Abstract
Recent advances in the study of the tumor microenvironment have revealed significant interaction between tumor cells and their surrounding stroma in model systems. We have previously shown that two distinct stromal signatures derived from a macrophage (CSF1) response and a fibroblastic (DTF-like) response are present in subsets of invasive breast cancers and show a correlation with clinical outcome [1–3]. In the present study we explore whether these signatures also exist in the stroma of ductal carcinoma in situ (DCIS). We studied the signatures by both gene expression profile analysis of a publically available data set of DCIS and by immunohistochemistry (IHC) on a tissue microarray of DCIS and invasive breast cancer cases. Both the gene expression and immunohistochemical data show that the macrophage response and fibroblast expression signatures are present in the stroma of subsets of DCIS cases. The incidence of the stromal signatures in DCIS is similar to the incidence in invasive breast cancer that we have previously reported. We also find that the macrophage response signature is associated with higher grade DCIS and cases which are ER and PR negative, whereas the fibroblast signature was not associated with any clinicopathologic features in DCIS. A comparison of 115 matched cases of DCIS and invasive breast cancer found a correlation between the type of stromal response in DCIS and invasive ductal carcinoma (IDC) within the same patient for both the macrophage response and the fibroblast stromal signatures (P = 0.03 and 0.08, respectively). This study is a first characterization of these signatures in DCIS. These signatures have significant clinicopathologic associations and tend to be conserved as the tumor progresses from DCIS to invasive breast cancer.
Keywords: Ductal carcinoma in situ, Tumor microenvironment, Cancer stroma, Fibroblast, Macrophage
Introduction
Breast cancer remains a leading cause of death amongst women worldwide. There has been a dramatic rise in the incidence of ductal carcinoma in situ (DCIS) in the US which is at least partially due to the use of screening mammography [4]. Currently, therapy for DCIS involves surgical resection with or without radiation and sometimes adjuvant hormone therapy [5–7]. Although Page et al. [8] have shown that overall there is a ninefold increased likelihood for women with DCIS to develop invasive breast carcinoma, there are few biomarkers to predict behavior in individual patients with DCIS [9, 10].
The connective tissue surrounding breast carcinoma cells (tumor microenvironment, TME) is a poorly characterized structure that consists of a wide variety of cell types (endothelial cells, inflammatory cells, fibroblasts, etc.) many of which are not well understood. Studies have shown that the TME is more than merely a scaffold on which the malignant cells rest but in fact plays an important interactive role with growth enhancing properties for the tumor [11–14]. One emerging area in TME research suggests that invasive cancer cells actively recruit stromal cells and interact with them to create a TME that promotes tumor growth and possibly metastasis. Less well studied is the impact of the surrounding stroma on pre-invasive (in situ) breast cancer. There have been a number of studies performed in animal model systems that demonstrate that stroma can induce pre-invasive cancer [15]. However, only a few papers have examined the expression patterns of genes within the stroma of DCIS [16–18].
In prior work, we have defined three stromal gene expression profiles in invasive breast cancer [1–3]. While most studies have treated the TME as a uniform entity, we have been able to distinguish different types of TME that occur in different cancers. One stromal gene expression profile is consistent with a fibroblast (DTF-like) response pattern and is composed of genes expressed during scar formation (e.g., type I and II collagen) and pro-fibrotic signaling proteins such as TGFB. Another represents a macrophage response to colony stimulating factor (CSF1). Analysis of these stromal signatures on large invasive breast cancer gene expression profiling datasets demonstrated that the signatures are present in distinct groups of invasive breast cancer, and the signatures are correlated with clinicopathologic features, such as survival and hormonal status. To date, we have focused on studying these stromal signatures in invasive breast carcinoma. Here we characterize both the fibroblast and macrophage response stromal reaction patterns in DCIS and show that these signatures also exist in non-invasive breast cancer.
Materials and methods
Gene expression data analysis
For an analysis of the stromal gene expression signatures in DCIS, we used a publicly available data set that includes 40 cases of DCIS [19]. The data was downloaded and imported into Excel. Stromal signatures were derived from our previous studies on invasive breast cancer [1, 2]. For the fibroblast (DTF) signature a list of 66 genes was used while the macrophage (CSF1) response signature contained 112 genes; these were mapped to the downloaded dataset. Expression values for genes were standardized by subtracting the mean expression level for this gene across all samples from each gene expression value. Unsupervised hierarchical clustering was done in each data set with the Cluster 3.0 software using the uncentered Pearson correlation as the distance metric and average linkage clustering. The resulting heat map and dendrogram were visualized on Java TreeView.
Evaluation of the fibroblast and macrophage response stromal signatures in breast cancer tumor microenvironment
To determine the patterns of macrophage (CSF1) and fibroblast (DTF) core protein expression in the breast cancer TME, we performed immunohistochemistry on two breast cancer TMAs (TA239 and TA241) containing samples from a total of 285 cases of DCIS and 115 cases of invasive ductal cancer obtained from Stanford University Medical Center from 2001 to 2007.
The primary antibodies used were FCGR3A (CD16, AbD Serotec, MCA1816, 1:40 dilution), CTSL1 (AbD Serotec, MCA2374, 1:25 dilution), FCGR2A (CD32; Ab-cam, AB45143, 1:200 dilution), CD163 (Novocastra, NCL-CD163, 1:200 dilution), SPARC (Zymed, 1:1000 dilution), CDH11 (Invitogen, Cat# 32-1700, 1:10 dilution), SDC1 (CD138, Serotec, cat# MCA681H, 1:400 dilution), and MMP11 (Calbiochem, Cat# IM86, 1:200 dilution). CD163 and CD138 were detected with the Ventana Benchmark autostainer while the remaining stains were manually applied and were visualized using mouse and rabbit versions of the EnVision + system (DAKO) using diam-inobenzidine. The immunohistochemical studies were interpreted by histopathologic evaluation by M.S., I.E., and R.W.
Cores were considered strongly positive if >30% of the stromal compartment stained positive and weakly positive if 5–30% of the stromal compartment was positive, and negative if <5% of the stromal compartment was positive. Scores were summed for both the fibroblast and macrophage response signatures and a case was deemed positive for the signature if two of three or three of four replicate cores were at least weakly positive.
Analysis of clinicopathologic variables
Fisher’s exact test was used to compute P values for association testing.
Results
The activated fibroblast and macrophage signatures are present in a subset of DCIS cases both by gene expression profiling and immunohistochemistry
In previous studies we defined core gene signatures for fibroblast (DTF-like) and macrophage (CSF1 response) stromal signature patterns in invasive breast cancer [1, 2]. The signatures are defined by expression levels for 2 distinct sets of genes, consisting of 66 genes for the fibroblast signature and 112 genes for the macrophage response. Each of the signatures was consistently found to be coordinately expressed by distinct subsets of invasive breast cancers in 5 publically available breast cancer datasets. We now examine a publically available published gene expression profiling dataset of 40 cases of DCIS [19] for the presence of these stromal signatures. From our original core gene signatures, 53 out of 66 genes were present and well measured in the DCIS dataset for the fibroblast signature while 100 out of 112 genes from the macrophage signature were well measured. Hierarchical clustering of the tumors based on the macrophage and fibroblast cores genes (Fig. 1a, c) show that there is a subset of tumors which express the fibroblast signature (16 of 40, 40%) and a subset which express the macrophage signature (17 of 40, 42.5%). The analysis on this relatively small dataset demonstrates that both stromal signatures previously identified in a subset of invasive breast carcinoma are likewise present in a subset of DCIS cases.
Fig. 1.
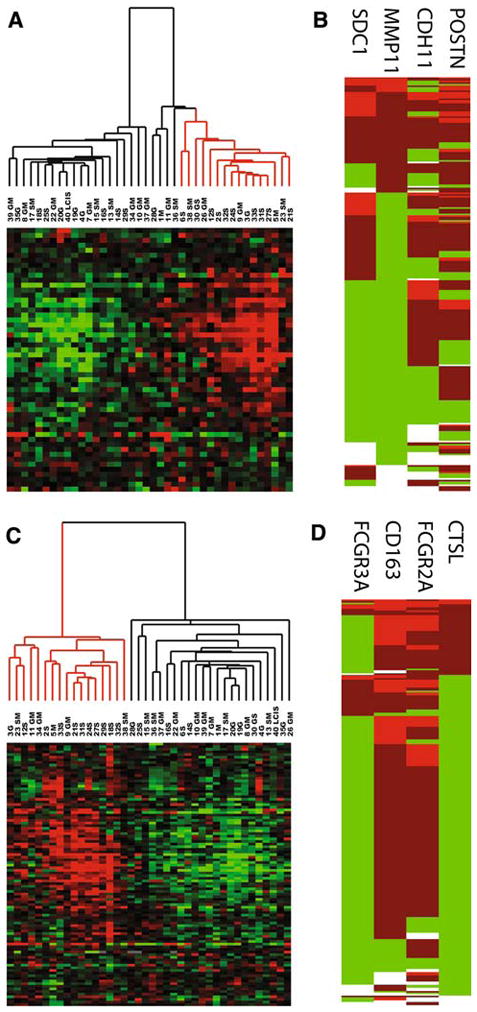
a Unsupervised hierarchical clustering of gene expression microarray data with fibroblast response core stromal genes in a DCIS data set. Cases are arranged along the X-axis and genes are arranged along the Y-axis. Within the heat map, red represents high levels of mRNA expression, black represents median expression, and green represents low expression. b Coordinate expression of fibroblast response as measured by immunohistochemistry. Unsupervised hierarchical clustering of 272 breast carcinomas based on TMA staining for four fibroblast (CDH11, POSTN, MMP11, and SDC1) proteins by immunohistochemistry. The breast cancer cases are arranged along the Y-axis, and fibroblast markers are arranged along the X-axis. Within the heat map, red represents high expression, green represents low expression, and white represents missing data. c Unsupervised hierarchical clustering of microarray data for macrophage response core stromal genes in a DCIS data set. d Coordinate expression of macrophage response proteins in breast cancer. Unsupervised hierarchical clustering of 272 breast carcinomas based on TMA staining for four macrophage response markers (CD163, FCGR3a, FCGR2a, and CTSL1) by immunohistochemistry
In order to confirm and expand on the results generated by analysis of the gene microarray dataset, we used immunohistochemistry on a DCIS tissue microarray to examine a large series of cases (Fig. 1b, d). We used a panel of four antibodies to represent each gene signature (8 antibodies total). Both sets of antibodies were obtained from the core gene lists with a preference for genes that were very highly and differentially expressed in DTF (fibroblast signature) or CSF1 (macrophage signature) with well-characterized antibodies. The biomarker panel for the macrophage response was previously defined and assessed on an invasive breast cancer array [1]. Each panel of antibodies was assessed for staining in the stromal compartments and scored as outlined in the “Materials and methods” section; a case was deemed positive for the signature if two of three or three of four replicate cores were at least weakly positive. 30% of all DCIS cases (55/236) were positive for the fibroblast signature and 19% (39/247) of cases were positive for the macrophage signature. This is similar to the fraction of positive cases seen in invasive breast cancer [1, 2] in which 25–35% were positive for the fibroblast signature and 17–25% were positive for the macrophage response signature. The protein expression pattern within the stroma varied between antibodies. Some genes (Cadherin 11, SPARC, CD163, and FCGF2B) were expressed diffusely within the stroma but were largely expressed in spindled stromal cells (Cadherin 11 and SPARC) or leukocytes (CD163 and FCGF2B). Other genes (SDC1, MMP11, CD16, and CTSL1) had restricted expression within specific cells within the TME. SDC1 and MMP11 were confined to a subset of spindled stromal cells while CD16 and CTSL1 were expressed in a subset of leukocytes. This is illustrated in Fig. 2a, which shows an example of a case which was diffusely positive for both fibroblast and macrophage response markers. The cell to cell variability of protein expression within the stromal compartment is illustrated by this case.
Fig. 2.
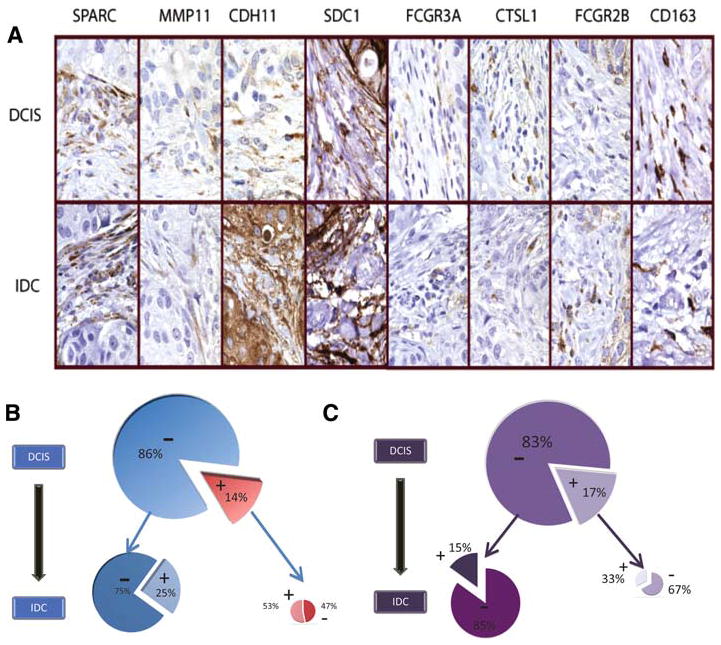
a Immunohistochemistry of four fibroblast response markers (SPARC, MMP11, CDH11, SDC1) and four macrophage response markers (FCGR3A, CTSL1, FCGR2B, CD163) showing an example of a single case of diffuse positivity for all four macrophage and four fibroblast stromal response markers in a patient who had both DCIS and invasive ductal carcinoma (IDC). b, c Pie chart representation of stability of stromal signatures from DCIS to IDC. b Fibroblast signature in DCIS (large pie) with positive (red, 14%) and negative (blue, 86%) cases progressing to IDC (smaller pies). c Macrophage response signature in DCIS (large pie) with positive (lavender, 17%) and negative (purple, 83%) cases progressing to IDC (smaller pies). For both the fibroblast and macrophage signature, there is an association of stromal signature expression in DCIS with the stromal signature expression in IDC (P = 0.03 for fibroblast signature; P = 0.09 for macrophage signature)
Correlation of stromal signatures with clinicopathologic features of DCIS
The fibroblast and macrophage response stromal signatures were correlated with the clinicopathologic features of grade and hormonal status, for the cases represented in the TMA. Histologic evaluation of all DCIS cases for tumor grade was conducted by two breast pathologists (KJ and RW) and tumors were placed into two categories: low/intermediate grade and high grade (Table 1). Cases that expressed the macrophage signature were associated with high grade DCIS in 24/39 (62%) of the cases while only 53/205 (26%) cases without macrophage response signature were high grade (P = 0.0001; Table 1). Additionally all DCIS cases were stained for ER, PR, and HER2 and evaluated based on ASCO scoring criteria (Table 2). Expression of the macrophage response signature in DCIS was significantly associated with PR negativity (25/39 (64%) of macrophage signature positive DCIS cases were PR negative compared with only 82/197 (42%) of macrophage signature negative DCIS cases; P = 0.01, Table 2). There was a tendency for macrophage response signature positive DCIS to be ER negative, but this did not reach statistical significance (P = 0.15, Table 2). These results are similar to our previous studies where we found that the macrophage response signature is associated with higher grade and negative hormone receptor status in invasive breast cancer. In contrast to the findings for the DTF stroma in invasive breast cancer, we were unable to identify a significant association of the fibroblast signature with grade or hormonal receptor status in DCIS.
Table 1.
Correlation of stromal signatures with DCIS grade
| Negative | Positive | P value | |
|---|---|---|---|
| Fibroblast response signature | |||
| DCIS grade | |||
| Low/Int | 125 (70%) | 34 (62%) | 0.32 |
| High | 54 (30%) | 21 (38%) | |
| Macrophage response signature | |||
| DCIS grade | |||
| Low/Int | 152 (74%) | 15 (38%) | 0.0001 |
| High | 53 (26%) | 24 (62%) | |
Table 2.
Correlation of stromal signatures ER, PR, and Her2 status in DCIS
| Fibroblast response signature |
P value | Macrophage response signature |
P value | |||
|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | |||
| ER | ||||||
| Negative | 77 (46%) | 21 (38%) | 0.35 | 79 (41%) | 20 (54%) | 0.15 |
| Positive | 90 (54%) | 34 (62%) | 113 (59%) | 17 (46%) | ||
| PR | ||||||
| Negative | 76 (44%) | 28 (51%) | 0.44 | 82 (42%) | 25 (64%) | 0.01 |
| Positive | 95 (56%) | 27 (49%) | 115 (58%) | 14 (36%) | ||
| Her2 | ||||||
| Negative | 107 (62%) | 34 (63%) | 1.0 | 127 (64%) | 21 (54%) | 0.28 |
| Positive | 66 (38%) | 20 (37%) | 71 (36%) | 18 (46%) | ||
Stromal signatures in DCIS are conserved in progression from DCIS to IDC
We considered that there might be an association between the stromal signatures and the progression to invasive breast cancer. To address this issue a TMA was created with 148 cases of DCIS without IDC present at the time of resection and 115 cases of DCIS with IDC present in the resection specimen at the time of resection. Cases of DCIS with and without IDC were analyzed for fibroblast and macrophage response signature positivity. We find that there was no significant association of either the fibroblast or macrophage signature in DCIS with the presence of IDC at time of resection.
The 115 IDC cases associated with DCIS were also scored for the two stromal signatures using the four fibroblast and four macrophage response markers (Fig. 2a). A schematic representation of how the matched DCIS and IDC cases either remained positive, gained positivity, lost positivity or stayed negative for the fibroblast and macrophage response signatures is shown in Fig. 2b and c. We found that as compared with fibroblast signature negative DCIS, DCIS with fibroblast signature positivity were more likely to be associated with a matched IDC case with fibroblast signature positivity (53% of cases with fibroblast signature positive DCIS showed fibroblast signature positivity in matched IBC, while only 25% of cases with fibroblast signature negative DCIS showed fibroblast signature positivity in matched IDC; P = 0.03). Likewise, as seen in Fig. 2c, there is a similar trend in the preservation of macrophage response from DCIS cases to invasive cancer (33% of cases with macrophage response positive DCIS showed the macrophage response signature in matched IDC, while only 15% of macrophage response signature negative DCIS showed the macrophage response signature in matched IDC; P = 0.09).
Table 3 shows the correlation of the four subgroups of matched case relationships (cases which remained positive from DCIS to IDC, gained positivity, lost positivity, and remained negative) with clinicopathologic variables. Cases of IDC that were macrophage response negative in both DCIS and IDC tended to be ER positive (47/69; 68%) compared with cases that expressed macrophage response in DCIS and/or IDC (12/28; 42%) (p = 0.04). No significant associations were found between fibroblast signature expression changes and clinicopathological features.
Table 3.
Correlation of the four subgroups of matched case relationships in DCIS and IDC with ER, PR, and Her2 status
| Fibroblast response signature |
P value | Macrophage response signature |
P value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stayed pos | Gained Pos | Lost Pos | Stayed Neg | Stayed Pos | Gained Pos | Lost Pos | Stayed Neg | |||
| ER | ||||||||||
| Neg | 5 | 11 | 1 | 21 | 0.096 | 4 | 6 | 6 | 22 | 0.025 |
| 24% | 17% | 5% | 11% | 22% | 16% | 18% | 11% | |||
| Pos | 2 | 10 | 7 | 39 | 2 | 6 | 4 | 47 | ||
| 10% | 16% | 31% | 21% | 11% | 16% | 12% | 22% | |||
| PR | ||||||||||
| Neg | 3 | 10 | 4 | 26 | 0.248 | 4 | 7 | 7 | 25 | 0.008 |
| 14% | 16% | 17% | 14% | 22% | 19% | 20% | 12% | |||
| Pos | 4 | 12 | 4 | 36 | 2 | 5 | 5 | 45 | ||
| 19% | 19% | 17% | 19% | 11% | 14% | 15% | 22% | |||
| HER2 | ||||||||||
| Neg | 4 | 15 | 4 | 44 | 0.508 | 2 | 10 | 7 | 48 | 0.151 |
| 19% | 23% | 17% | 24% | 11% | 27% | 20% | 23% | |||
| Pos | 3 | 6 | 3 | 21 | 4 | 3 | 5 | 22 | ||
| 14% | 19% | 13% | 11% | 22% | 8% | 15% | 10% | |||
Discussion
Invasive breast cancer is often associated with a stromal response that includes proliferating stromal cells and new matrix deposition. At least two, not necessarily mutually exclusive, views exist. On the one hand, researchers have proposed that the stromal reaction to tumor could be due to tumor cells invade through the tissue and thus induce a “wound response” [20, 21]. In this view the stroma reacts to the presence of the tumor by expressing a defined set of genes associated with healing. On the other hand, researchers have found evidence that the stroma can play a more active role in the induction or promotion of oncogenesis [14, 22–25]. For example, tumor cells can actively recruit stromal cells (inflammatory cells, vascular cells, and fibroblasts) into the tumor and this recruitment is essential for tumor growth. In vitro and in vivo studies have also shown that stromal angiogenesis and aberrant apoptotic signals in myoepithelial cells and stromal cells can actually precede invasion, further evidence supporting a more causative role of the stromal compartment in breast cancer [26, 27].
An even more pronounced role of stroma in tumor progression was suggested by Olumi et al. [14], who showed that fibroblasts associated with carcinomas stimulate tumor progression of initiated non-tumorigenic epithelial cells both in an in vivo tissue recombination system and in an in vitro co-culture system. Others have found that primary, phenotypically normal fibroblasts associated with a human epithelial malignancy can stimulate progression of a non-tumorigenic epithelial cell into a neoplastic one [15]. These studies reveal a complex interaction with cross-talk between tumor and stromal cells that plays an important role in promoting the oncogenic process [11]. Many studies approach the stromal response as a relatively uniform entity with little variation between different patients’ tumors. In previous studies, we have characterized two different stromal signatures in invasive breast cancer that are present at different levels in different patients [1–3]. The signatures represent a fibroblast and macrophage derived response, and they are obtained through measurements of several dozen genes by gene expression profiling. Expression of these genes was found to be largely restricted to stromal cells as demonstrated by immunohistochemistry and in situ hybridization. The presence of these stromal signatures varies between invasive breast cancers in a way that correlates with clinicopathologic features of the tumor that include hormone status, tumor grade, and clinical outcome. In addition, in prior studies [1–3], we found that prognostic performance of the fibroblast stromal signature was independent in multivariate analysis for clinical risk factors including tumor size and lymph node status.
In the current study, we evaluated whether these stromal signatures are expressed at variable levels in pre-invasive ductal breast cancer, DCIS. Our hypothesis was that the study of stromal signatures in DCIS could shed light on the two competing views of tumor–stromal interaction as outlined above. If the stroma is responding to the invasive nature of the tumor as a “wound response” we might expect that the stromal signatures we have observed in invasive breast cancer would not be present in DCIS, which has yet to develop an invasive phenotype. However, if the stroma plays an important role in inducing and promoting oncogenesis then we might expect to see the same stromal signatures in DCIS as we do in invasive breast cancer.
Our results demonstrate that distinct subsets of DCIS have the fibroblast and macrophage response signatures that can also be found in invasive breast cancer. We have shown this with two different methodologies: by analysis of a published gene expression profiling dataset, which provides measurements of all the gene transcripts in the stromal signatures, and by immunohistochemistry, which looks at a limited number of protein products from the genes but allows for localization within specific compartments of the TME. The incidences of DCIS cases positive for both the stromal signatures, 30% for the fibroblast signature and 19% for the macrophage response signature, are similar to the incidences in invasive breast cancer that we have previously reported: 25–35% for the fibroblast signature [2] and 17–25% for the macrophage response signature [1]. Importantly, we find that both signatures are more likely than not to be conserved during progression from DCIS to IDC.
The macrophage response signature in DCIS is associated with higher grade tumors and cases that lack ER and PR expression. This is consistent with our study on the macrophage response in invasive breast cancer which found that the stromal macrophage response signature correlated with higher tumor grade, decreased expression of ER, decreased expression of PR, and increased p53 mutations [1]. It is possible that this stromal signature may be associated with the inherent biology of a specific subset of ER and PR negative breast cancers and could be related to the basal-like phenotype. Some studies have suggested that a small subset of breast cancers with an intense inflammatory response (e.g., medullary carcinoma) are also associated with the basal-like breast cancer signature [28, 29]. A recent study from the Pollard group suggests that CSF1 responsive macrophages are important in breast cancer metastasis [30]. This study focused on the effects of macrophages in the metastatic TME. Our study suggests that for a subset of breast cancers, the recruitment of this type of macrophage begins at a much earlier stage in breast cancer progression.
The biologic and clinicopathologic aspects of the fibroblast associated stromal signature are less clear. While the stromal signature involves scar and wound fibroblastic responses, it is unknown whether this provides a benefit to the tumor or represents a host response that interferes with tumor growth. Likewise, while we have termed this stromal response the “fibroblast signature” because it some of the genes are expressed in fibroblastic cells and the initial gene studies were derived from desmoid-type fibromatosis (a soft tissue tumor with fibroblastic origins), it is unclear what cell initiates this response pattern. In separate studies on invasive cancer, we have found that there is some association between histologic appearance of desmoplasia and the DTF fibroblast signature. Likewise there is a correlation between the presence of inflammatory cells and the CSF1 macrophage response signature in breast cancer. However, this neither of the signatures is tightly correlated with histology and there are cases with desmoplasia and/or inflammation that are not positive for either of the stromal signatures and vice versa.
This study is one of the first to attempts to classify DCIS based on the expression of groups of functionally related genes involved in stromal response. Others have previously examined the vascularity in smaller sets of DCIS [16–18]. These studies have looked at markers for vessels and a number of genes that are likely involved in angiogenesis. These studies found that increases in vascularity occurred in DCIS and preceded invasion. But the low number of cases studies (18 DCIS) precluded the determination of discrete groups of DCIS based on these markers.
While we cannot determine from these studies whether either stromal signature is due to a stimulus in the neoplastic cells or originates in the host stroma, it is clear that both the fibroblast and macrophage stromal signatures are present in pre-invasive breast cancer. It is even possible that these signatures may first emerge at a stage of breast cancer development that precedes DCIS. As others have suggested before, the stromal response might prove to be helpful in clinical practice as it may lead to a better prognostication of tumors or may even provide additional targets for therapy [11].
Ultimately these findings will improve the chance of identifying tumors that will respond to a specific stroma-targeted therapy that might be developed in the TME field. The majority of cancer research has focused on the neoplastic cells and as a result, the vast majority of therapies available to oncologists are therapeutic agents that act on these cancer cells. Cancers, however, often develop resistance to these therapies, in large part due to their inherent genomic instability [31]. An alternative, emerging avenue of therapy focuses on targeting various microenvironmental processes [11, 32] Since stromal cells within the tumor are thought to be “normal” and less genetically labile than the neoplastic cells, development of acquired resistance to therapy is thought to be less likely. As such, the tumor stroma may be an excellent target for directed therapy. Our findings suggest that stroma targeted therapies could also treat pre-invasive disease.
Acknowledgments
Supported in part by research grants from the California Breast Cancer Research Program 15NB-0156 and the National Cancer Institute R01 CA129927.
Contributor Information
M. Sharma, Department of Pathology, Stanford University Hospital, Room L235, 300 Pasteur Drive, Stanford, CA 94305, USA
A. H. Beck, Department of Pathology, Stanford University Hospital, Room L235, 300 Pasteur Drive, Stanford, CA 94305, USA
J. A. Webster, Department of Pathology, Stanford University Hospital, Room L235, 300 Pasteur Drive, Stanford, CA 94305, USA
I. Espinosa, Department of Pathology, Stanford University Hospital, Room L235, 300 Pasteur Drive, Stanford, CA 94305, USA
K. Montgomery, Department of Pathology, Stanford University Hospital, Room L235, 300 Pasteur Drive, Stanford, CA 94305, USA
S. Varma, Veterans Affairs Palo Alto Health Care System, Palo Alto, CA, USA
M. van de Rijn, Department of Pathology, Stanford University Hospital, Room L235, 300 Pasteur Drive, Stanford, CA 94305, USA
K. C. Jensen, Department of Pathology, Stanford University Hospital, Room L235, 300 Pasteur Drive, Stanford, CA 94305, USA. Veterans Affairs Palo Alto Health Care System, Palo Alto, CA, USA
R. B. West, Email: rbwest@stanford.edu, Department of Pathology, Stanford University Hospital, Room L235, 300 Pasteur Drive, Stanford, CA 94305, USA. Veterans Affairs Palo Alto Health Care System, Palo Alto, CA, USA
References
- 1.Beck AH, Espinosa I, Edris B, Li R, Montgomery K, Zhu S, Varma S, Marinelli RJ, van de Rijn M, West RB. The macrophage colony-stimulating factor 1 response signature in breast carcinoma. Clin Cancer Res. 2009;15(3):778–787. doi: 10.1158/1078-0432.CCR-08-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beck AH, Espinosa I, Gilks CB, van de Rijn M, West RB. The fibromatosis signature defines a robust stromal response in breast carcinoma. Lab Invest. 2008;88(6):591–601. doi: 10.1038/labinvest.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West RB, Nuyten DS, Subramanian S, Nielsen TO, Corless CL, Rubin BP, Montgomery K, Zhu S, Patel R, Hernandez-Boussard T, Goldblum JR, Brown PO, van de Vijver M, van de Rijn M. Determination of stromal signatures in breast carcinoma. PLoS Biol. 2005;3(6):e187. doi: 10.1371/journal.pbio.0030187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ernster VL, Barclay J, Kerlikowske K, Grady D, Henderson C. Incidence of and treatment for ductal carcinoma in situ of the breast. JAMA. 1996;275(12):913–918. [PubMed] [Google Scholar]
- 5.Czerniecki BJ, Koski GK, Koldovsky U, Xu S, Cohen PA, Mick R, Nisenbaum H, Pasha T, Xu M, Fox KR, Weinstein S, Orel SG, Vonderheide R, Coukos G, DeMichele A, Araujo L, Spitz FR, Rosen M, Levine BL, June C, Zhang PJ. Targeting HER-2/neu in early breast cancer development using dendritic cells with staged interleukin-12 burst secretion. Cancer Res. 2007;67(4):1842–1852. doi: 10.1158/0008-5472.CAN-06-4038. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez RJ, Buzdar AU, Fraser Symmans W, Yen TW, Broglio KR, Lucci A, Esteva FJ, Yin G, Kuerer HM. Novel clinical trial designs for treatment of ductal carcinoma in situ of the breast with trastuzumab (herceptin) Breast J. 2007;13(1):72–75. doi: 10.1111/j.1524-4741.2006.00366.x. [DOI] [PubMed] [Google Scholar]
- 7.Yen TW, Kuerer HM, Ottesen RA, Rouse L, Niland JC, Edge SB, Theriault RL, Weeks JC. Impact of randomized clinical trial results in the national comprehensive cancer network on the use of tamoxifen after breast surgery for ductal carcinoma in situ. J Clin Oncol. 2007;25(22):3251–3258. doi: 10.1200/JCO.2006.10.2699. [DOI] [PubMed] [Google Scholar]
- 8.Page DL, Dupont WD, Rogers LW, Jensen RA, Schuyler PA. Continued local recurrence of carcinoma 15–25 years after a diagnosis of low grade ductal carcinoma in situ of the breast treated only by biopsy. Cancer. 1995;76(7):1197–1200. doi: 10.1002/1097-0142(19951001)76:7<1197::aid-cncr2820760715>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 9.Gauthier ML, Berman HK, Miller C, Kozakeiwicz K, Chew K, Moore D, Rabban J, Chen YY, Kerlikowske K, Tlsty TD. Abrogated response to cellular stress identifies DCIS associated with subsequent tumor events and defines basal-like breast tumors. Cancer Cell. 2007;12(5):479–491. doi: 10.1016/j.ccr.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu M, Peluffo G, Chen H, Gelman R, Schnitt S, Polyak K. Role of COX-2 in epithelial-stromal cell interactions and progression of ductal carcinoma in situ of the breast. Proc Natl Acad Sci USA. 2009;106(9):3372–3377. doi: 10.1073/pnas.0813306106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bissell MJ, Radisky D. Putting tumours in context. Nat Rev Cancer. 2001;1(1):46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elenbaas B, Weinberg RA. Heterotypic signaling between epithelial tumor cells and fibroblasts in carcinoma formation. Exp Cell Res. 2001;264(1):169–184. doi: 10.1006/excr.2000.5133. [DOI] [PubMed] [Google Scholar]
- 13.Mueller MM, Fusenig NE. Friends or foes—bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4(11):839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 14.Olumi AF, Grossfeld GD, Hayward SW, Carroll PR, Tlsty TD, Cunha GR. Carcinoma-associated fibroblasts direct tumor progression of initiated human prostatic epithelium. Cancer Res. 1999;59(19):5002–5011. doi: 10.1186/bcr138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orimo A, Gupta PB, Sgroi DC, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey VJ, Richardson AL, Weinberg RA. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121(3):335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 16.Brown LF, Guidi AJ, Schnitt SJ, Van De Water L, Iruela-Arispe ML, Yeo TK, Tognazzi K, Dvorak HF. Vascular stroma formation in carcinoma in situ, invasive carcinoma, and meta-static carcinoma of the breast. Clin Cancer Res. 1999;5(5):1041–1056. [PubMed] [Google Scholar]
- 17.Guidi AJ, Fischer L, Harris JR, Schnitt SJ. Microvessel density and distribution in ductal carcinoma in situ of the breast. J Natl Cancer Inst. 1994;86(8):614–619. doi: 10.1093/jnci/86.8.614. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs TW, Schnitt SJ, Tan X, Brown LF. Radial scars of the breast and breast carcinomas have similar alterations in expression of factors involved in vascular stroma formation. Hum Pathol. 2002;33(1):29–38. doi: 10.1053/hupa.2002.30190. [DOI] [PubMed] [Google Scholar]
- 19.Hannemann J, Kristel P, van Tinteren H, Bontenbal M, van Hoesel QG, Smit WM, Nooij MA, Voest EE, van der Wall E, Hupperets P, de Vries EG, Rodenhuis S, van de Vijver MJ. Molecular subtypes of breast cancer and amplification of topoi-somerase II alpha: predictive role in dose intensive adjuvant chemotherapy. Br J Cancer. 2006;95(10):1334–1341. doi: 10.1038/sj.bjc.6603449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang HY, Sneddon JB, Alizadeh AA, Sood R, West RB, Montgomery K, Chi JT, van de Rijn M, Botstein D, Brown PO. Gene expression signature of fibroblast serum response predicts human cancer progression: similarities between tumors and wounds. PLoS Biol. 2004;2(2):E7. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315(26):1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 22.Bhowmick NA, Moses HL. Tumor-stroma interactions. Curr Opin Genet Dev. 2005;15(1):97–101. doi: 10.1016/j.gde.2004.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunha GR, Hayward SW, Wang YZ, Ricke WA. Role of the stromal microenvironment in carcinogenesis of the prostate. Int J Cancer. 2003;107(1):1–10. doi: 10.1002/ijc.11335. [DOI] [PubMed] [Google Scholar]
- 24.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 25.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 26.Pavlakis K, Messini I, Vrekoussis T, Yiannou P, Keramopoullos D, Louvrou N, Liakakos T, Stathopoulos EN. The assessment of angiogenesis and fibroblastic stromagenesis in hyperplastic and pre-invasive breast lesions. BMC Cancer. 2008;8:88. doi: 10.1186/1471-2407-8-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shekhar MP, Tait L, Pauley RJ, Wu GS, Santner SJ, Nangia-Makker P, Shekhar V, Nassar H, Visscher DW, Heppner GH, Miller FR. Comedo-ductal carcinoma in situ: A paradoxical role for programmed cell death. Cancer Biol Ther. 2008;7(11):1774–1782. doi: 10.4161/cbt.7.11.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Pinilla SM, Rodriguez-Gil Y, Moreno-Bueno G, Sarrio D, Martin-Guijarro Mdel C, Hernandez L, Palacios J. Sporadic invasive breast carcinomas with medullary features display a basal-like phenotype: an immunohistochemical and gene amplification study. Am J Surg Pathol. 2007;31(4):501–508. doi: 10.1097/01.pas.0000213427.84245.92. [DOI] [PubMed] [Google Scholar]
- 29.Vincent-Salomon A, Gruel N, Lucchesi C, MacGrogan G, Dendale R, Sigal-Zafrani B, Longy M, Raynal V, Pierron G, de Mascarel I, Taris C, Stoppa-Lyonnet D, Pierga JY, Salmon R, Sastre-Garau X, Fourquet A, Delattre O, de Cremoux P, Aurias A. Identification of typical medullary breast carcinoma as a genomic sub-group of basal-like carcinomas, a heterogeneous new molecular entity. Breast Cancer Res. 2007;9(2):R24. doi: 10.1186/bcr1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian B, Deng Y, Im JM, Muschel RJ, Zou Y, Li J, Lang RA, Pollard JW. A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLoS ONE. 2009;4(8):e6562. doi: 10.1371/journal.pone.0006562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akli S, Zheng PJ, Multani AS, Wingate HF, Pathak S, Zhang N, Tucker SL, Chang S, Keyomarsi K. Tumor-specific low molecular weight forms of cyclin E induce genomic instability and resistance to p21, p27, and antiestrogens in breast cancer. Cancer Res. 2004;64(9):3198–3208. doi: 10.1158/0008-5472.can-03-3672. [DOI] [PubMed] [Google Scholar]
- 32.Kenny PA, Lee GY, Bissell MJ. Targeting the tumor microenvironment. Front Biosci. 2007;12:3468–3474. doi: 10.2741/2327. [DOI] [PMC free article] [PubMed] [Google Scholar]


