Abstract
BACKGROUND
Ovarian clear-cell and endometrioid carcinomas may arise from endometriosis, but the molecular events involved in this transformation have not been described.
METHODS
We sequenced the whole transcriptomes of 18 ovarian clear-cell carcinomas and 1 ovarian clear-cell carcinoma cell line and found somatic mutations in ARID1A (the AT-rich interactive domain 1A [SWI-like] gene) in 6 of the samples. ARID1A encodes BAF250a, a key component of the SWI–SNF chromatin remodeling complex. We sequenced ARID1A in an additional 210 ovarian carcinomas and a second ovarian clear-cell carcinoma cell line and measured BAF250a expression by means of immunohistochemical analysis in an additional 455 ovarian carcinomas.
RESULTS
ARID1A mutations were seen in 55 of 119 ovarian clear-cell carcinomas (46%), 10 of 33 endometrioid carcinomas (30%), and none of the 76 high-grade serous ovarian carcinomas. Seventeen carcinomas had two somatic mutations each. Loss of the BAF250a protein correlated strongly with the ovarian clear-cell carcinoma and endometrioid carcinoma subtypes and the presence of ARID1A mutations. In two patients, ARID1A mutations and loss of BAF250a expression were evident in the tumor and contiguous atypical endometriosis but not in distant endometriotic lesions.
CONCLUSIONS
These data implicate ARID1A as a tumor-suppressor gene frequently disrupted in ovarian clear-cell and endometrioid carcinomas. Since ARID1A mutation and loss of BAF250a can be seen in the preneoplastic lesions, we speculate that this is an early event in the transformation of endometriosis into cancer. (Funded by the British Columbia Cancer Foundation and the Vancouver General Hospital–University of British Columbia Hospital Foundation.)
In the united states, ovarian cancer ranks as the fifth deadliest cancer among women.1 Of the several subtypes of epithelial ovarian cancer, high-grade serous carcinomas are the most common, accounting for approximately 70% of all cases of epithelial ovarian cancer in North America.2 Although ovarian clear-cell carcinoma is the second most common subtype in North America (accounting for 12% of cases and an even higher percentage in Japan3) and is the second leading cause of death from ovarian cancer,2 it is relatively understudied. Ovarian clear-cell carcinoma is defined on the basis of histopathological findings, including a predominance of clear cells and “hobnail” cells.4 Ovarian clear-cell carcinomas have a low mitotic rate,5,6 are genetically stable, and do not exhibit the complex karyotypes or chromosomal instability of high-grade serous carcinomas5,7–9 that may contribute to their lack of sensitivity to platinum-based chemotherapy.10–12 Although ovarian clear-cell carcinoma does not respond well to conventional platinum–taxane chemotherapy for ovarian carcinoma, this remains the adjuvant treatment of choice, because effective alternatives have not been identified. Both ovarian clear-cell and endometrioid carcinomas are associated with endometriosis.13,14 The genetic events associated with the transformation of endometriosis into ovarian clear-cell carcinoma and endometrioid carcinoma are unknown.
The SWI–SNF chromatin remodeling complex, present in all eukaryotes, is involved in the regulation of many cellular processes, including development, differentiation, proliferation, DNA repair, and tumor suppression.15 The complex uses ATP to mobilize nucleosomes, thereby modulating the accessibility of promoters to transcriptional activation or repression. BAF250a, the protein encoded by ARID1A (the AT-rich interactive domain 1A [SWI-like] gene),16,17 is one of the accessory subunits of the SWI–SNF complex believed to confer specificity in regulation of gene expression. Mutations or other aberrations in ARID1A have not been described in ovarian carcinomas; however, an ARID1A rearrangement has been found in a breast-cancer cell line, an ARID1A deletion has been identified in a lung-cancer cell line, and it has been suggested that ARID1A is a tumor-suppressor gene.18
We used data derived from the whole-transcriptome sequencing (RNA sequencing) of 18 ovarian clear-cell carcinomas and an ovarian clear-cell carcinoma cell line to identify variants in ARID1A, as previously described.19,20 We then studied this gene in a larger cohort of patients with ovarian carcinoma and associated endometriosis. The results suggest that ARID1A is a tumor suppressor in ovarian clear-cell and endometrioid carcinomas.
METHODS
PATIENTS AND SAMPLES
Ovarian clear-cell carcinomas from 18 patients, obtained from the OvCaRe (Ovarian Cancer Research) frozen-tumor bank, and 1 ovarian clear-cell carcinoma–derived cell line (TOV21G)21 were selected as the discovery cohort for RNA sequencing. Approval from the hospital’s institutional review board was obtained to permit the use of these samples for RNA-sequencing experiments.
To determine the frequency of ARID1A mutations in ovarian clear-cell carcinoma and other subtypes of ovarian cancer, we performed targeted exon resequencing in the discovery cohort, as well as in a mutation-validation cohort, consisting of 210 samples: samples of ovarian clear-cell carcinoma from 101 patients (independent of the 19 samples used in RNA sequencing for the discovery cohort), samples of endometrioid carcinoma from 33 patients, samples of high-grade serous carcinoma from 76 patients, and the ovarian clear-cell carcinoma–derived cell line ES2.22 Ten specimens of ovarian clear-cell carcinoma came from Johns Hopkins University, 29 from the Centre Hospitalier de l’Université de Montréal–Hôpital Notre-Dame, and 42 from the Australian Ovarian Cancer Study (AOCS); all others were obtained from the OvCaRe frozen-tumor bank. All patients from both the discovery and mutation-validation cohorts provided written informed consent to have their tumors and germ-line DNA used for research, including genomic studies. Details regarding the consents and other approvals by the institutional review boards are supplied in the Supplementary Appendix (available with the full text of this article at NEJM.org).
PATHOLOGICAL REVIEW
All tumor samples were reviewed independently by a gynecologic pathologist before mutational analysis was performed. In cases in which the review diagnosis differed from the diagnosis at the source institution, the samples were further reviewed by another gynecologic pathologist, who acted as an arbiter. Both review pathologists were unaware of the results of the genomic studies.
PAIRED-END RNA SEQUENCING AND ANALYSIS
RNA sequencing and analysis were performed as previously described.19,20 For details, see the Methods section in the Supplementary Appendix.
TARGETED EXON RESEQUENCING OF ARID1A AND MUTATION VALIDATION
Genomic DNA from samples in both the discovery and mutation-validation cohorts were subjected to targeted exon resequencing. Selected ARID1A variants (those with truncating changes or radical missense mutations23 with an allele frequency of >10%) detected by means of exon resequencing were validated in tumor DNA by means of Sanger sequencing. In most cases, germ-line DNA (from formalin-fixed paraffin-embedded sections, blood samples, or cultured fibroblasts) was also analyzed by means of Sanger sequencing (see Table 3 in the Supplementary Appendix). Full details are provided in the Supplementary Appendix.
IMMUNOHISTOCHEMICAL ANALYSIS OF BAF250A
Immunohistochemical staining for BAF250a was performed in all samples except 42 ovarian clear-cell carcinoma specimens from the AOCS and 4 from Johns Hopkins University. A total of 455 additional ovarian-carcinoma samples — including 132 ovarian clear-cell carcinomas, 125 endometrioid carcinomas, and 198 high-grade serous carcinomas — from a previously described tissue microarray6 were used for an immunohistochemical validation cohort and were analyzed for BAF250a expression. All normal gynecologic tissues showed moderate or intense nuclear immunoreactivity for BAF250a. Tumors were scored positive for BAF250a if tumor cells showed definite nuclear staining and negative if tumor nuclei had no immunoreactivity but endothelial and other nontumor cells from the same samples showed immunoreactivity. Cases in which neither normal cells in the stroma nor tumor cells were immunoreactive were considered to be the result of technical failure. Details of the staining protocol are provided in the Supplementary Appendix. Additional immunohistochemical staining for hepatocyte nuclear factor 1β (HNF-1β) and estrogen receptor was performed on whole sections for two tumors with contiguous atypical endometriosis, as previously described.24
LASER-CAPTURE MICRODISSECTION, DNA ISOLATION, AND CLONING
In two tumors with identified ARID1A mutations, sections of atypical (contiguous) and distant endometriosis were identified by a gynecologic pathologist. Laser-capture microdissection was used to isolate endometriotic epithelium. DNA extracted from these cells was analyzed by means of sequencing for the mutations seen in the tumor (see the Supplementary Appendix).
RESULTS
ARID1A MUTATIONS
The RNA-sequencing data, including the number of mapped sequencing reads and potential non-synonymous sequence variants, are summarized in Table 1 in the Supplementary Appendix. RNA sequencing of the 19 samples in the discovery cohort resulted in the detection of the following nucleotide mutations (and corresponding amino acid mutations) (also shown in Table 1 and Fig. 1): three somatic nonsense mutations — C4201T (Q1401*), C5164T (R1722*), and C1680A (Y560*) (stars denote a stop codon); two somatic indels (insertion–deletion) — 6018-6020delGCT and 5541insG; one somatic missense mutation — T5953C (S1989P) (found in the same sample as the 5541insG mutation); and one gene rearrangement involving ARID1A and the neighboring gene ZDHHC18 (encoding the zinc-finger DHHC domain-containing protein 18). The fusion ends of this rearrangement map to a homozygous deletion involving most of the ARID1A gene (Fig. 1 in the Supplementary Appendix).
Table 1.
Results of RNA Sequencing and Exon Resequencing of the Discovery Cohort of 19 Specimens of Ovarian Clear-Cell Carcinoma (Including a Cell Line).*
| Sample No. | ARID1A Mutation | Exon Resequencing | RNA Sequencing | Immunohistochemical Test for BAF250a Expression | Predicted ARID1A Status | Other Somatic Mutations in Oncogenes or Tumor Suppressors | |
|---|---|---|---|---|---|---|---|
| no. of reads containing mutation/total no. of reads at mutation position (%) | |||||||
| CCC01 | 6018-6020delGCT | 223/1529 (15) | 10/40 (25) | Positive | Heterozygous† | ||
| CCC02 | 404delC‡ | Not applicable§ | No coverage | Negative | Mutation with loss of heterozygosity¶ | CTNNB1 C110G (S37C) | |
| CCC03 | 5518delG‡ | 395/1725 (23) | 1/2 (50) | Positive | Heterozygous† | ||
| CCC04 | Deletion and rearrangement | Not applicable | 6/6 (100) | Negative | Deletion and rearrangement (homozygous) | ||
| CCC06 | C4201T (Q1401*) | 100/914 (11) | 8/26 (31) | Positive | Heterozygous† | ||
| CCC09 | C5164T (R1722*) | 1132/1513 (75) | 30/30 (100) | Negative | Mutation with loss of heterozygosity | ||
| CCC10 | 3948delG‡ | 166/758 (22) | No coverage | Negative | Heterozygous† | ||
| CCC13 | 5541insG | 395/1518 (26) | 23/97 (24) | Negative | Heterozygous|| | CTNNB1 C110G (S37C) | |
| CCC13 | T5953C (S1985P) | 339/1093 (31) | 25/60 (42) | Negative | Heterozygous|| | CTNNB1 C110G (S37C) | |
| CCC14 | C1680A (Y560*) | 1411/2651 (53) | 3/6 (50) | Negative | Heterozygous | ||
| CCC05 | None | Positive | KRAS G38A (G13D) | ||||
| CCC67 | None | Positive | |||||
| CCC68 | None | Positive | |||||
| CCC66 | None | Positive | |||||
| CCC69 | None | Positive | |||||
| CCC70 | None | Negative | |||||
| CCC71 | None | Positive | |||||
| CCC72 | None | Positive | |||||
| CCC73 | None | Positive | |||||
| TOV21G cell line | 1645insC‡ | 484/1821 (27) | 5/34 (15) | Negative | Heterozygous† | PIK3CA C3139T (H1047Y), KRAS G37T (G13C) | |
The mutations listed are nucleotide mutations, followed by amino acid mutations (if known) in parentheses, with a star indicating a stop codon. CTNNB1 denotes the catenin beta-1 gene, KRAS the v-Ki-ras2 Kirsten rat sarcoma viral oncogene homologue gene, and PIK3CA the phosphoinositide-3-kinase, catalytic, alpha polypeptide gene (Entrez Gene record numbers NM_001904.3, NM_004985.3, and NM_006218.2, respectively).
The percentage of reads containing a mutation in samples CCC01, CCC03, CCC06, and CCC10 and the TOV21G cell line suggests some form of allelic imbalance, with extra copies of the wild-type allele.
The ARID1A mutations in samples CCC02, CCC03, CCC10, and the TOV21G cell line were not initially identified or discovered through RNA sequencing.
For sample CCC02, no reads were available from the exon resequencing of exon 1, but Sanger sequencing showed a dominant peak from the mutation.
For sample CCC02, the predicted ARID1A status is based on microarray data (Affymetrix SNP 6.0).
For sample CCC13, the two somatic mutations in ARID1A can be found both in the trans configuration (on independent alleles) and in the cis configuration (on the same alleles). For details, see Figure 4 in the Supplementary Appendix.
Figure 1. Mutations Found in ARID1A and the BAF250a Protein It Encodes.
The 20 exons of ARID1A are represented (as numbered gray boxes) above a schematic of the BAF250a protein (the blue segment, with the ARID [AT-rich interactive domain] DNA-binding domain in pink, the HIC1 [hypermethylated in cancer 1] binding domain in green, and the three C-terminal leucine-rich LXXLL motifs that facilitate interaction with gluco-corticoid receptor in yellow). The nucleotide mutations (with corresponding amino acid mutations in parentheses) listed above the schematic are those identified by means of transcriptome sequencing (RNA sequencing) of the 18 samples of ovarian clear-cell carcinoma and the TOV21G cell line in the discovery cohort, and those listed below the schematic were identified in subsequent validation efforts with the use of targeted exon resequencing and Sanger sequencing of genomic DNA from the 210 ovarian-cancer samples in the mutation-validation cohort. All unique somatic mutations detected in samples of ovarian clear-cell carcinoma, endometrioid carcinoma, and high-grade serous carcinoma are shown. Numbers 1 through 6858 below the schematic indicate the nucleotide (nt) position, starting with the A in the ATG start codon for ARID1A in position 1 (based on the sequence given in record number NM_006015.4 in Entrez Gene; also see Table 1 in the Supplementary Appendix). UTR denotes untranslated region.
All predicted variants of ARID1A were validated with the use of Sanger sequencing of DNA from the source tumors, except for the deletion–rearrangement, which was validated with the use of microarray data (Affymetrix SNP 6.0) (Table 3 in the Supplementary Appendix). The finding of multiple types of mutations in a single gene, ARID1A, in ovarian clear-cell carcinoma led us to further explore ARID1A in this cancer type. Since mutations in PIK3CA (the phosphoinositide-3-kinase, catalytic, alpha polypeptide gene), CTNNB1 (the catenin beta-1 gene), KRAS (the v-Ki-ras2 Kirsten rat sarcoma viral oncogene homologue gene), and TP53 (the tumor protein p53 gene) are recurrent in ovarian clear-cell carcinoma,25 we analyzed the RNA-sequencing data and performed a polymerase-chain-reaction assay for the presence of variants in these genes (Table 1). Whole-transcriptome sequence data for the 19 samples of the discovery cohort have been deposited at the European Genome–Phenome Archive (accession number, EGAS00000000075).
ARID1A mutation frequency in ovarian clear-cell carcinomas and other ovarian-cancer subtypes was established through targeted exon resequencing of the mutation-validation cohort of 210 samples of various subtypes of ovarian carcinomas and 1 ovarian clear-cell carcinoma cell line, along with the original discovery cohort of 18 samples of ovarian clear-cell carcinoma and 1 ovarian clear-cell carcinoma cell line. ARID1A mutations were identified in 55 of 119 (46%) ovarian clear-cell carcinomas, 10 of the 33 (30%) endometrioid carcinomas, and none of the 76 high-grade serous carcinomas (Table 2, and Table 3 in the Supplementary Appendix). A total of 17 samples (12 of ovarian clear-cell carcinoma and 5 of endometrioid carcinoma) each had two validated ARID1A mutations. In addition, the ovarian clear-cell carcinoma cell line TOV21G had a truncating mutation in ARID1A (1645insC).
Table 2.
Mutational Status in the Discovery and Mutation-Validation Cohorts (Excluding the Two Cell Lines), According to Carcinoma Type.
| Mutational Status | Ovarian Clear-Cell Carcinoma | Endometrioid Carcinoma | High-Grade Serous Carcinoma | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Immunohistochemical Test for BAF250a Expression | Total | Immunohistochemical Test for BAF250a Expression | Total | Immunohistochemical Test for BAF250a Expression | |||||||
| negative | positive | not available | negative | positive | not available | negative | positive | not available | ||||
| ARID1A mutation | ||||||||||||
| One somatic nonsense or indel mutation | 41 | 19 | 9 | 13 | 5 | 1 | 4 | 0 | 0 | 0 | 0 | 0 |
| Two somatic nonsense or indel mutations | 10 | 5 | 0 | 5 | 4 | 4 | 0 | 0 | 0 | 0 | 0 | 0 |
| One somatic nonsense or indel mutation, one missense mutation | 2 | 2 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| One missense mutation | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Other mutation (deletion and rearrangement) | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 55 | 27 | 10 | 18 | 10 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| Normal ARID1A | 64 | 4 | 32 | 28 | 23 | 2 | 21 | 0 | 76 | 1 | 75 | 0 |
| Overall total | 119 | 31 | 42 | 46 | 33 | 7 | 26 | 0 | 76 | 1 | 75 | 0 |
We analyzed germ-line DNA from 55 samples (47 ovarian clear-cell carcinomas and 8 endometrioid carcinomas) in the discovery and mutation- validation cohorts for the presence of 65 truncating mutations (53 found in ovarian clear-cell carcinomas and 12 found in endometrioid carcinomas). In all 55, the mutations were found to be somatic. On this basis, we made the assumption that 12 subsequent truncating mutations (10 in ovarian clear-cell carcinoma and 2 in endometrioid carcinoma) would be somatic (i.e., predicted to be somatic without germ-line DNA testing) (Table 3 in the Supplementary Appendix). The presence of ARID1A mutations showed a strong association (P<0.001 by Fisher’s exact test) with the two ovarian-cancer subtypes associated with endometriosis (ovarian clear-cell carcinoma and endometrioid carcinoma).
BAF250A PROTEIN EXPRESSION
The correlation between ARID1A mutations and BAF250a expression was evaluated by means of immunohistochemical staining for BAF250a in 182 tumors for which formalin-fixed, paraffin-embedded sections were available in the discovery cohort and the mutation-validation cohort: 73 ovarian clear-cell carcinomas, 33 endometrioid carcinomas, and 76 high-grade serous carcinomas. The presence of mutations was significantly associated with BAF250a loss in endometriosis-associated cancers (P<0.001 by Fisher’s exact test). A total of 27 of 37 samples (73%) and 5 of 10 samples (50%) of ovarian clear-cell carcinoma and endometrioid carcinoma, respectively, with an ARID1A mutation showed a loss of BAF250a expression, as compared with 4 of 36 samples (11%) and 2 of 23 samples (9%), respectively, without an ARID1A mutation (Fig. 2A and Table 2). Loss of BAF250a expression was strongly associated with the endometriosis-related ovarian cancers — with 31 of 73 samples (42%) of ovarian clear-cell carcinoma and 7 of 33 samples (21%) of endometrioid carcinoma showing a loss of expression — as compared with high-grade serous carcinomas, for which 1 of the 76 samples (1%) had loss of expression (P<0.001 by Fisher’s exact test) (Fig. 2B). ARID1A mutations were not significantly associated with the presence of endometriosis in 86 ovarian clear-cell carcinomas and 33 endometrioid carcinomas (Table 5 in the Supplementary Appendix).
Figure 2. Results of Immunohistochemical Analyses of BAF250a Expression.
The percentages of tumors (with number and total number in parentheses) from three subtypes of ovarian cancer — clear-cell carcinoma (CCC), endometrioid carcinoma (EC), and high-grade serous (HGS) carcinoma — from the discovery and mutation-validation cohorts that showed loss of BAF250a expression are shown in Panel A for samples with and samples without ARID1A mutations and in Panel B for samples in the discovery and mutation-validation cohorts and samples in the immunohistochemical validation cohort. The rate of BAF250a loss was higher among CCC specimens with an ARID1A mutation than among those without an ARID1A mutation (P<0.001); the same was true for EC specimens (P = 0.02). The loss of expression was also consistently more common in CCC and EC (the two endometriosis-associated carcinomas) than in HGS carcinoma when assessed in the discovery and mutation-validation cohorts and again in the immunohistochemical validation cohort (Panel B), with P<0.001 for all comparisons. All P values were calculated with the use of Fisher’s exact test.
The immunohistochemical validation cohort was also assessed for BAF250a expression (Fig. 2B). This analysis revealed that 55 of the 132 samples (42%) of ovarian clear-cell carcinoma, 39 of the 125 samples (31%) of endometrioid carcinoma, and 12 of the 198 samples (6%) of high-grade serous carcinoma lacked BAF250a expression. These findings are in agreement with the proportions observed in the discovery and mutation-validation cohorts. No significant associations with absence of BAF250a expression were noted on the basis of age of presentation, stage of disease (low or high), or disease-specific survival within any of the cancer subtypes, as assessed by means of Welch’s analysis of variance, Fisher’s exact test, and the log-rank statistic, respectively (P>0.05 for all analyses).
ANALYSIS OF ARID1A IN ENDOMETRIOSIS ASSOCIATED WITH OVARIAN CANCER
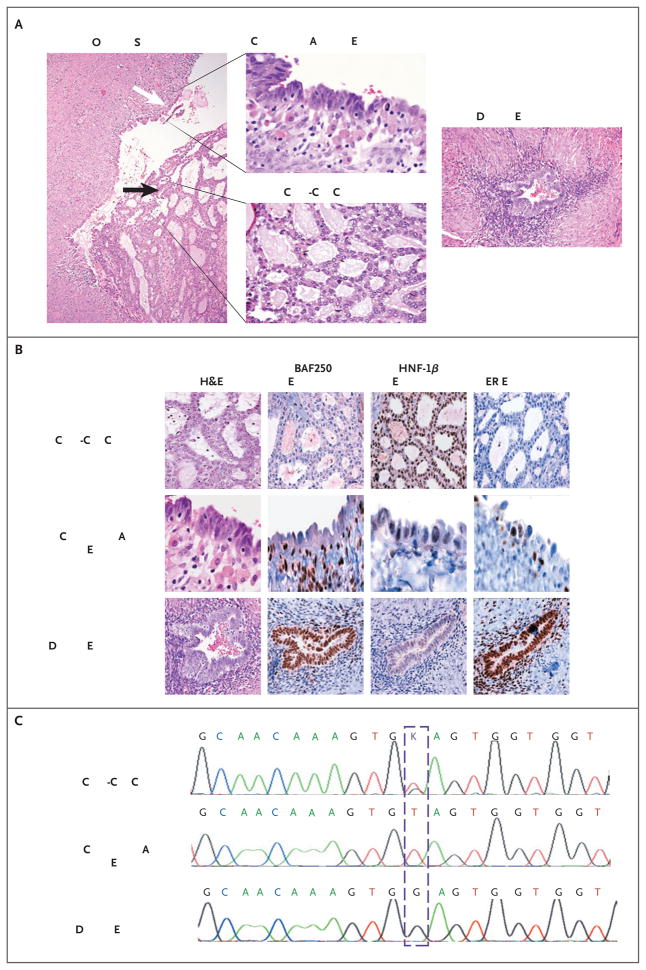
Two patients with ovarian clear-cell carcinomas (samples CCC13 and CCC23) carrying ARID1A mutations had contiguous atypical endometriosis (Fig. 3, and Fig. 3 in the Supplementary Appendix). For one of the two patients, the specimen was heterozygous for an ARID1A truncating mutation (G6139T [E2047*]) in exon 20. This mutation was also found in 17 of 42 clones derived from atypical endometriosis but in none of 52 clones from a distant endometriotic lesion (P<0.001 by Fisher’s exact test) (Fig. 3C). Epithelial samples of both the ovarian clear-cell carcinoma and atypical endometriosis had loss of BAF250a expression, whereas expression was maintained in the distant endometriotic lesion (Fig. 3B). HNF-1β was expressed in the ovarian clear-cell carcinoma but not in the contiguous atypical or distant endometriosis, and estrogen receptor was expressed in both the contiguous and distant endometriosis but not in the ovarian clear-cell carcinoma, as was expected.24 Thus, atypical endometrium could be distinguished from the distant endometrium only on the basis of loss of BAF250a expression, which correlated with the presence of an ARID1A mutation.
Figure 3. Analysis of Ovarian Clear-Cell arcinoma and Associated Endometriosis in a Study Patient.
Panel A shows a section (hematoxylin and eosin [H&E]) on which a clear-cell carcinoma (black arrow) has arisen in an endometriotic cyst (white arrow). The same section, viewed at a higher magnification, shows regions of the clear-cell carcinoma and contiguous atypical endometriosis. A region of distant endometriosis from the same patient is also shown. Panel B shows the results of immunohistochemical staining of the epithelial portions of tissue specimens shown in Panel A for expression of BAF250a, hepatocyte nuclear factor 1β (HNF-1β), and estrogen receptor (ER). BAF250a immunoreactivity is lost in both the clear-cell carcinoma and the contiguous atypical endometriosis but is maintained in the distant endometriosis. Both regions of endometriosis differ from the carcinoma in their lack of HNF-1β expression (with weak expression in the contiguous atypical endometriosis) and maintenance of estrogen-receptor expression. Panel C shows sequencing chromatograms for the clear-cell carcinoma and polymerase-chain-reaction (PCR) clones of microdissected material from the contiguous atypical endometriosis and distant endometriosis, from which DNA was extracted. The carcinoma and contiguous atypical endometriosis show nucleotide variation corresponding to G6139T (as indicated with the dashed box); the tumor shows a heterozygous peak at that location, whereas the atypical endometriosis is homozygous for the substitution (in 17 of 42 clones). In contrast, the distant endometriosis shows wild-type sequence (in all 52 clones analyzed). None of the PCR clones from the distant endometriosis showed variation from the wild-type sequence.
For the other patient, the sample of ovarian clear-cell carcinoma had two somatic mutations in ARID1A (and loss of BAF250a expression): both these mutations, along with a CTNNB1 missense mutation, were present in the tumor and the adjacent atypical endometriosis but not in a distant endometriotic lesion (Fig. 3B in the Supplementary Appendix).
DISCUSSION
Overall, 46% of patients with ovarian clear-cell carcinoma and 30% of those with endometrioid carcinoma had somatic truncating or missense mutations in ARID1A; no ARID1A mutations were found in any of the 76 specimens of high-grade serous carcinoma analyzed. Loss of ARID1A expression was also specific to the subtype of ovarian cancer, with loss of nuclear BAF250a expression seen in 36% of ovarian clear-cell carcinomas and endometrioid carcinomas, but only 1% of high-grade serous carcinomas. Our initial mutation-screening assays involving RNA sequencing in the discovery cohort identified seven somatic mutations in ARID1A in the 19 samples; four additional mutations were subsequently identified when these samples were analyzed by means of amplicon-exon resequencing. Most likely, the additional mutations had not been seen in the RNA-sequencing data owing to their transcripts being rapidly targeted for nonsense-mediated decay26 or the inherently decreased sensitivity of the assay to mutations at the 5 end of transcripts. Thus, although RNA sequencing is a useful tool for discovery, targeted exon resequencing may be more appropriate for the determination of true mutation frequency.
ARID1A is located at 1p36.11.27 This chromosomal region is commonly deleted in tumors, and it has been suggested that deletion regions encompassing 1p36 could contain tumor-suppressor genes.28,29 Rearrangements and deletions in ARID1A have been identified in a primary breast-cancer cell line and a lung-cancer cell line, respectively,18 and the loss of BAF250a has also been observed in cervical- and breast-carcinoma cell lines.30 In a study by Wang and colleagues,31 the screening of 241 tumors revealed that ARID1A transcript levels are decreased in approximately 6% of cancers in general and in 30% of renal carcinomas and 10% of breast carcinomas, specifically; however, none of the 14 ovarian cancers showed loss of expression, probably because they were predominantly the high-grade serous subtype.
The ARID1A mutations identified in our study were mostly truncating mutations, which were evenly distributed across the gene. The presence of mutations is strongly correlated with the loss of BAF250a protein (Table 2 and Fig. 2A). Loss of BAF250a expression was seen in 73% and 50% of samples of ovarian clear-cell carcinoma and endometrioid carcinoma with an ARID1A mutation, respectively, and in only 11% and 9% of samples without an ARID1A mutation, respectively. Seventeen of the mutation-positive samples had two ARID1A mutations; in all but one of the specimens with two mutations for which immunohistochemical data were available, BA-F250a expression was not detected. That single exception (an endometrioid carcinoma) had both a C-terminal truncating mutation and a mis-sense mutation; either of these changes could produce a detectable protein. A single sample of ovarian clear-cell carcinoma had ARID1A loss and rearrangement resulting in the homozygous deletion of the gene. Three other cases of ovarian clear-cell carcinoma also appear to be characterized by loss of heterozygosity, on the basis of the frequency of mutant alleles and wild-type alleles (Table 3 in the Supplementary Appendix) and subsequent loss of BAF250a expression. However, the majority of cancers with somatic ARID1A mutations and loss of BAF250a expression appear to have a wild-type allele present. Data from exon resequencing and RNA sequencing show excellent agreement between the fraction of mutant and wild-type alleles at both the DNA and RNA levels (Table 1), suggesting that epigenetic silencing is not a significant factor. Post-transcriptional or post-translational regulation or dominant negative effects of the mutations are possible, albeit untested, explanations for the lack of protein expression in these heterozygous cases.
The presence of BAF250a immunoreactivity in 15 samples positive for an ARID1A mutation (all but 1 of which had truncating mutations) may indicate that haploinsufficiency is pathogenic, as has been reported in mice.32 Alternatively, immunohistochemical detection of a truncated but nonfunctional BAF250a protein may account for the immunostaining results. The antibody used in the assay targets a region of 111 amino acids (amino acids 1216 through 1326) in the middle of the protein, and 7 of the 15 specimens that were positive for loss of BAF250a expression had mutations that would result in truncation distal to the epitope.
The mutations are common in ovarian carcinomas that are associated with endometriosis (ovarian clear-cell carcinoma and endometrioid carcinoma) but not in the unrelated high-grade serous carcinoma. This suggests that the mutations may be pathogenic, rather than random, events. Mutations in the PTEN gene (encoding the phosphatase and tensin homologue) have been described in 20% of endometriotic cysts,33 and conditional expression of either oncogenic Kras or deletion of the Pten tumor suppressor in the ovarian surface epithelium in mice was found to induce endometriosis.34 Expression of oncogenic Kras accompanied by simultaneous loss of Pten resulted in widely metastatic ovarian carcinoma; however, KRAS mutations are not seen in human cases of endometriosis and are uncommon in endometriosis-associated ovarian cancers in humans. By comparing ovarian clear-cell carcinomas to their contiguous atypical endometriotic lesions in two patients, we show that the same mutations may be present in the putative precursor lesions and in the tumors. In contrast, the distant endometriotic lesions do not have ARID1A mutations. In the case of ovarian clear-cell carcinoma described in Figure 3, the mutation (G6139T [E2047*]) was present before the atypical endometriosis resulted in the development of the immunophenotype associated with the cancer (estrogen-receptor–negative, HNF-1β–positive24), suggesting that the mutation is an early event in neoplastic transformation. Taken together, these data suggest that ARID1A is a classic tumor-suppressor gene. Unlike BRCA or TP53 mutations, which can be found in the germ-line DNA, all truncating ARID1A mutations were somatic. Deletion of ARID1A on one allele results in embryonic lethality in mice.32
Mutations in ARID1A and loss of BAF250a expression were seen preferentially in ovarian clear-cell carcinomas and endometrioid carcinomas, cancers that do not feature the chromosomal instability, nearly ubiquitous TP53 mutations, and frequent abnormalities in BRCA (associated with early breast cancer) seen in high-grade serous carcinomas.5,35 It is possible that defects in genes that alter the accessibility of transcription factors to chromatin, such as ARID1A, in addition to mutations in the WNT and PI3 kinase pathways,25 will help to define ovarian clear-cell carcinomas and endometrioid carcinomas. If such a model is correct, other abnormalities affecting the ARIDIA locus or dysregulation of other chromatin-remodeling genes may be found in ovarian clear-cell and endometrioid carcinomas that are negative for an ARID1A mutation. This idea is supported by the clinical similarities between ovarian clear-cell carcinomas positive for and those negative for an ARID1A mutation.
The mechanism by which somatic mutations in ARID1A enable the progression of benign endometriosis to carcinoma is unclear; however, our findings are consistent with a critical role for ARID1A mutations in the genesis of a substantial fraction of ovarian clear-cell and endometrioid carcinomas.
Supplementary Material
Acknowledgments
Supported by grants from the British Columbia (BC) Cancer Foundation and the Vancouver General Hospital (VGH)–University of British Columbia Hospital Foundation (to the OvCaRe ovarian cancer research team in Vancouver) and the Canadian Institutes of Health Research (CIHR). The Michael Smith Genome Sciences Centre (MSGSC), with which many of the authors are affiliated, is funded by Genome Canada, and OvCaRe and the MSGSC are also funded by the Michael Smith Foundation for Health Research (MSFHR). Salary support is provided by MSFHR to Drs. Shah, Marra, Jones, and Huntsman; by the CIHR Training Program for Clinician Scientists in Molecular Oncologic Pathology to Drs. Al-Agha and Turashvili (STP-53912); by the CIHR Bioinformatics Training Program for Health Research to Mr. McPherson and Mr. Ha; by the Canadian Breast Cancer Foundation to Dr. Shah; and a Canada Research Chair in Molecular Oncology to Dr. Aparicio. The Genetic Pathology Evaluation Centre, which constructed the tissue microarrays, has received nondirected research grants from Sanofi-Aventis, Canada. The contributing tumor banks were supported by OvCaRe and Ovarian Cancer Canada (VGH, Banque de Tissus et de Données of the Réseau de Recherche sur le Cancer of the Fonds de la Recherche en Santé du Québec, affiliated with the Canadian Tumor Repository Network), and by a grant (DAMD17-O1-1-0729) from the U.S. Army Medical Research and Materiel Command; grants from the Cancer Council Tasmania, the Cancer Foundation of Western Australia, and the National Health and Medical Research Council of Australia to AOCS; and grants from the National Cancer Institute, National Institutes of Health (RO1CA103937 and RO1CA129080).
We thank all members of the OvCaRe ovarian cancer research team (www.ovcare.ca) for their enthusiastic support of this project. We also thank the AOCS management group (D. Bowtell, G. Chenevix-Trench, A. Green, P. Webb, A. deFazio, and D. Gertig) and the AOCS study nurses and research assistants for their contributions. The full AOCS study group is listed at www.aocstudy.org. We thank all the women who donated the samples used in this study. In addition, we thank Christian Steidl for technical consultation; Coco Yu for help in preparation of the manuscript; Robert Bartusiak, Sylvia Lee, and Julie Lorette for technical assistance; and the fellows of the University of British Columbia Gynecologic Oncology Program for obtaining consent from patients for data in our tumor bank.
This article is dedicated to the memory of OvCaRe research administrator Cecelia Suragh.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer Statistics, 2010. CA Cancer J Clin. 2010 July 7; doi: 10.3322/caac.20073. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 2.Köbel M, Kalloger SE, Huntsman DG, et al. Differences in tumor type in low-stage versus high-stage ovarian carcinomas. Int J Gynecol Pathol. 2010;29:203–11. doi: 10.1097/PGP.0b013e3181c042b6. [DOI] [PubMed] [Google Scholar]
- 3.Itamochi H, Kigawa J, Terakawa N. Mechanisms of chemoresistance and poor prognosis in ovarian clear cell carcinoma. Cancer Sci. 2008;99:653–8. doi: 10.1111/j.1349-7006.2008.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tavassoli FA, Devilee P, editors. World Health Organization of classification of tumours. Vol. 4. Lyon, France: IARC Press; 2003. Pathology and genetics of tumours of the breast and female genital organs. [Google Scholar]
- 5.Press JZ, De Luca A, Boyd N, et al. Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer. 2008;8:17. doi: 10.1186/1471-2407-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Köbel M, Kalloger SE, Boyd N, et al. Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med. 2008;5(12):e232. doi: 10.1371/journal.pmed.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dent J, Hall GD, Wilkinson N, et al. Cytogenetic alterations in ovarian clear cell carcinoma detected by comparative genomic hybridisation. Br J Cancer. 2003;88:1578–83. doi: 10.1038/sj.bjc.6600896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilks CB. Molecular abnormalities in ovarian cancer subtypes other than high-grade serous carcinoma. J Oncol. 2009 December 30; doi: 10.1155/2010/740968. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suehiro Y, Sakamoto M, Umayahara K, et al. Genetic aberrations detected by comparative genomic hybridization in ovarian clear cell adenocarcinomas. Oncology. 2000;59:50–6. doi: 10.1159/000012137. [DOI] [PubMed] [Google Scholar]
- 10.Crotzer DR, Sun CC, Coleman RL, Wolf JK, Levenback CF, Gershenson DM. Lack of effective systemic therapy for recurrent clear cell carcinoma of the ovary. Gynecol Oncol. 2007;105:404–8. doi: 10.1016/j.ygyno.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 11.Goff BA, Sainz de la Cuesta R, Muntz HG, et al. Clear cell carcinoma of the ovary: a distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy in stage III disease. Gynecol Oncol. 1996;60:412–7. doi: 10.1006/gyno.1996.0065. [DOI] [PubMed] [Google Scholar]
- 12.Sugiyama T, Kamura T, Kigawa J, et al. Clinical characteristics of clear cell carcinoma of the ovary: a distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy. Cancer. 2000;88:2584–9. [PubMed] [Google Scholar]
- 13.Ness RB. Endometriosis and ovarian cancer: thoughts on shared pathophysiology. Am J Obstet Gynecol. 2003;189:280–94. doi: 10.1067/mob.2003.408. [DOI] [PubMed] [Google Scholar]
- 14.Viganó P, Somigliana E, Chiodo I, Abbiati A, Vercellini P. Molecular mechanisms and biological plausibility underlying the malignant transformation of endometriosis: a critical analysis. Hum Reprod Update. 2006;12:77–89. doi: 10.1093/humupd/dmi037. [DOI] [PubMed] [Google Scholar]
- 15.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–68. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 16.Sif S, Saurin AJ, Imbalzano AN, Kingston RE. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–18. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–30. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 18.Huang J, Zhao YL, Li Y, Fletcher JA, Xiao S. Genomic and functional evidence for an ARID1A tumor suppressor role. Genes Chromosomes Cancer. 2007;46:745–50. doi: 10.1002/gcc.20459. [DOI] [PubMed] [Google Scholar]
- 19.Shah SP, Köbel M, Senz J, et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med. 2009;360:2719–29. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 20.Goya R, Sun MG, Morin RD, et al. SNVMix: predicting single nucleotide variants from next-generation sequencing of tumors. Bioinformatics. 2010;26:730–6. doi: 10.1093/bioinformatics/btq040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Provencher DM, Lounis H, Champoux L, et al. Characterization of four novel epithelial ovarian cancer cell lines. In Vitro Cell Dev Biol Anim. 2000;36:357–61. doi: 10.1290/1071-2690(2000)036<0357:COFNEO>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 22.Lau DH, Lewis AD, Ehsan MN, Sikic BI. Multifactorial mechanisms associated with broad cross-resistance of ovarian carcinoma cells selected by cyanomorpholino doxorubicin. Cancer Res. 1991;51:5181–7. [PubMed] [Google Scholar]
- 23.Dagan T, Talmor Y, Graur D. Ratios of radical to conservative amino acid replacement are affected by mutational and compositional factors and may not be indicative of positive Darwinian selection. Mol Biol Evol. 2002;19:1022–5. doi: 10.1093/oxfordjournals.molbev.a004161. [DOI] [PubMed] [Google Scholar]
- 24.Köbel M, Kalloger SE, Carrick J, et al. A limited panel of immunomarkers can reliably distinguish between clear cell and high-grade serous carcinoma of the ovary. Am J Surg Pathol. 2009;33:14–21. doi: 10.1097/PAS.0b013e3181788546. [DOI] [PubMed] [Google Scholar]
- 25.Kuo KT, Mao TL, Jones S, et al. Frequent activating mutations of PIK3CA in ovarian clear cell carcinoma. Am J Pathol. 2009;174:1597–601. doi: 10.2353/ajpath.2009.081000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 27.Kozmik Z, Machon O, Králová J, Kreslová J, Paces J, Vlcek C. Characterization of mammalian orthologues of the Drosophila osa gene: cDNA cloning, expression, chromosomal localization, and direct physical interaction with Brahma chromatin-remodeling complex. Genomics. 2001;73:140–8. doi: 10.1006/geno.2001.6477. [DOI] [PubMed] [Google Scholar]
- 28.Mitelman F, Johansson B, Mandahl N, Mertens F. Clinical significance of cytogenetic findings in solid tumors. Cancer Genet Cytogenet. 1997;95:1–8. doi: 10.1016/s0165-4608(96)00252-x. [DOI] [PubMed] [Google Scholar]
- 29.Mitelman F, Mertens F, Johansson B. A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet. 1997;15(Special no):417–74. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- 30.Decristofaro MF, Betz BL, Rorie CJ, Reisman DN, Wang W, Weissman BE. Characterization of SWI/SNF protein expression in human breast cancer cell lines and other malignancies. J Cell Physiol. 2001;186:136–45. doi: 10.1002/1097-4652(200101)186:1<136::AID-JCP1010>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Nagl NG, Jr, Flowers S, Zweitzig D, Dallas PB, Moran E. Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumors. Int J Cancer. 2004;112:636. doi: 10.1002/ijc.20450. [DOI] [PubMed] [Google Scholar]
- 32.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–61. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato N, Tsunoda H, Nishida M, et al. Loss of heterozygosity on 10q23.3 and mutation of the tumor suppressor gene PTEN in benign endometrial cyst of the ovary: possible sequence progression from benign endometrial cyst to endometrioid carcinoma and clear cell carcinoma of the ovary. Cancer Res. 2000;60:7052–6. [PubMed] [Google Scholar]
- 34.Dinulescu DM, Ince TA, Quade BJ, Shafer SA, Crowley D, Jacks T. Role of K-ras and Pten in the development of mouse models of endometriosis and endometrioid ovarian cancer. Nat Med. 2005;11:63–70. doi: 10.1038/nm1173. [DOI] [PubMed] [Google Scholar]
- 35.Ahmed AA, Etemadmoghadam D, Temple J, et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol. 2010;221:49–56. doi: 10.1002/path.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.