this may be driven by an increased frequency of HLA DRB1*0803.
Methods
Patient population
This study was approved by our respective (Institutional Review Boards and all subjects, parents, or guardians gave informed consent and for minors, assent. We enrolled subjects with severe peanut allergy who had a peanut-tolerant full sibling who would participate in the study. Subjects were recruited from within our own allergy practices, referred from our allergy colleagues, referred by other subjects, and identified by our attendance at activities sponsored by the Food Allergy and Anaphylaxis Network (FAAN). The peanut-allergic subjects were from the greater Denver, Colorado (n=34) and Ann Arbor, Michigan (n=19) areas.
These were discordant sibling-pairs for whom we had high confidence that one of the siblings was clinically allergic to peanuts with an excellent history and either a peanut-specific IgE >14 U/ml (ImmunoCap, Pharmacia, Upsula, Sweden), a skin prick test >5mm larger than negative control, or a positive double-blind, placebo-controlled food challenge as these findings are associated with a high confidence of reproducible reactions [25-27]. The paired sibling had eaten peanuts within 3 months and without a clinical reaction. In addition, all of the siblings were tested for peanut-specific IgE and none had values >14 U/ml (ImmunoCap, Pharmacia, Upsula, Sweden) (Figure 1). Fifty-nine of the 63 peanut-tolerant siblings had skin prick tests for peanut and none of these tests were >5mm larger than the negative control (data not shown). Blood was drawn from the peanut-allergic subjects and from the peanut-tolerant siblings on the same day and handled the same way. For the analysis of sibling pairs, when there was more than one peanut-tolerant sibling, we used the data from the sibling who was first to volunteer. To identify the maximal number of sibling pairs with identical HLA Class II, we examined the entire set of 64 siblings. If two or more peanut-tolerant siblings were identical to the peanut-allergic subject at HLA Class II we used the data from the one who volunteered first.
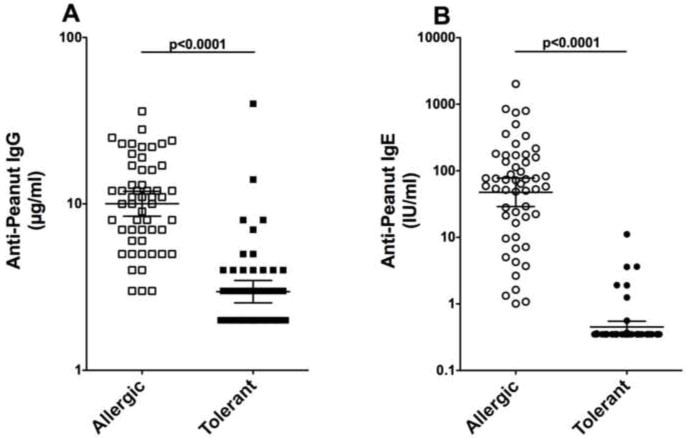
Figure 1. Anti-peanut IgG and IgE.
A) Anti-peanut IgG and B) anti-peanut IgE were determined by ImmunoCap. Log data of 53 sibling pairs were compared with unpaired t-tests (p<0.0001).
Immunoglobulin levels
In vitro peanut-specific IgG, IgG4, and IgE determinations were performed on sera by the ImmunoCap Method (Pharmacia; Uppsala, Sweden). For samples with anti-peanut IgE >100, sera were diluted 1:10, reassayed, and the data corrected for dilution. IgG specific for Ara h 1, Ara h 2, and Ara h 6 were measured in 49 or our 53 subjects by ELISA assays similar to those described by Tay using methodology developed by Rieben and Blaser except that HRP-labeled mouse anti-human IgG (Clone JDC-10; Southern Biotechnology, Birmingham, AL) was the detecting antibody [10, 28].
HLA Class II
DNA was isolated using the BioRobot EZ1 Workstation (Qiagen; Valencia, CA) and HLA class II alleles were determined by high resolution DNA methodology (Atria Genetics; San Francisco, CA). HLA class II DR supertypes were determined as initially described by Ou et al. and refined by others [15, 18-20].
Statistical Analysis
Based on published data examining HLA supertypes, we determined that this population size gave us an 85% power to find significant differences that would withstand a Bonferonni correction for 7 DRB1 supertypes [15, 19]. We compared the proportion of siblings having the supertype of interest using a McNemar’s test to account for correlation between the siblings. We used a Chi-square test or Fishers exact test (as appropriate) to test differences in the proportion of peanut allergic sibs vs controls having the supertypes of interest. The Bonferroni correction was applied to account for multiple comparisons. Unpaired two-tailed t-tests and Pearson correlations were performed on natural logs of data (Prism v 5.0a for the MacIntosh, GraphPad Software, La Jolla, CA). Graphs show geometric means and 95% confidence intervals.
Results
Clinical and laboratory data for our subjects are shown in Table 1. Our sib-pairs were predominantly of European descent (n=44; 83%). Most of the peanut-allergic subjects had reactions involving more than one organ system (89%), and most had anti-peanut IgE >14 IU/ml (81%).
Table 1.
Clinical and Laboratory data
| Sequential number | Race | Age | Most Severe Reaction* | Total IgE (IU/ml)** | Anti-Pnut IgE (IU/ml)*** | Skin Test wheal (mm) or DBPCFC**** |
|---|---|---|---|---|---|---|
| 1 | C | 14 | AE, GI | 1312 | 58 | |
| 2 | C | 69 | S,AE | 587 | 16.4 | |
| 3 | C+H | 11 | S,AE,R,GI | 727 | 77.3 | |
| 4 | C | 11 | S,AE,R,GI | 1185 | 74.7 | |
| 5 | C | 29 | S,R | 136 | 59 | |
| 6 | C | 10 | S,AE,R,GI | 138 | 9.6 | 28 |
| 7 | C | 3 | S, R, GI | 341 | 96.8 | |
| 8 | C | 6 | S,GI | 298 | 2.65 | Positive DBPCFC |
| 9 | C | 10 | S, AE, R, GI | 295 | 50.1 | |
| 10 | C | 16 | S,AE,R | 454 | 159 | |
| 11 | C | 7 | S,AE | 161 | 63.8 | |
| 12 | C | 16 | S,AE,R,GI | 293 | 51.6 | |
| 13 | C+H | 8 | S,GI | 303 | 1.08 | >5 |
| 14 | C | 16 | S,AE,R | 142 | 10.1 | >5 |
| 15 | C | 14 | S,AE,R,GI | 56 | 21.3 | |
| 16 | C | 9 | S,AE,R | 568 | 137 | |
| 17 | C+H | 15 | S, AE, R | 202 | 20.3 | |
| 18 | C | 9 | S, AE, R | 1639 | 748 | |
| 19 | A | 19 | S,AE,R,GI | 2416 | 113 | |
| 20 | C | 19 | S,AE,R | 82 | 1.01 | >5 |
| 21 | C | 7 | S,AE,R,GI | 245 | 52.7 | |
| 22 | C | 12 | S,AE,R | 560 | 216 | |
| 23 | C | 11 | R | 1386 | 24 | |
| 24 | C | 5 | S, GI | 415 | 5.03 | 10 |
| 25 | C | 13 | GI | 625 | 6.78 | 9 |
| 26 | C | 8 | S,GI | 599 | 175 | |
| 27 | A | 6 | S,GI,R | 127 | 3.7 | 15 |
| 28 | C | 12 | S,AE | 91 | 29.2 | |
| 29 | C | 5 | S, GI | 216 | 76.5 | |
| 30 | C | 6 | S | 697 | 334 | |
| 31 | C | 9 | S,AE,R | 2439 | 848 | |
| 32 | C | 6 | S,GI | 493 | 171 | |
| 33 | C+H | 6 | S, AE | 619 | 171 | |
| 34 | C | 11 | S,R | 505 | 48.7 | |
| 35 | C | 6 | S,AE,R | 51 | 1.63 | 7 |
| 36 | C+H | 11 | S,AE,R | 2421 | 180 | |
| 37 | C | 11 | S,AE,R | 83 | 1.33 | 5 |
| 38 | C | 7 | S,AE, R, GI | 671 | 73.3 | |
| 39 | C | 9 | S,R,AE | 708 | 4.25 | 25 |
| 40 | C | 7 | S, GI | 816 | 357 | |
| 41 | C | 4 | S,AE,GI | 1418 | 254 | |
| 42 | C | 42 | S,AE,R | 3680 | 7.17 | >5 |
| 43 | C | 13 | S,AE,R | 1423 | 500 | |
| 44 | PI | 7 | S,GI | 4939 | 88.4 | |
| 45 | C | 22 | S,AE,R,GI | 3060 | 796 | |
| 46 | C | 10 | S,AE | 297 | 28 | |
| 47 | C | 17 | S,GI | 171 | 53.6 | |
| 48 | C | 10 | GI | 338 | 82.8 | |
| 49 | AA | 3 | S, AE, R | 6490 | 2020 | |
| 50 | C | 6 | S, AE, R | 357 | 134 | |
| 51 | C | 9 | S,AE,R,GI | 166 | 72 | |
| 52 | C | 8 | S, AE, R | 292 | 22.3 | |
| 53 | C | 7 | R,GI | 186 | 77 |
Symptoms associated with each subject’s most severe reaction. S, skin (erythema and/or hives); R, wheezing; A, anaphylaxis; GI, gastrointestinal symptoms; AE, angioedema.
Total IgE: geom. mean (95% confidence intervals); allergic, 464 (388, 637) IU/ml; tolerant, 95 (65, 166) IU/ml.
Anti-peanut IgE: geom. mean (95% confidence intervals); allergic, 48 (29, 77) IU/ml; tolerant, 0.45 (0.37, 0.54) IU/ml. Values of <0.35 were set at 0.35 for this calculation.
For those subjects with anti-peanut IgE <14 IU/ml, mm wheal greater than the negative control. DBPCFC, double blind, placebo-controlled food challenge.
Similar to the findings of both Howell and Shreffler [22, 23] who examined HLA at the serologic level, we found no significant differences in HLA class II between these discordant sib-pairs when high-resolution DNA techniques were applied. Data for DRB1 are shown in Table 2. Data for DPB1 and DQB1 did not reveal any significant findings (data not shown). Furthermore, analysis of these data using published supertypes of DRB1 similarly did not reveal any differences between these sibling pairs (data not shown) [15, 18, 20, 21]. An unexpected finding from this study is that there appears to be an increased frequency of DRB1*0803 in both our peanut-allergic (3 out of 44; 6.8%) and peanut-tolerant subjects (also 3 out of 44; 6.8%) of European descent compared to a large control group of bone marrow donors of European descent in which DRB1*0803 is a very rare allele in (0.27%; n=7,870) [24]. After correction for the 59 DRB1, DQB1, and DPB1 alleles identified in this study, this is a statistically significant finding both for the peanut-allergic and tolerant siblings (pc=4.5 × 10-9) (Table 2). In addition to its unexpected frequency in our individual subjects, DRB1*0803 was found in 5 out of 44 families of European descent (11.4%) as there was only 1 family in which it occurred in both the peanut-allergic and peanut-tolerant. Subjects with this allele did not have distinguishing clinical characteristics or statistically different values for either total or peanut-specific IgE (data not shown).
Table 2.
Population Frequency of DRB1 Alleles in Subjects of European Descent
| DRB1 | Peanut-Allergic (n=44) | Peanut-Tolerant Sibling (n=44) | Large database* n=7,870 | P/Pc** allergic and tolerant c/w large database |
|---|---|---|---|---|
| 0101 | 4 (9.1) | 4 (9.1) | (17.5) | ns/ns |
| 0301 | 16 (36.4) | 9 (20.4) | (24.2) | ns/ns |
| 0401 | 7 (15.9) | 9 (20.4) | (17.4) | ns/ns |
| 0701 | 9 (20.4) | 7 (15.9) | (25.6) | ns/ns |
| 0801 | 4 (9.1) | 4 (9.1) | (4.7) | ns/ns |
| 0803 | 3 (6.8) | 3 (6.8) | (0.3) | p=7.7×10-11/pc=4.5×10-9 |
| 1101 | 5 (11.3) | 4 (9.1) | (11.0) | ns/ns |
| 1301 | 6 (13.6) | 6 (13.6) | (12.2) | ns/ns |
| 1302 | 6 (13.6) | 6 (13.6) | (7.9) | ns/ns |
| 1401 | 0(0) | 0(0) | (4.9) | ns/ns |
| 1501 | 12 (27.2) | 10 (22.7) | (26.8) | ns/ns |
National Marrow Donor Program® (NMDP) (http://bioinformatics.nmdp.org). This database contains data from 7,870 normal donors. Population frequency was calculated using the formula P=1-(1-G).
Corrected for 59 DR, DQ, and DP alleles identified.
Symptoms are those associated with each subject’s worse reaction. Resp., wheezing; Anaph., anaphylaxis; GI, gastrointestinal symptoms; AE, angioedema; LOC, loss of consciousness; BP, blood pressure (hypotension).
As expected, the anti-peanut IgG and anti-peanut IgE were higher in the peanut-allergic subjects compared to the peanut-tolerant siblings (Figure 1; n=53 pairs; p<0.0001) and were correlated in the allergic (r=0.628; P<0.0001) (Figure 2). We then asked if there was any relationship between IgG or IgE levels and HLA. To address this question, we examined levels of IgG and IgE in sibling pairs with identical HLA Class II (Figure 3). As can be seen the dramatic differences in both peanut-specific IgG and IgE between the peanut-allergic and peanut-tolerant siblings shown in Figure 1 are also present in a subset of subjects in which each pair has identical HLA class II (n=14 pair; p<0.0001) (Figure 3). Again a clear correlation was seen between anti-peanut IgG and anti-peanut IgE in the allergic subjects (r=0.642; p<0.01) (data not shown). We also examined total IgG and IgG4 values for total peanut protein using ImmunoCaps and total IgG for Ara h 1, Ara h 2 IgG, and anti-Ara h 6 by ELISA. Although these correlated well with each other (r values between 0.6 and 0.9; p<0.001) none of these measurements were correlated with HLA alleles, supertypes, or haplotypes (data not shown).
Figure 2. Correlations between anti-peanut IgG and IgE.
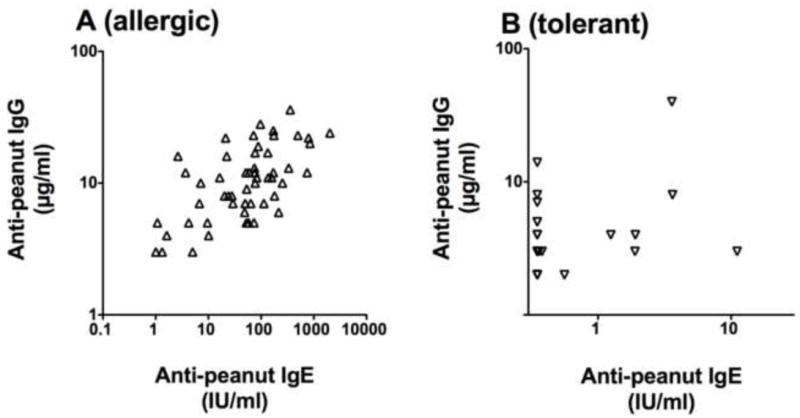
A) Anti-peanut IgG is correlated with anti-peanut IgE in peanut-allergic subjects (p<0.0001; Pearson r=0.628; rˆ2=0.394). B) For the peanut-tolerant siblings, anti-peanut IgG was compared with anti-peanut IgE. Much of the data were below the lower limits of detection so a correlation cannot be accurately determined. The data for the peanut-tolerant subjects (B) is driven by 44 samples that had anti-peanut IgE at the lower limits of detection (<0.35). Excluding these points, the correlation between anti-peanut IgG and anti-peanut IgE for the remaining 9 peanut-tolerant subjects was not significant (data not shown).
Figure 3. Anti-peanut IgG and IgE in subjects with identical HLA Class II.
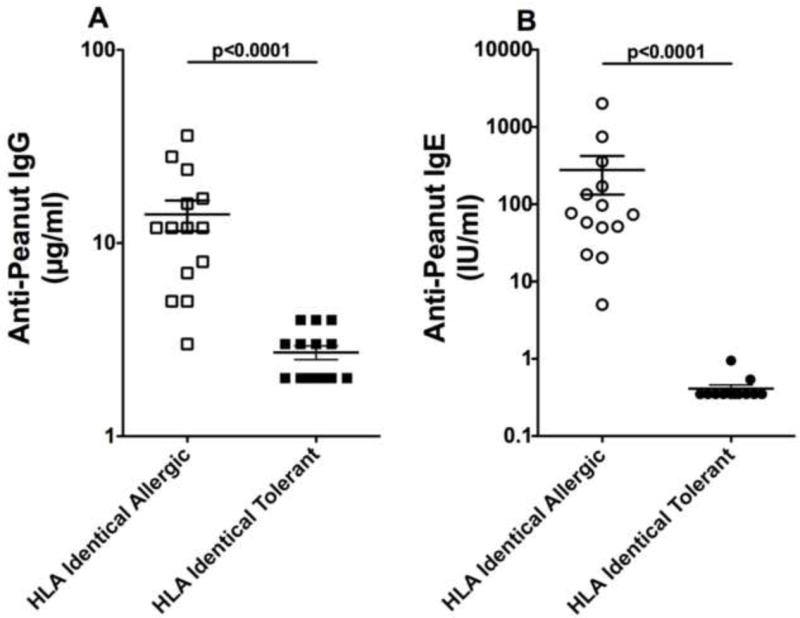
A) Anti-peanut IgG and B) anti-peanut IgE. Log data for 14 sibling pairs with identical HLA Class II were compared with unpaired t-tests.
Since two previous studies of HLA in peanut allergy have been published with very similar designs the data were examined together in an separate analysis (Table 3). To do this, our high-resolution DRB1 data were contracted into the appropriate serotypes. Again, there were no significant differences between the peanut-allergic and peanut-tolerant siblings. However, compared to a large public database there is increased prevalence of the DRB1*08 serologic allele in the peanut-allergic subjects (pc=0.008) but not in the peanut-tolerant siblings (pc=ns) (Table 3). In our cohort, 57% of the DRB1*08 alleles are DRB1*0801 and 43% are DRB1*0803 (Table 2). Since the DRB1*0803 allele is more prevalent in subjects of Asian descent, it is possible that the 5% of the Shreffler subjects and the 4% of the subjects in the current study who were of Asian descent could bias this analysis. However, the increased prevalence still holds when the peanut-allergic subjects known to be of European descent from the current study are pooled with only those from the Howell study in which there were no Asian subjects (pc<0.001; data not shown).
Table 3.
Combined analysis of 3 studies*
| Locus (13 DR serologic groups identified) | Peanut allergic n=176 | Peanut tolerant siblings n=161 | Large database** n=7,870 | P/Pc c/w sibs | P/Pc*** allergic c/w large database |
|---|---|---|---|---|---|
| DRB1*03 | 54 (31%) | 35 (22%) | (24%) | 0.06/ns | 0.04/ns |
| DRB1*08 | 20 (11%) | 14 (9%) | (5.2%) | ns/ns | 0.0006/0.00785 |
| DQB1*04 | 17 (10%) | 10 (7%) | (5.0%) | ns/ns | ns/ns |
Discussion
These data demonstrate unequivocally that peanut allergy, at both clinical and immunologic levels, is entirely independent of differences in HLA class II. We have shown this in our population of full siblings who are discordant for clinically important peanut allergy by examining HLA class II with high resolution DNA-based technology, by looking for epitope sharing with the DRB1 alleles using published “supertypes”, and by measuring peanut-specific IgG. Of note, 8 peanut-tolerant subjects had measurable levels of peanut-specific IgE (0.36-11 IU/ml; geometric mean 1.75 IU/ml) so the peanut-tolerant population is not absolutely non-allergic. The sibling pairs that included siblings with detectable anti-peanut IgE did not contain any dominant HLA class II alleles (data not shown). Of the 14 sibling pairs with identical HLA (Figure 3), only 2 peanut-tolerant siblings had detectable IgE (0.54 and 0.95 IU/ml). Our finding of large differences in peanut-specific IgG in peanut-allergic subjects compared with controls corroborates the report of Tay et al. [10] and extends them in that we find this disparity in full siblings discordant for peanut allergy (n=53 pairs) as well as a smaller cohort in which the paired siblings discordant for peanut allergy have identical HLA class II genotypes (n=14 pairs) (Figure 2). This underscores the concept that the aberrant immune response to peanuts in peanut-allergic subjects occurs at steps earlier than switching to IgE and is independent of the ability to present antigen. In further support of this finding, we measured IgG specific for Ara h 1, Ara h 2, and Ara h 3 in our peanut-allergic subjects. As found for total IgG, there was no association of allergen-specific IgG with any HLA allele, supertype, or haplotype (data not shown). Perhaps HLA-related specificity could be found if specific linear epitopes were examined.
A secondary finding is that, as described by Howell and colleagues, HLA DRB1*08, appears to be more prevalent in patients with peanut allergy compared with a large public database [22]. It is important to point out that all DRB1*08 alleles were present only in 11% of subjects (Table 3) and were found in similar numbers for both the allergic and the tolerant siblings. Furthermore, based on its increased frequency in our subjects, DRB1*0803 which differs from DRB1*0801 only at amino acid #67 with an isoleucine (I) for *0803 compared with a phenylalanine (F) for *0801 [29], appears to account for this increased prevalence of DRB1*08. So, DRB1*08 and particularly DRB1*0803 appears to be a marker of families with a propensity toward peanut allergy and not a marker of peanut allergy per se. Alternatively, this may be a statistical aberration of no biological consequence. In a review of the current literature, we were not able to find association of DRB1*08, DRB1*0801, or DRB1*0803 with any other immunologic diseases.
In summary, we have shown that elevated anti-peanut IgG in peanut-allergic subjects compared to their peanut-tolerant siblings occurs independently of HLA class II and offer evidence that HLA DRB1*0803 is may be a risk factor for peanut allergy in families of European descent.
Acknowledgments
The authors thank study coordinators, Darcy G. Schlichting and Hanna Talwar for their efforts in recruiting these subjects and collecting parts of the data. We thank Drs. Laurent Pons, Michael Kulis, Wesley Burks, and Soheila Maleki for providing purified Ara h 1 and Dr. Xueni Chen for supplying purified Ara h 2 and Ara h 6. We also thank Drs. Allen Adinoff, Karen Andrews, Sandy Avner, Alan Bock, Leon Greos, John James, Grant Olson, Jerry Koepke, Jeffery Rumbyrt, and Katherine VanKerkhove for referring patients and our study subjects for their participation.
Supported by grant from the Food Allergy and Anaphylaxis Network and RO1-AI052164 (National Institute of Allergy and Infectious Diseases) to Dr. Dreskin, and by institutional funds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sicherer SH, Sampson HA. Peanut allergy: emerging concepts and approaches for an apparent epidemic. J Allergy Clin Immunol. 2007;120:491–503. doi: 10.1016/j.jaci.2007.07.015. quiz 504-495. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Shoshan M, Kagan RS, Alizadehfar R, Joseph L, Turnbull E, St Pierre Y, Clarke AE. Is the prevalence of peanut allergy increasing? A 5-year follow-up study in children in Montreal. J Allergy Clin Immunol. 2009;123:783–788. doi: 10.1016/j.jaci.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 3.Burks AW. Peanut allergy. Lancet. 2008;371:1538–1546. doi: 10.1016/S0140-6736(08)60659-5. [DOI] [PubMed] [Google Scholar]
- 4.Shreffler WG, Beyer K, Chu TH, Burks AW, Sampson HA. Microarray immunoassay: association of clinical history, in vitro IgE function, and heterogeneity of allergenic peanut epitopes. J Allergy Clin Immunol. 2004;113:776–782. doi: 10.1016/j.jaci.2003.12.588. [DOI] [PubMed] [Google Scholar]
- 5.Thomas WR, Hales BJ. Immune Responses to Inhalant Allergens. WAO Journal. 2008:89–95. doi: 10.1097/WOX.0b013e3181788324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Platts-Mills TA. Local production of IgG, IgA and IgE antibodies in grass pollen hay fever. J Immunol. 1979;122:2218–2225. [PubMed] [Google Scholar]
- 7.Platts-Mills TA, von Maur RK, Ishizaka K, Norman PS, Lichtenstein LM. IgA and IgG anti-ragweed antibodies in nasal secretions. Quantitative measurements of antibodies and correlation with inhibition of histamine release. J Clin Invest. 1976;57:1041–1050. doi: 10.1172/JCI108346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson M, Reinholdt J, Cardell LO. Allergen-reactive antibodies are found in nasal fluids from patients with birch pollen-induced intermittent allergic rhinitis, but not in healthy controls. Allergy. 2003;58:386–392. doi: 10.1034/j.1398-9995.2003.00113.x. [DOI] [PubMed] [Google Scholar]
- 9.Kolopp-Sarda MN, Moneret-Vautrin DA, Gobert B, Kanny G, Guerin L, Faure GC, Bene MC. Polyisotypic antipeanut-specific humoral responses in peanut-allergic individuals. Clin Exp Allergy. 2001;31:47–53. [PubMed] [Google Scholar]
- 10.Tay SS, Clark AT, Deighton J, King Y, Ewan PW. Patterns of immunoglobulin G responses to egg and peanut allergens are distinct: ovalbumin-specific immunoglobulin responses are ubiquitous, but peanut-specific immunoglobulin responses are up-regulated in peanut allergy. Clin Exp Allergy. 2007;37:1512–1518. doi: 10.1111/j.1365-2222.2007.02802.x. [DOI] [PubMed] [Google Scholar]
- 11.Jarvis D, Zock JP, Heinrich J, Svanes C, Verlato G, Olivieri M, Villani S, Ponzio M, Leynaert B, Sunyer J, Dahlman-Hoglund A, Chinn S, Luczynska C, Norback D, Burney P. Cat and dust mite allergen levels, specific IgG and IgG4, and respiratory symptoms in adults. J Allergy Clin Immunol. 2007;119:697–704. doi: 10.1016/j.jaci.2006.10.042. [DOI] [PubMed] [Google Scholar]
- 12.Jones SM, Pons L, Roberts JL, Scurlock AM, Perry TT, Kulis M, Shreffler WG, Steele P, Henry KA, Adair M, Francis JM, Durham S, Vickery BP, Zhong X, Burks AW. Clinical efficacy and immune regulation with peanut oral immunotherapy. J Allergy Clin Immunol. 2009;124:292–300. 300 e291–297. doi: 10.1016/j.jaci.2009.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapsenberg ML, Jansen HM. Antigen Presentation and Immunoregulation. In: Y J, Adkinson NF, Busse WW, Bochner BS, Holgate ST, Simons FER, editors. Middleton’s Allergy: Principles and Practice. Mosby; Philadelphia, PA: 2003. pp. 177–188. [Google Scholar]
- 14.Robinson JH, Delvig AA. Diversity in MHC class II antigen presentation. Immunology. 2002;105:252–262. doi: 10.1046/j.0019-2805.2001.01358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ou D, Mitchell LA, Tingle AJ. A new categorization of HLA DR alleles on a functional basis. Hum Immunol. 1998;59:665–676. doi: 10.1016/s0198-8859(98)00067-6. [DOI] [PubMed] [Google Scholar]
- 16.Castelli FA, Buhot C, Sanson A, Zarour H, Pouvelle-Moratille S, Nonn C, Gahery-Segard H, Guillet JG, Menez A, Georges B, Maillere B. HLA-DP4, the most frequent HLA II molecule, defines a new supertype of peptide-binding specificity. J Immunol. 2002;169:6928–6934. doi: 10.4049/jimmunol.169.12.6928. [DOI] [PubMed] [Google Scholar]
- 17.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 18.Pavoni DP, Roxo VM, Marquart Filho A, Petzl-Erler ML. Dissecting the associations of endemic pemphigus foliaceus (Fogo Selvagem) with HLA-DRB1 alleles and genotypes. Genes Immun. 2003;4:110–116. doi: 10.1038/sj.gene.6363939. [DOI] [PubMed] [Google Scholar]
- 19.Blanco C, Sanchez-Garcia F, Torres-Galvan MJ, Dumpierrez AG, Almeida L, Figueroa J, Ortega N, Castillo R, Gallego MD, Carrillo T. Genetic basis of the latex-fruit syndrome: association with HLA class II alleles in a Spanish population. J Allergy Clin Immunol. 2004;114:1070–1076. doi: 10.1016/j.jaci.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 20.du Montcel ST, Michou L, Petit-Teixeira E, Osorio J, Lemaire I, Lasbleiz S, Pierlot C, Quillet P, Bardin T, Prum B, Cornelis F, Clerget-Darpoux F. New classification of HLA-DRB1 alleles supports the shared epitope hypothesis of rheumatoid arthritis susceptibility. Arthritis Rheum. 2005;52:1063–1068. doi: 10.1002/art.20989. [DOI] [PubMed] [Google Scholar]
- 21.Jones EY, Fugger L, Strominger JL, Siebold C. MHC class II proteins and disease: a structural perspective. Nat Rev Immunol. 2006;6:271–282. doi: 10.1038/nri1805. [DOI] [PubMed] [Google Scholar]
- 22.Howell WM, Turner SJ, Hourihane JO, Dean TP, Warner JO. HLA class II DRB1, DQB1 and DPB1 genotypic associations with peanut allergy: evidence from a family-based and case-control study. Clin Exp Allergy. 1998;28:156–162. doi: 10.1046/j.1365-2222.1998.00224.x. [DOI] [PubMed] [Google Scholar]
- 23.Shreffler WG, Charlop-Powers Z, Sicherer SH. Lack of Association of HLA Class II alleles with Peanut Allergy. Annals of Allergy Asthma & Immunology. 2006;96:865–869. doi: 10.1016/S1081-1206(10)61351-8. [DOI] [PubMed] [Google Scholar]
- 24.Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol. 2007;68:779–788. doi: 10.1016/j.humimm.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 25.Maloney JM, Rudengren M, Ahlstedt S, Bock SA, Sampson HA. The use of serum-specific IgE measurements for the diagnosis of peanut, tree nut, and seed allergy. J Allergy Clin Immunol. 2008;122:145–151. doi: 10.1016/j.jaci.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 26.Nolan RC, Richmond P, Prescott SL, Mallon DF, Gong G, Franzmann AM, Naidoo R, Loh RK. Skin prick testing predicts peanut challenge outcome in previously allergic or sensitized children with low serum peanut-specific IgE antibody concentration. Pediatr Allergy Immunol. 2007;18:224–230. doi: 10.1111/j.1399-3038.2007.00519.x. [DOI] [PubMed] [Google Scholar]
- 27.Poznik GD, Adamska K, Xu X, Krolewski AS, Rogus JJ. A novel framework for sib pair linkage analysis. Am J Hum Genet. 2006;78:222–230. doi: 10.1086/499827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rieben R, Blaser K. Quantification of IgG and IgG4 antibodies to bee venom phospholipase A2 by competitive inhibition in ELISA. J Immunol Methods. 1989;119:1–8. doi: 10.1016/0022-1759(89)90374-8. [DOI] [PubMed] [Google Scholar]
- 29.Eberle M, Baxter-Lowe LA. Molecular analysis of HLA-DRB1*08/12 alleles: identification of two additional alleles. Hum Immunol. 1992;34:24–30. doi: 10.1016/0198-8859(92)90081-w. [DOI] [PubMed] [Google Scholar]