Abstract
Across all major vertebrate groups, androgen receptors (ARs) have been identified in neural circuits that shape reproductive-related behaviors, including vocalization. The vocal control network of teleost fishes presents an archetypal example of how a vertebrate nervous system produces social, context-dependent sounds. We cloned a partial cDNA of AR that was used to generate specific probes to localize AR expression throughout the central nervous system of the vocal plainfin midshipman fish (Porichthys notatus). In the forebrain, AR mRNA is abundant in proposed homologs of the mammalian striatum and amygdala, and in anterior and posterior parvocellular and magnocellular nuclei of the preoptic area, nucleus preglomerulosus, and posterior, ventral and anterior tuberal nuclei of the hypothalamus. Many of these nuclei are part of the known vocal and auditory circuitry in midshipman. The midbrain periaqueductal gray, an essential link between forebrain and hindbrain vocal circuitry, and the lateral line recipient nucleus medialis in the rostral hindbrain also express abundant AR mRNA. In the caudal hindbrain-spinal vocal circuit, high AR mRNA is found in the vocal prepacemaker nucleus and along the dorsal periphery of the vocal motor nucleus congruent with the known pattern of expression of aromatase-containing glial cells. Additionally, abundant AR mRNA expression is shown for the first time in the inner ear of a vertebrate. The distribution of AR mRNA strongly supports the role of androgens as modulators of behaviorally defined vocal, auditory, and neuroendocrine circuits in teleost fish and vertebrates in general.
Keywords: vocal control system, hearing, inner ear, hypothalamus, estrogen receptor, aromatase
Androgen receptors (ARs) and androgen-concentrating cells have been consistently identified in neural circuits that shape reproductive-related behaviors, including sexually dimorphic nuclei that control vocalization, across most vertebrate taxa: amphibians (Kelley, 1986; Perez et al., 1996), reptiles (Tang et al., 2001), and birds (Gahr, 2001; Ball et al., 2002; Schlinger and Brenowitz, 2002; Kim et al., 2004) (for review, see Bass, 2008). While androgen-concentrating neurons have long been known in the central nervous system (CNS) of fish as well (e.g., Morrell et al., 1975; Fine et al., 1996), recent studies have shown that androgens can strongly modulate the vocal pattern generating circuitry in teleost fish (Remage-Healey and Bass, 2004, 2006a, 2007), although the loci for possible direct androgen actions on the vocal system remain unknown. The current study delineates the position of ARs in the plainfin midshipman fish, Porichthys notatus, that has become a well-established model system for studying the neural and hormonal mechanisms of vocal-acoustic communication among vertebrates (Bass and McKibben, 2003).
The plainfin midshipman has two male morphs that exhibit alternative reproductive tactics including divergent vocal behaviors (Brantley and Bass, 1994; Lee and Bass, 2006). Territorial, type I males defend nests and court females acoustically with territorial and advertisement calls generated via the contraction of paired muscles attached to the gas-filled swim bladder (Cohen and Winn, 1967; Bass, 1996). Unlike type I males, type II males are nonterritorial, reach sexual maturity early, and invest in large testes size rather than body size and vocal muscle, and either “sneak” or “satellite” spawn in competition with type I males to fertilize eggs of which the type I will invest in parental behavior (Brantley and Bass, 1994; Bass, 1996). In general, type II males and females are similar to each other but differ from type I males in a suite of somatic, endocrinological, and neurological traits, including differing levels of circulating androgens (Bass, 1996). Sexual polymorphisms for this species are best exemplified in the vocal motor system where a hindbrain-spinal vocal pattern generator and sound-producing muscle is hypertrophied in type I males (Bass and Marchaterre, 1989; Bass et al., 1996). The capacity for multiple types of vocalizations (advertisement and agonistic calls) of widely divergent duration (msec to >1 hour) is reflected in the expanded vocal motor system in type I males, whereas females and type II males are limited to producing only short duration (msec) agonistic calls (Brantley and Bass, 1994).
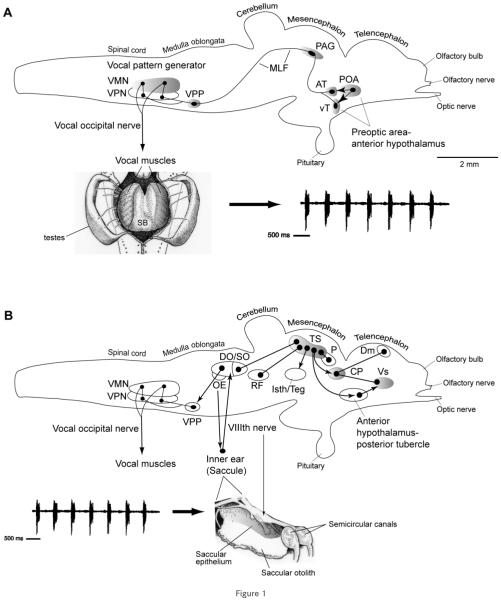
The vocal and auditory systems in midshipman and the closely related toadfish (same family: Batrachoididae; Nelson, 2006) show a conserved pattern of organization that shares many traits with tetrapods (Bass and Baker, 1997; Goodson and Bass, 2002; Bass et al., 2005, 2008; Kittelberger et al., 2006). An expansive vocal motor system (Fig. 1) includes a midline motor nucleus (VMN) that spans the caudal hindbrain-rostral spinal cord boundary and ipsilaterally innervates each vocal swim bladder muscle via paired occipital nerve roots (Bass and Baker, 1990; Bass et al., 1994). Each VMN receives bilateral input from adjacent columns of vocal pacemaker neurons (VPN) that, in turn, are innervated by a more rostral prepacemaker nucleus (VPP, formerly ventral medullary nucleus). Neurophysiological studies support the hypothesis that the VPP-VPN-VMN circuit collectively comprises a vocal pattern generator (VPG) that directly determines the temporal pattern of natural calls (Bass and Baker, 1990; Remage-Healey and Bass, 2004; Chagnaud et al., 2009) and compares closely to vocal patterning circuits in tetrapods (Bass et al., 2008). Other studies show that auditory inputs interface with vocal nuclei in the forebrain, midbrain, and hindbrain (Bass et al., 2000; 2001; Goodson and Bass, 2002; Kittelberger et al., 2006).
Figure 1.
Side views of the brain showing vocal motor (A) and central auditory (B) systems in batrachoidid fish (midshipman and toadfish) (modified from Bass and McKibben, 2003; Kittelberger et al., 2006). Solid dots represent somata, and lines represent axonal projection pathways. Two connected dots indicate reciprocal connections. Shaded areas indicate localization of AR expression. A: Descending vocal motor pathways (see Bass and Baker, 1990; Bass et al., 1994; Goodson and Bass, 2002; Kittelberger et al., 2006). Preoptic (POA) and ventral (vT) and anterior (AT) tuberal nuclei in the hypothalamic forebrain project to the periaqueductal gray (PAG) in the midbrain which then connects to the vocal pattern generator (VPG) in the hindbrain-spinal cord. The VPG consists of vocal prepacemaker (VPP), pacemaker (VPN), and motor (VMN) nuclei. The VMN projects directly via occipital nerve roots to sound-producing muscles on the swim bladder (shown in ventral, dissected view). The arrow points to a vocalization produced by the swim bladder, a typical “grunt train” often produced during nest defense by a type I male (Brantley and Bass, 1994). B: Central auditory system (see Bass et al., 2000, 2001). Natural sounds, represented by a “grunt train,” are detected by the inner ear which projects via the VIIIth nerve to octaval nuclei in the hindbrain and further to the auditory midbrain torus semicircularis (TS). Shown are nuclei interconnected with TS. The dorsal thalamic central posterior nucleus (CP) contains reciprocal connections to the dorsomedial (Dm) and ventral supracommissural (Vs) telencephalon which also receives input from the anterior hypothalamus-posterior tubercle (for nomenclature, see Braford and Northcutt, 1983). TS and CP also connect to vocal motor nuclei in the forebrain (anterior hypothalamus and POA) and midbrain (PAG and isthmal/tegmentum), while auditory-recipient octaval nuclei in the hindbrain connect to the VPG (also see Bass et al., 1994; Goodson and Bass, 2002).
Midshipman fish and toadfish are ideal model systems to investigate the effects of androgens on defined neural circuitry which directly impacts vocal-acoustic communication during social behavior. Androgens masculinize vocal motor neurons and muscle (reviewed in Bass and Forlano, 2008) and rapidly modulate vocal patterning (reviewed in Remage-Healey and Bass, 2006a; Bass and Remage-Healey, 2008). Androgens are elevated during nesting and gonadal recrudescence in males and females (Brantley et al., 1993a; Knapp et al., 1999; Modesto and Canario, 2003; Sisneros et al., 2004a; Fine et al., 2004), during advertisement calling in males (Knapp et al., 2001), and during acoustic playback challenge to nesting males (Remage-Healey and Bass, 2005). Androgens also induce changes in vocal behavior in nesting males in their natural habitat (Remage-Healey and Bass, 2006b). Seasonal peaks in circulating androgens correlate with high brain aromatase (estrogen synthase) mRNA expression (Forlano and Bass, 2005a) in both males and females. Testosterone upregulates aromatase expression in glial cells throughout the brain (Forlano and Bass, 2005b; Bass and Forlano, 2008) and induces natural, seasonal-like changes in the temporal encoding of the frequency content of male calls in the peripheral auditory system (Sisneros et al., 2004b).
The main goal of this study was to test the hypothesis that central integration sites for vocal, auditory, and neuroendocrine functions in the midshipman fish would show high levels of AR expression, consistent with the known role of androgens in modulating vocal-acoustic and reproductive behaviors.
MATERIALS AND METHODS
Animals
Midshipman fish were collected from field sites in northern California during the spawning season or in Monterey Bay, CA at other times of the year and maintained in seawater tanks until sacrificed shortly afterwards (see Sisneros et al., 2004a, for a description of the reproductive and nonreproductive time periods). The animals chosen for study were from the spring and summer months when plasma levels of steroids in general are elevated in females and males (Sisneros et al., 2004a). All experimental protocols were approved by the Cornell University Institutional Animal Care and Use Committee.
Cloning of partial cDNA of midshipman AR
Methods were previously reported for cloning of partial cDNA for midshipman ERα (Forlano et al., 2005) and were similar to those used to clone a partial cDNA for the aromatase gene from this species (Forlano et al., 2001). Degenerate primers (forward: GARGCNTCNGGNTGYCAYTAYGG; reverse: NCCYTTNGCCCAYTTNACNACYTT) were designed based on highly conserved regions in the DNA and ligand binding domains of six AR sequences from four teleost species (Halichoeres trimaculatus, Oncorhynchus mykiss [alpha and beta isoforms], Anguilla japonica [alpha and beta isoforms], and Chrysophrys major). Partial cDNA clones (462 bp) of AR were amplified from vocal muscle, brain, and the sensory epithelium from the saccule division of the inner ear of two type I males in reproductive condition.
Reverse-transcription polymerase chain reaction (RT-PCR)
Internal, midshipman-specific primers (forward: GCTGGAATGACCCTTGGAGCA; reverse: TCAACTCCCACGTGGTCTTCCTC) were employed to identify AR mRNA transcripts (150 bp) from brain, sonic muscle, and saccular epithelium of three type I males, two females and a type II male. Brains were dissected into four areas: 1) a large forebrain region including the olfactory bulbs, telencephalon, and preoptic area; 2) a middle region including the caudal diencephalon, midbrain tectum, and tegmentum; 3) the corpus of the cerebellum; and 4) remaining hindbrain and rostral spinal cord. Total RNA was isolated from various tissues and DNase-treated (Ambion, Austin, TX) according to the manufacturer’s instructions, after which 2.5 μg of RNA was reverse-transcribed as previously reported (Forlano et al., 2001). PCR was performed in a final volume of 50 μL containing 1× reaction buffer, 1.5 mM MgCl2, 200 μM each dNTP, 0.5 μM each primer, 0.5 μL Tfl DNA polymerase, and 1 μL of first-strand cDNA reaction. The reaction was run under the following conditions: 94°C for 4 minutes followed by 35 cycles of 94°C for 45 seconds, 56°C for 45 seconds, 72°C for 1 minute, and a final extension step at 72°C for 10 minutes. Negative controls included a no template control, and samples were verified as negative for genomic DNA after DNAse treatment. Reactions were run on a 2% agarose gel stained with ethidium bromide to detect the presence of transcripts in the various tissues.
In situ hybridization (ISH)
The procedure for ISH detection of AR mRNA was similar to our previous study for ERα mRNA (Forlano et al., 2005). Adult midshipman fish were deeply anesthetized in 0.025% benzocaine (Sigma Chemical, St. Louis, MO) in seawater and perfused transcardially with teleost ringers followed by 4% paraformaldehyde in 0.1M phosphate buffer (PB; pH 7.2). Brains (five type I males, three type II males, three females) and saccular epithelium of the inner ear (two females) were removed and postfixed in the same fixative for 1 hour and stored in PB. Prior to sectioning, various tissues were cryoprotected overnight in 30% sucrose in PB and frozen in Cryo-M-bed (Hacker Instruments, Huntington, UK). Sections were made at 30 μm in the transverse plane for brain tissue and at 20 μm in the oblique plane for the inner ear; alternate sections were collected onto Super-frost Plus slides (Erie Scientific, Portsmouth, NH) and stored at −80°C until processed. Prior to hybridization, slides were equilibrated to room temperature for 10 minutes, washed 2 × in 0.1 M potassium phosphate-buffered saline (KPBS; pH 7.2) and placed into freshly prepared 0.25% acetic anhydride in 0.1M triethanolamine 2 × 5 minutes, dehydrated in a series of ethanol washes and chloroform, and air-dried.
Hybridization was carried out with a mixture of three oligo probes antisense to nucleotides 160-186 (AAATGTTTTGAAGCTGGAATGACCC), 240–265 (CCTCTCCAGGAGTCTGTTGAGATCA), 401–425 (CTCTGCTCACCAGCCTCAATGAGC) of the 462-bp sequence of midshipman AR. A mixture of sense oligos from which the antisense sequences were derived were used as negative controls. Oligonucleotides were labeled with a terminal deoxynucleotidyl transferase reaction using α-33P d-ATP (SA, 1,000-3,000 Ci/mmole; NEN, Boston, MA). Hybridization solution (4× SSC [1× SSC = 0.15 M sodium chloride; 0.015 M sodium citrate, pH 7.2], 40% deionized formamide, 500 μg/mL denatured calf thymus DNA, 250 μg/mL transfer RNA, 4× Denhardt’s solution, 4 mM EDTA, 5 mM sodium phosphate [pH 6.5], 10% [wt/vol] Dextran sulfate and 3 × 106 cpm total radiolabeled probe [1.0 × 106 cpm each probe]) was placed on each slide (300 μL), coverslipped with parafilm, and incubated overnight (at least 15 hours) in a humidified 37°C chamber. Following hybridization, slides were briefly washed twice at 23°C in 1× SCC, washed twice for 30 minutes at 55°C in 1× SCC in a shaking water bath, and washed once in 1× SCC in 0.1% Triton X-100 at 23°C, briefly rinsed in distilled water, 70% ethanol, and air-dried. Slides were then exposed to Biomax MR film (Eastman Kodak, Rochester, NY) for 3 days at 4°C to confirm signal, and subsequently dipped in nuclear emulsion (NTB, Kodak) and exposed for 4 weeks at 4°C. Slides were developed in Kodak D-19 (4 minutes at 14°C), rinsed in distilled water (10 seconds at 14°C), and fixed (Kodak GBX fixer, 5 minutes at 14°C), rinsed in running distilled water (5 minutes), counter-stained in cresyl violet, dehydrated, and coverslipped with Permount (Fisher Scientific, Fair Lawn, NJ). The neuroanatomical nomenclature of Braford (1995), Braford and Northcutt (1983), and Northcutt (1995) was adopted for this study.
Photomicroscopy
Darkfield optics were used to best visualize the overall mRNA signal pattern throughout the brain at low magnification, while brightfield optics were used to best visualize ISH signal at higher magnification over Nissl-stained tissue. Photographs were taken using Kodak Gold color film (iso 400 for darkfield; iso 100 for brightfield) and a 35 mm Nikon camera mounted on a Nikon ES800 compound microscope. Negatives were then scanned at 1800 dpi, compiled into plates, converted to grayscale, contrast-enhanced, and labeled in Adobe Photoshop CS4 (San Jose, CA).
RESULTS
Cloning of partial cDNA for midshipman AR
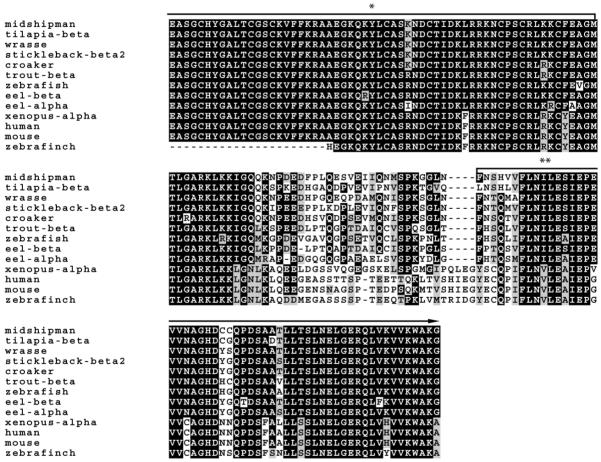
A predicted 462-bp sequence was amplified from adult male vocal muscle cDNA and was verified from 12 independent clones, as well as four brain clones, and five clones from the saccular epithelium of the inner ear of males. Midshipman vocal muscle is a known androgen-sensitive target (Brantley et al., 1993b). Figure 2 shows the partial cDNA sequence with translated amino acid sequence below. The sequence spans conserved regions of the DNA and ligand binding regions of the AR gene across vertebrates. Figure 3 shows a comparison of midshipman AR translated amino acid sequence to other vertebrate ARs. A BLAST (National Center for Biotechnical Information) search using the midshipman translated protein suggests a close match to other teleost AR beta (ARβ) subtypes. Multiple clones from different tissues support the likelihood of a single AR gene in midshipman. A sequence alignment revealed an 89% identity to three spot wrasse (Halichoeres trimaculatus) AR (GenBank Access. No. AF326200.1), 88% sequence identity to Nile Tilapia (Oreochromis niloticus) ARβ (GenBank Access. No. AB045212.1), stickleback (Gasterosteus aculeatus) ARβ1 and β2 (GenBank Access. No. AY247206.1 and AY247207.1, respectively) and Atlantic croaker (Micropogonias undulatus) AR (GenBank Access. No. AY701761.1), 85% identity to rainbow trout (Oncorhynchus mykiss) ARβ (GenBank Access. No. AB012096.1), 82% identity to zebrafish (Danio rerio) AR (GenBank Access. No. EF440290.1), 84% identity to Japanese eel (Anguilla japonica) ARβ and 80% to ARα (GenBank Access. No. AB025361.1 and AB023960.1, respectively), 69% to African clawed frog (Xenopus laevis) ARα (GenBank Access. No. NM_001090884.1), 68% identity to zebra finch (Taeniopygia guttata) AR (GenBank Access. No. NM_001076688.1), 66% to human (Homo sapiens) AR (GenBank Access. No. L29496.1), and 65% identity to mouse (Mus musculus) AR (GenBank Access. No. NM_013476.3).
Figure 2.
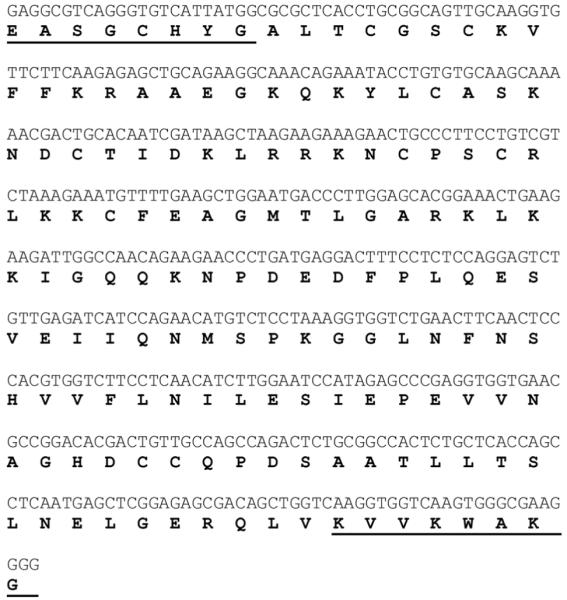
Nucleotide and deduced amino acid sequence of AR partial cDNA clone isolated from midshipman brain. Underlined amino acids and basepairs correspond to sequence derived from degenerate primers.
Figure 3.
Alignment of deduced amino acid sequences of midshipman with other representative vertebrate ARs: teleost (tilapia, wrasse, stickleback, croaker, trout, zebrafish, and eel), amphibian (Xenopus), mammalian (human and mouse) and avian (zebra finch). Black and gray shading indicate identical and similar amino acids to midshipman AR, respectively. The first and last eight amino acids are conserved regions to which degenerate primers were made for RT-PCR. The entire sequence falls within the DNA (*) and ligand binding domain (**) and is up to 90% conserved with other teleosts.
RT-PCR
AR transcripts were identified in four CNS regions in adult individuals (Fig. 4): a large forebrain region including the olfactory bulbs, telencephalon, and preoptic area; a middle region including the caudal diencephalon, midbrain tectum, and midbrain tegmentum; the corpus of the cerebellum; and remaining hindbrain and rostral spinal cord. AR mRNA was also found in saccular epithelium of the inner ear and vocal muscle of both adult females and type I males. No transcripts were detected when DNase-treated RNA was used as a template (no RT) and in no template controls for PCR reactions.
Figure 4.

Using midshipman-specific primers, a predicted 150-bp product containing AR transcripts was identified by RT-PCR in a forebrain region (F), a middle CNS region including midbrain and caudal diencephalon (M), the cerebellum (C), hindbrain and rostral spinal cord (H), vocal muscle (VM), and saccular epithelium of the inner ear (E) of individual adult midshipman. Shown is a representative type I male.
In situ hybridization
A neuroanatomical atlas of AR mRNA expression is represented in Figures 6-12 at 13 transverse levels shown in Figure 5 from a type II male brain. For each representative level of the atlas, the left half of the Nissl-stained hybridized section is presented in darkfield (left) at low magnification and then in brightfield (middle). Higher-magnification views of individual nuclei are displayed in brightfield (right) to demonstrate the most abundant AR hybridization patterns. The general patterns of hybridization are representative of all animals used in the study (type I, II males and females). No specific hybridization signal was seen with negative controls (sense probes). Table 1 also lists the relative abundance of AR expression in nuclei throughout the CNS in midshipman and highlights sites identified as part of the auditory and/or vocal system (also see Discussion).
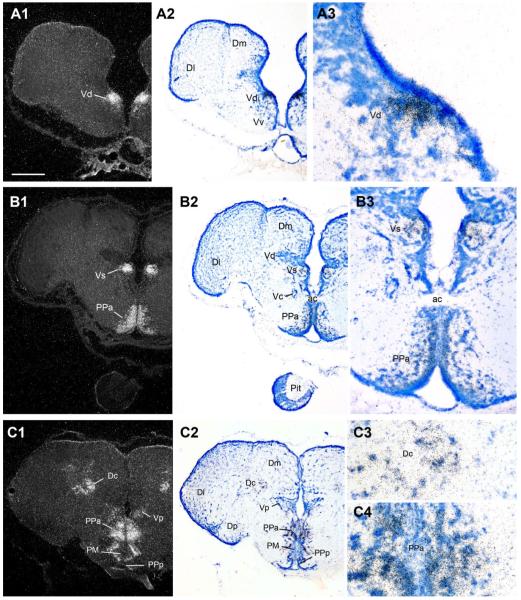
Figure 6.
AR mRNA expression in the ventral telencephalon and rostral preoptic area. In this figure and in Figures 6-12, neuroanatomical localization of in situ hybridization signal is shown for the same section in low-magnification darkfield (left) and brightfield Nissl-stained sections (middle) and higher-magnification brightfield (right). A1–3: Robust AR expression in dorsal nucleus of area ventralis (Vd). B1–B3: High AR mRNA expression in the supracommissural nucleus of area ventralis (Vs) and anterior parvocellular preoptic area (PPa), part of the forebrain vocal-acoustic complex (see Fig. 1A,B; Table 1). C1–C4: Strong AR expression in the dorsal pallium in the central zone of area dorsalis (Dc) as well as throughout the extent of the PPa and the postcommissural nucleus of area ventralis (Vp). Scale bar = 500 μm for panels 1, 2 in A–C; 100 μm, 200 μm, and 100 μm panels in A3, B3, and C3–C4, respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
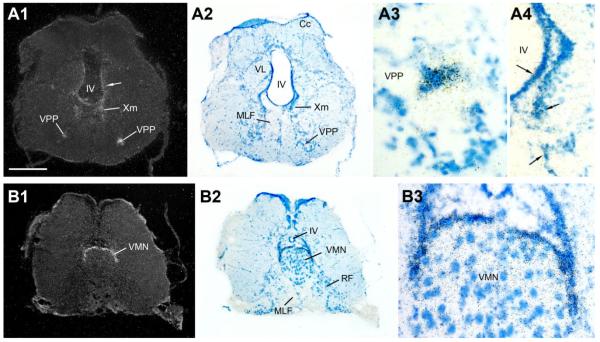
Figure 12.
AR mRNA expression in the vocal, hindbrain-spinal cord pattern generator. A1–A4: The vocal prepacemaker nucleus (VPP) shows robust AR expression; also consistent but diffuse mRNA signal found along the ventricle (IV). B1–B3: Strong AR mRNA expression forms a cap around the dorsal periphery of the vocal motor nucleus (VMN). Scale bar = 500 μm for panels 1, 2 in A,B; 100 μm in A3; 200 μm in A4,B3. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 5.
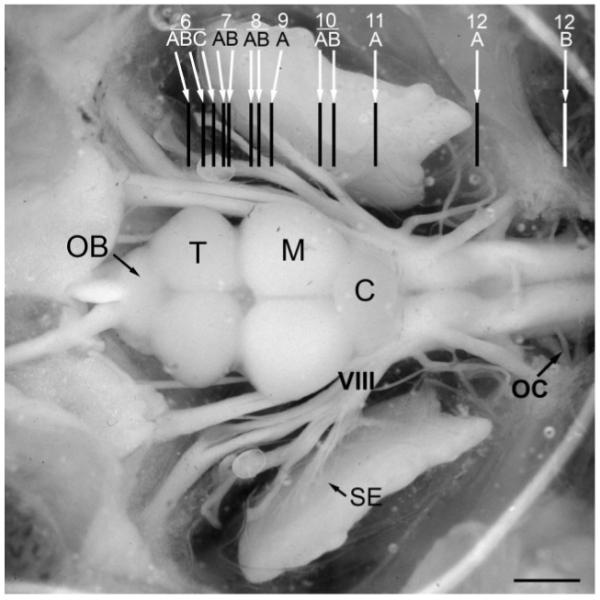
Dorsal view of exposed brain and inner ear of a midshipman indicating levels (rostral-caudal) of transverse sections for the brain atlas of androgen receptor distribution shown in Figures 6-12. C, cerebellum; M, midbrain; OB, olfactory bulb; OC, occipital nerve roots; T, telencephalon; SE, saccular epithelium of the inner ear; VIII, eighth nerve. Modified from Forlano et al. (2005). Scale bar = 1.5 mm.
TABLE 1.
Comparative Distribution of Androgen Receptor (AR), Aromatase (Estrogen Synthase), and Estrogen Receptor alpha (ERα) in the Brain of the Plainfin Midshipman, Porichthys notatus
| Brain area | AR mRNA | Aromatase mRNA/ protein1† |
ERα mRNA2 |
|---|---|---|---|
| Ventral telencephalon | |||
| Olfactory bulb (OB) | − | +++ | + |
| Ventral nucleus (Vv) AUDITORY | − | +++ | + |
| Supracommissural nucleus (Vs) AUDITORY, VOCAL | +++ | +++ | + |
| Postcommissural nucleus (Vp) AUDITORY | ++ | +++ | + |
| Dorsal nucleus (Vd) | +++ | +++ | − |
| Intermediate nucleus (Vi) | +++ | +++ | − |
| Dorsal telencephalon | |||
| Central zone (Dc) | ++ | ++ | + |
| Central medial division of medial zone (Dm-cm) | +++ | ++ | ++ |
| Posterior division of medial zone (Dm-p) AUDITORY, VOCAL | + | ++ | + |
| Dorsal posterior zone (Dp) | − | ++ | + |
| Preoptic area | |||
| Anterior parvocellular (PPa) AUDITORY, VOCAL | +++ | +++ | +++ |
| Posterior parvocellular (PPp) AUDITORY, VOCAL | ++ | +++ | − |
| Magnocellular (PM/PMg) | +++ | +++ | ++ |
| Ventral hypothalamus | |||
| Anterior tuberal (AT) AUDITORY, VOCAL | ++ | +++ | ++ |
| Ventral tuberal (vT) AUDITORY, VOCAL | ++ | +++ | − |
| Periventricular (Hv/Hd) | +++ | +++ | +++ |
| Thalamus | |||
| Central posterior nucleus (CP) AUDITORY | +*† | +++ | + |
| Dorsal posterior (DPo) | +*† | +++ | + |
| Nucleus preglomerulosus (PGl/m) AUDITORY | + | + | − |
| Periventricular nucleus of posterior tuberal (TPp) | ++*† | +++ | + |
| Posterior tuberal nucleus (TP) | +++ | +++ | − |
| Pineal | − | ++ | +++ |
| Brainstem | |||
| Periacqueductal gray (PAG) VOCAL | + | +++ | − |
| Mesencephalic tectum (TeM) | + | − | + |
| Medial longitudinal fasiculus (MLF) (peri) | +*† | ++ | − |
| Nucleus of medial longitudinal fasiculus (nMLF) | + | +++ | + |
| Periventricular cell layer of TS (Pe) AUDITORY | ++ | − | − |
| Griseum central (GC) | +*† | +++ | − |
| Medial octavolateralis nucleus (MED) LATERAL LINE | + | − | − |
| Vagal motor nucleus (Xm) | +*† | + | − |
| Vocal prepacemaker (VPP) VOCAL | ++ | − | − |
| Vocal motor nucleus (VMN) VOCAL | ++*† | +++ | + |
| Peripheral tissues | |||
| Saccular epithelium (inner ear) AUDITORY | + | ++ | + |
| Vocal muscle VOCAL | ++ | − | − |
Forlano et al. (2005). See Discussion for auditory, vocal ,and lateral line references.
Glial expression
similar pattern to glial aromatase expression; +++, ++, + = relative levels of hybridization signal.
Forebrain
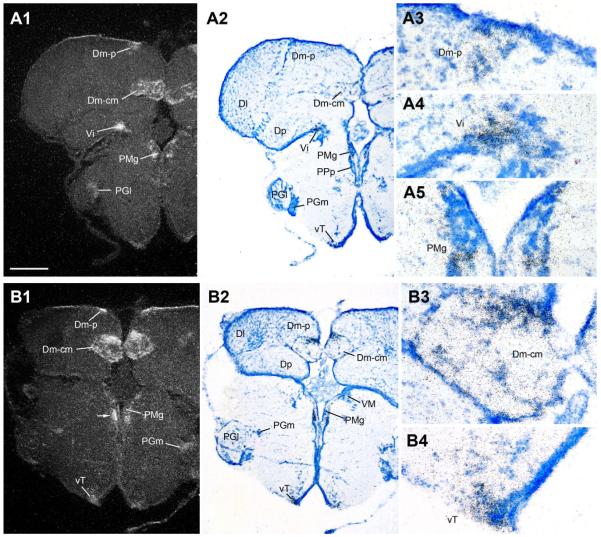
Robust AR mRNA expression is found in discrete nuclei in the ventral telencephalon, including the dorsal, supracommissural, postcommissural, and intermediate nuclei of area ventralis (Vd, Vs, Vp, and Vi, respectively) (Figs. 6A–C, 7A). Similarly, prominent AR mRNA is seen in very specific regions of the dorsal telencephalon. Rostrally, the central zone of area dorsalis (Dc) is comprised of two distinct clusters of cells well-defined by AR mRNA expression: the slightly more medial group adjoins Dm while the ventrolateral group sits above Dp (Fig. 6C). In the caudal telencephalon, the posterior (Dm-p) and the central medial (Dm-cm) divisions of the medial zone of area dorsalis are sites of strong AR mRNA signal (Fig. 7A,B); and AR expression in Dc (Fig. 6C) is contiguous with Dm-cm.
Figure 7.
AR mRNA expression in the dorsal telencephalon and caudal preoptic area. A1–A5: The posterior (p) and central medial (cm) divisions of the medial zone of area dorsalis (Dm-p and Dm-cm), the intermediate nucleus of area ventralis (Vi), and the gigantocellular division of the magnocellular preoptic area (PMg) show strong AR mRNA expression. B1–B4: In addition to strong hybridization signal in the dorsal pallium (Dm-p, Dm-cm) and ventricular zone adjacent to PMg, the ventral tuberal hypothalamus (vT), part of the descending vocal motor pathway (see Fig. 1), shows robust AR expression. Scale bar = 500 μm for panels 1, 2 in A,B; 100 μm for A3–A5 and B3–B4. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
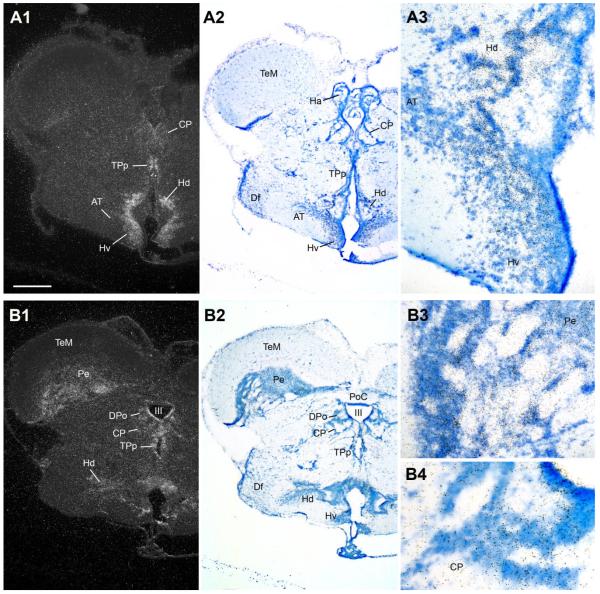
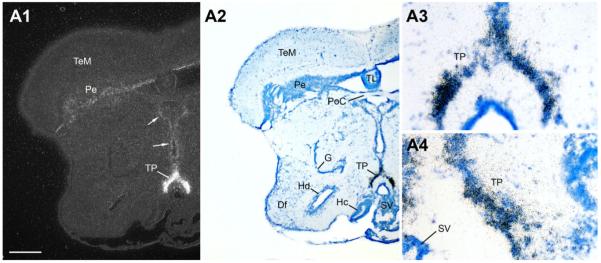
Abundant AR mRNA is found throughout the preoptic area. Robust signal covers the extent of the anterior parvocellular preoptic area (PPa) (Fig. 6B,C) and the peripheral margin of the anterior magnocellular preoptic area (PM) (Fig. 6C), and is as also over the more caudal gigantocellular division of PM (PMg) (Fig. 7A,B). Less intense expression is found in lateral and medial subdivisions of nucleus preglomerulosus (PGl/m) along the lateral aspect of the diencephalon (Fig. 7A,B). Medial nuclei of the anterior ventral hypothalamus contain abundant AR mRNA, including the ventral and anterior tuberal nuclei (vT, Fig. 7B; AT, Fig. 8A), and dorsal and ventral periventricular areas (Hd and Hv, Fig. 8A,B). The caudal extent of Hd and Hc, however, are devoid of AR mRNA expression (Fig. 9). Throughout its extent, the periventricular posterior tuberal nucleus (TPp, Fig. 8A,B) shows dense AR mRNA signal that appears contiguous with expression along the third ventricle and within the central and dorsal posterior nuclei of the thalamus (CP and DPo, Fig. 8B). Caudal to TPp, the posterior tuberal nucleus (TP) likely contains the greatest concentration of AR mRNA in the brain (Fig. 9); the ventricular midline just dorsal to TP contains lower but clear AR mRNA signal (arrows, Fig. 9A1).
Figure 8.
AR mRNA expression in the diencephalon and rostral hypothalamus. A1–A3: Dorsal (Hd) and ventrolateral periventricular (Hv) nuclei of the hypothalamus show abundant AR; lower levels are found in the anterior tuberal nucleus (AT), part of the forebrain vocal-acoustic complex (see Fig. 1). B1–B4: Diffuse AR mRNA expression is found in the auditory central posterior nucleus (CP, see Fig. 1), the dorsal posterior (DPo) nucleus of the thalamus, and the periventricular layer of the midbrain tectum (Pe), as well as along the third ventricle (III) including the periventricular posterior tuberal nucleus (TPp, also see A). Scale bar = 500 μm for panels 1, 2 in A,B; 200 μm and 100 μm for A3 and B3–B4, respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Figure 9.
A1–A4: The posterior tuberal nucleus (TP), shown in high magnification at two levels, has perhaps the highest abundance of AR mRNA in the central nervous system. Also note consistent signal (arrows) along the ventricular midline in the thalamus (midline) and in the periventricular cell layer of the tectum (Pe). Scale bar = 500 μm for panels A1–A2; 100 μm for A3–A4. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Midbrain
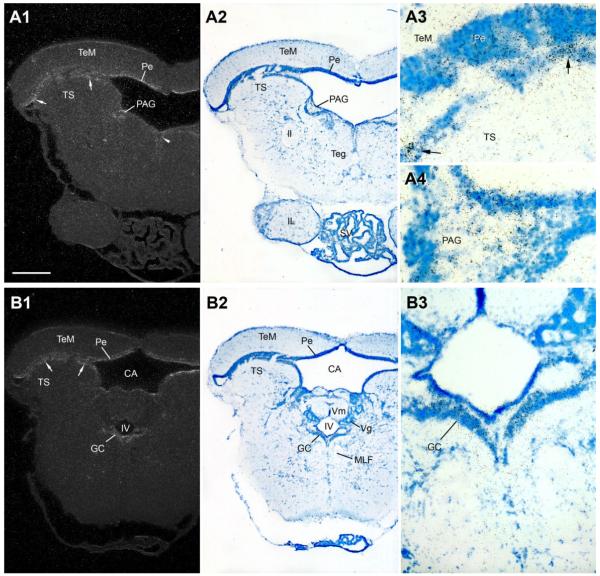
In general, hybridization signal in the midbrain is lower and more diffuse compared to forebrain nuclei. Consistent AR mRNA expression is seen throughout the periventricular cell layer of the midbrain tectum (Pe, Figs. 8 - 10; see Brantley and Bass, 1988, for cytoarchitecture) and the periventricular cell layer of the torus semicircularis (arrows in TS, Fig. 10A1,A3,B1; see Bass et al., 2000, for cytoarchitecture). Hybridization signal is apparent at rostral levels along the dorsal tegmental midline (midline arrowhead, Fig. 10A1) and a region of the central gray previously identified as the periaqueductal gray (PAG) in midshipman (Fig. 10A) because of its shared neuroanatomical and neurophysiological traits with the same-named region in mammals (Goodson and Bass, 2002; Kittelberger et al., 2006). In the caudal midbrain, AR mRNA expression is consistently found over the band of cells that comprise the griseum centrale (GC) bordering the fourth ventricle (Fig. 10B).
Figure 10.
AR mRNA expression at two midbrain levels. A1–A4: Hybridization signal in the periaqueductal gray (PAG), part of the descending vocal motor pathway (see Fig. 1), and the periventricular torus semicircularis (TS, arrows), tectum (Pe) and dorsal tegmental midline (arrowhead). B1–B3: AR mRNA expression in a more caudal section of the midbrain which also demonstrates signal in periventricular TS (arrows), tectum’s Pe, and griseum centrale (GC). Scale bar = 500 μm for panels 1, 2 in A,B; 100 μm and 200 μm for A3–A4 and B3, respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Hindbrain and spinal cord
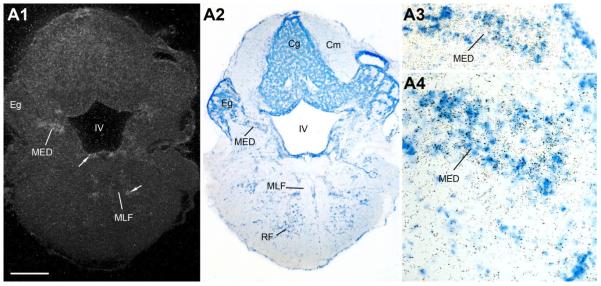
Similar to the midbrain, the rostral hindbrain shows diffuse AR mRNA signal along the fourth (IV) ventricle (arrows, Figs. 11, 12A). A distinct patch of AR expression is found over the cell plate of the medial octavolateralis nucleus (MED, Fig. 11), with additional AR signal at this level between the medial longitudinal fasiculus (MLF) and lateral lemniscus (ll) (arrow, Fig. 11A1). More caudally, at the level of the vagal lobe (VL), AR mRNA lines the fourth ventricle and continues ventrally into the vagal motor nucleus (Xm) and along the lateral edge of the MLF (Fig. 12A). Robust AR mRNA expression in the caudal hindbrain, comparable in density to that found in forebrain nuclei, is found over the vocal prepacemaker nucleus (VPP) within the ventrolateral reticular formation (Fig. 12A). Equally prominent AR expression is found overlying the VMN spanning the hindbrain-spinal cord boundary (Fig. 12B).
Figure 11.
A1–A4: AR mRNA expression in the lateral line recipient nucleus medialis (MED). Also notice diffuse signal along the fourth ventricle (IV) and lateral to the medial longitudinal fasciculus (MLF) (arrows). Scale bar = 500 μm for A1–A2; 200 μm and 100 μm for A3 and A4, respectively. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Inner ear
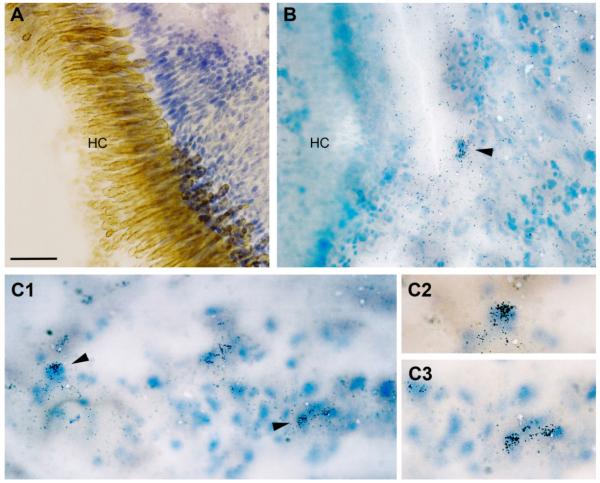
AR gene expression in the inner ear was initially demonstrated by RT-PCR (Fig. 4). The main end organ of hearing in midshipman and many other teleosts is the saccule (Cohen and Winn, 1967; Bass and McKibben, 2003). Using anatomical landmarks of the saccule (see Forlano et al., 2005; Fig. 13A), we determined that the expression for AR mRNA was concentrated over discrete patches of cells, likely support cells, just outside the hair cell layer (Fig. 13).
Figure 13.
AR mRNA expression in the inner ear of female midshipman. A: The hair cell layer (HC) of the sensory epithelium of the saccule is delineated by a hair cell-specific antibody (brown), overlaying Nissl stained support cells (see Forlano et al., 2005, for methods). B: Arrowhead indicates AR mRNA (clusters of silver grains) over cells just outside the HC layer. C1–C3: Examples of hybridization specificity in the inner ear in a similar location as B. C2–C3: Higher magnification of cells indicated by arrowheads in C1. Scale bar = 100 μm in A,B; 50 μm in C1; 30 μm in C2–C3. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
DISCUSSION
The current study demonstrates AR mRNA hybridization signal at all brain levels, providing anatomical support for androgen actions on vocal, auditory, and neuroendocrine circuits in midshipman fish and the closely related toadfish, and in teleosts in general. Abundant AR mRNA expression was observed at forebrain, midbrain, and hindbrain-spinal cord levels of the descending vocal motor pathway as well as in sites of vocal-acoustic integration. Furthermore, our findings show the first evidence of AR mRNA in the inner ear of a vertebrate, in this case in the saccule, which is the main auditory division of the inner ear in many teleosts including midshipman fish (Bass and McKibben, 2003). Table 1 lists the relative abundance of AR expression in nuclei throughout the CNS in midshipman and highlights sites identified as part of the auditory and/or vocal system. Figure 1 summarizes AR mRNA expression in sites identified as part of the auditory and/or vocal system.
AR subtypes
Aside from the present study, the neuroanatomical distribution of AR mRNA expression in teleosts has been documented in the cichlid Astatotilapia burtoni (Harbott et al., 2007), and the zebrafish Danio rerio (Gorelick et al., 2008). In A. burtoni, the distribution of two AR subtypes, ARα and ARβ, was documented mainly throughout the telencephalon and ventral hypothalamus. While both subtypes were highly expressed in the preoptic area, ARβ was more widely distributed in the brain, notably in dorsal and central divisions of area dorsalis in the telencephalon, and found at higher expression levels in the forebrain and midbrain by quantitative (Q)RT-PCR. Although localization was not described in the hindbrain by ISH, transcripts of both subtypes were identified by QRT-PCR and expression of ARα was found to be significantly greater in the hindbrain and the pituitary gland. The sequence comprising the partial cDNA for midshipman AR shares highest identities with other teleost ARβs (Fig. 3), and its robust distribution in the dorsal telencephalon and widespread expression throughout the brain further supports that the midshipman AR described here is an ARβ subtype. Also, the AR in the present study was first isolated from vocal muscle which is responsive to both testosterone and the nonaromatizable androgen 11-ketotestosterone (Brantley et al., 1993b; Lee and Bass, 2005), and therefore may contain one AR sensitive to different androgens, again similar to the broad binding affinity of ARβ/AR2 described in other teleosts (Sperry and Thomas, 1999, 2000). Recent studies indicate a single AR gene in zebrafish that is also closest to ARβ in sequence, has binding affinities to both testosterone and 11-ketotestosterone, and likely encodes a functional ortholog of the mammalian AR (Jorgensen et al., 2007; Gorelick et al., 2008; Hossain et al., 2008). Another cyprinid fish, Spinibarbus denticulatus, also expresses only one AR isoform; RT-PCR analysis showed AR in the olfactory bulb, telencephalon, and hypothalamus of males and females, but only in posterior brain regions (optic tectum, medulla, and spinal cord) in males (Liu et al., 2009).
The AR isoform most similar to ARβ in fishes appears closest to that of other vertebrates in sequence homology and androgen binding and is the probable ancestral vertebrate AR (Sperry and Thomas, 1999; Harbott et al., 2007; Gorelick et al., 2008). A recent analysis proposes that basal teleosts such as Anguilla have retained both AR subtypes, while intermediate groups (e.g., salmonids and ostariophysians that includes zebrafish) secondarily lost one subtype and derived teleosts (percomorphs that include cichlids) have retained both subtypes (Douard et al., 2008). Given this pattern, midshipman would likely fall into the intermediate groups that express a single AR gene. However, there may be an 11-ketotestosterone-specific receptor yet to be identified in midshipman equivalent to ARβ2 identified in male stickleback kidney (Olsson et al., 2005), although this receptor may only be expressed in androgen-sensitive peripheral tissues.
Comparative neuroanatomical AR distribution with other teleosts
The widespread distribution of AR mRNA documented in the present study overlaps, in part, that described in cichlids and zebrafish (Harbott et al., 2007; Gorelick et al., 2008) and using steroid autoradiography in earlier studies of other teleosts, including toadfish, (Morrell et al., 1975; Davis et al., 1977; Bass et al., 1986; Fine et al., 1996). Like A. burtoni (Harbott et al., 2007), midshipman show AR expression in Dc, but in addition have robust signal in the central medial (cm) and posterior (p) divisions of Dm. In contrast, zebrafish alone shows AR mRNA signal in the dorsolateral telencephalon (Gorelick et al., 2008). The expression in Dm-cm and Dm-p of midshipman borders a larger medial division of Dm that is retrogradely labeled following neurobiotin injection into the ventral tuberal region of the anterior hypothalamus (vT; Fig. 7A), a vocal site in midshipman that receives input from the ascending auditory system (Bass et al., 2000; Goodson and Bass, 2000a, 2002). A review of that original material (A. Bass, unpubl. obs.) shows retrogradely filled cells in Dm-cm and Dm-p, although very modest in comparison to the larger Dm region. Thus, AR mRNA expression may include vocal and/or auditory related regions of Dm.
Compared to studies in other teleosts, AR mRNA expression in supracommisural (Vs), postcommissural (Vp), and intermediate (Vi) nuclei of the ventral telencephalon has only been reported here in midshipman. Androgen-concentrating cells were also documented in the Vs of toadfish, but not other species (review in Fine et al., 1996). Fine et al. (1996) report androgen-concentrating cells in a restricted area of Dp that is most likely equivalent to Vi in the current study. Unlike A. burtoni (Harbott et al., 2007) and paradise fish (Davis et al., 1977), AR mRNA was not found in Vv in midshipman (this study) and toadfish (Fine et al., 1996). We also found robust AR expression in periventricular and posterior tuberal nuclei and along the periventricular optic tectum, like zebrafish (Gorelick et al., 2008) but unlike A. burtoni (Harbott et al., 2007). Consistent with all other teleosts so far studied (see above), midshipman have abundant AR expression in several divisions of the preoptic area and the ventral hypothalamus, including proposed homologs to mammalian arcuate (Hv; Cerda-Reverter et al., 2003; Forlano and Cone, 2007), para-ventricular (PM; Gilchriest et al., 2000; Kapsimali et al., 2001) (also see Forlano and Cone, 2007, and references therein), and ventromedial (AT; Goodson, 2005; Forlano et al., 2005; Forlano and Cone, 2007) hypothalamic nuclei. Similarly, AR mRNA is found highly expressed in amygdala homologs in the ventral telencephalon (Vi, Vp, Vs) (Northcutt and Braford, 1980; Northcutt, 1995), and a striatal homolog (Vd) (Wullimann and Mueller, 2004; Northcutt, 2006). Regions of Dm and Dc that show abundant AR mRNA may compare to either ventral or dorsal pallium (see Braford, 1995; Wullimann and Mueller, 2004; Yamamoto and Ito, 2005; Northcutt, 2006; Yamamoto et al., 2007). This is consistent with findings across vertebrate groups, where AR expression is located in reproductive and limbic brain areas, including the preoptic area-anterior hypothalamus, as this conserved brain region is involved in sexual behavior and control of gonadotropin release (Goodson and Bass, 2001; De Vries and Simerly, 2002).
In the present study, additional sites of AR mRNA were reported for the first time in a teleost in the diencephalon (ventral and anterior tuber, central and dorsal posterior thalamus, lateral and medial divisions of nucleus preglomerulosus), midbrain (torus semicircularis, periaqueductal gray, griseum centrale), hindbrain (nucleus medialis, vagal motor nucleus, vocal prepacemaker and motor nuclei, reticular and periventricular areas) and in the saccule division of the inner ear. Androgen-concentrating cells labeled in the inferior reticular formation in toadfish (Fine et al., 1996) are probably equivalent to AR mRNA signal in midshipman vocal prepacemaker (A. Bass, unpubl. obs.).
Androgen-dependent plasticity in teleost vocal motor system
Testosterone and 11-ketotestosterone are elevated in the circulation during prespawning and spawning seasons in midshipman and toadfish (Modesto and Canario, 2003; Fine et al., 2004; Sisneros et al., 2004a), and influence the vocal motor system and behavior of midshipman and toadfish (see Introduction; Remage-Healey and Bass, 2006b; Forlano et al., 2007; Bass, 2008; Bass and Remage-Healey, 2008). AR expression in one or more vocal sites (Fig. 1A) likely contributes to the known effects of androgens on the vocal system and vocal behavior (see Introduction). The mechanism by which androgens rapidly modulate the vocal pattern generator is unknown (Remage-Healey and Bass, 2004, 2006, 2007); however, these effects are blocked by cyproterone acetate (CA), a selective AR antagonist in fishes (Wells and Van Der Kraak, 2000). This implies that the mode of action involves a receptor or membrane binding site that contains a similar structure to a nuclear-like AR receptor (Remage-Healey and Bass, 2004). Interestingly, CA similarly blocks the rapid action of both testosterone and 11-ketotestosterone on vocal output, and may indicate that the same receptor is responsible for the effects of both androgens, although their sensitivities are tuned to a specific androgen in each morph/sex (Remage-Healey and Bass, 2007; Bass and Remage-Healey, 2008).
Comparisons with AR distribution in vocal tetrapods
Our results are consistent with evidence for expression of AR in vocal forebrain and brainstem nuclei, and vocal muscles across vertebrates (for review, see Bass and Remage-Healey, 2008). AR expression is well documented in vocal neurons in amphibians (Kelley, 1986; Perez et al., 1996), songbirds (Nastiuk and Clayton, 1995; Gahr and Wild, 1997; Gahr and Metzdorf, 1997; Bernard et al., 1999; Metzdorf et al., 1999; Gahr, 2001; Kim et al., 2004) non-songbirds (Shaw and Kennedy, 2002; Matsunaga and Okanoya, 2008), and reptiles (Tang et al., 2001). In primates, although extensive mapping of vocal pathways and associated motor nuclei is well documented (see Jurgens, 2002), AR expression associated with these areas has not been specifically investigated. However, extensive AR expression is found in vocal-related motor nuclei in rats (i.e., nucleus ambiguus, trigeminal motor, facial, hypoglossal) (Simerly et al., 1990) (see Jurgens, 2002, for vocal motor overview). Like midshipman fish (Goodson and Bass, 2002; Kittelberger et al., 2006), the midbrain PAG that activates hindbrain vocal nuclei in birds and mammals are abundant with ARs (see references above). Also like midshipman, amphibian preoptic and hypothalamic nuclei that project to brainstem vocal nuclei express high amounts of AR (Kelley, 1986). Lastly, like vocal motor and prepacemaker nuclei in midshipman, the vocal motor and premotor nuclei in songbirds express AR (see Gahr and Wild, 1997). Thus, in addition to brainstem-spinal vocal motor circuitry itself (Bass et al., 2008), AR expression within vocal nuclei also appears to be an ancestral, conserved phenotype of vertebrates (also see Bass and Remage-Healey, 2008).
Steroid-dependent plasticity in the auditory system
Improved temporal encoding of the frequency components of male calls have been recorded from eighth nerve, saccular afferents in female midshipman fish during reproductive periods, paralleling seasonal increases in circulating testosterone and 17β-estradiol (Sisneros and Bass, 2003; Sisneros et al., 2004a). Both testosterone and 17β-estradiol treatments induce a reproductive-like auditory phenotype in nonreproductive females, and the testosterone effect could be due to its local conversion to 17β-estradiol by aromatase (Sisneros et al., 2004b). In support of this hypothesis, estrogen receptor (ER)α mRNA was identified just outside the hair cell layer and aromatase-immunoreactive ganglion cells were found in the branch of the eighth nerve proximal to the saccule’s sensory epithelium (Forlano et al., 2005). Similar to ERα, we show AR mRNA expressed in cells just outside the hair cell layer of the sensory epithelium, in support of androgen-dependent plasticity in the inner ear. While these AR-positive cells, likely support cells, could indirectly affect the properties of the hair cells, androgens may also act directly on hair cells; mRNA levels may either be expressed below the threshold of detection of ISH or fluctuate seasonally and therefore have been potentially missed in our sampling. It is likely, however, that androgens function, in part, to upregulate local aromatase in eighth nerve ganglion cells, as found in the brain (Forlano and Bass, 2005b), thereby increasing local 17β-estradiol production proximal to ERs (also see Forlano et al., 2005).
Several nuclei in the central auditory system (Bass et al., 2000) consistently expressed AR mRNA (Table 1; Fig. 1B), including the torus semicircularis (TS) that is the homolog of the tetrapod inferior colliculus (Bass et al., 2005), and several sites in the diencephalon (CP, AT, PGl) that receive direct input from the auditory torus (Bass et al., 2000, 2001). AR mRNA is also highly expressed in preoptic-anterior hypothalamic (PPp, PPa, vT) and ventral telencephalic (Vd, Vp, Vs) nuclei that receive input from the auditory thalamus (CP) (Goodson and Bass, 2002). AR mRNA in the lateral line recipient hindbrain nucleus medialis suggests the potential influence of androgens on the auditory-like vocal encoding abilities of the lateral line system (Weeg and Bass, 2000, 2002). With the exception of the auditory-recipient telencephalic nuclei, AR mRNA is expressed more robustly than ERα (Forlano et al., 2005) throughout central auditory circuitry, which suggests that androgens have an important role for modulation of auditory function in midshipman (also see next section).
AR mRNA has been documented in the central auditory system of a vocal lizard, Geckko gecko (Tang et al., 2001) and in rats (Simerly et al., 1990), while the TS of the frog Xenopus concentrates androgens (Kelley, 1980), but in all cases the functional significance has not been investigated. Interestingly, humans with Kennedy’s disease (also known as X-linked bulbospinal muscular atrophy) that is caused by a mutation on the AR gene (Palazzolo et al., 2008) show impaired hearing accompanying abnormal brainstem auditory evoked potentials (Lai et al., 2007).
AR in relation to aromatase and estrogen receptor alpha expression
Compared to mammals, teleosts have exceptionally high aromatase and AR levels in brain tissue (Pasmanik and Callard, 1988; Callard et al., 1990). Testosterone has a high affinity for aromatase and therefore high levels of brain aromatase may significantly reduce brain androgen levels that may, in turn, necessitate high AR levels to activate AR signaling pathways (Pasmanik and Callard, 1988).
Table 1 compares the relative abundance of AR mRNA (this report) with aromatase mRNA/protein (Forlano et al., 2001) and ERα mRNA (Forlano et al., 2005) in midshipman fish, although we fully recognize the need for double-labeling experiments to demonstrate cellular colocalization. In general, AR and aromatase are abundantly coexpressed in forebrain nuclei in the telencephalon, preoptic area, hypothalamus, and thalamus (aromatase-positive glial cells along ventricles are adjacent to and have processes extending into CP, DPo and TPp; see fig. 2 in Forlano et al., 2001, and fig. 12 in Forlano et al., 2005). Similar to songbirds (Bernard et al., 1999; Metzdorf et al., 1999), AR expression is comparatively more widespread and found in greater abundance than ER in vocal nuclei (indicated in Table 1). ARs are also more prominent in the central auditory system (Table 1). Thus, there appears to be a greater relative abundance of “classical” androgen to estrogen receptors throughout vocal-acoustic nuclei, although the distribution of other ER subtypes have yet to be defined in midshipman.
Our previous neuroanatomical studies show that testosterone upregulates brain aromatase expression, particularly in the VMN (Forlano and Bass, 2005b; Bass and Forlano, 2008). This is consistent with abundant AR expression along the dorsal rim of the VMN (Fig. 12B) where aromatase-positive glial cell bodies are similarly abundant (see Forlano et al., 2001, for aromatase), suggesting colocalization of AR and aromatase. ARs have been identified in glial cells in mammals (for review, see DonCarlos et al., 2006; Sarkey et al., 2008), although it is not known whether these same cells also express aromatase. The results are also consistent with the hypothesis that local estrogen production in the VMN, dependent on aromatization of testosterone by glial cells, contributes to estrogen modulation of the vocal hindbrain-spinal cord (Remage-Healey and Bass, 2004, 2007). The robust expression of AR over VMN glial cells contrasts sharply with the more rostral VPP that shows AR expression over neuronal somata themselves and not in an aromatase-glial-like pattern (P. Forlano and A. Bass, unpubl. obs.). Given VPP’s important role in vocal patterning (Chagnaud et al., 2009), the VPP is a likely a specific site underlying androgen action in the vocal system (Remage-Healey et al., 2004, 2007).
Androgen receptor expression in relation to neuropeptides
While androgens and steroid hormones in general increase vocal motor output (Remage-Healey and Bass, 2004), the neuropeptides arginine vasotocin (AVT) and isotocin (IT) (homologs to mammalian vasopressin and oxytocin, respectively) inhibit evoked fictive vocalizations in midshipman (Goodson and Bass, 2000a). Due to the opposing actions of androgens and AVT/IT on vocal motor output, androgens may function, in part, to inhibit AVT/IT release (Bass and Remage-Healey, 2008). Indeed, AR is highly expressed in all vocal-related nuclei that contain AVT/IT-ir neurons (POA, Fig. 1A) and in hypothalamic vocal stimulation sites that are responsive to AVT application and contain AVT/IT-ir terminals (AT, vT; Fig. 1A) (Goodson and Bass, 2000a,b; Goodson et al., 2003).
CONCLUSIONS
It is only through the recent delineation of the functional signature of discrete neuronal populations, for example, GnRH preoptic neurons regulating gonadal size (see Harbott et al., 2007) and vocal-auditory circuits (see Introduction), that AR localization in the CNS of teleosts becomes especially salient in the context of identifying the mechanisms underlying steroid-dependent behaviors. In addition to localizing abundant AR mRNA in preoptic-hypothalamic regions (POA-AH) with ubiquitous involvement in vertebrate reproduction (Goodson and Bass, 2001; Remage-Healey and Bass, 2006b; Forlano et al., 2007; Bass and Forlano, 2008), the current study also demonstrates AR mRNA localization in vocal and auditory circuits that include the POA-AH (Goodson and Bass, 2002). Studies of vocal fish provide the opportunity to show how androgens influence vocal-acoustic networks that are conserved between fish and tetrapods (Bass et al., 2005, 2008) and include sites of AR mRNA expression (this report). Thus, seasonal breeding fish and tetrapods alike that rely on vocal-acoustic communication show an intricate link between reproductive steroid cycling, audition, and vocalization via steroid, including androgen, receptors acting in the central and peripheral nervous system.
Acknowledgments
Grant sponsor: National Institutes of Health; Grant number: DC00092 (to A.H.B.).
Abbreviations
- ac
Anterior commissure
- AT
Anterior tuberal nucleus
- C
Cerebellum
- CA
Cerebral aqueduct
- Cc
Cerebellar crest
- Cg
Granular layer of the corpus of the cerebellum
- Cm
Molecular layer of the corpus of the cerebellum
- CP
Central posterior nucleus of the thalamus
- D
Area dorsalis of the telencephalon
- Dc
Central zone of D
- Df
Diffuse nucleus of the hypothalamus
- Dl
Lateral zone of D
- Dm
Medial zone of D
- Dm-cm
Central medial division of Dm
- Dm-p
Posterior division of Dm
- DO
Descending octaval nucleus
- Dp
Posterior zone of D
- DPo
Dorsal posterior nucleus of the thalamus
- Eg
Eminentia granularis
- F
Forebrain
- G
Nucleus glomerulosus
- GC
Griseum centrale
- H
Hindbrain
- Ha
Habenula
- HC
Hair cell layer
- Hd
Dorsal periventricular hypothalamus
- HoCo
Horizontal commissure
- Hv
Ventral periventricular hypothalamus
- III
Third ventricle
- IL
Inferior lobe of the hypothalamus
- Isth/Teg
Isthmal and midbrain tegmental region
- IV
Fourth ventricle
- LH
Lateral hypothalamus
- ll
Lateral lemniscus
- M
Midbrain
- MED
Cell plate of medial octavolateralis nucleus
- MLF
Medial longitudinal fasciculus
- nMLF
Nucleus of the medial longitudinal fasciculus
- OB
Olfactory bulb
- OE
Octavolateral efferent nucleus
- oc
Occipital nerves
- ot
Optic tract
- P
Medial pretoral nucleus of the pretectum
- PAG
Periaqueductal gray
- Pe
Periventricular cell layer of the midbrain tectum
- PGl
Lateral division of nucleus preglomerulosus
- PGm
Medial division of nucleus preglomerulosus
- Pit
Pituitary
- PM
Magnocellular preoptic nucleus
- PMg
Gigantocellular division of PM
- PPa
Anterior parvocellular preoptic nucleus
- PPp
Posterior parvocellular preoptic nucleus
- PoC
Posterior commissure
- RF
Reticular formation
- SE
Sensory cell epithelium of the saccule
- SO
Secondary octaval nucleus
- SV
Saccus vasculosus
- T
Telencephalon
- TEG
Tegmentum
- TeM
Midbrain tectum
- TL
Torus longitudinalis
- TP
Posterior tuberal nucleus
- TPp
Periventricular posterior tuberal nucleus
- TS
Torus semicircularis
- v
Ventricle
- V
Area ventralis of the telencephalon
- Vc
Central nucleus of V
- Vd
Dorsal nucleus of V
- Vg
Granular layer of the valvula
- Vi
Intermediate nucleus of V
- Vl
Ventrolateral nucleus of the thalamus
- VL
Vagal lobe
- Vm
Molecular layer of the valvula
- VM
Nucleus ventromedialis
- VMN
Vocal motor nucleus
- Vp
Postcommissural nucleus of V
- VPG
Vocal pattern generator
- VPN
Vocal pacemaker neurons
- VPP
Vocal prepacemaker nucleus
- Vs
Supracommissural nucleus of V
- vT
Ventral tuberal hypothalamus
- Vv
Ventral nucleus of V
- VIII
Eighth nerve
LITERATURE CITED
- Ball GF, Riters LV, Balthazart J. Neuroendocrinology of song behavior and avian brain plasticity: multiple sites of action of sex steroid hormones. Front Neuroendocrinol. 2002;23:137–178. doi: 10.1006/frne.2002.0230. [DOI] [PubMed] [Google Scholar]
- Bass AH. Shaping brain sexuality. Am Sci. 1996;84:352–363. [Google Scholar]
- Bass AH. Steroid-dependent plasticity of vocal motor systems: novel insights from teleost fish. Brain Res Rev. 2008;57:299–308. doi: 10.1016/j.brainresrev.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Bass AH, Baker R. Sexual dimorphisms in the vocal control system of a teleost fish: morphology of physiologically identified neurons. J Neurobiol. 1990;21:1155–1168. doi: 10.1002/neu.480210802. [DOI] [PubMed] [Google Scholar]
- Bass AH, Baker R. Phenotypic specification of hindbrain rhombomeres and the origins of rhythmic circuits in vertebrates. Brain Behav Evol. 1997;50:3–16. doi: 10.1159/000113351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AH, Forlano PM. Neuroendocrine mechanisms of alternative reproductive tactics: the chemical language of social plasticity. In: Oliveira R, Taborsky M, Brockmann J, editors. Alternative reproductive reproductivetactics: an integrative approach. Cambridge University Press; Cambridge, UK: 2008. [Google Scholar]
- Bass AH, Marchaterre MA. Sound-generating (sonic) motor system in a teleost fish (Porichthys notatus): sexual polymorphisms and general synaptology of sonic motor nucleus. J Comp Neurol. 1989;286:154–169. doi: 10.1002/cne.902860203. [DOI] [PubMed] [Google Scholar]
- Bass AH, McKibben JR. Neural mechanisms and behaviors for acoustic communication in teleost fish. Prog Neurobiol. 2003;69:1–26. doi: 10.1016/s0301-0082(03)00004-2. [DOI] [PubMed] [Google Scholar]
- Bass AH, Remage-Healey L. Central pattern generators for social vocalization: androgen-dependent neurophysiological mechanisms. Horm Behav. 2008;53:659–672. doi: 10.1016/j.yhbeh.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AH, Segil N, Kelley DB. Androgen binding in the brain and electric organ of a mormyrid fish. J Comp Physiol [A] 1986;159:535–544. doi: 10.1007/BF00604173. [DOI] [PubMed] [Google Scholar]
- Bass AH, Marchaterre MA, Baker R. Vocal-acoustic pathways in a teleost fish. J Neurosci. 1994;14:4025–4039. doi: 10.1523/JNEUROSCI.14-07-04025.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AH, Horvath BJ, Brothers EB. Nonsequential developmental trajectories lead to dimorphic vocal circuitry for males with alternative reproductive tactics. J Neurobiol. 1996;30:493–504. doi: 10.1002/(SICI)1097-4695(199608)30:4<493::AID-NEU5>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Bass AH, Bodnar DA, Marchaterre MA. Midbrain acoustic circuitry in a vocalizing fish. J Comp Neurol. 2000;419:505–531. doi: 10.1002/(sici)1096-9861(20000417)419:4<505::aid-cne7>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Bass AH, Bodnar DA, Marchaterre MA. Acoustic nuclei in the medulla and midbrain of the vocalizing gulf toadfish (Opsanus beta) Brain Behav Evol. 2001;57:63–79. doi: 10.1159/000047226. [DOI] [PubMed] [Google Scholar]
- Bass AH, Rose GJ, Pritz MB. Auditory midbrain of fish, amphibians and reptiles: models systems for understanding auditory function. In: Winer JA, Schreiner CE, editors. The inferior colliculus. Springer; New York: 2005. pp. 459–492. [Google Scholar]
- Bass AH, Gilland EH, Baker R. Evolutionary origins for social vocalization in a vertebrate hindbrain-spinal compartment. Science. 2008;321:417–421. doi: 10.1126/science.1157632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard DJ, Bentley GE, Balthazart J, Turek FW, Ball GF. Androgen receptor, estrogen receptor alpha, and estrogen receptor beta show distinct patterns of expression in fore-brain song control nuclei of European starlings. Endocrinology. 1999;140:4633–4643. doi: 10.1210/endo.140.10.7024. [DOI] [PubMed] [Google Scholar]
- Braford MR., Jr. Comparative aspects of forebrain organization in the ray-finned fishes: touchstones or not? Brain Behav Evol. 1995;46:259–274. doi: 10.1159/000113278. [DOI] [PubMed] [Google Scholar]
- Braford MR, Jr, Northcutt RG. Organization of the diencephalon and pretectum of the ray-finned fishes. In: Davis RE, Northcutt RG, editors. Fish neurobiology. Vol. 2. University of Michigan Press; Ann Arbor: 1983. pp. 117–164. [Google Scholar]
- Brantley RK, Bass AH. Cholinergic neurons in the brain of a teleost fish (Porichthys notatus) located with a monoclonal antibody to choline acetyltransferase. J Comp Neurol. 1988;275:87–105. doi: 10.1002/cne.902750108. [DOI] [PubMed] [Google Scholar]
- Brantley RK, Bass AH. Alternative male spawning tactics and acoustic-signals in the plainfin midshipman fish Porichthys notatus Girard (Teleostei, Batrachoididae) Ethology. 1994;96:213–232. [Google Scholar]
- Brantley RK, Wingfield JC, Bass AH. Sex steroid levels in Porichthys notatus, a fish with alternative reproductive tactics, and a review of the hormonal bases for male dimorphism among teleost fishes. Horm Behav. 1993a;27:332–347. doi: 10.1006/hbeh.1993.1025. [DOI] [PubMed] [Google Scholar]
- Brantley RK, Marchaterre MA, Bass AH. Androgen effects on vocal muscle structure in a teleost fish with inter-sexual and intra-sexual dimorphism. J Morphol. 1993b;216:305–318. doi: 10.1002/jmor.1052160306. [DOI] [PubMed] [Google Scholar]
- Callard G, Schlinger B, Pasmanik M. Nonmammalian vertebrate models in studies of brain-steroid interactions. J Exp Zool Suppl. 1990;4:6–16. doi: 10.1002/jez.1402560404. [DOI] [PubMed] [Google Scholar]
- Cerda-Reverter JM, Ringholm A, Schioth HB, Peter RE. Molecular cloning, pharmacological characterization, and brain mapping of the melanocortin 4 receptor in the goldfish: involvement in the control of food intake. Endocrinology. 2003;144:2336–2349. doi: 10.1210/en.2002-0213. [DOI] [PubMed] [Google Scholar]
- Chagnaud B, Baker R, Bass AH. Temporal patterning of vocalizations by a hindbrain network. Abstract Viewer/Itinerary Planner. Soc Neurosci Online; Washington, DC: 2009. in press. [Google Scholar]
- Cohen MJ, Winn HE. Electrophysiological observations on hearing and sound production in the fish, Porichthys notatus. J Exp Zool. 1967;165:355–370. doi: 10.1002/jez.1401650305. [DOI] [PubMed] [Google Scholar]
- Davis RE, Morrell JI, Pfaff DW. Autoradiographic localization of sex steroid-concentrating cells in brain of teleost Macropodus opercularis (Osteichthyes Belontiidae) Gen Comp Endocrinol. 1977;33:496–505. doi: 10.1016/0016-6480(77)90108-3. [DOI] [PubMed] [Google Scholar]
- De Vries GJ, Simerly RB. Anatomy, development, and function of sexually dimorphic neural circuits in the mammalian brain. In: Pfaff DW, Arnold AP, Etgen AM, Fahrbach SE, Rubin RT, editors. Hormones, brain and behavior. Elsevier; Amsterdam: 2002. [Google Scholar]
- DonCarlos LL, Sarkey S, Lorenz B, Azcoitia I, Garcia-Ovejero D, Huppenbauer C, Garcia-Segura LM. Novel cellular phenotypes and subcellular sites for androgen action in the forebrain. Neuroscience. 2006;138:801–807. doi: 10.1016/j.neuroscience.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Douard V, Brunet F, Boussau B, Ahrens I, Vlaeminck-Guillem V, Haendler B, Laudet V, Guiguen Y. The fate of the duplicated androgen receptor in fishes: a late neofunctionalization event? BMC Evol Biol. 2008;8:336. doi: 10.1186/1471-2148-8-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine ML, Chen FA, Keefer DA. Autoradiographic localization of dihydrotestosterone and testosterone concentrating neurons in the brain of the oyster toadfish. Brain Res. 1996;709:65–80. doi: 10.1016/0006-8993(95)01275-3. [DOI] [PubMed] [Google Scholar]
- Fine ML, Johnson MS, Matt DW. Seasonal variation in androgen levels in the oyster toadfish. Copeia. 2004;2004:235–244. [Google Scholar]
- Forlano PM, Bass AH. Seasonal plasticity of brain aromatase mRNA expression in glia: Divergence across sex and vocal phenotypes. J Neurobiol. 2005a;65:37–49. doi: 10.1002/neu.20179. [DOI] [PubMed] [Google Scholar]
- Forlano PM, Bass AH. Steroid regulation of brain aromatase expression in glia: female preoptic and vocal motor nuclei. J Neurobiol. 2005b;65:50–58. doi: 10.1002/neu.20178. [DOI] [PubMed] [Google Scholar]
- Forlano PM, Cone RD. Conserved neurochemical pathways involved in hypothalamic control of energy homeostasis. J Comp Neurol. 2007;505:235–248. doi: 10.1002/cne.21447. [DOI] [PubMed] [Google Scholar]
- Forlano PM, Deitcher DL, Myers DA, Bass AH. Anatomical distribution and cellular basis for high levels of aromatase activity in the brain of teleost fish: aromatase enzyme and mRNA expression identify glia as source. J Neurosci. 2001;21:8943–8955. doi: 10.1523/JNEUROSCI.21-22-08943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlano PM, Deitcher DL, Bass AH. Distribution of estrogen receptor alpha mRNA in the brain and inner ear of a vocal fish with comparisons to sites of aromatase expression. J Comp Neurol. 2005;483:91–113. doi: 10.1002/cne.20397. [DOI] [PubMed] [Google Scholar]
- Forlano PM, Schlinger BA, Bass AH. Brain aromatase: new lessons from non-mammalian model systems. Front Neuroendocrinol. 2006;27:247–274. doi: 10.1016/j.yfrne.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Forlano PM, Remage-Healey L, Sisneros JA, Bass AH. Steroid-induced plasticity in the auditory and vocal motor system: recent studies in a teleost fish. In: Canonaco M, Facciolo RM, editors. Evolutionary molecular strategies and plasticity. Research Signpost; Kerala, India: 2007. pp. 25–50. [Google Scholar]
- Gahr M. Distribution of sex steroid hormone receptors in the avian brain: functional implications for neural sex differences and sexual behaviors. Microsc Res Tech. 2001;55:1–11. doi: 10.1002/jemt.1151. [DOI] [PubMed] [Google Scholar]
- Gahr M, Metzdorf R. Distribution and dynamics in the expression of androgen and estrogen receptors in vocal control systems of songbirds. Brain Res Bull. 1997;44:509–517. doi: 10.1016/s0361-9230(97)00233-5. [DOI] [PubMed] [Google Scholar]
- Gahr M, Wild JM. Localization of androgen receptor mRNA-containing cells in avian respiratory-vocal nuclei: an in situ hybridization study. J Neurobiol. 1997;33:865–876. [PubMed] [Google Scholar]
- Gilchriest BJ, Tipping DR, Hake L, Levy A, Baker BI. The effects of acute and chronic stresses on vasotocin gene transcripts in the brain of the rainbow trout (Oncorhynchus mykiss) J Neuroendocrinol. 2000;12:795–801. doi: 10.1046/j.1365-2826.2000.00522.x. [DOI] [PubMed] [Google Scholar]
- Goodson JL. The vertebrate social behavior network: evolutionary themes and variations. Horm Behav. 2005;48:11–22. doi: 10.1016/j.yhbeh.2005.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson JL, Bass AH. Forebrain peptides modulate sexually polymorphic vocal circuitry. Nature. 2000a;403:769–772. doi: 10.1038/35001581. [DOI] [PubMed] [Google Scholar]
- Goodson JL, Bass AH. Vasotocin innervation and modulation of vocal-acoustic circuitry in the teleost Porichthys notatus. J Comp Neurol. 2000b;422:363–379. doi: 10.1002/1096-9861(20000703)422:3<363::aid-cne4>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Goodson JL, Bass AH. Social behavior functions and related anatomical characteristics of vasotocin/vasopressin systems in vertebrates. Brain Res Rev. 2001;35:246–265. doi: 10.1016/s0165-0173(01)00043-1. 36:91–94. [DOI] [PubMed] [Google Scholar]
- Goodson JL, Bass AH. Vocal-acoustic circuitry and descending vocal pathways in teleost fish: convergence with terrestrial vertebrates reveals conserved traits. J Comp Neurol. 2002;448:298–322. doi: 10.1002/cne.10258. [DOI] [PubMed] [Google Scholar]
- Goodson JL, Evans AK, Bass AH. Putative isotocin distributions in sonic fish: relation to vasotocin and vocal-acoustic circuitry. J Comp Neurol. 2003;462:1–14. doi: 10.1002/cne.10679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelick DA, Watson W, Halpern ME. Androgen receptor gene expression in the developing and adult zebrafish brain. Dev Dyn. 2008;237:2987–2995. doi: 10.1002/dvdy.21700. [DOI] [PubMed] [Google Scholar]
- Harbott LK, Burmeister SS, White RB, Vagell M, Fernald RD. Androgen receptors in a cichlid fish, Astatotilapia burtoni: structure, localization, and expression levels. J Comp Neurol. 2007;504:57–73. doi: 10.1002/cne.21435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MS, Larsson A, Scherbak N, Olsson PE, Orban L. Zebrafish androgen receptor: isolation, molecular, and biochemical characterization. Biol Reprod. 2008;78:361–369. doi: 10.1095/biolreprod.107.062018. [DOI] [PubMed] [Google Scholar]
- Jorgensen A, Andersen O, Bjerregaard P, Rasmussen LJ. Identification and characterisation of an androgen receptor from zebrafish Danio rerio. Comp Biochem Physiol C Toxicol Pharmacol. 2007;146:561–568. doi: 10.1016/j.cbpc.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Jurgens U. Neural pathways underlying vocal control. Neurosci Biobehav Rev. 2002;26:235–258. doi: 10.1016/s0149-7634(01)00068-9. [DOI] [PubMed] [Google Scholar]
- Kapsimali M, Bourrat F, Vernier P. Distribution of the orphan nuclear receptor Nurr1 in medaka (Oryzias latipes): cues to the definition of homologous cell groups in the vertebrate brain. J Comp Neurol. 2001;431:276–292. doi: 10.1002/1096-9861(20010312)431:3<276::aid-cne1070>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Kelley DB. Auditory and vocal nuclei in the frog brain concentrate sex hormones. Science. 1980;207:553–555. doi: 10.1126/science.7352269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley DB. Neuroeffectors for vocalization in Xenopus laevis: hormonal regulation of sexual dimorphism. J Neurobiol. 1986;17:231–248. doi: 10.1002/neu.480170307. [DOI] [PubMed] [Google Scholar]
- Kim YH, Perlman WR, Arnold AP. Expression of androgen receptor mRNA in zebra finch song system: developmental regulation by estrogen. J Comp Neurol. 2004;469:535–547. doi: 10.1002/cne.11033. [DOI] [PubMed] [Google Scholar]
- Kittelberger M, Land BR, Bass AH. Midbrain periaqueductal gray and vocal pattering in a teleost fish. J Neurophysiol. 2006;96:71–85. doi: 10.1152/jn.00067.2006. [DOI] [PubMed] [Google Scholar]
- Knapp R, Wingfield JC, Bass AH. Steroid hormones and paternal care in the plainfin midshipman fish (Porichthys notatus) Horm Behav. 1999;35:81–89. doi: 10.1006/hbeh.1998.1499. [DOI] [PubMed] [Google Scholar]
- Knapp R, Marchaterre MA, Bass AH. Relationship between courtship behavior and steroid hormone levels in parental male plainfin midshipman fish. Horm Behav. 2001;39:335. [Google Scholar]
- Lai TH, Soong BW, Chen JT, Chen YY, Lai KL, Wu ZA, Liao KK. Multimodal evoked potentials of Kennedy’s disease. Can J Neurol Sci. 2007;34:328–332. doi: 10.1017/s0317167100006764. [DOI] [PubMed] [Google Scholar]
- Lee JSF, Bass AH. Differential effects of 11-ketotestosterone on dimorphic traits in a teleost with alternative male reproductive morphs. Horm Behav. 2005;47:523–531. doi: 10.1016/j.yhbeh.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Lee JSF, Bass AH. Dimorphic male midshipman fish: reduced sexual selection or sexual selection for reduced characters? Behav Ecol. 2006;17:670–675. [Google Scholar]
- Liu X, Su H, Zhu P, Zhang Y, Huang J, Lin H. Molecular cloning, characterization and expression pattern of androgen receptor in Spinibarbus denticulatus. Gen Comp Endocrinol. 2009;160:93–101. doi: 10.1016/j.ygcen.2008.10.026. [DOI] [PubMed] [Google Scholar]
- Matsunaga E, Okanoya K. Vocal area-related expression of the androgen receptor in the budgerigar (Melopsittacus undulatus) brain. Brain Res. 2008;1208:87–94. doi: 10.1016/j.brainres.2008.02.076. [DOI] [PubMed] [Google Scholar]
- Metzdorf R, Gahr M, Fusani L. Distribution of aromatase, estrogen receptor, and androgen receptor mRNA in the forebrain of songbirds and nonsongbirds. J Comp Neurol. 1999;407:115–129. [PubMed] [Google Scholar]
- Modesto T, Canario AV. Morphometric changes and sex steroid levels during the annual reproductive cycle of the Lusitanian toadfish, Halobatrachus didactylus. Gen Comp Endocrinol. 2003;131:220–231. doi: 10.1016/s0016-6480(03)00027-3. [DOI] [PubMed] [Google Scholar]
- Morrell JI, Kelley DB, Pfaff DW. Sex steroid binding in the brains of vertebrates. In: Knigge KM, Scott DE, Kobayashi H, Miura-shi S, editors. Brain-endocrine interaction II. The ventricular system. S. Karger; Basel: 1975. pp. 230–256. [Google Scholar]
- Nastiuk KL, Clayton DF. The canary androgen receptor mRNA is localized in the song control nuclei of the brain and is rapidly regulated by testosterone. J Neurobiol. 1995;26:213–224. doi: 10.1002/neu.480260206. [DOI] [PubMed] [Google Scholar]
- Nelson JS. Fishes of the world. John Wiley & Sons; New York: 2006. [Google Scholar]
- Northcutt RG. The forebrain of gnathostomes: in search of a morphotype. Brain Behav Evol. 1995;46:275–318. doi: 10.1159/000113279. [DOI] [PubMed] [Google Scholar]
- Northcutt RG. Connections of the lateral and medial divisions of the goldfish telencephalic pallium. J Comp Neurol. 2006;494:903–943. doi: 10.1002/cne.20853. [DOI] [PubMed] [Google Scholar]
- Northcutt RG, Braford MR., Jr. New observations on the organization and evolution of the telencephalon of actinopterygian fishes. In: Ebbesson SOE, editor. Comparative neurology of the telencephalon. Plenum; New York: 1980. pp. 41–98. [Google Scholar]
- Olsson PE, Berg AH, von Hofsten J, Grahn B, Hellqvist A, Larsson A, Karlsson J, Modig C, Borg B, Thomas P. Molecular cloning and characterization of a nuclear androgen receptor activated by 11-ketotestosterone. Reprod Biol Endocrin. 2005;3:37–54. doi: 10.1186/1477-7827-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzolo I, Gliozzi A, Rusmini P, Sau D, Crippa V, Simonini F, Onesto E, Bolzoni E, Poletti A. The role of the polyglutamine tract in androgen receptor. J Steroid Biochem Mol Biol. 2008;108:245–253. doi: 10.1016/j.jsbmb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Pasmanik M, Callard GV. A high abundance androgen receptor in goldfish brain: characteristics and seasonal changes. Endocrinology. 1988;123:1162–1171. doi: 10.1210/endo-123-2-1162. [DOI] [PubMed] [Google Scholar]
- Perez J, Cohen MA, Kelley DB. Androgen receptor mRNA expression in Xenopus laevis CNS: sexual dimorphism and regulation in laryngeal motor nucleus. J Neurobiol. 1996;30:556–568. doi: 10.1002/(SICI)1097-4695(199608)30:4<556::AID-NEU10>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Remage-Healey L, Bass AH. Rapid, hierarchical modulation of vocal patterning by steroid hormones. J Neurosci. 2004;24:5892–5900. doi: 10.1523/JNEUROSCI.1220-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remage-Healey L, Bass AH. Rapid elevations in both steroid hormones and vocal signaling during playback challenge: a field experiment in Gulf toadfish. Horm Behav. 2005;47:297–305. doi: 10.1016/j.yhbeh.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Remage-Healey L, Bass AH. A rapid neuromodulatory role for steroid hormones in the control of reproductive behavior. Brain Res. 2006a;1126:27–35. doi: 10.1016/j.brainres.2006.06.049. [DOI] [PubMed] [Google Scholar]
- Remage-Healey L, Bass AH. From social behavior to neural circuitry: steroid hormones rapidly modulate advertisement calling via a vocal pattern generator. Horm Behav. 2006b;50:432–441. doi: 10.1016/j.yhbeh.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Remage-Healey L, Bass AH. Plasticity in brain sexuality is revealed by the rapid actions of steroid hormones. J Neurosci. 2007;27:1114–1122. doi: 10.1523/JNEUROSCI.4282-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkey S, Azcoitia I, Garcia-Segura LM, Garcia-Ovejero D, Don-Carlos LL. Classical androgen receptors in nonclassical sites in the brain. Horm Behav. 2008;53:753–764. doi: 10.1016/j.yhbeh.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlinger BA, Brenowitz EA. Neural and hormonal control of birdsong. In: Pfaff DW, Arnold AP, Etgen AM, Fahrbach SE, Moss R, Rubin R, editors. Hormones, brain, and behavior. Academic Press; New York: 2002. pp. 799–839. [Google Scholar]
- Shaw BK, Kennedy GG. Evidence for species differences in the pattern of androgen receptor distribution in relation to species differences in an androgen-dependent behavior. J Neurobiol. 2002;52:203–220. doi: 10.1002/neu.10079. [DOI] [PubMed] [Google Scholar]
- Simerly RB, Chang C, Muramatsu M, Swanson LW. Distribution of androgen and estrogen receptor messenger RNA-containing cells in the rat brain — an in situ hybridization study. J Comp Neurol. 1990;294:76–95. doi: 10.1002/cne.902940107. [DOI] [PubMed] [Google Scholar]
- Sisneros JA, Bass AH. Seasonal plasticity of peripheral auditory frequency sensitivity. J Neurosci. 2003;23:1049–1058. doi: 10.1523/JNEUROSCI.23-03-01049.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisneros JA, Forlano PM, Knapp R, Bass AH. Seasonal variation of steroid hormone levels in an intertidal-nesting fish, the vocal plainfin midshipman. Gen Comp Endocrinol. 2004a;136:101–116. doi: 10.1016/j.ygcen.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Sisneros JA, Forlano PM, Deitcher DL, Bass AH. Steroid-dependent auditory plasticity leads to adaptive coupling of sender and receiver. Science. 2004b;305:404–407. doi: 10.1126/science.1097218. [DOI] [PubMed] [Google Scholar]
- Sperry TS, Thomas P. Characterization of two nuclear androgen receptors in Atlantic croaker: comparison of their biochemical properties and binding specificities. Endocrinology. 1999;140:1602–1611. doi: 10.1210/endo.140.4.6631. [DOI] [PubMed] [Google Scholar]
- Sperry TS, Thomas P. Androgen binding profiles of two distinct nuclear androgen receptors in Atlantic croaker (Micropogonias undulatus) J Steroid Biochem. 2000;73:93–103. doi: 10.1016/s0960-0760(00)00069-8. [DOI] [PubMed] [Google Scholar]
- Tang YZ, Piao YS, Zhuang LZ, Wang ZW. Expression of androgen receptor mRNA in the brain of Gekko gecko: implications for understanding the role of androgens in controlling auditory and vocal processes. J Comp Neurol. 2001;438:136–147. doi: 10.1002/cne.1305. [DOI] [PubMed] [Google Scholar]
- Weeg MS, Bass AH. Central lateral line pathways in a vocalizing fish. J Comp Neurol. 2000;418:41–64. [PubMed] [Google Scholar]
- Weeg MS, Bass AH. Frequency response properties of lateral line superficial neuromasts in a vocal fish, with evidence for acoustic sensitivity. J Neurophysiol. 2002;88:1252–1262. doi: 10.1152/jn.2002.88.3.1252. [DOI] [PubMed] [Google Scholar]
- Wells K, Van Der Kraak G. Differential binding of endogenous steroids and chemicals to androgen receptors in rainbow trout and goldfish. Environ Toxicol Chem. 2000;19:2059–2065. [Google Scholar]
- Wullimann MF, Mueller T. Teleostean and mammalian forebrains contrasted: Evidence from genes to behavior. J Comp Neurol. 2004;475:143–162. doi: 10.1002/cne.20183. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Ito H. Fiber connections of the anterior preglomerular nucleus in cyprinids with notes on telencephalic connections of the preglomerular complex. J Comp Neurol. 2005;491:212–233. doi: 10.1002/cne.20681. [DOI] [PubMed] [Google Scholar]
- Yamamoto N, Ishikawa Y, Yoshimoto M, Xue HG, Bahaxar N, Sawai N, Yang CY, Ozawa H, Ito H. A new interpretation on the homology of the teleostean telencephalon based on hodology and a new eversion model. Brain Behav Evol. 2007;69:96–104. doi: 10.1159/000095198. [DOI] [PubMed] [Google Scholar]