Abstract
Background
Non-tuberculous mycobacteria (NTM) are increasingly important as opportunistic infections after major and minor surgical procedures, likely because they are ubiquitous and not effectively killed by many commonly used disinfectants. Outbreaks of soft tissue infections with NTM appeared related to the use of commercial disinfectants based on quaternary ammonium compounds (QACs).
Methods
We studied the survival of clinical and environmental isolates of Mycobacterium abscessus, Mycobacterium massiliense, Mycobacterium chelonae and Mycobacterium fortuitum after 20 min, 60 min or 24 h exposures to different QACs, and the surviving bacteria were then re-exposed to QACs to see if the percentage of surviving bacteria had increased. The bacteria were labelled with a dnaA–gfp fusion and their level of QAC resistance monitored as increasing fluorescence. The QAC-resistant bacteria were then serially restreaked onto non-selective medium and retested for QAC survival.
Results
The frequency of survivors was <1 in 105 bacteria with Mycobacterium smegmatis, but >1 in 100 with the other mycobacteria studied. Different environmental and clinical isolates had similar QAC MICs, but QAC survivors of each strain were resistant. The QAC-surviving strains reverted to the original, non-resistant phenotype after several passages on non-selective medium.
Conclusions
QACs should not be used in settings where even minimally invasive procedures are performed, as they select for a non-genetically determined reversible resistant phenotype that appears at high frequency with several rapidly growing mycobacterial species associated with healthcare-related infections. M. smegmatis behaves differently and is not an adequate model for testing the activity of disinfectants against NTM.
Keywords: quaternary ammonium compounds, disinfectants, NTM, resistance, tolerance, persistence
Introduction
The recent popularity of conventional and non-conventional cosmetic procedures, often performed by diverse practitioners in less than optimal settings, has been associated with outbreaks of non-tuberculous mycobacteria (NTM) infections of the skin and soft tissues.1–3 Infections with the Mycobacterium abscessus–chelonae group and Mycobacterium fortuitum are among the most common and most difficult to treat.4 While some of these cases were associated with the injection of contaminated local anaesthetics and mixtures purported to reduce adiposity, many cases were apparently related to the use of surgical instruments that were inadequately sterilized or incomplete disinfection of the skin prior to the procedure.5 Some outbreaks appeared to be associated with the use of disinfectants whose active ingredient is a quaternary ammonium compound (QAC), a class of disinfectants regarded as having only mycobacteriostatic activity.6 In order to explain more precisely how the use of a disinfectant might be associated with infections,7 we investigated the activity of QACs against several species of NTM.
Materials and methods
Bacterial strains and plasmids
Mycobacterium smegmatis mc2155 and Mycobacterium bovis BCG are strains commonly used in the laboratory (Laboratorio de Genética Molecular, IVIC). Mycobacterium terrae was obtained from the ATCC (ATCC 15755). The other mycobacterial strains were isolated in Caracas (Lab. TB, Inst. Biomedicina) from patient material or environmental sources (Table 1). The PRA method was used for initial strain identification (polymerase chain reaction–restriction enzyme analysis) (http://app.chuv.ch/prasite/index.html), but definitive species assignment was obtained by sequencing a fragment of the rpoB gene amplified with primers MycoF (5′-GGCAAGGTCACCCCGAAGGG-3′) and MycoR (5′-AGCGGCTGCTGGGTGATCATC-3′).8
Table 1.
Strains used in this study: mycobacterial isolates, their resistant derivatives selected after exposure to the indicated disinfectants and revertants obtained by restreaking the resistant strains on medium without disinfectant
| Strain | Origin |
|---|---|
| M. massiliense WT 1.0 | M. massiliense clinical isolate LTE3550 |
| M. massiliense 1.2 | M. massiliense WT 1.0 selected in 9% CTAB |
| M. abscessus WT 2.0 | M. abscessus clinical isolate LTE3514 |
| M. abscessus 2.1 | M. abscessus WT 2.0 selected in 9% CTAB |
| M. abscessus 2.1.3 | M. abscessus 2.1 restreaked 3 times |
| M. abscessus 2.1.6 | M. abscessus 2.1 restreaked 6 times |
| M. abscessus 2.2 | M. abscessus WT 2.0 selected in 9% DBLAB |
| M. abscessus 2.2.3 | M. abscessus 2.2 restreaked 3 times |
| M. abscessus 2.2.6 | M. abscessus 2.2 restreaked 6 times |
| M. abscessus 2.3 | M. abscessus WT 2.0 selected in glutaraldehyde 2% for 5 min |
| M. abscessus 2.3.3 | M. abscessus 2.3 restreaked 3 times |
| M. abscessus 2.3.6 | M. abscessus 2.3 restreaked 6 times |
| M. fortuitum WT 1.0 | M. fortuitum clinical isolate LTE1428 |
| M. fortuitum 1.2 | M. fortuitum WT 1.0 selected in 9% DBLAB |
| M. chelonae WT 2.0 | M. chelonae clinical isolate LTE2714 |
| M. chelonae 2.1 | M. chelonae WT 2.0 selected in 9% CTAB |
| M. chelonae 2.2 | M. chelonae WT 2.0 selected in 9% DBLAB |
| M. fortuitum WT 3.0 | M. fortuitum environmental isolate LTE2297 |
| M. fortuitum 3.2 | M. fortuitum WT 3.0 selected in 9% DBLAB |
| M. terrae | ATCC 15755 |
| M. smegmatis mc2155 | laboratory strain (Laboratorio de Genética Molecular, IVIC) |
| M. bovis BCG | laboratory strain (Laboratorio de Genética Molecular, IVIC) |
Resistant survivors and revertants were obtained as noted in Table 1. The strains were grown at 37°C in Middlebrook 7H9 liquid medium supplemented with 10% OAD (oleic acid, albumin, dextrose), 0.2% glycerol and 0.05% Tween 80 (7H9-OAD-Tw) or on Petri dishes containing Middlebrook 7H10 supplemented with 10% OAD and 0.2% glycerol. Plasmids pFPV279 and pYUB41210 were gifts from Dr Lalita Ramakrishnan (University of Washington, Seattle, WA, USA) and Drs Martin Pavelka (University of Rochester, NY, USA) and William R. Jacobs Jr (Albert Einstein College of Medicine, Bronx, NY, USA), respectively.
Disinfectants
The disinfectants evaluated were: hexadecyl trimethyl ammonium bromide (centrimonium bromide; C19H42NBr; CTAB); dodecyl trimethyl ammonium bromide (C15H34NBr; DTAB); cetylpyridinium chloride (C21H38NCl; CPC); tetradecyl trimethyl ammonium bromide (C17H38NBr; TTAB); and dimethyl benzyl lauryl ammonium bromide [C13H27NBr; DBLAB, marketed under a commercial product name (http://www.pharmcast.com/Patents/Yr2002/August2002/082702/6441045_Disinfectant082702.htm)]. The disinfectants were prepared by dissolving CTAB, CPC, DTAB and TTAB in 50 mL volumes of 7H9-OAD-Tw to obtain 4% or 10% solutions of each. For the marketed disinfectant, the 10% commercial product solution was used directly in some studies or diluted to 4% in 7H9-OAD-Tw as the starting concentration in the 96-well plate assay. All disinfectant solutions were kept at 4°C until used. The highest concentrations tested were 9%, made up of 9 parts of the 10% solution and one part of bacterial inoculum.
Determination of mycobactericidal activity and selection of resistant survivors
The mycobacterial strains were grown in 50 mL tubes to an OD600 of 1.0 (∼109 cfu/mL). Ten 1.0 mm glass beads were added and the tube was vortexed for 20 s and then left to stand for 10 min to allow for clumps to sediment. When necessary, the resulting suspension was diluted serially, 1 : 10, in 7H9. An aliquot of 1 mL of the final suspension was then added to 9 mL of 10% DBLAB (the undiluted commercial preparation) or 10% CTAB and left to incubate with agitation for 20 min, 1 h or 24 h. Subsequently, after vortexing for 1 min, 1 mL of the disinfectant containing bacterial suspension was added to 9 mL of neutralizing solution (10% Tween 80, 0.5% sodium thiosulphate, 0.1% histidine in 0.25 M phosphate buffer, pH 7.0), which was then centrifuged at 3000 rpm for 10 min. The supernatant was discarded and the pellet resuspended in ∼300 µL of the residual medium. Three ∼100 µL aliquots of this pellet were then spread onto three 7H10 OAD plates without detergent and incubated at 37°C for 4–8 days, at which point surviving colonies were counted. Assays were performed in triplicate. Controls were treated identically, but incubated without disinfectant.
The selection for glutaraldehyde-resistant colonies was slightly different. After vortexing and allowing for clumps to settle, as above, the resultant supernatant was then transferred to a new 50 mL tube that was placed into a bath sonicator for 5 min. An aliquot of 100 µL of this suspension was added to 900 µL of 2% glutaraldehyde. After 5, 20 and 60 min, 100 µL of the glutaraldehyde suspension was added to 900 µL of neutralizing solution (6% Tween 80, 1% sodium thiosulphate, 0.5% sodium thioglycolate and 0.15% cysteine). From the neutralized suspension, three 100 µL aliquots were plated onto 7H10 plates without OAD and incubated at 37°C for 4–8 days, when surviving colonies were counted. Negative controls were performed with sterile water in place of glutaraldehyde.
Cloning of the dnaA–gfp fusion
Reagents for DNA manipulations were obtained from commercial sources and used according to routine protocols. For the construction of plasmid pdnaA–gfp, the dnaA promoter region from M. tuberculosis (582 bp) was amplified from a cosmid11 containing the rpmH–dnaA region, using primers GLRA3 (5′-CTA TGT CTG GCA ACA T) and GLRA4 (5′-TAT CTC CCT GGT TCT CGT TA), and ligated into the BamHI and EcoRV sites upstream of gfp on plasmid pFPV27. The chromosomal integrating version of the dnaA–gfp fusion, on plasmid pintdnaA–gfp, was obtained by cloning a BamHI–BclI fragment from pdnaA–gfp into the BclI site of pYUB412.10 Plasmids pdnaA–gfp and pintdnaA–gfp were electroporated into the mycobacterial strains and plated onto medium containing kanamycin (pdnaA–gfp) or hygromycin (pintdnaA–gfp). GFP-expressing mycobacteria were visualized for fluorescence using a Nikon Eclipse 600 fluorescence microscope.
Monitoring bacterial growth by fluorescence
A colony of mycobacteria from solid medium was inoculated into 5 mL of 7H9-OAD-Tw (with kanamycin when the strains contained plasmid pdnaA–gfp or hygromycin for strains with integrating plasmid pintdnaA–gfp) in a 50 mL conical tube and grown with agitation at 37°C until reaching exponential phase. The colony was then transferred to 50 mL of the same medium and grown in 500 mL flasks. For the initial studies comparing fluorescence versus growth, samples were taken periodically to measure the optical density (OD) and fluorescence: every 3 h for M. smegmatis; every 6 h for M. abscessus; and every 24 h for M. chelonae. OD600 was measured in a 96-well spectrophotometer (Molecular Devices SpectraMax 340) and fluorescence (485 nm excitation and 535 nm emission) was measured in a 96-well plate fluorimeter (Molecular Devices SpectraMax, GeminisXS).
Assay of disinfectant activity using fluorescent mycobacteria
The different bacteria were grown in 50 mL tubes to an OD600 of 1.0 (∼109 cfu/mL). Then, 5 mL of each was transferred to a new 50 mL conical tube, 10 glass beads (1.0 mm) were added, and the tube was vortexed for 20 s and left to stand for 10 min to allow for clumps to sediment. An aliquot of 100 µL of the suspension was added to 9.9 mL of 7H9-OAD-TW and, then, 1 mL of this was added to 9 mL of the same medium, which was then mixed and used as the inoculum in the 96-well plate assay. For some assays, a second 1 : 10 dilution in medium was performed, so that 105 or 104 bacteria, in 100 µL, were added to each well.
The assay was performed using black 96-well plates, one disinfectant per plate. An aliquot of 200 μL of the highest concentration of the disinfectant to be tested was placed into the first column and 100 µL of 7H9-OAD-Tw was added to the other columns. Using a multichannel pipette, 100 µL from the wells with the highest disinfectant concentration was diluted into the 100 μL of 7H9-OAD-Tw in the next column, and the dilution process serially repeated to obtain final concentrations of 4%, 2%, 1%, 0.5%, 0.25%, 0.125% and 0.06%. Each concentration was evaluated in triplicate. An aliquot of 100 μL of the mycobacterial suspension was added into each well, and positive controls without disinfectants and negative controls with medium alone were included on each plate. Peripheral wells were filled with 200 mL of sterile distilled water. The plates were incubated with agitation at 37°C and the fluorescence read periodically over 4 days in a Molecular Dynamics Spectramax GeminisXS Fluorimeter/Luminometer.
Results
QACs kill most mycobacteria, but survivors appear at high frequencies in M. chelonae and M. abscessus
Although the QAC disinfectants are cidal for most bacteria, they have been generally regarded as only mycobacteriostatic. To test this, M. smegmatis, M. chelonae and M. abscessus were exposed to CTAB or to a commercial disinfectant commonly used in Venezuela, which will be referred to as DBLAB, for dimethyl benzyl lauryl ammonium bromide (C13H27NBr), its active QAC ingredient. Both CTAB and DBLAB were used as 9% concentrations, close to the 10% commercial presentation of DBLAB.
The mycobacteria were exposed to the QACs for 20 min, 60 min or 24 h, after which the QACs were neutralized and the mycobacteria plated on non-selective medium. After 20 or 60 min exposures, both QACs appeared to effectively kill M. smegmatis, with resistant colonies appearing at a frequency of <1 in 105 (Table 2). The disinfectants also appeared to kill most M. abscessus and M. chelonae, but surviving colonies appeared at frequencies of >5 in 100 after 20 and 60 min exposures, with lower frequencies of survival after a 24 h exposure.
Table 2.
The number of colonies of different strains surviving exposures to CTMB, DBLAB or glutaraldehyde
|
M. abscessus |
M. chelonae |
M. smegmatis |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Exposure | 2.0 WT ∼102 cfu plated | 2.1 Select. CTAB ∼102 cfu plated | 2.2 Select. DBLAB ∼102 cfu plated | 2.3 Select. GTA ∼102 cfu plated | 2.0 WT ∼105 cfu plated | 2.3 Select. GTA ∼105 cfu plated | 2.0 WT ∼102 cfu plated | 2.1 Select. CTAB ∼102 cfu plated | 2.2 Select. DBLAB ∼102 cfu plated | 2.0 WT ∼105 cfu plated | mc2155 WT ∼105 cfu plated |
| 5 min | |||||||||||
| 2% GTA | 0 | 0 | 0 | 0 | 1 | 4 | 0 | 0 | 0 | 2 | 0 |
| 20 min | |||||||||||
| 9% CTAB | 12 | 72 | 61 | 78 | NT | NT | 14 | 86 | 76 | NT | 0 |
| 9% DBLAB | 15 | 68 | 58 | 92 | NT | NT | 4 | NT | NT | NT | 0 |
| 2% GTA | 0 | 0 | 0 | 0 | 1 | 2 | 0 | NT | NT | 0 | 0 |
| 60 min | |||||||||||
| 9% CTAB | 6 | 47 | 57 | 52 | NT | NT | 8 | 53 | 73 | NT | 0 |
| 9% DBLAB | 6 | 63 | 48 | 70 | NT | NT | 3 | NT | NT | NT | 0 |
| 2% GTA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NT | NT | 0 | 0 |
| 24 h | |||||||||||
| 9% CTAB | 1 | 0 | 0 | 0 | NT | NT | 0 | 0 | 0 | NT | 0 |
| 9% DBLAB | 0 | 0 | 1 | 0 | NT | NT | 0 | 0 | 0 | NT | 0 |
The strains tested were the parental isolates and their resistant derivatives, selected as survivors of previous exposures to the indicated disinfectants. NT, not tested; GTA, glutaraldehyde; Select., the strain was selected as a survivor after exposure to the indicated disinfectant.
The surviving colonies were then selected, grown in liquid medium without QACs and again subjected to 20 min, 60 min or 24 h exposures to 9% CTAB or 9% DBLAB. The survival rates were now ≥∼50% after 20 and 60 min exposures to both agents, but remained at <1 in 100 after 24 h exposures (Table 2). This was true even for those strains obtained from colonies surviving a 24 h exposure. Strains obtained from colonies surviving a 5 min or 20 min exposure to 2% glutaraldehyde behaved similarly, but were only minimally more resistant to glutaraldehyde than the parent strain. No survivors were obtained with longer exposures to 2% glutaraldehyde.
Equivalent results were obtained when the experiment was repeated with clinical or environmental isolates of M. massiliense and M. fortuitum. Some of these original, parental isolates displayed the rough colony morphotype, while others had smooth colonies. The original colony morphotype tended to persist in the QAC surviving colonies, but some smooth QAC surviving strains began to produce a minority of rough colonies when restreaked. Smooth strains showed greater motility (data not shown), but neither motility nor colony morphotype appeared to be correlated with QAC resistance.
Susceptibility to QACs determined by fluorescence
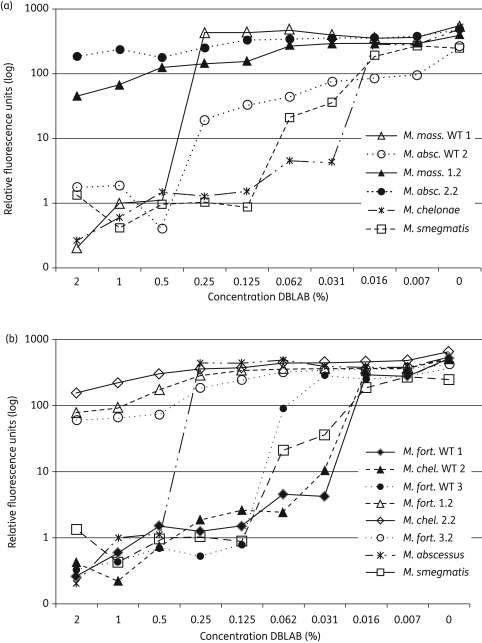
To assess QAC susceptibility in another format, the mycobacteria were labelled with a dnaA–gfp fusion, which permitted bacterial growth to be monitored as fluorescence in a 96-well plate assay (Figure S1, available as Supplementary data at JAC Online). Using this method, clinical and environmental isolates of M. abscessus and M. massiliense were less susceptible to DBLAB than M. chelonae and M. fortuitum, but the strains grown from colonies surviving exposure to DBLAB and CTAB were resistant to all concentrations tested (Figure 1).
Figure 1.
Different isolates of the M. abscessus-chelonae group and M. fortuitum show similar susceptibilities to DBLAB, while their derivatives, selected after surviving a previous exposure, are resistant to at least 2% DBLAB. The strains were labelled with plasmid pdnaA–gfp and grown in 96-well plates in medium containing DBLAB at the indicated concentrations. Mycobacterial growth was measured as fluorescence after 4 days of incubation at 37°C. All results are from representative experiments that were repeated at least three times. (a) Comparison of the growth in DBLAB of M. massiliense (M. mass. WT 1), M. abscessus (M. absc. WT 2), derivatives of each that were selected as survivors of a previous exposure of DBLAB (M. mass. 1.2 and M. absc. 2.2), M. smegmatis and M. chelonae. (b) Comparison of the growth in DBLAB of M. chelonae (M. chel. WT 2), two isolates of M. fortuitum (M. fort. WT 1 and M. fort. WT 2), derivatives of each that were selected as survivors of a previous exposure to DBLAB (M. fort. 1.2, M. chel. 2.2, and M. fort. 3.2), M. smegmatis and M. abscessus.
To learn more about the innate susceptibility of different mycobacteria to QACs, we compared the growth of M. smegmatis, M. chelonae, M. abscessus, M. bovis BCG and M. terrae in the presence of increasing concentrations of CTAB, DBLAB and three other QACs (Figure S2, available as Supplementary data at JAC Online). The susceptibilities varied somewhat with the different QACs, but M. abscessus tended to be the most resistant and M. smegmatis the most susceptible. Notably though, the susceptibility of M. smegmatis to DBLAB was very similar to that of M. chelonae and M. fortuitum (Figure 1b).
Reversibility of QAC resistance
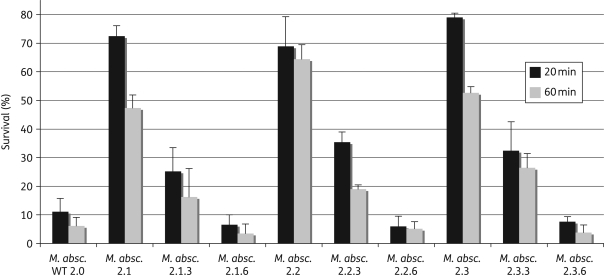
The very high frequency of QAC-resistant survivors of M. abscessus, M. massiliense, M. chelonae and M. fortuitum seemed inconsistent with a genetic alteration, so we tested whether the resistant phenotype was reversible. Three M. abscessus survivor strains, derived from the same parent strain after exposure to CTAB (2.1), DBLAB (2.2) or a 5 min exposure to glutaraldehyde (2.3), were serially restreaked to non-selective medium. After three and six passages, they were again subjected to 20 or 60 min exposures to CTAB and DBLAB. After three passages, the survival of all three strains had fallen to ∼30% and after six passages only ∼5% of the plated bacteria survived, similar to the original parent strain (Figure 2). Resistant M. chelonae survivor strains showed similar reductions in QAC survival after serial restreaking on non-selective medium (data not shown).
Figure 2.
The QAC-resistant phenotype reverts to normal when passaged on medium without disinfectants. An original clinical M. abscessus isolate—M. absc. WT 2.0 (LTE3514), as well as its resistant derivatives and revertants, were plated after 20 or 60 min exposures to 9% CTAB. Survival is expressed as the number of colonies appearing after the CTAB exposures, compared with the number of colonies appearing after a parallel, control treatment without the presence of CTAB. The derivatives of clinical isolate WT 2.0 were selected as survivors after exposures to CTAB (2.1), DBLAB (2.2) or glutaraldehyde (2.3). The revertants were obtained after either three (2.1.3, 2.2.3 and 2.3.3) or six (2.1.6, 2.2.6 and 2.3.6) serial passages on solid medium without disinfectants.
Discussion
Provoked by the occurrence of NTM infections that appeared to be associated with the use of commercial QAC disinfectants,12,13 we investigated the effectiveness of QACs against mycobacteria. QACs are described in the literature as only mycobacteriostatic, so we were surprised to find that a 20 min exposure to QACs appeared to be cidal against M. smegmatis, with at least 5 logs of killing;14 although their effectiveness would likely be less if assayed in a carrier test in the presence of organic material.15 Against M. fortuitum, M. chelonae, M. massiliense and M. abscessus, however, even with the simple suspension test employed here, we found that QACs kill only ∼90%–95% of bacteria. This suggests that the use of QACs will not prevent, and may even promote, the occurrence of infections with M. fortuitum and the M. chelonae–abscessus group of NTM.
From these results, the QACs cannot accurately be called mycobacteriostatic, because when the agent is removed, the majority of the bacteria are not viable. Neither can they be termed effectively mycobactericidal, at least for M. fortuitum and the M. chelonae–abscessus group, because there are <2 logs of killing after 1 h of exposure. This high percentage of survival has been previously reported,14,15 but the nature of the survivors apparently was not investigated. It is not clear why this high frequency of persisters is not seen with M. smegmatis, but it cannot simply depend upon the level of innate resistance, as some of the original parental M. fortuitum and M. chelonae isolates were as susceptible to DBLAB as was M. smegmatis (Figure 1b).
The QAC survivors of M. fortuitum and the M. chelonae–abscessus group appeared at frequencies between 1% and 10%, which seems too high to be caused by genomic mutations. Although these survivors showed QAC resistance, when they were serially restreaked in the absence of QACs, the frequency of QAC survivors returned to that of the original strains. This reversible resistance, although it persists for several generations, suggests a phenotype dependent upon non-genetic changes in gene expression. After a 5 min or 20 min exposure to 2% glutaraldehyde, there also appeared, at a lower frequency, glutaraldehyde-tolerant survivors that showed only minimally increased resistance to glutaraldehyde, but had 60% QAC survival, suggesting that they had also acquired the QAC-resistant phenotype. This phenotype could therefore reflect some sort of general stress or global resistance response.16,17 Escherichia coli tolerant to QACs have been described, but were selected by adaptation to serially increasing concentrations of the disinfectant,18 much different from the one-step selection in high QAC concentrations described here.
The QAC resistance seen in the survivors is not complete, as there was no meaningful increase in survival after 24 h exposures to the QACs. The M. abscessus and M. chelonae bacteria that survived 24 h exposures to QACs appear to represent a second, additional form of QAC persisters. While they survive a 1 h exposure to QACs, they are no more resistant to a 24 h QAC exposure than the parent strains, which is similar to the previously described antibiotic persistence of non-replicating bacteria.19,20 However, the frequency of the 24 h survivors seen here (∼1/100–1/1000) is higher than has been reported for non-replicating persisters and suggests that mycobacteria could perhaps survive in QAC disinfectant stocks.
Like many other NTM, the M. chelonae–abscessus group have been observed in two morphotypes, smooth and rough. While the rough morphotype is more virulent in pulmonary infection models,21 the smooth morphotype has a higher production of glycopeptidolipids22,23 that is thought to cause their increased motility and the increased biofilm formation that has been associated with decreased drug susceptibility.24 We found no association of QAC resistance and colony morphotype, as QAC-resistant strains were selected from both rough and smooth isolates, and tended to maintain their morphotype (data not shown).
M. abscessus and M. massiliense belong to a family of closely related mycobacteria.25 They are among the NTM most commonly isolated from healthcare-associated skin and soft-tissue infections,26 although M. chelonae is closely related and also frequently encountered. M. fortuitum is more phylogenetically distant, but is also found in healthcare-related infections. The dramatic recent increase in articles describing human infections with NTM, especially with the M. abscessus family, suggests that their incidence is increasing, and M. abscessus may have greater pathogenic capacity than other NTM.27 The innocuous M. smegmatis would seem an inadequate model to study the effectiveness of disinfectants against mycobacteria.
We are currently investigating possible mechanisms for the QAC-resistant phenotype, such as decreased cell wall permeability28 or the extrusion of the QACs7,29,30 by efflux pumps,31 similar to LfrA32 or Tap.33 However, while the molecular mechanisms responsible for the resistant phenotype may yield insights into the pathogenic potential of these bacteria, the principal conclusion from this work is that QAC disinfectants are insufficient to prevent, and may actually promote, healthcare-associated infections with M. fortuitum and the M. abscessus–chelonae group of NTM.
Funding
This work was funded by FONACIT (Fondo Nacional de Ciencia, Tecnología y Innovación) Proyecto G-2005000393 and by Helmerich & Paine de Venezuela, C.A. through LOCTI (Ley Orgánica de Ciencia, Tecnología y Innovación) project ‘Las Cepas de Tuberculosis Mas Virulentas de Venezuela’. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Transparency declarations
None to declare.
Author contributions
C. C. performed the experiments, analysed results and helped write the manuscript, G. J. L. constructed reagents and developed methods employed, J. H. de W. helped to conceive the study, provided strains and analysed the results, and H. E. T. conceived the study, analysed the results and wrote the manuscript.
Supplementary data
Figures S1 and S2 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Supplementary Material
Acknowledgements
We thank Donald Court for helpful discussions and Drs Sylvia Leaó, Elba Guerrero, Glenda Vílchez, Donald Court and Stewart Cole for critically reading the manuscript.
References
- 1.Cooksey RC, de Waard JH, Yakrus MA, et al. Mycobacterium cosmeticum sp. nov., a novel rapidly growing species isolated from a cosmetic infection and from a nail salon. Int J Syst Evol Microbiol. 2004;54:2385–91. doi: 10.1099/ijs.0.63238-0. doi:10.1099/ijs.0.63238-0. [DOI] [PubMed] [Google Scholar]
- 2.Piquero J, Casals VP, Higuera EL, et al. Iatrogenic Mycobacterium simiae skin infection in an immunocompetent patient. Emerg Infect Dis. 2004;10:969–70. doi: 10.3201/eid1005.030681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivera-Olivero IA, Guevara A, Escalona A, et al. Soft-tissue infections due to non-tuberculous mycobacteria following mesotherapy. What is the price of beauty. Enferm Infecc Microbiol Clin. 2006;24:302–6. doi: 10.1157/13089664. doi:10.1157/13089664. [DOI] [PubMed] [Google Scholar]
- 4.van Ingen J, van der Laan T, Dekhuijzen R, et al. In vitro drug susceptibility of 2275 clinical non-tuberculous Mycobacterium isolates of 49 species in The Netherlands. Int J Antimicrob Agents. 2010;35:169–73. doi: 10.1016/j.ijantimicag.2009.09.023. doi:10.1016/j.ijantimicag.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 5.van Ingen J, Boeree MJ, Dekhuijzen PN, et al. Environmental sources of rapid growing nontuberculous mycobacteria causing disease in humans. Clin Microbiol Infect. 2009;15:888–93. doi: 10.1111/j.1469-0691.2009.03013.x. doi:10.1111/j.1469-0691.2009.03013.x. [DOI] [PubMed] [Google Scholar]
- 6.McDonnell G, Russell AD. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev. 1999;12:147–79. doi: 10.1128/cmr.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegstad K, Langsrud S, Lunestad BT, et al. Does the wide use of quaternary ammonium compounds enhance the selection and spread of antimicrobial resistance and thus threaten our health? Microb Drug Resist. 2010;16:91–104. doi: 10.1089/mdr.2009.0120. [DOI] [PubMed] [Google Scholar]
- 8.Adekambi T, Colson P, Drancourt M. rpoB-based identification of nonpigmented and late-pigmenting rapidly growing mycobacteria. J Clin Microbiol. 2003;41:5699–708. doi: 10.1128/JCM.41.12.5699-5708.2003. doi:10.1128/JCM.41.12.5699-5708.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valdivia RH, Hromockyj AE, Monack D, et al. Applications for green fluorescent protein (GFP) in the study of host–pathogen interactions. Gene. 1996;173:47–52. doi: 10.1016/0378-1119(95)00706-7. doi:10.1016/0378-1119(95)00706-7. [DOI] [PubMed] [Google Scholar]
- 10.Pavelka MS, Jr, Jacobs WR., Jr Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. J Bacteriol. 1996;178:6496–507. doi: 10.1128/jb.178.22.6496-6507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salazar L, Fsihi H, de Rossi E, et al. Organization of the origins of replication of the chromosomes of Mycobacterium smegmatis, Mycobacterium leprae and Mycobacterium tuberculosis and isolation of a functional origin from M. smegmatis. Mol Microbiol. 1996;20:283–93. doi: 10.1111/j.1365-2958.1996.tb02617.x. doi:10.1111/j.1365-2958.1996.tb02617.x. [DOI] [PubMed] [Google Scholar]
- 12.Tiwari TS, Ray B, Jost KC, Jr, et al. Forty years of disinfectant failure: outbreak of postinjection Mycobacterium abscessus infection caused by contamination of benzalkonium chloride. Clin Infect Dis. 2003;36:954–62. doi: 10.1086/368192. doi:10.1086/368192. [DOI] [PubMed] [Google Scholar]
- 13.Murillo J, Torres J, Bofill L, et al. Skin and wound infection by rapidly growing mycobacteria: an unexpected complication of liposuction and liposculpture. The Venezuelan Collaborative Infectious and Tropical Diseases Study Group. Arch Dermatol. 2000;136:1347–52. doi: 10.1001/archderm.136.11.1347. doi:10.1001/archderm.136.11.1347. [DOI] [PubMed] [Google Scholar]
- 14.Bello T, Rivera-Olivero IA, de Waard JH. Inactivation of mycobacteria by disinfectants with a tuberculocidal label. Enferm Infecc Microbiol Clin. 2006;24:319–21. doi: 10.1157/13089667. doi:10.1157/13089667. [DOI] [PubMed] [Google Scholar]
- 15.Best M, Sattar SA, Springthorpe VS, et al. Comparative mycobactericidal efficacy of chemical disinfectants in suspension and carrier tests. Appl Environ Microbiol. 1988;54:2856–8. doi: 10.1128/aem.54.11.2856-2858.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grkovic S, Brown MH, Skurray RA. Regulation of bacterial drug export systems. Microbiol Mol Biol Rev. 2002;66:671–701. doi: 10.1128/MMBR.66.4.671-701.2002. table of contents doi:10.1128/MMBR.66.4.671-701.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hecker M, Pane-Farre J, Volker U. SigB-dependent general stress response in Bacillus subtilis and related Gram-positive bacteria. Annu Rev Microbiol. 2007;61:215–36. doi: 10.1146/annurev.micro.61.080706.093445. doi:10.1146/annurev.micro.61.080706.093445. [DOI] [PubMed] [Google Scholar]
- 18.Langsrud S, Sundheim G, Holck AL. Cross-resistance to antibiotics of Escherichia coli adapted to benzalkonium chloride or exposed to stress-inducers. J Appl Microbiol. 2004;96:201–8. doi: 10.1046/j.1365-2672.2003.02140.x. doi:10.1046/j.1365-2672.2003.02140.x. [DOI] [PubMed] [Google Scholar]
- 19.Singh R, Barry CE, III, Boshoff HI. The three RelE homologs of Mycobacterium tuberculosis have individual, drug-specific effects on bacterial antibiotic tolerance. J Bacteriol. 2010;192:1279–91. doi: 10.1128/JB.01285-09. doi:10.1128/JB.01285-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiuff C, Zappala RM, Regoes RR, et al. Phenotypic tolerance: antibiotic enrichment of noninherited resistance in bacterial populations. Antimicrob Agents Chemother. 2005;49:1483–94. doi: 10.1128/AAC.49.4.1483-1494.2005. doi:10.1128/AAC.49.4.1483-1494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ordway D, Henao-Tamayo M, Smith E, et al. Animal model of Mycobacterium abscessus lung infection. J Leukoc Biol. 2008;83:1502–1. doi: 10.1189/jlb.1007696. doi:10.1189/jlb.1007696. [DOI] [PubMed] [Google Scholar]
- 22.Byrd TF, Lyons CR. Preliminary characterization of a Mycobacterium abscessus mutant in human and murine models of infection. Infect Immun. 1999;67:4700–7. doi: 10.1128/iai.67.9.4700-4707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howard ST, Rhoades E, Recht J, et al. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology. 2006;152:1581–90. doi: 10.1099/mic.0.28625-0. doi:10.1099/mic.0.28625-0. [DOI] [PubMed] [Google Scholar]
- 24.Greendyke R, Byrd TF. Differential antibiotic susceptibility of Mycobacterium abscessus variants in biofilms and macrophages compared to that of planktonic bacteria. Antimicrob Agents Chemother. 2008;52:2019–6. doi: 10.1128/AAC.00986-07. doi:10.1128/AAC.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leao SC, Tortoli E, Viana-Niero C, et al. Characterization of mycobacteria from a major Brazilian outbreak suggests that revision of the taxonomic status of members of the Mycobacterium chelonae–M. abscessus group is needed. J Clin Microbiol. 2009;47:2691–8. doi: 10.1128/JCM.00808-09. doi:10.1128/JCM.00808-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medjahed H, Gaillard JL, Reyrat JM. Mycobacterium abscessus: a new player in the mycobacterial field. Trends Microbiol. 2010;18:117–23. doi: 10.1016/j.tim.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 27.Ripoll F, Pasek S, Schenowitz C, et al. Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS One. 2009;4:e5660. doi: 10.1371/journal.pone.0005660. doi:10.1371/journal.pone.0005660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svetlikova Z, Skovierova H, Niederweis M, et al. Role of porins in the susceptibility of Mycobacterium smegmatis and Mycobacterium chelonae to aldehyde-based disinfectants and drugs. Antimicrob Agents Chemother. 2009;53:4015–8. doi: 10.1128/AAC.00590-09. doi:10.1128/AAC.00590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar R, Bibi E. MdfA, an Escherichia coli multidrug resistance protein with an extraordinarily broad spectrum of drug recognition. J Bacteriol. 1997;179:2274–80. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bore E, Hebraud M, Chafsey I, et al. Adapted tolerance to benzalkonium chloride in Escherichia coli K-12 studied by transcriptome and proteome analyses. Microbiology. 2007;153:935–46. doi: 10.1099/mic.0.29288-0. doi:10.1099/mic.0.29288-0. [DOI] [PubMed] [Google Scholar]
- 31.Esteban J, Martin-de-Hijas NZ, Ortiz A, et al. Detection of lfrA and tap efflux pump genes among clinical isolates of non-pigmented rapidly growing mycobacteria. Int J Antimicrob Agents. 2009;34:454–6. doi: 10.1016/j.ijantimicag.2009.06.026. doi:10.1016/j.ijantimicag.2009.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Takiff HE, Cimino M, Musso MC, et al. Efflux pump of the proton antiporter family confers low-level fluoroquinolone resistance in Mycobacterium smegmatis. Proc Natl Acad Sci USA. 1996;93:362–6. doi: 10.1073/pnas.93.1.362. doi:10.1073/pnas.93.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ainsa JA, Blokpoel MC, Otal I, et al. Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacterium fortuitum and Mycobacterium tuberculosis. J Bacteriol. 1998;180:5836–43. doi: 10.1128/jb.180.22.5836-5843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.