Abstract
The stability and structure of nylon nucleic acid duplexes with complementary DNA and RNA strands was examined. Thermal denaturing studies of a series of oligonucleotides containing nylon nucleic acids (1 to 5 amide linkages) revealed that the amide linkage enhanced significantly the binding affinity of nylon nucleic acids towards both complementary DNA (up to 26°C increase in the thermal transition temperature (Tm) for 5 linkages) and RNA (around 15°C increase in Tm for 5 linkages) when compared with non-amide linked precursor strands. For both DNA and RNA complements, increasing derivatization decreased the melting temperatures of uncoupled molecules relative to unmodified strands; by contrast, increasing lengths of coupled copolymer raised Tm from less to slightly greater than Tm of unmodified strands. Thermodynamic data extracted from melting curves and CD spectra of nylon nucleic acid duplexes were consistent with loss of stability due to incorporation of pendent groups on the 2′ position of ribose, and recovery of stability upon linkage of the side chains.
Keywords: circular dichroism, conformational restriction, DNA analogs, template synthesis, thermodynamics
Introduction
Several examples of DNA-templated organic reactions have been reported,[1–3] but far fewer stereo- and regio-specific DNA-mediated polymerization reactions are known.[4] In previous work, we have reported the attachment of dicarboxyl and diamino groups to the 2′ position of uridine nucleoside analogs.[5] These side chains were condensed to form short segments of a nylon-like polymer pendent from the nucleic acid backbone. The long-term goal of this work is to use the topological control afforded by nucleic acids[6] to direct the topology of polymers of industrial importance. Our synthesis of nylon nucleic acid oligomers is markedly more robust than the previous version, so we are now able to isolate enough material for a study of the effect of structural restrictions from the nylon-like polymer on thermal stability. In the design of the strands reported in our earlier work, the modified nucleosides were flanked on both sides by short stretches of oligo-dT. The disadvantage of an oligonucleotide containing exclusively rU or dT nucleobases is that there is no control over the position or manner in which it binds its oligo-dA complement, including the possibility of triplex formation.[7] Furthermore, the melting temperatures of homopolymer duplexes containing only A and T are lower than those that also contain G and C, so to carry out thermodynamic studies, higher strand concentrations are required; however, this leads to a more complicated system, which may contain aggregated concatamers consisting of overlapping oligomers.[8]
In this study, the modified nucleosides in the oligonucleotide are flanked by general DNA sequences containing all four bases, not only oligo-dT. This is one step further toward the goal of controlling the topology of industrial polymers by nucleic acids, as more bases must be used for such a system to be effective. In the work reported here, two to six modified nucleosides are located in the middle region of 18-mer oligonucleotides. After condensation between carboxyl and amino groups, short nylon-like polymers with one to five amide linkages result. Thermodynamic properties of nylon nucleic acid strands with their DNA or RNA complements were investigated by thermal denaturing experiments with precursor strands (prior to amide coupling) and unmodified DNA for comparison. Thermodynamic parameters were extracted from melting curves based on van’t Hoff analysis. Duplex formation was also detected and analyzed by circular dichroism.
Results and Discussion
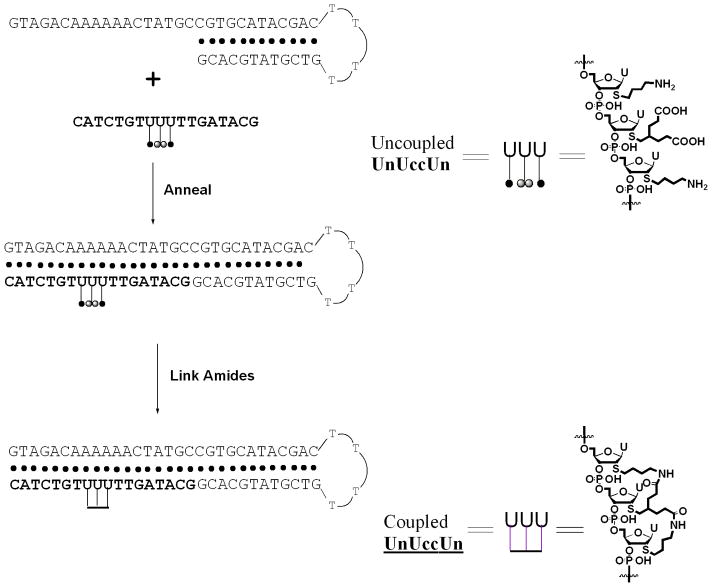
Strands were designed to place a contiguous stretch of modified uridine nucleotides in the middle region of a heterobase sequence. The number of modified nucleotides ranged from two to six. The sequences are shown and the strands are named in Table 1. We denote uridine nucleotides containing single amino or carboxyl modifications as Un or Uc, respectively, and diamino or dicarboxyl modifications are labeled as Unn or Ucc, respectively (Fig. 1). The nylon nucleic acids are indicated by an underline at the coupled nucleotides (e.g., UnUc). Oligonucleotide strands with free carboxyl and amino groups were synthesized by automated phosphoramidite synthesis, cleaved from the solid support and deprotected using procedures similar to those described previously, although the incubation temperature for deprotection was 50°C, rather than room temperature.[5a] After purification by denaturing gel electrophoresis, all strands were characterized by MALDI-TOF mass spectrometry. A new amide formation procedure was developed in which the precursor strand was hybridized with a complementary hairpin DNA molecule before the coupling reaction.[9] Schematic drawings in Fig. 1 illustrate the templated synthesis of nylon nucleic acid molecule 3b. After annealing with a 46-mer hairpin molecule, a strand containing pendent groups forms a duplex with complementary DNA. Duplex formation prevents various potential side reactions, such as coupling with amine groups on nucleobases, or reaction with remote pendent groups. Amide formation catalyzed by DMT-MM also followed previous procedures, except that the reaction mixtures were shaken gently during the coupling reaction. The use of a hairpin with a longer sequence facilitated electrophoretic separation of the modified molecule from its complement after coupling (Supporting Information S1).
Table 1.
Oligonucleotides used in this study and MALDI-TOF MS analysis. Strand 1 is unmodified DNA, used as a control; 2a – 6a are uncoupled precursor strands with 2, 3, 4, 5 or 6 modified nucleotides incorporated; and 2b – 6b contain corresponding nylon nucleic acid sequences.
| ODNs | Sequence | Calculated | Found |
|---|---|---|---|
| 1 | 5′-GCATAGTTTTTTGTCTAC | -- | -- |
| 2a | 5′-GCATAGTTUnUcTTGTCTAC | 5678.9 | 5678.8 |
| 2b | 5′-GCATAGTTUnUcTTGTCTAC | 5660.9 | 5661.3 |
| 3a | 5′-GCATAGTTUnUccUnTGTCTAC | 5840.8 | 5839.5 |
| 3b | 5′-GCATAGTTUnUccUnTGTCTAC | 5804.8 | 5803.7 |
| 4a | 5′-GCATAGTUcUnnUccUnTGTCTAC | 6000.4 | 6000.3 |
| 4b | 5′-GCATAGTUcUnnUccUnTGTCTAC | 5946.4 | 5947.0 |
| 5a | 5′-GCATAGTUcUnnUccUnnUcGTCTAC | 6161.7 | 6161.9 |
| 5b | 5′-GCATAGTUcUnnUccUnnUcGTCTAC | 6089.7 | 6090.9 |
| 6a | 5′-GCATAGUcUnnUccUnnUccUnGTCTAC | 6322.9 | 6323.0 |
| 6b | 5′-GCATAGUcUnnUccUnnUccUnGTCTAC | 6232.9 | 6233.9 |
Figure 1.
Schematic depiction of the templated synthesis of nylon nucleic acid molecule 3b. A DNA hairpin template strand with a sequence complementary to strand 3a forms a stable duplex. Proximal amines and carboxylates are then connected using chemical ligation. The nylon nucleic acid product is separated from the hairpin template by denaturing gel electrophoresis.
Thermal Denaturing Studies
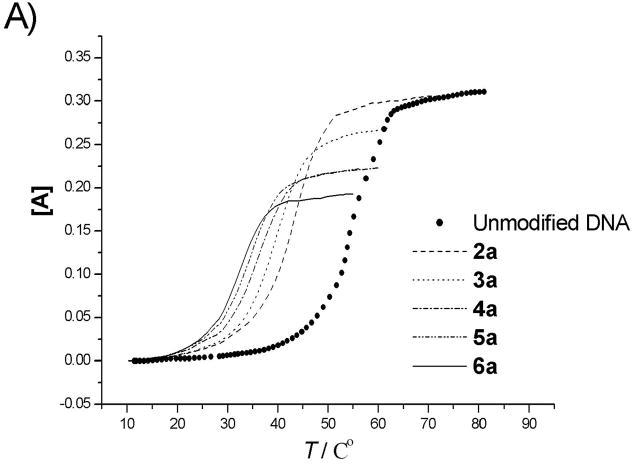
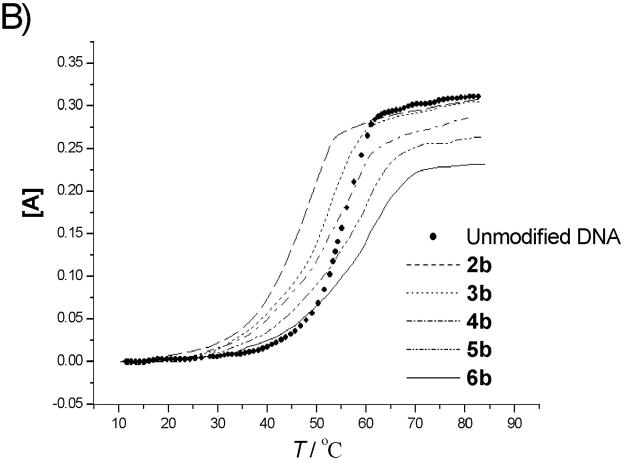
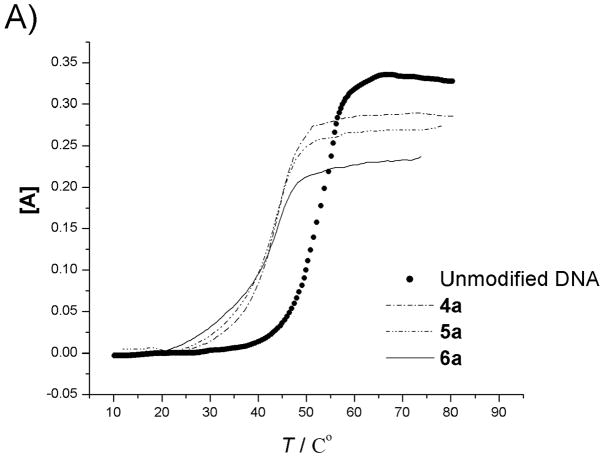
The duplex stabilities of nylon nucleic acid-containing strands hybridized with unmodified DNA and RNA complements were determined by thermal denaturing experiments that monitored absorbance at 260 nm. Fig. 2 (DNA) and Fig. 3 (RNA) show thermal transition profiles plotted as the change in absorbance (AT − AT0)/AT0 vs. temperature. In all cases, at least two consecutive heating-annealing cycles were applied. Superimposable heating and annealing profiles indicate that transition processes are kinetically reversible. The effect of extra polyamide linkages was examined by comparing the melting temperatures of 1 : DNA duplex or 1 : RNA duplex (using the unmodified DNA 1 as control). Melting temperatures (Tm), determined as that temperature at which half of the double-strand complexes had dissociated,[10] are summarized in Table 2. Uncoupled duplexes showed cooperative melting behaviour, but coupled oligomers melted over a relatively broad temperature range. Nevertheless, single transition behaviour was assumed for the thermodynamic analysis.
Figure 2.
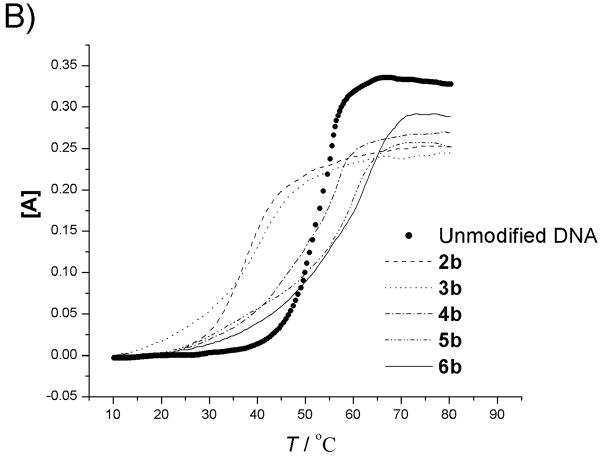
Melting curves for modified oligonucleotides before A) and after B) coupling with complementary DNA. [A] = (AT−AT0)/AT0. (AT0 is the absorbance at 10°C). UV absorption was monitored at 260 nm.
Figure 3.
Melting curves for modified oligonucleotides before A) and after B) coupling with complementary RNA. [A] = (AT−AT0)/AT0. (AT0 is the absorbance at 10°C). UV absorption was monitored at 260 nm.
Table 2.
Tm value and thermodynamic data for nylon nucleic acid duplexes with DNA complement (a) and RNA complement (b).[a], [b]
| (a) | Nylon nucleic acid precursor strands | Coupled nylon nucleic acids |
ΔTm[c] (°C) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Tm (°C) |
ΔHo (kJ·mol−1) |
ΔSo (J·mol−1K−1) |
T·ΔSo (kJ·mol−1) (298K) |
ΔG (kJ·mol−1) (298K) |
Tm (°C) |
ΔHo (kJ·mol−1) |
ΔSo (J·mol−1K−1) |
T·ΔSo (kJ·mol−1) (298K) |
ΔG (kJ·mol−1) (298K) |
|||
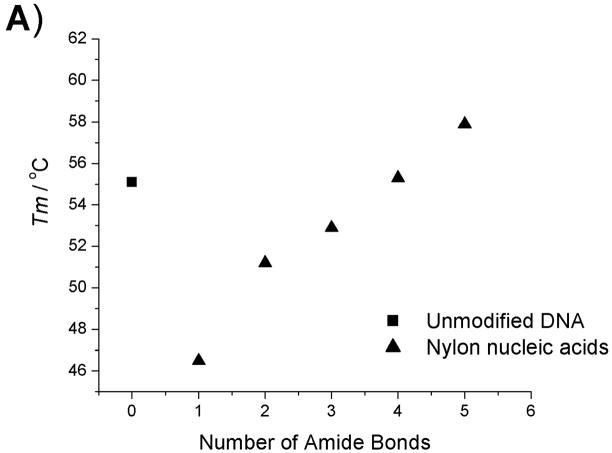
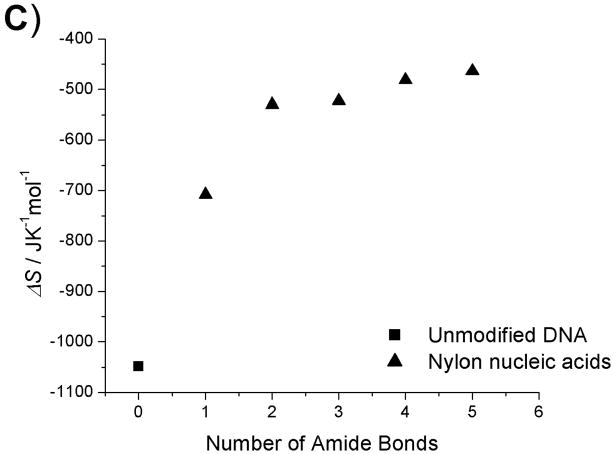
| 1 | 55.1 | −384 | −1048 | −312 | −71.3 | 1 | 55.1 | −384 | −1048 | −312 | −71.3 | -- |
| 2a | 42.8 | −381 | −1086 | −324 | −57.6 | 2b | 46.5 | −264 | −707.9 | −211 | −53.5 | 3.7 |
| 3a | 39.5 | −376 | −1084 | −323 | −53.3 | 3b | 51.2 | −210 | −529.7 | −158 | −52.6 | 11.7 |
| 4a | 35.8 | −366 | −1065 | −317 | −48.7 | 4b | 52.9 | −209 | −522.6 | −156 | −52.9 | 17.1 |
| 5a | 34.0 | −341 | −988.7 | −295 | −45.9 | 5b | 55.3 | −197 | −481.2 | −143 | −53.6 | 21.3 |
| 6a | 31.9 | −317 | −916.3 | −273 | −43.6 | 6b | 57.9 | −193 | −463.6 | −138 | −54.7 | 26.0 |
| (b) | Nylon nucleic acid precursor strands | Coupled nylon nucleic acids |
ΔTm[c] (°C) |
|||||||||
|
Tm (°C) |
ΔHo (kJ·mol−1) |
ΔSo (J·mol−1K−1) |
T·ΔSo (kJ·mol−1) (298K) |
ΔG (kJ·mol−1) (298K) |
Tm (°C) |
ΔHo (kJ·mol−1) |
ΔSo (J·mol−1K−1) |
T·ΔSo (kJ·mol−1) (298K) |
ΔG (kJ·mol−1) (298K) |
|||
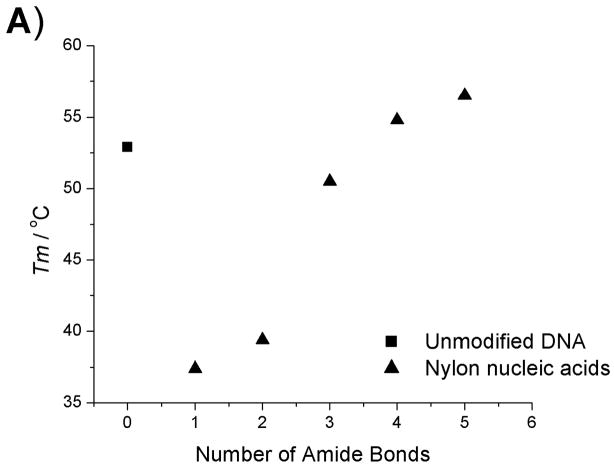
| 1 | 52.9 | −467 | −1312 | −391 | −75.6 | 1 | 52.9 | −467 | −1312 | −391 | −75.6 | -- |
| 2a | -- | -- | -- | -- | -- | 2b | 37.4 | −262 | −719.0 | −214 | −47.8 | -- |
| 3a | -- | -- | -- | -- | -- | 3b | 39.4 | −199 | −508.0 | −151 | −47.4 | -- |
| 4a | 43.1 | −377 | −1071 | −319 | −57.8 | 4b | 50.5 | −196 | −495.0 | −148 | −48.9 | 7.4 |
| 5a | 42.4 | −313 | −873.5 | −260 | −53.0 | 5b | 54.8 | −186 | −448.0 | −134 | −52.8 | 12.4 |
| 6a | 41.8 | −242 | −651.3 | −194 | −48.3 | 6b | 56.5 | −177 | −416.0 | −124 | −53.0 | 14.7 |
Complementary DNA sequence: 5′-GTAGACAAAAAACTATGC; Complementary RNA sequence: 5′-GUAGACAAAAAACUAUGC.
The melting curves (A260 vs. temperature) were recorded in medium salt hybridization buffer (40 mM sodium cacodylate and 100 mM sodium chloride, pH 7.3) containing the two complementary strands at 1μM concentration.
ΔTm are the Tm difference between nylon nucleic acids and corresponding uncoupled precursor strands.
The uncoupled precursor strands form duplexes with DNA and RNA that are less stable than analogous duplexes of unmodified DNA. Thus, introduction of 2′-pendent thioalkyl diamino or dicarboxyl groups destabilizes the duplex. In the case of DNA, a trend is observed that the melting temperature decreases as the number of modified nucleotides is increased. This destabilization may be attributed to steric effects exerted by the bulky 2′ moieties. Although 2′-O-methyl and 2′-O-methoxymethyl substituents are known to stabilize duplex with DNA, 2′-O-alkyl modification lowers binding affinity towards complementary DNA strands with increasing size of the alkyl chain.[11] Destabilization could also result from altered sugar puckering of the 2′-deoxy-2′-alkylthio ribose, as the less electronegative[12] 2′-alkylmercapto modification should lead to a decrease in the preponderance of the 3′-endo conformer.[13] The higher ratio of 2′-endo conformer may perturb the backbone, which may lead to further distortions, in addition to the steric hindrance exerted by bulky 2′-S pendent groups. Furthermore, this destabilization effect may also be associated with reports that oligonucleotides containing dT are more stable than analogous strands containing dU.[14] In addition, the charges on the pendent groups could also have a local effect on melting, although the exact nature of the effect is unclear; overall, the number of added positive and negative charges is equal in all molecules. For duplex molecules hybridizing the uncoupled precursor strands with RNA, destabilization of the duplex was also observed. However, the decrease of melting temperature with increase in the number of modified nucleotides is not as large (Table 2b, Fig. 3A).
In contrast with the uncoupled strands, coupling of the amino and carboxyl groups led to higher stability in duplex molecules. Coupled strand 2b, containing one amide bond, gives a 3.7°C increase in Tm for the duplex with complementary DNA as compared with the uncoupled precursor 2a, but still much lower than control strand 1. However, the enhancement of duplex stability to natural DNA continues with the number of amide bonds introduced. For strand 3b (two amide linkages) Tm increases to 51.2°C and is 11.7°C higher than its uncoupled counterpart. This trend continues, so that strands with four or five linkages in fact form more stable duplexes with their natural DNA complement than the unmodified control. Coupled strand 6b has a Tm 2.8°C higher than unmodified DNA and 26°C higher than its uncoupled analog. The situation for the RNA duplexes is similar. Higher melting temperatures were obtained with more amide linkages, so that coupled strand 6b has a Tm 3.6°C higher than the unmodified DNA : RNA control. This enhanced thermostability probably can be attributed to conformational restriction, imposed by the presence of a second polyamide backbone that links adjacent nucleotides in nylon nucleic acids. Conformational restriction has been reported to account for increasing the stability of duplexes for other nucleic acid analogs[15] including peptide nucleic acid analogs.[16] Most such restrictions have been reported previously within a single nucleotide and rarely involved links between adjacent nucleotides as done in the present case. Nielsen reported covalent links between nucleotides and observed stabilization of nucleic acid three-way junctions.[17]
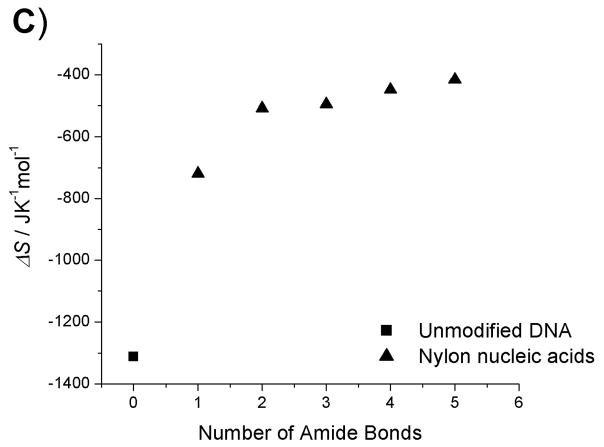
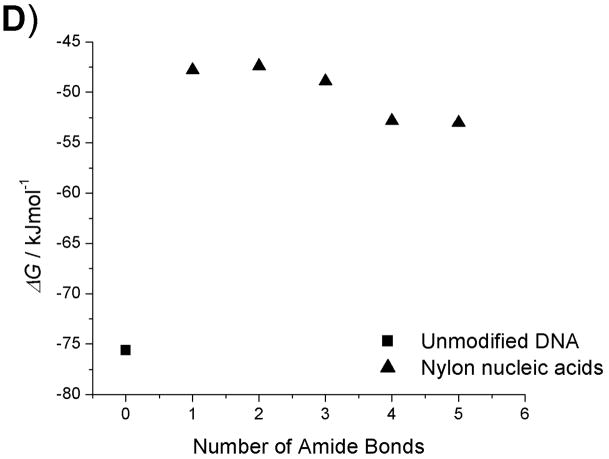
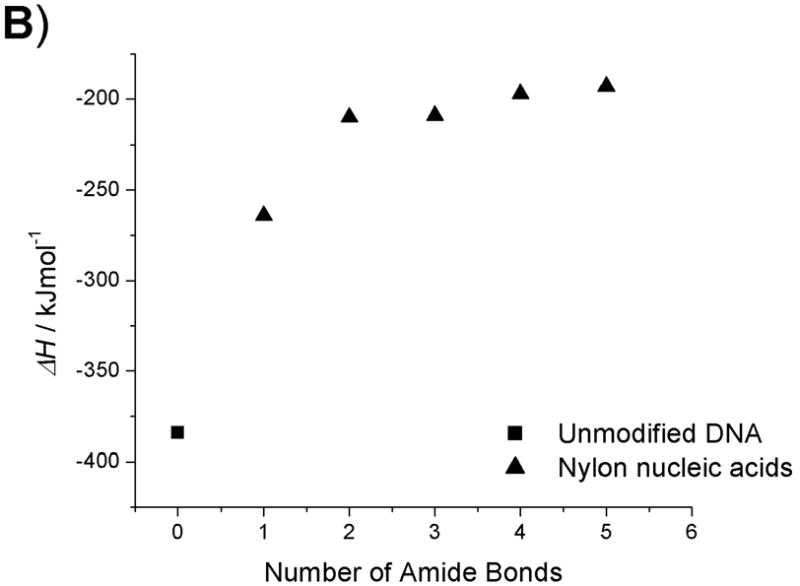
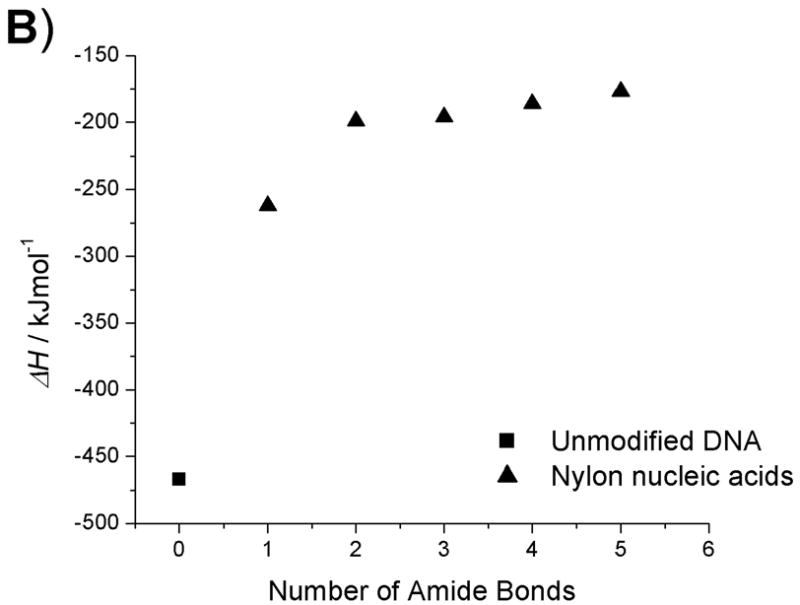
The enthalpies and entropies of the thermal transition were determined using van’t Hoff analysis (Table 2, Fig. 4–5).[18] Compared with unmodified DNA : DNA and DNA : RNA duplexes, there is a significant difference in both enthalpy and entropy for all modified strands, including the fully coupled strand 6b, which shows higher Tm for both types of duplexes. This reflects the broader temperature range of the transition observed for the duplexes containing modified strands. Although the latter portions of the Tm curves for strands 5b and 6b are observed at clearly higher temperatures than for the controls, initiation of the curve occurs at temperatures well below the control. This broad range suggests lower cooperativity in the melting of duplexes containing modified strands, which is reflected in the enthalpy and entropy terms. The less sharp dependence upon temperature may result from distortions in the duplex structure that arise from the macrobicyclic rings associated with the nylon nucleic acid ladder polymer structure. Another contribution to this behaviour may arise from the heterogeneity of the nylon nucleic acid region of each strand, which is flanked by unmodified DNA sequences.
Figure 4.
Thermodynamic data plots of nylon nucleic acid : DNA duplexes. A) Tm change as a function of the number of amide bonds formed. B) ΔH change as a function of the number of amide bonds formed. C) ΔS change as a function of the number of amide bonds formed. D) ΔG (298K) change as a function of the number of amide bonds formed.
Figure 5.
Thermodynamic data plots of nylon nucleic acid : RNA duplexes. A) Tm change as a function of the number of amide bonds formed. B) ΔH change as a function of the number of amide bonds formed. C) ΔS change as a function of the number of amide bonds formed. D) ΔG (298K) change as a function of the number of amide bonds formed.
Within the series of duplexes with DNA (2–6), the thermodynamic analysis reveals that nylon nucleic acids and precursor strands exhibit a trend in that the enthalpy of duplex formation becomes less favorable, but entropy becomes more favorable with more modified nucleotides incorporated. However, the values are very different from those from unmodified DNA 1: The values of enthalpy and entropy for the melting of strand 6b are only about half those of unmodified DNA 1. The strongly favorable entropy is probably attributable to restricted conformation exerted by 2′ amide linkage.[15f] A similar trend was also obtained for thermodynamic parameters of the nylon nucleic acid complementary with RNA strands (Table 2b). It is likely that nylon nucleic acids impose similar restrictions on the duplex formed with RNA.
Circular Dichroism Spectroscopy
Circular dichroism (CD) measurements were undertaken to compare the secondary structures of hybridized uncoupled precursor strands and coupled nylon nucleic acids with natural DNA and RNA duplex.[19] In Fig. 6, CD spectra of duplexes involving nylon nucleic acid 6b with A) DNA and B) RNA complements are shown and compared with the corresponding uncoupled precursor strand 6a and unmodified DNA 1. In Fig. 6A, the overall CD curves of both uncoupled and coupled strands are similar to DNA control strand 1, and consistent with the B-form structure. Both uncoupled and coupled strands show a less intense negative band around 250 nm and a more intense positive band around 270 – 280 nm, the latter being shifted toward shorter wavelengths compared with 1. Although the maximum number of modified nucleotides accounts only for one third of the strand, it is clear that perturbation by the modified nucleotides is present. In general, it appears that incorporation of the 2′-alkylthio substituent causes a small change in the spectra, and linking the substituents to form nylon nucleic acids brings the spectra back towards that of the unmodified strand 1. Similar effects were observed for strands with 2 – 5 modified nucleotides, for which CD data are shown in the Supporting Information section. In the series 2a–6a, as more nucleotides with pendent groups are incorporated into the sequence, the uncoupled precursor strands show increasing differences from the spectra of the control. However, the spectra of all of the duplexes formed between coupled molecules 2b–6b and DNA are nearly superimposable. Therefore, coupling of the amides fixes the conformation of the strands to preclude the continuous deviation from B-form observed in the uncoupled sequences.
Figure 6.
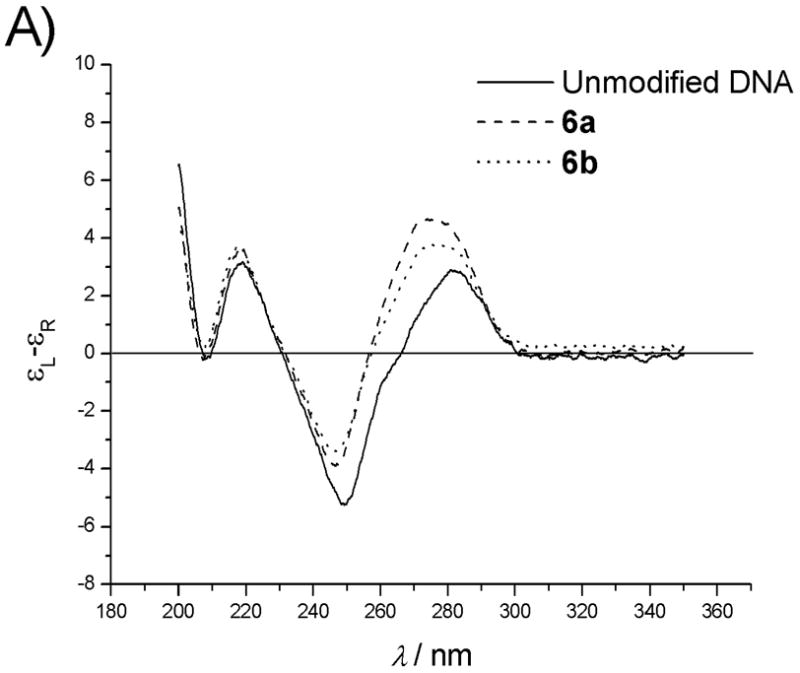
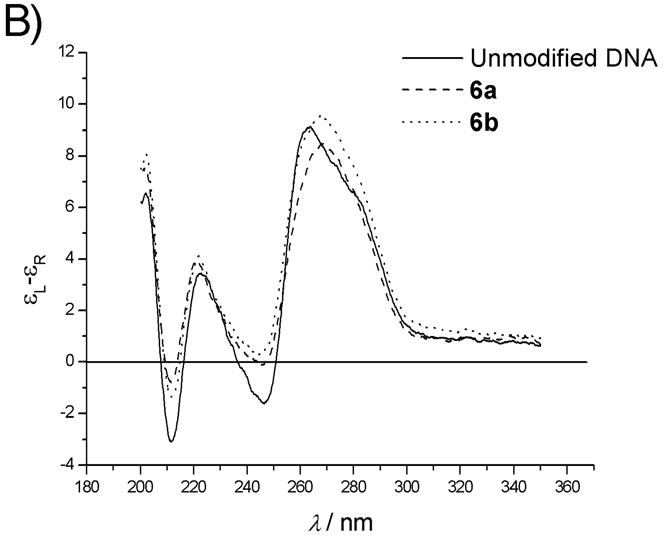
A) CD spectra of duplexes of unmodified DNA strand 1, uncoupled 6a and coupled nylon nucleic acid 6b with DNA complement. B) With RNA complement. Duplex concentration: 4μM. Buffer: 10mM sodium phosphate, 150mM sodium chloride and 0.1mM EDTA, pH 7.0.
Both uncoupled precursor strand 6a and nylon nucleic acid 6b show the A-form structure in the CD spectra when paired with their RNA complement (Fig. 6B). Similar to the DNA 1 : RNA control, there is an intense positive band at long wavelength centered around 265 nm. However, the two negative bands at 210 nm and 245 nm are less intense than the unmodified reference. In contrast to the control, the CD spectrum of the uncoupled precursor strand : RNA duplex for the series 2a–6a shows a decrease of intensity of the 265 nm band and is also accompanied by a small bathochromic shift. For coupled 2b–6b, the changes in CD spectra are much smaller (Supporting Information S3). The polyamide linkage of nylon nucleic acid presents less perturbation of the conformation of duplexes with RNA than the pendent groups of precursor strands. Thus, there is no major evidence from the CD spectroscopy to suggest a structural perturbation to explain the less sharp dependence on temperature seen in the melting data.
Overall, the effect of introducing 2′-alkylthio-substituted nucleotides into DNA sequences brings about a small distortion of the conformation with a significant reduction in the stability of duplexes. However, linking the pendent groups together to form nylon nucleic acids reverses this trend, even producing an increase in stability in the longer copolymer chains.
Given that many modified oligonucleotides have been studied for biomedical applications,[20] a natural question arises regarding the potential suitability of nylon nucleic acids for such a purpose. The increased stability observed with nylon nucleic acid : DNA duplexes is encouraging in this regard. The increase in the transition temperature per nucleotide for nylon nucleic acid bound to its complementary DNA strand is competitive with many modified oligonucleotides[21] although not as large as some, such as locked nucleic acids.[15c,d,22] Stabilization of duplexes in our system appears to originate from conformational restriction enforced by the amide links. Prior examples of conformational constraints have involved primarily those within a single nucleotide. A few examples of rigidifying linkers between remote nucleotides have been reported.[23] To our knowledge, rigidification of the oligonucleotide backbone via covalent connections between adjacent nucleotides has not been explored previously. It seems likely that further optimization of nylon nucleic acid linkers could lead to even more stable duplexes. Further work including examination of enzyme resistance, cell permeability, and other properties, will be needed to assess properly the potential of nylon nucleic acid for in vivo applications.
Conclusions
We have designed and synthesized a series of strands containing modified uridine nucleotides flanked by hetero-base sequences. The thermostability of nylon nucleic acid : DNA/RNA duplexes was studied by monitoring thermal denaturation via absorbance at 260 nm. It was found that fully coupled nylon nucleic acid strands display greater stability of duplex formation, compared with the analogous precursor strands. With an increase in the number of coupled units, nylon nucleic acid strands increase binding affinity to complementary DNA or RNA, while the precursor strands show a decreasing trend. At 1μM DNA concentration, after four or more amide linkages (strands 5b, 6b) were formed, nylon nucleic acid was even more stably hybridized with its complement than the unmodified DNA control. All these data suggest that the formation of the nylon linkage can stabilize the formation of duplex DNA or RNA by strands modified with nylon precursors. Analysis of thermodynamic data extracted from melting curves suggests that this stability results from a balance between lost enthalpy and less negative entropy as a function of modified nucleotide incorporation. Such differences can be attributed to conformational restriction exerted by the nylon linkages. Solution structural analysis by circular dichroism spectroscopy suggests that nylon nucleic acid hybridized with DNA has spectra more characteristic of B-form than A-form conformation, while nylon nucleic acid hybridized with RNA shows evidence of A-form conformation. However, in both cases the CD spectra of coupled polymers show less conformational change with increasing numbers of modified nucleotides incorporated than do precursor strands (i.e., with free pendent carboxylates and amines prior to amide bond formation). This observation is consistent with the conformational restriction hypothesis. These results provide a good model for understanding the factors affecting DNA binding ability.
Experimental Section
Materials
Buffer solutions were prepared from chemicals purchased from Aldrich (St. Louis, MO) and doubly distilled water and adjusted to desired pH values. For experiments involving RNA, all laboratory equipments were autoclaved prior to use and sterilized water and buffers were purchased from Ambion (Austin, TX). The preparation of the modified phosphoramidite uridine monomers and synthesis of nylon nucleic acid precursor strands followed the procedures previously reported, [5a] except that strands were cleaved from resin and deprotected at 50°C, instead of room temperature. All commercial nucleic acid strands and nylon nucleic acid uncoupled precursor strands were purified by denaturing gel electrophoresis (20% acrylamide; running buffer containing 89 mM Tris·HCl, pH 8.0, 89 mM Boric acid, and 2 mM EDTA). For purification of uncoupled strands, care was taken not to dissolve DNA strands in formamide before loading onto gels, to avoid a side reaction of amino groups with formamide upon heating. Concentrations of oligonucleotides were determined by UV spectroscopy (OD260).
MALDI-TOF MS analysis
MALDI-TOF mass spectra were recorded on a Bruker OmniFLEX MALDI-TOF spectrometer. A 3-HPA matrix solution was prepared by dissolving 3-HPA (18mg) in CH3CN (150μL) and H2O (150μL). An ammonium citrate co-matrix solution was prepared by dissolving ammonium citrate (35mg) in H2O (1mL). The working matrix solution was obtained by mixing 3-HPA matrix solution (40μL) and ammonium citrate co-matrix solution (10μL). An oligonucleotide sample (20~100μM, 2μL) was mixed with the working matrix solution (2.5μL) using a vortex and then centrifuged. The mixture was deposited on a target. DNA molecules of known masses were used as either external or internal calibrants in the measurements.
Thermal denaturing studies
Pairs of complementary strands (700 pmol each) were dissolved in a buffer (40 mM sodium cacodylate and 100 mM sodium chloride, pH 7.3) to a final volume of 700 μL, and annealed overnight from 9°C to room temperature. The samples were transferred to quartz cuvettes with 1 cm path length, and the same cacodylate buffer was used as a blank. Thermal denaturation was monitored at 260 nm on a Spectronic Genesys 5 spectrophotometer, using a Neslab RTE-111 programmable circulating bath. At least two consecutive heating-cooling cycles were applied with a linear temperature gradient of 0.1°C/min. Heating and cooling ramps were superimposable in all cases, indicating equilibrium conditions. Absorbance vs. temperature curves were converted into θ vs. temperature curves (where θ is the fraction of oligomers in the associated state) by subtracting upper and lower base lines. These upper and lower linear base lines define temperature-dependent extinction coefficients for associated and dissociated states. Tm is defined as the temperature at which half of the strands are in the associated form and half in the dissociated form, i.e. θ = 0.5.[10] Thermodynamic data were generated from van’t Hoff plots of the data (ln K vs. 1/T).[10]
CD experiments
The CD spectra were recorded from 350 nm to 200 nm on an AVIV Model 202SF Circular Dichroism Spectropolarimeter at room temperature in 0.2 cm cuvettes. Two complementary strands (2.8 nmol each) were dissolved in buffer (10mM sodium phosphate, 150mM sodium chloride, and 0.1mm EDTA) to a final volume of 700μL and annealed as described above. The duplex solution was filtered through a 0.2 um syringe filter (Millex) before recording CD spectra. At least three scans were collected for each sample and a buffer baseline was subtracted from the average of these scans to yield the CD plots. The single strand CD spectra were recorded under the same conditions except at 15 μM concentration.
Supplementary Material
Acknowledgments
We thank Akinori Kuzuya for valuable advice. We gratefully acknowledge support by NSF (CTS-0548774 and CTS-0608889) to N.C.S. and J.W.C., grant GM-076202 from NIGMS and CHE-0316589 from NSF to J.W.C., and by grant GM-29554 from NIGMS, grants DMI-0210844, EIA-0086015, CCF-0432009, CTS-0548774, CTS-0608889, CCF-0726378 and CCF-0523290 from the NSF, 48681-EL and MURI W911NF-07-1-0439 from ARO, grant DE-FG02-06ER64281 from DOE (Subcontract from the Research Foundation of SUNY), and a grant from the W.M. Keck Foundation to N.C.S. The instrument facility was supported by Research Facilities Improvement Grant Number C06 RR-16572-01 from the National Center for Research Resources, National Institutes of Health. Instrumentation in the facility was purchased with support from NSF grants CHE-0234863 and MRI-0116222.
Footnotes
Supporting information for this article is available on the WWW under http://www.chembiochem.org or from the author.
Contributor Information
Prof. Dr. James W. Canary, Email: james.canary@nyu.edu.
Prof. Dr. Nadrian C. Seeman, Email: ned.seeman@nyu.edu.
References
- 1.Silverman AP, Kool ET. Chem Rev. 2006;106:3775. doi: 10.1021/cr050057+. [DOI] [PubMed] [Google Scholar]
- 2.a) Li X, Liu DR. Angew Chem Int Ed. 2004;43:4848. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]; b) Calderone CT, Liu DR. Current Opinion in Chemical Biology. 2004;8:645. doi: 10.1016/j.cbpa.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 3.a) Zhan ZJ, Ye J, Li X, Lynn DG. Curr Org Chem. 2001;5:885. [Google Scholar]; b) Gat Y, DG . In: Lynn in Templated Organic Synthesis. Stang PJ, Diederich F, editors. Wiley-VCH Verlag GmbH; Weinheim: 1999. pp. 133–157. [Google Scholar]; c) Summerer D, Marx A. Angew Chem Int Ed. 2002;41:89. [PubMed] [Google Scholar]
- 4.Datta B, Schuster GB, McCook A, Harvey SC, Zakrzewska K. J Am Chem Soc. 2006;128:14428. doi: 10.1021/ja0648413. [DOI] [PubMed] [Google Scholar]
- 5.a) Zhu L, Lukeman PS, Canary JW, Seeman NC. J Am Chem Soc. 2003;125:10178. doi: 10.1021/ja035186r. [DOI] [PubMed] [Google Scholar]; b) Zhu L, dos Santos O, Seeman NC, Canary JW. Nucleotides, Nucleosides & Nucleic Acids. 2002;21:723. doi: 10.1081/NCN-120015728. [DOI] [PubMed] [Google Scholar]
- 6.Seeman NC. Mol Eng. 1992;2:297.Rothemund PWK. Nature. 2006;440:297. doi: 10.1038/nature04586.For recent review, see: Lund K, Williams B, Ke Y, Liu Y, Yan H. Current Nanoscience. 2006;2(2):113.Seeman NC. Trends Biochem Sci. 2005;30(3):119. doi: 10.1016/j.tibs.2005.01.007.Seeman NC. Rep Prog Phys. 2005;68(1):237. doi: 10.1088/0034-4885/68/1/R05.
- 7.Felsenfeld G, Davies DR, Rich A. J Am Chem Soc. 1957;79:2023. [Google Scholar]
- 8.Gennis RB, Cantor CR. Biochemistry. 1970;9:4714. doi: 10.1021/bi00826a014. [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Ding L, Sha R, Seeman NC, Canary JW. manuscript in preparation. [Google Scholar]
- 10.Mergny J, Lacroix L. Oligonucleotides. 2003;13:515. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 11.a) Werner D, Brunar H, Noe CR. Pharm Acta Helv. 1998;73:3. [Google Scholar]; b) Urban E, Noe CR. Il Farmaco. 2003:243. doi: 10.1016/S0014-827X(03)00022-3. [DOI] [PubMed] [Google Scholar]
- 12.a) Uesugi S, Miki H, Ikehara M, Iwahashi H, Kyogoku Y. Tetrahedron lett. 1979;42:4073. [Google Scholar]; b) Guschlbauer W, Janlowski K. Nucleic Acids Res. 1980;8:1421. doi: 10.1093/nar/8.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.a) Venkateswarlu D, Lind KE, Mohan V, Manoharan M, Ferguson DM. Nucleic Acids Res. 1999;27:2189. doi: 10.1093/nar/27.10.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ozaki H, Sato Y, Azuma S, Sawai H. Nucleosides, Nucleotides Nucleic acids. 2000;19:593. doi: 10.1080/15257770008035010. [DOI] [PubMed] [Google Scholar]
- 14.a) Ross PD, Howard FB. Biopolymers. 2003;68:210. doi: 10.1002/bip.10306. [DOI] [PubMed] [Google Scholar]; b) Howard FB. Biopolymers. 2005;78:221. doi: 10.1002/bip.20289. [DOI] [PubMed] [Google Scholar]
- 15.a) Herdewijn P. Liebigs Ann. 1996:1337. [Google Scholar]; b) Kool E. Chem Rev. 1997;97:1473. doi: 10.1021/cr9603791. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Singh SK, Nielsen P, Koshkin A, Wengel J. Chem Commun. 1998:455. [Google Scholar]; d) Vester B, Wengel J. Biochemistry. 2004;43:13233. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]; e) Morita K, Hasegawa C, Kaneko M, Tsutsumi S, Sone J, Ishikawa T, Imanishi T, Koizumi M. Bioorg Med Chem Lett. 2002;12:73. doi: 10.1016/s0960-894x(01)00683-7. [DOI] [PubMed] [Google Scholar]; f) Renneberg D, Leumann C. J Am Chem Soc. 2002;124:5993. doi: 10.1021/ja025569+. [DOI] [PubMed] [Google Scholar]; g) Dupouy C, Iche-Tarrat N, Durrieu M, Rodriguez F, Escudier J, Vigroux A. Angew Chem Int Ed. 2006;45:3623. doi: 10.1002/anie.200504475. [DOI] [PubMed] [Google Scholar]
- 16.Kumar VA, Ganesh KN. Acc Chem Res. 2005;38:400. doi: 10.1021/ar030277e. [DOI] [PubMed] [Google Scholar]
- 17.a) Borsting P, Nielsen KE, Nielsen P. Org Biomol Chem. 2005;3:2183. doi: 10.1039/b502720a. [DOI] [PubMed] [Google Scholar]; b) Sharma PK, Mikkelsen BH, Christensen MS, Nielsen KE, Kirchhoff C, Pedersen SL, Sorensen AM, Ostergaard K, Petersen M, Nielsen P. Org Biomol Chem. 2006;4:2433. doi: 10.1039/b603830a. [DOI] [PubMed] [Google Scholar]
- 18.a) Breslauer KJ. In: Thermodynamic Data for biochemistry and Biotechnology. Hinz H, editor. Academic Press; New York: 1986. pp. 402–427. [Google Scholar]; b) Marky LA, Breslauer KJ. Biopolymer. 1987;26:1601. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 19.Johnson WC. In: Circular Dichroism: Principles and Application. Berova N, Nakanishi K, Woody RW, editors. Wiley-VCH; New York: 2000. pp. 719–740. [Google Scholar]
- 20.a) Friere SM, Altmann KH. Nucleic Acids Res. 1997;25:4429. doi: 10.1093/nar/25.22.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Crooke ST. Antisense Nucleic Acid Drug Dev. 1998;8:vii–viii. doi: 10.1089/oli.1.1998.8.vii. [DOI] [PubMed] [Google Scholar]; c) Dean NM, Butler M, Monla BP, Manoharan M. Antisense Drug Technology. 2001:319. [Google Scholar]; d) Corey DR, Abrams JM. Genome Biology. 2001;2:5. doi: 10.1186/gb-2001-2-5-reviews1015. reviews 1015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Larsen HJ, Bentin T, Nielsen PE. Biochim Biophys Acta. 1999;1489:159. doi: 10.1016/s0167-4781(99)00145-1. [DOI] [PubMed] [Google Scholar]
- 21.a) Inoue H, Hayase Y, Imura A, Iwai S, Miura, Ohtsuka E. Nucleic Acid Res. 1987;15:6131. doi: 10.1093/nar/15.15.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Majlessi M, Nelson NC, Becker MM. Nucleic Acid Res. 1998;26:2224. doi: 10.1093/nar/26.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Cramer H, Pfleiderer W. Nucleosides, Nucleotides Nucleic acids. 2000;19:1765. doi: 10.1080/15257770008045458. [DOI] [PubMed] [Google Scholar]; d) Prakash TP, Kawasaki AM, Lesnik EA, Sioufi N, Manoharan M. Tetrahedron. 2003:7413. [Google Scholar]; e) Marjia Prhavc, Prakash TP, Minasov G, Cook PD, Egli M, Manoharan M. Org Lett. 2003:2017. doi: 10.1021/ol0340991. [DOI] [PubMed] [Google Scholar]; f) Honcharenko D, Varghese OP, Plashkevych O, Barman J, Chattopadhyaya J. J Org Chem. 2006;71:299. doi: 10.1021/jo052115x. [DOI] [PubMed] [Google Scholar]
- 22.a) Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. Tetrahedron. 1998;54:3607. [Google Scholar]; b) Koshkin AA, Rajwanshi VK, Wengel J. Tetrahedron Lett. 1998;39:4381. [Google Scholar]; c) Singh SK, Wengel J. Chem Commun. 1998:1247. [Google Scholar]
- 23.a) Glick GD. Biopolymers. 1998;48:83. doi: 10.1002/(SICI)1097-0282(1998)48:1<83::AID-BIP8>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]; b) Alefelder S, Sigurdsson STh. Bioorg Med Chem. 2000;8:269. doi: 10.1016/s0968-0896(99)00280-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.