Abstract
Human multidrug and toxin extrusion member 1, MATE1 (SLC47A1), plays an important role in the renal and biliary excretion of endogenous and exogenous organic cations including many therapeutic drugs. In this study, we characterized the transcriptional effects of five polymorphic variants and six common haplotypes in the basal promoter region of MATE1 that were identified in 272 DNA samples from ethnically diverse U.S. populations. We measured luciferase activities of the six common promoter haplotypes of MATE1 using in vitro and in vivo reporter assays. Haplotypes that contain the most common variant (mean allele frequency in four ethnics: 0.322), g.−66T>C, showed a significant decrease in reporter activities compared to the reference. Two transcription factors, AP-1 and AP-2rep, were predicted to bind to the promoter in the region of g.−66T>C. Results from electrophoretic mobility shift assays showed that the g.−66T allele, exhibited greater binding to AP-1 than the g.−66C allele. AP-2rep inhibited the binding of AP-1 to the MATE1 basal promoter region, and the effect was considerably greater for the g.−66T>C. These data suggest that the reduced transcriptional activity of g.−66T>C results from a reduction in the binding potency of the transcriptional activator, AP-1, and an enhanced binding potency of the repressor, AP-2rep to the MATE1 basal promoter region. Consistent with the reporter assays, MATE1 mRNA expression levels were significantly lower in kidney samples from individuals who were homozygous or heterozygous for g.−66T>C in comparison to samples from individuals who were homozygous for the g.−66T allele. Our study suggests that the rate of transcription of MATE1 is regulated by AP-1 and AP-2rep and that a common promoter variant, g.−66T>C may affect the expression level of MATE1 in human kidney, and ultimately result in variation in drug disposition and response.
Keywords: MATE1, Haplotype, Promoter, AP-1, AP-2rep, Transcriptional activity, Pharmacogenomics, Membrane transporters, SLC47A1, Polymorphisms
Introduction
The liver and kidney are the major organs responsible for the elimination of drugs and other xenobiotics from the body [1]. Transporters localized to the sinusoidal membrane of the hepatocyte, take up drugs from the portal blood into the hepatocyte. Subsequently, the drugs can be transported back into the blood or excreted into the bile in their original chemical forms or as metabolic products [2]. Human multidrug and toxin extrusion member 1 (MATE1, SLC47A1, or also known as H+/organic cation antiporter member), is present on the canalicular membrane of the hepatocyte and appears to work in concert with organic cation transporter 1 (OCT1) to mediate the biliary excretion of many cationic drugs and their metabolites [3]. Previous studies have demonstrated that various drugs and environmental toxins including cimetidine, metformin, and paraquat are substrates of MATE1 [4, 5]. In the kidney, MATE1 is highly expressed on the apical membrane of the proximal tubule and may work with the basolateral membrane transporter, human organic cation transporter 2 (OCT2), in the renal secretion of organic cations [3, 6]. In addition to the liver and kidney, other tissues that express MATE1 in high abundance include the adrenal gland, testis, and skeletal muscle [7].
Recently, we identified four synonymous and six non-synonymous coding region variants of MATE1 in DNA samples from a large ethnically diverse cohort of healthy volunteers (68 individuals from each population: European Americans, African Americans, Chinese Americans and Mexican Americans). Using a mammalian expression system, we observed that several singleton non-synonymous single nucleotide polymorphisms (SNPs) of MATE1 showed significant loss of function for all substrates tested and that several common non-synonymous SNPs (> 5% minor allele frequency) showed selectively reduced transport for some substrates [8]. These findings suggested that genetic variants in MATE1 may alter drug disposition and ultimately affect clinical drug response.
Genetic variation in the promoter region of MATE1 has also been described. In particular, Kajiwara M, et al [9] identified a SNP, g.−32G>A (from translational start site, allele frequency: 3.7%) in DNA samples from a Japanese population within a specificity protein 1 (Sp-1) binding site of the proximal promoter region of MATE1. This polymorphism had reduced expression activity in reporter assays [9].
The goal of the current study was to identify and functionally characterize basal promoter region polymorphisms in MATE1. We screened DNA samples from a large ethnically diverse U.S. population for variants in the basal promoter of MATE1 and investigated the function of the variants using both in vivo and in vitro reporter assays. Our study provides information about the mechanisms responsible for the transcriptional regulation of MATE1 and identifies a functionally important genetic polymorphism that may contribute to variation in the expression level of this transporter.
Materials and Methods
Genetic analysis
Genomic DNA samples were collected from unrelated healthy individuals from four major ethnic groups (68 each from European Americans, African Americans, Chinese Americans, and Mexican Americans) as a part of the Study Of PHarmacogenetics In Ethnically-diverse populations (SOPHIE). To identify polymorphisms in the promoter region of MATE1, a polymerase chain reaction (PCR) fragment (−636 to +114 bp from the translational start site) was amplified and directly sequenced by an automated genetic analyzer using the primers listed in Table 3. Haplotype assembly was performed using the Haploview 4.1 program (Broad Institute, Cambridge, MA, USA), based on a standard expectation-maximization algorithm [10], to reconstruct individual haplotypes from population genotype data. Nucleotide location number was assigned from the translational start site according to the MATE1 mRNA sequence (GenBank accession number; NM_018242). We followed the nomenclature system that was recommended by Antonarakis SE, et al [11, 12].
Table 3.
Oligonucleotide primers used in the construction of MATE1 reporter plasmids or AP-2rep plasmid and the electrophoretic mobility shift analysis
| Primers for MATE1 promoter sequencing | |
| Sense | 5’-GGG AGC ATG TTG CTC TAT CC-3’ |
| Antisense | 5’-AGG AGC TTC CAT GTG ACT CG-3’ |
| Primers for MATE1 promoter cloning | |
| First PCR (−543 to +15) | |
| Sense | 5’-AGT ATG CAA CAC CCT AAC CAG CA-3’ |
| Antisense | 5’-CCT CAG GAG CTT CCA TGT GAC T-3’ |
| Second PCR (−234 to −12) | |
| Sense (NheI site)1 | 5’-CTA GCT AGC GGT GCA GAG AGA GGT GCA A-3’ |
| Antisense (HindIII site)1 | 5’-CCC AAG CTT GCT GCG GCC GGG TAG-3’ |
| Primers for AP-2rep cloning | |
| Sense (NheI site)1 | 5’-GCT AGC ATG AAT ATC CAT ATG AAG AGA-3’ |
| Antisense (KpnI site)1 | 5’-GGT ACC GCT GGA CAG GTA GCA TTC CTC AC-3’ |
| Primers for EMSA | |
| Reference (g.−66T)2 | 5’-TGC GCG GTA CTC ACT GCC GGC C-3’ |
| Variant (g.−66C)2 | 5’-TGC GCG GTA CCC ACT GCC GGC C-3’ |
| Consensus AP-13 | 5’-AAG CCT GTG ACT CAG GAC CTG-3’ |
| Consensus AP-2rep4 | 5’-CTG GGG CAG TGG GAC TGG CA-3’ |
Construction of MATE1 variants in the promoter region
To construct the reporter plasmids containing the MATE1 promoter region, 559 bp of the MATE1 gene was amplified from genomic DNA samples from individuals having each haplotype listed in Table 2. The amplified products were inserted into pCR2.1-TOPO vectors (Invitrogen Corporation) and were directly sequenced. The second PCR was performed using the primers containing the restriction endonuclease sites, NheI and HindIII (Table 3). The amplified 223 bp products were inserted into the pGL4.11b [luc2] vectors (Promega Corporation) and DNA sequences were confirmed by direct sequencing.
Table 2.
Frequencies of the common haplotypes in the MATE1 promoter region
| ID | g.−66T>C | g.−53C>G | g.−48_−26del | g.−44C>T | g.−27_−26ins | Minor Allele Frequency | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| African American |
European American |
Chinese American |
Mexican American |
Total | ||||||
| H1 | T | C | G | C | G | 0.540 | 0.679 | 0.743 | 0.694 | 0.661 |
| H2 | C | C | G | C | G | 0.365 | 0.254 | 0.218 | 0.210 | 0.265 |
| H3 | C | G | G | C | ins | 0.016 | 0.045 | 0.000 | 0.081 | 0.035 |
| H4 | C | C | del | C | G | 0.048 | 0.007 | 0.000 | 0.008 | 0.016 |
| H5 | T | G | G | T | ins | 0.000 | 0.000 | 0.026 | 0.000 | 0.008 |
| H6 | C | G | G | T | ins | 0.016 | 0.000 | 0.012 | 0.000 | 0.006 |
The minor alleles are marked in bold-faced letter with underlines.
Data were obtained from a DNA samples from 272 unrelated individuals including 68 from each of four major ethnic groups.
Position of the variant is based upon the translational start site.
Measurement of MATE1 promoter activity in vitro
The reporter plasmids containing the reference or variants of MATE1 were transfected into HCT-116 (human colon carcinoma), ACHN (human kidney adenocarcinoma), HepG2 (human liver, carcinoma), and HeLa (human uterus carcinoma) cell lines using Lipofectamine LTX and Plus reagents (Invitrogen) according to the manufacturers’ protocol. HCT-116 cells were cultured in high glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) while ACHN cells were cultured in Roswell Park Memorial Institute (RPMI) medium 1640 supplemented with 10% FBS. HepG2 and HeLa cells were cultured in minimum essential medium (MEM) supplemented with 10% FBS. Thirty hours after transfection, the reporter activity was measured using the Dual-luciferase® reporter assay system (Promega) according to the manufacturers’ protocol and quantified using the Glomax 96-well plate luminometer (Promega). The firefly luciferase to renilla luciferase ratios were determined and expressed as relative luciferase activity. To examine the effect of AP-2rep on the promoter activity of MATE1, reference or variant reporter plasmids were co-transfected with increasing amounts of AP-2rep-pcDNA5-FRT vector (3 ng to 50 ng). The AP-2rep gene (BC_019680) was amplified from human kidney cDNA by PCR using primers listed in Table 3 and inserted into pcDNA5-FRT vector. The amount of transfected plasmid was equalized by adding pcDNA5-FRT vector.
Measurement of MATE1 promoter activity in vivo
For the in vivo functional assay, hydrodynamic tail vein injection experiments were performed on at least 10 mice per construct using the TransIT® In Vivo gene delivery system (Mirus Bio Corporation) following the manufacturers’ protocol. Briefly, 10 µg of pGL4.11b [luc2] vector containing either no promoter or the MATE1 promoter constructs (reference haplotype H1 or variant haplotype H2) alongside 2 µg of pGL4.74 [hRluc/TK] vector, to correct for injection efficiency, was injected into the tail vein of CD1 mice (Charles River) weighing 22–24 g. After 24 hours, animals were euthanized, livers were dissected and homogenized in passive lysis buffer (Promega) followed by centrifugation for 30 min at 14,000 rpm. Firefly luciferase and renilla luciferase activity in the supernatant (diluted 1:20 with buffer) were measured six times for each liver using the Dual-luciferase® reporter assay system, and quantified using the Synergy 2 (BioTek Instruments, Inc.). The firefly luciferase to renilla luciferase ratios were determined and expressed as relative luciferase activity.
Electrophoretic mobility shift assay (EMSA) of MATE1
Nuclear protein extracts were obtained from naïve HCT-116 cells, from HCT-116 cells treated with 10 ng/ml of TNF-α, or from HCT-116 cells transfected with the AP-2rep-pcDNA-FRT plasmid. The nuclear protein extraction procedure was performed according to a previously described method with some modifications [13]. Oligonucleotides used in the EMSA are listed in Table 3. The method used for the EMSA has been described previously [13]. Nuclear protein extracts (20 or 30 µg) were incubated with 2 µl of 32P-labeled oligonucleotide (1×105 counts/min or 2×105 counts/min or 4×105 counts/min) in a buffer containing 7% glycerol, 10 mM HEPES (pH 7.9), 1 mM EDTA (pH 8.0), 1 mM DTT, and 0.2 µg poly(deoxyinosinate-deoxycytidylate) in a final volume of 20 µl for 30 minutes at room temperature. The reaction mixtures were loaded on 6% nondenaturing polyacrylamide gel and electrophoresed for 2 hours at 200 V. The dried gel was exposed to Kodak XR-5 film (Eastman Kodak Corporation) on an intensifying screen for 10–24 hours at room temperature or −80°C. For the shift assay, 2 µl of phospho-c-jun (9164, Cell Signaling Technology Inc.) or 1 µl of c-fos (sc-253X, Santa Cruz Biotechnology Inc.) antibody was incubated with the nuclear extract for 20 minutes at room temperature prior to the binding reaction. The intensity of each band on the film was measured using Scion image (Scion Corporation).
Quantification of MATE1 gene expression using quantitative real-time RT-PCR
Using TRIZOL reagent (Invitrogen), total RNA was isolated from 38 surgical sections of human kidney (23 normal renal cortex adjacent to tumor section of renal cell carcinoma and 15 renal cortex from tissues of individuals with various disease diagnoses) and 34 human liver samples were normal postmortem tissue. The age of the donors ranges from 36 to 83 years. The clinical diagnoses of the kidney donors were renal cell carcinoma, chronic obstructive pulmonary disease, ischemic heart disease or chronic heart failure; whereas the clinical diagnoses of the donors of the post-mortem liver samples were pneumonia, ischemic heart disease and Non-Hodgkin Lymphoma. These samples were purchased from Asterand plc and Capital Biosciences. All samples collected were stored frozen at −80 °C from collection until processing for RNA and DNA. These samples were all from individual with European ancestry background. Single strand cDNAs were synthesized using a Superscript III first-strand synthesis system for RT-PCR (Invitrogen). Quantitative real-time RT-PCR was performed using a 7500 Fast RT-PCR system (Applied Biosystems) with Taqman gene expression assays. The total reaction volume in each well was 10 µl, including 50 ng of cDNA, 0.5 µl of 20-fold primers (Hs00217320_m1, Applied Biosystems), and 5 µl of 2-fold Taqman Fast Universal PCR Master Mix (Applied Biosystems). The RT-PCR was started with 95°C for 20 seconds, and continued with 40 cycles of 95°for 3 seconds and 60°C for 30 seconds. Relative quantitative analysis was performed with 7500 Fast System SDS software 1.4 and the expression of endogenous human GAPDH gene was used as an internal standard to correct for the amount of added cDNA. The genomic DNA of these tissue samples were isolated, and a 559 bp PCR fragment was amplified using the MATE1 promoter cloning primers and directly sequenced by an automated genetic analyzer.
RNA mediated interference
Validated small interfering RNA (siRNA) for c-jun (VHS40918, Invitrogen) was reverse-transfected into HCT-116 cells using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturers’ protocol. Stealth RNAi negative control high GC (Invitrogen) was used as a negative control. Forty-two hours after transfection, the reporter plasmids containing the reference or variant MATE1 were transfected into the cells using Lipofectamine LTX and Plus reagents according to the manufacturer’s protocol. Thirty hours after transfection, the reporter activity was measured using the Dual-luciferase® reporter assay system according to the manufacturer’s protocol and quantified using the Glomax 96-well plate luminometer. Firefly luciferase to renilla luciferase ratios were determined and expressed as relative luciferase activity.
Statistical analysis
Results of luciferase assays presented were derived from two or three independent experiments and were shown as the mean ± SD. All P values except for those obtained in the in vivo luciferase assays were calculated using Dunnett's multiple comparison test. The P value for the in vivo luciferase assay was calculated using Student’s t test. P values less than 0.05 were considered statistically significant. A D’Agostino and Pearson omnibus normality test (GraphPad Prism 5) was used to determine whether the expression levels of MATE1 in kidney and in liver were normally distributed.
Results
Genetic polymorphisms of MATE1 in the promoter region
We identified eight variants in the promoter region of MATE1 in 272 DNA samples from ethnically diverse U.S. populations. Five of the variants were polymorphic (minor allele frequency > 1%) in at least one of the four ethnic groups (Table 1). The g.−66T>C was present in all four populations and the frequency of this variant was the highest in all ethnic groups. In our study, we did not observe the g.−32G>A (from transcriptional start site) polymorphism identified previously in a Japanese population [9]. Table 2 shows the frequency distributions of the common haplotypes in the MATE1 promoter region that occurred at minor allele frequencies > 1% in at least one of the four ethnic groups. Haplotype 1, which consists of all major alleles, showed the highest frequency, followed by Haplotype 2, which contains g.−66T>C.
Table 1.
Allele frequencies of MATE1 variants in the proximal promoter
| Identification | Variant | Allele Frequency | |||
|---|---|---|---|---|---|
| African American |
European American |
Chinese American |
Mexican American |
||
| V1 | g.−66T>C | 0.445 | 0.321 | 0.231 | 0.289 |
| V2 | g.−53C>G | 0.017 | 0.054 | 0.031 | 0.078 |
| V3 | g.−48_−26delGTACCCACTGCCGGCCTCCGCG | 0.025 | 0.000 | 0.000 | 0.000 |
| V4 | g.−44C>T | 0.015 | 0.015 | 0.029 | 0.008 |
| V5 | g.−27_−26insGTACCCACTGCCGGCCTCCGCG | 0.037 | 0.059 | 0.044 | 0.068 |
Data were obtained from a DNA samples from 272 unrelated individuals including 68 from each of four major ethnic groups.
Position of the variant is based upon the translational start site.
Promoter activity of MATE1 variants in vitro
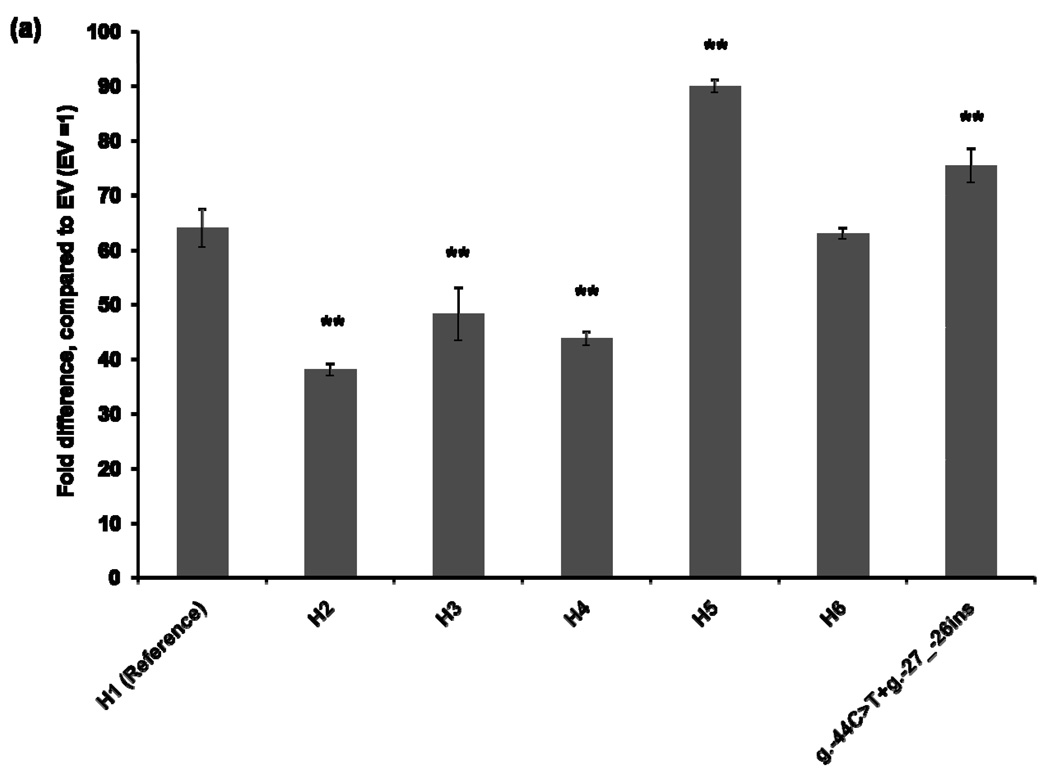
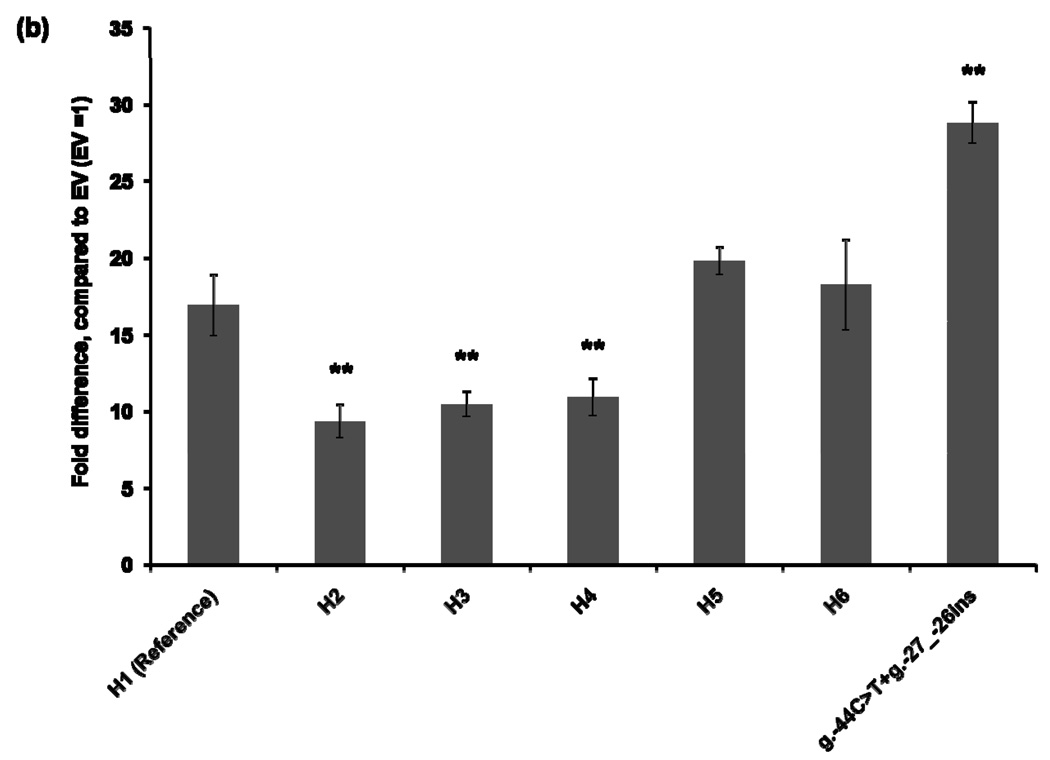
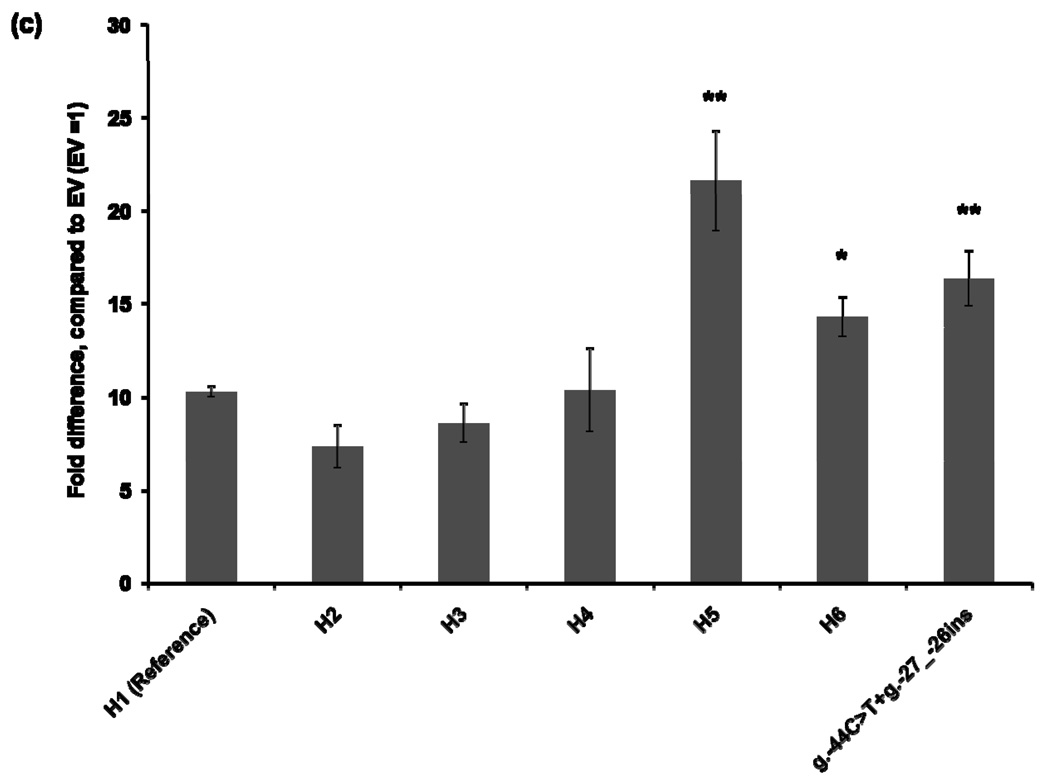
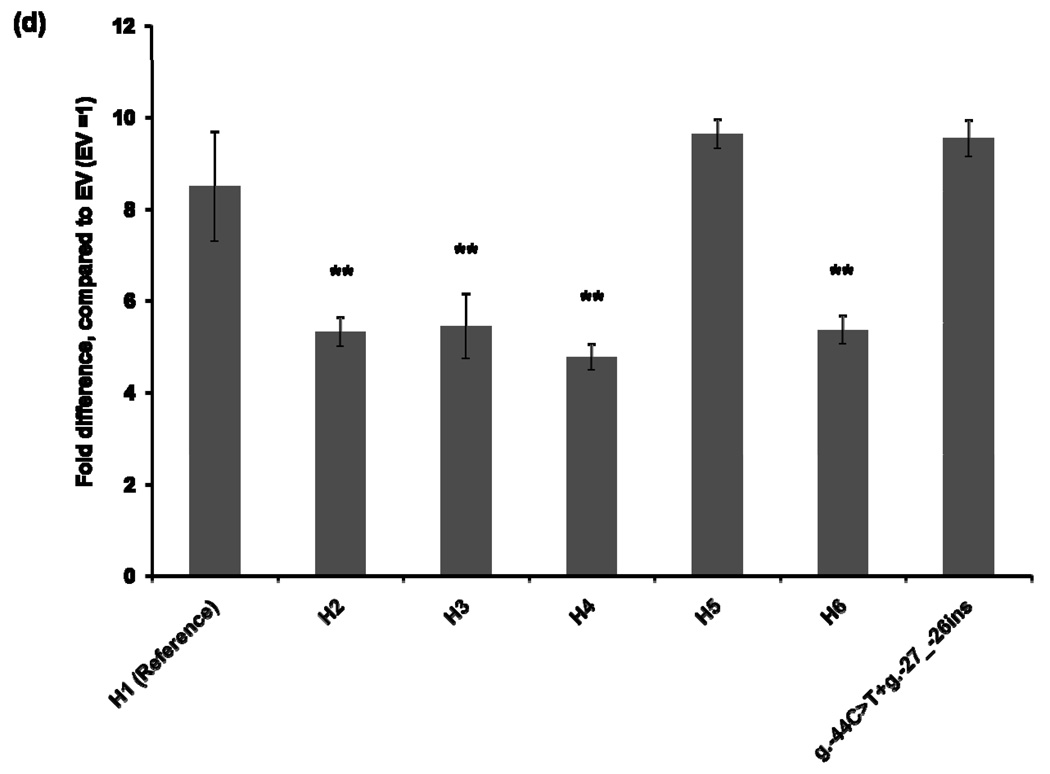
To characterize the functional effects of the promoter variants, we constructed MATE1-pGL4.11b [luc2] reporter plasmids that included the essential region for the basal transcriptional activity of MATE1 [9]. The reporter assay was performed 30 hours after transfection of the reporter plasmids into four different cell lines: HCT-116, ACHN, HepG2, and HeLa. Haplotype 1 was used as a reference for the functional analysis. The promoter activities of Haplotypes 2, 3, and 4, all of which contained the g.−66T>C variant showed decreased reporter gene expression by 16–44% compared to that of the reference in all cell lines except for HepG2 cells in which haplotype 3 and 4 showed similar activity to the reference haplotype 1 (Fig. 1). Haplotype 5, which contains variants, g.−53C>G, g.−44C>T, and g.−27_−26ins, showed 13–110% increased promoter activity, compared to that of the reference haplotype in all cell lines. These results appear to be explained by the presence of the variants g.−44C>T, and g.−27_−26ins, which tended to have higher reporter activities in these cell lines. There was no individual having a haplotype that contained only the variants g.−44C>T, and g.−27_−26ins in this study. Interestingly, the promoter activity of Haplotype 6, which contained both the reduced function variant, g.−66T>C, and the increased function variants, g.−44C>T and g.−27_−26ins, was comparable to that of the reference haplotype in HCT-116 and ACHN cell lines (Fig. 1).
Figure 1.
Luciferase activities in cell lines expressing reporter constructs containing the common haplotypes of the basal promoter of MATE1. Luciferase activities were measured 30 hours after transfection of the reporter plasmids into HCT-116 (a), ACHN (b), HepG2 (c), and HeLa (d) cell lines. The reporter activity of each construct was compared with that of empty vector (pGL4.11b [luc2]). Data shown represent mean ± SD from triplicate wells in a representative experiment. *P < 0.05, **P < 0.01 vs. the reference. Haplotypes (H1–H6) are defined in Tables 2. The variant g.66T>C is contained in H2, H3, H4 and H6. H6 also contains g.−44C>T, present in H5.
Promoter activity of a MATE1 variant in vivo
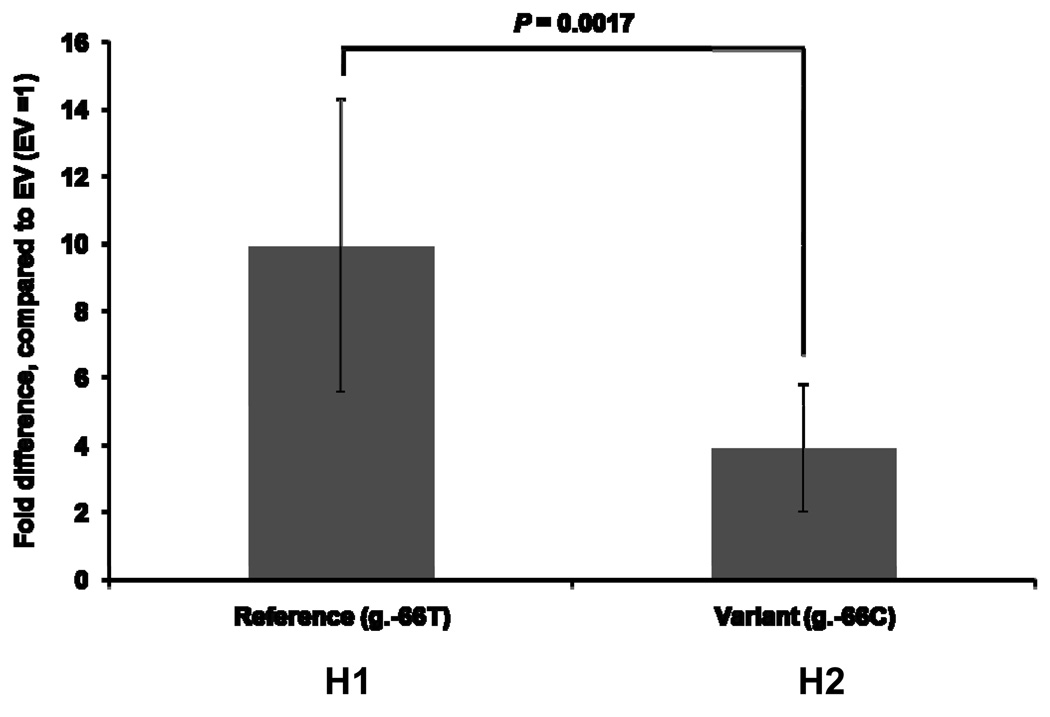
To confirm the findings obtained in the reporter assays in cells with respect to g.−66T>C, we measured the promoter activity of MATE1 in vivo in mouse liver. We took advantage of the hydrodynamic tail vein injection technique in order to express our reporter assay constructs in the mouse liver [14]. Consistent with our in vitro results, we observed that the construct containing the variant g.−66T>C showed a significant decrease in luciferase activity in comparison to the reference allele (Fig. 2).
Figure 2.
The activity of a common promoter region variant, g.−66T>C, of MATE1 in mouse liver. Activity was assessed after in vivo hydrodynamic injection of reporter constructs into mouse tail vein. Luciferase activities were measured 24 hours after the injection of the reporter plasmids into the tail vein of mice (n=10 per each group). The luciferase activity of each construct was normalized to the luciferase activity measured in liver samples from mice injected with constructs containing the empty vector (pGL4.11b [luc2]). Data shown represent mean ± SD from reporter assays from livers of 10 mice. A t-test was used to measure statistical significance.
Effect of variant, g.−66T>C on the binding affinity of AP-1 and AP-2rep within the MATE1 promoter region
Due to the high frequency and reduced function of the g.−66T>C, we investigated the mechanisms by which this variant may cause a reduction in transcriptional activity. First, we identified the transcription factors that could bind to the basal promoter region of MATE1 in the vicinity of g.−66T>C. Two different transcription factors, namely activating protein-1 (AP-1) and activating protein-2repressor (AP-2rep) were identified using TFSearch 3.1 (Real-World Company Partnership) and the Delta-Match query tool (developed by Dr. David W. Williamson (personal communication)). AP-1 is a strong oncogene that mediates tumor cell invasion and AP-2rep belongs to a family of Krüppel-like factor (KLF) and works as a repressor of AP-2α [15, 16]. TFSearch 3.1 predicted that AP-1 would have a higher binding affinity to the g.−66T allele, than the g.−66C allele, whereas the Delta-Match query tool predicted that the g.−66C allele, would bind with a higher affinity to AP-2rep than the g.−66T allele. These predictions, if validated, would explain the reduced transcriptional activity of the variant, i.e., in the presence of the variant, AP-1 would activate transcription less potently whereas AP-2rep would repress transcription more potently.
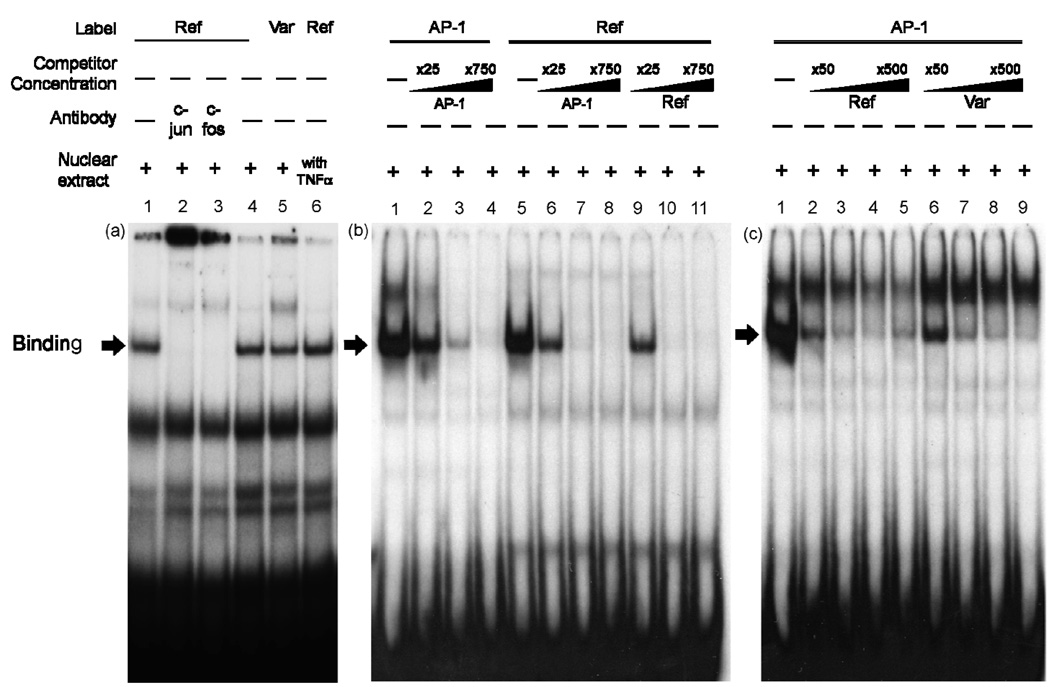
To functionally validate these predictions, we first determined whether AP-1 could bind to the promoter region containing the g.−66T>C site through electrophoretic mobility shift assay (EMSA). Oligonucleotides (2×105 counts/min, reference g.−66T: lanes 1–4 and 6; variant g.−66C: lane 5, Fig. 3 (a)) were incubated with nuclear protein extracts (20 µg) obtained from HCT-116 (lanes 1–5) or tumor necrosis factor-α (TNF-α) treated HCT-116 cells (lane 6), because TNF-α is known to induce the expression of AP-1 [17]. Both reference and variant probes formed DNA-protein complexes, although the intensity of the complex with the variant probe was decreased by 12%, compared to the reference (lanes 4–5, Fig. 3 (a)). AP-1 is found either as a homodimer of the Jun family or in heterodimers between members of the Fos and Jun families [16]. The presence of the supershift with antibodies against phosphorylated c-jun and c-fos confirmed that AP-1 was present in the complex (lanes 2–3, Fig. 3 (a)). Consistent with a previous study indicating that TNF-α could induce the expression of AP-1, we confirmed that the binding of the reference probe with AP-1 was increased by 17% after exposure to TNF-α (lane 6, Fig. 3 (a)) [17]. To support the result from the supershift assay, we performed a competition assay using the AP-1 consensus probe (Fig. 3 (b) and (c)) [15]. The AP-1 consensus probes (1×105 counts/min, lanes 1–4, Fig. 3 (b)) or MATE1 reference probes (1×105 counts/min, lanes 5–11, Fig. 3 (b)) were incubated with nuclear protein extracts (20 µg) obtained from HCT-116 cells. In the competition assay, various concentrations of unlabelled AP-1 consensus oligonucleotides competed with AP-1 consensus (lanes 2–4, Fig. 3 (b)) or MATE1 reference probes (lanes 6–8, Fig. 3 (b)) in a dose dependent manner. Similarly, the unlabelled MATE1 reference oligonucleotides competed in a dose-dependent manner with the labeled MATE1 reference probe as shown in lanes 9–11 (Fig. 3 (b)). The binding affinity between reference, g.−66T and variant, g.−66C for AP-1 consensus sequence were examined in a competition assay using AP-1 consensus probes (1×105 counts/min) with various concentrations of the unlabeled MATE1 reference (lanes 2–5, Fig. 3 (c)) or variant (lanes 6–9, Fig. 3 (c)) oligonucleotides and HCT-116 nuclear protein extracts (20 µg). The reference oligonucleotide appeared to be a stronger competitor than the variant, suggesting that the MATE1 reference, g.−66T has a higher affinity for the AP-1 consensus than the MATE1 variant, g.−66C.
Figure 3.
Electrophoretic mobility shift analysis of MATE1 reference and variant g.−66T>C oligonucleotides. (a) Oligonucleotides (reference g.−66T: lanes 1–4 and 6; variant g.−66C: lane 5) were incubated with nuclear protein extracts (20 µg) obtained from HCT-116 or HCT-116 cells that were pre-incubated with 10 ng/ml of TNF-α for 1 hour (lane 6). The arrow indicates the position of the DNA protein complex. Supershift experiments were performed with the antibodies against phosphorylated c-jun (lane 2) and c-fos (lane 3) added separately to each gel shift reaction. (b) Oligonucleotides (1×105 counts/min, AP-1 consensus: lanes 1–4, reference g.−66T: lanes 5–11) were incubated with nuclear protein extracts (20 µg) obtained from HCT-116. Competition reactions were performed using different folds (25 (lanes 2, 6, 9), 250 (lanes 3, 7, 10), 750 (lanes 4, 8, 11)) of molar excess of unlabeled consensus AP-1 (lanes 2–4 and 6–8) or reference g.−66T (lanes 9–11) oligonucleotides. (c) AP-1 consensus oligonucleotides (1×105 counts/min) were incubated with nuclear protein extracts (20 µg) obtained from HCT-116. Competition reactions were performed using different folds (50 (lanes 2, 6), 125 (lanes 3, 7), 250 (lanes 4, 8), 500 (lanes 5, 9)) of molar excess of unlabeled reference g.−66T (lanes 2–5) or variant g.−66C (lanes 6–9) oligonucleotides. The arrow indicates the position of the DNA protein complex.
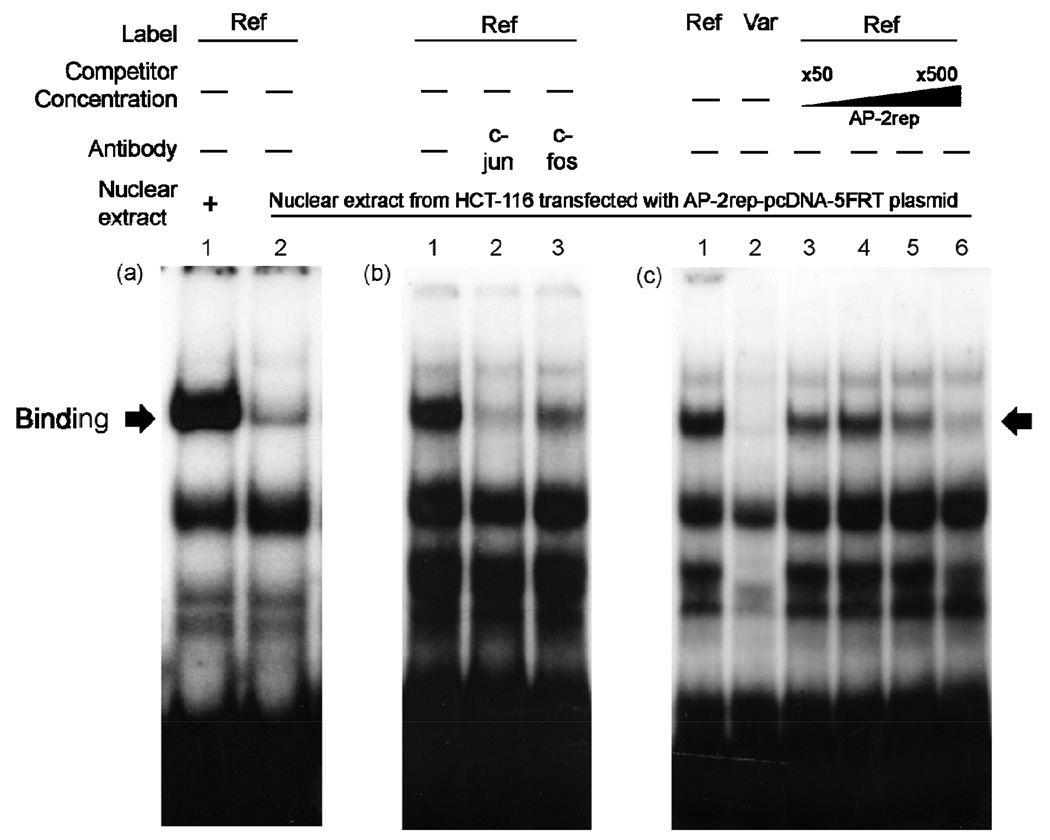
Next, we conducted EMSAs with nuclear protein extracts obtained from AP-2rep-over-expressed HCT-116 cells (Fig. 4). The reference probes (2×105 counts/min) were incubated with nuclear protein extracts (20 µg) obtained from HCT-116 (lane 1, Fig. 4 (a)) or AP-2rep-overexpressed HCT-116 cells (lane 2, Fig. 4 (a)). We found that the intensity of DNA-protein complex was much weaker in nuclear protein extracts from AP-2rep-over-expressed HCT-116 cells, compared to extracts from control HCT-116 cells. We hypothesized that AP-2rep can inhibit the binding of AP-1 to the promoter region of MATE1. The DNA-protein complexes were supershifted with phosphorylated c-jun and c-fos antibodies indicating that these bands are complexes of DNA with AP-1 (lanes 2–3, Fig. 4 (b)). We also examined the inhibitory effect of AP-2rep on the binding of DNA with AP-1 through EMSA using the variant probe (Fig. 4 (c)). Oligonucleotides (4×105 counts/min, reference g.−66T: lanes 1, 3–6, variant g.−66C: lane 2, Fig. 4 (c)) were incubated with nuclear protein extracts (30 µg) obtained from AP-2rep-over-expressed HCT-116 cells. The intensity of DNA-protein complex was much weaker in lane 2, in which the variant probe was used, compared to lane 1, which was probed with the reference probe. These data suggest that the inhibitory effect of AP-2rep on DNA-AP-1 binding is much stronger in the presence of the variant, g.−66C, compared to the reference. A dose-dependent competition of DNA-AP-1 binding was observed after addition of AP-2rep oligonucleotides (lanes 3–6, Fig. 4 (c)) indicating that AP-1 and AP-2rep can bind competitively to the region of the MATE1 promoter containing variant, g.−66T>C.
Figure 4.
Effect of AP-2rep on the binding of DNA with AP-1. (a) Reference oligonucleotides (2×105 counts/min) were incubated with nuclear protein extracts (20 µg) obtained from HCT-116 (lane 1) or AP-2rep-over-expressed HCT-116 cells (lane 2). (b) Reference oligonucleotides (4×105 counts/min) were incubated with nuclear protein extracts (30 µg) obtained from AP-2rep-overexpressed HCT-116 cells (lanes 1–3). Supershift experiments were performed with the antibodies against phosphorylated c-jun (lane 2) and c-fos (lane 3) added separately to each gel shift reaction. (c) Oligonucleotides (4×105 counts/min, reference g.−66T: lane 1 and lanes 3–6, variant g.−66C: lane 2) were incubated with nuclear protein extracts (30 µg) obtained from AP-2rep-over-expressed HCT-116 cells. Competition reactions were performed using different folds (25 (lane 3), 50 (lane 4), 100 (lane 5), and 250 (lane 6)) of molar excess of unlabeled consensus AP-2rep oligonucleotides. The arrow indicates the position of the DNA protein complex.
Effect of AP-1 or AP-2rep on the MATE1 promoter activity
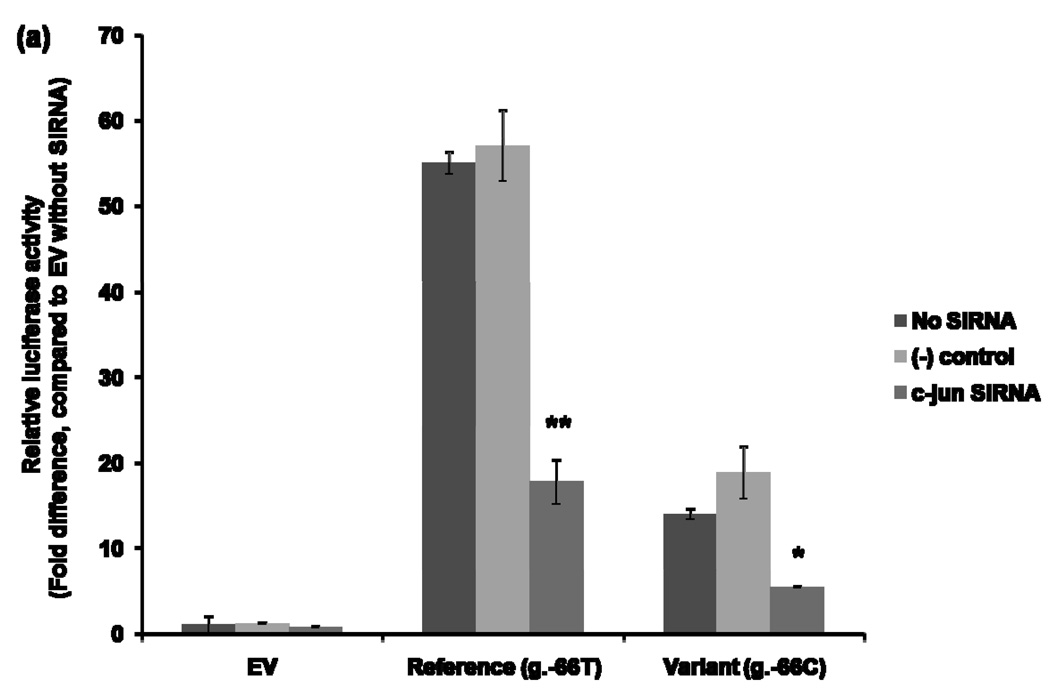
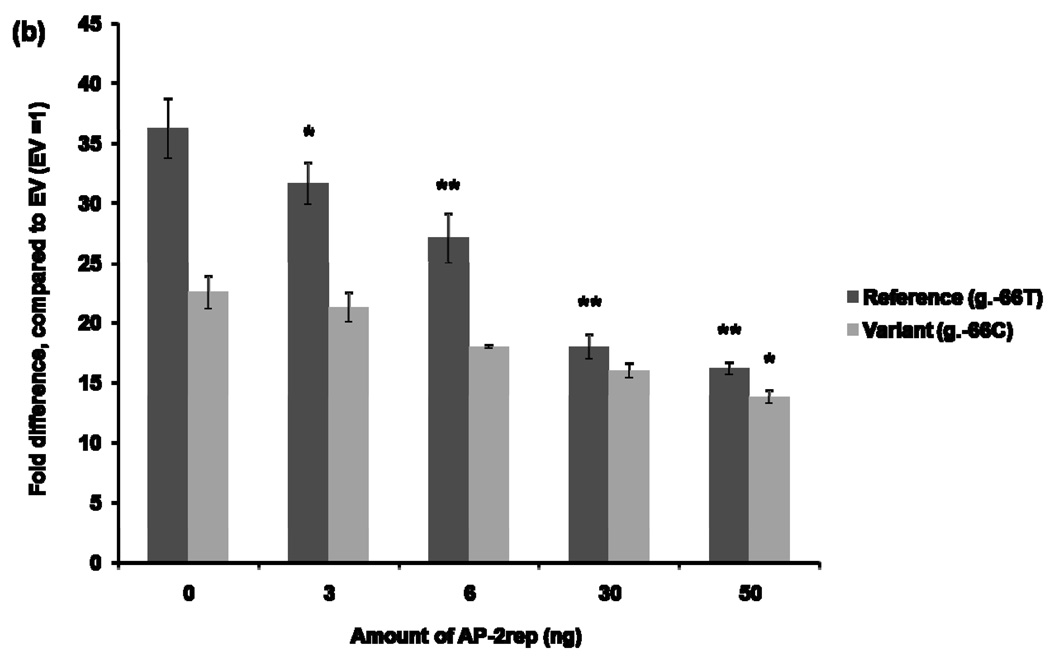
The effect of AP-1 on the MATE1 promoter activity was examined by using siRNA to knockdown c-jun. After 42 hours of siRNA transfection to HCT-116, the reference (H1) or variant (H2) reporter constructs were transfected into the cells. Reporter activities were determined after 30 hours. Figure 5 (a) showed a significant reduction in luciferase activities of the H1 (3.4 fold reduced) and H2 (3.5 fold reduced) reporter constructs in the presence of c-jun siRNA compared to the luciferase activities with the negative control siRNA. This result suggests that AP-1 works as a transcription factor that binds to the MATE1 promoter to regulate its expression. The effect of AP-2rep on the promoter activity of MATE1 was examined by conducting a luciferase assay after over-expression of AP-2rep transcription factor. To examine the effect of AP-2rep on the promoter activity of MATE1, reference (H1) or variant (H2) reporter constructs with increasing amounts of plasmids containing AP-2rep were co-transfected in HCT-116. We observed that AP-2rep produced a dose-dependent repression of the MATE1 promoter activity (Fig. 5 (b)).
Figure 5.
Effects of AP-1 and AP-2rep on the promoter activity of MATE1. (a) 42 hours after reverse–transfection using validated small RNA interfering for c-jun, reference or variant reporter constructs were co-transfected in HCT-116. Thirty hours after the transfection, promoter activities were measured. The reporter activity of each construct was compared with that of empty vector (pGL4.11b [luc2]) without small RNA interfering. Significant differences were observed in promoter activity after knockdown of c-jun. Data shown represent mean ± SD from triplicate wells in a representative experiment. *P < 0.05, **P < 0.01 vs. the naïve empty vector promoter activity. (b) Promoter activities were measured 30 hours after the co-transfection of the reference or variant reporters and various amounts of AP-2rep plasmids into HCT-116 cell. The reporter activity of each construct was compared with that of empty vector (pGL4.11b [luc2]). Data shown represent mean ± SD from triplicate wells in a representative experiment. *P < 0.05, **P < 0.01 vs. the naïve promoter activity.
Effect of g.−66T>C variant on MATE1 expression in human kidney samples
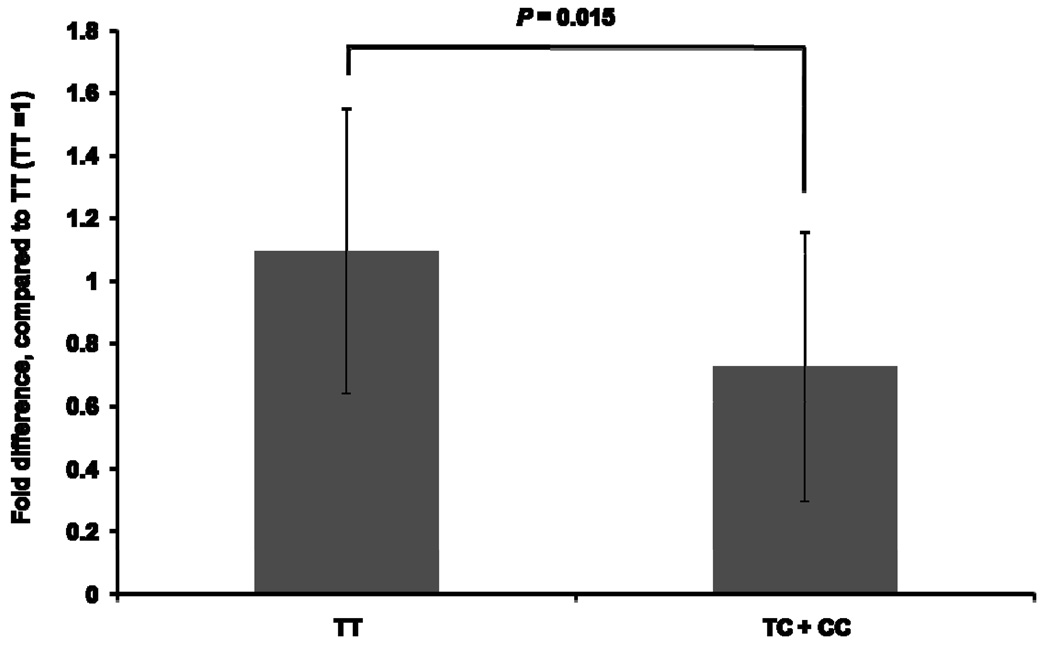
MATE1 expression levels in human kidney samples, determined by a quantitative real-time RT-PCR, was associated with the g.−66T>C genotype. In particular, MATE1 expression in the samples with the T/C (n=21) or C/C (n=5) genotypes were significantly lower (34%), compared to that of samples with the T/T genotype (n=12) (Fig. 6). The MATE1 gene expression in kidney tissue was normally distributed. This was tested based on D’Agostino and Pearson omnibus normality test, which demonstrated that the distribution was not significantly different from normality (P = 0.4734).
Figure 6.
The effect of genotype, g.−66T>C on MATE1 expression levels in human kidney. Expression of MATE1 in the human kidney was determined by a quantitative real-time RT-PCR. The expression levels of MATE1 in the T/C or C/C groups were compared with that in the T/T group. Data shown represent mean ± SD. The P value was calculated using Student’s t test.
Discussion
MATE1 is highly expressed on the apical membrane of the hepatocytes and renal proximal tubule cells, and mediates the transport of a broad array of drugs [4]. Functional changes in the activity or expression level of MATE1 caused by genetic variants in the coding or non-coding region of this transporter could result in changes in the levels of drugs substrates for the transporter. For example, reduction in the expression level of Mate1 in rats has been associated with reduced tubular secretion and high blood levels of the anti-ulcer drug, cimetidine [18].
This study was conducted to identify and functionally characterize genetic variants in the proximal promoter region of MATE1. By screening a large number of samples, we identified five variants that are polymorphic in four major ethnic groups. Recently Kajiwara M, et al [9] described a variant, g.−32G>A, in the MATE1 promoter region that showed markedly decreased promoter activity. Their study indicated that the Sp1 transcription factor has a critical role in the basal promoter activity of MATE1, and that the variant, g.−32G>A reduced the binding affinity of Sp1. Though the allelic frequency of g.−32G>A in the Japanese sample was 3.7%, it was not observed in our study, which included DNA samples from 68 individuals with ancestries in China. Chinese and Japanese share most alleles, but also have alleles that are population specific. Our results suggest that g.−32G>A may be a variant not shared between these two closely related populations.
The most common variant in this study, g.−66T>C showed a significant decrease in promoter activity in reporter assays in four cell lines (Fig. 1) and in mouse liver (Fig. 2). Transcription factor binding site (TFBS) analyses suggested that two transcription factors could bind to the region encompassing g.−66T>C and that the binding affinity of these transcription factors was variant-dependent. In particular, AP-1 would have a higher binding affinity for the g.−66T allele whereas AP-2rep would have a higher binding affinity for the g.−66C allele.
AP-1 is a strong oncogene that mediates tumor cell invasion. The expression of AP-1 is highly elevated in many kinds of tumors such as squamous cell carcinoma, colon carcinoma, breast carcinoma and fibrosarcoma, and it has a critical role in tumor invasion [15]. Previous studies reported that AP-1 induces the transcription of transporter genes including the human glucose transporter 1, the sodium dependent glutamate/aspartate transporter, and the sodium dependent bile acid transporter [19–21]. In some cases, AP-1 may also act as a repressor; it enhances tumor cell invasion by the repression of genes that function as invasion suppressors. AP-1 is thought to bind to the DNA sequence, TGAg/cTCA [15]. We observed that MATE1 contains the sequence, GTACTCA, which is similar to the consensus sequence recognized by AP-1. The variant g.−66T>C results in the sequence, GTACCCA, which is not as good of a match as the reference. Therefore we predicted that AP-1 would have binding preference to the reference DNA sequence as compared to the variant sequence. The results of EMSAs supported our hypothesis. The intensity of the DNA-AP-1 complex was decreased in the presence of g.−66T>C (Fig. 3 (a)). From the competition assay, we observed that g.−66T was a stronger competitor than g.−66C (Fig. 3 (c)), suggesting that g.−66T has a higher affinity for AP-1 than g.−66C. We also demonstrated that AP-1 regulates MATE1 transcription (Fig. 5 (a)), establishing that this transcription factor may be important in regulating the expression of this transporter.
In contrast to AP-1, there are few reports about AP-2rep. AP-2rep belongs to the KLFs family, which are characterized by three highly conserved classical Cys2/His2 zinc fingers at the C-terminus. These motifs enable KLFs to bind to related GC- and CACCC-boxes of DNA. The N-termini of KLFs contain activation or repression domains. To date, a total of 17 KLFs have been identified. AP-2rep, also known as KLF 12, has a repressor domain (PVDLS motif) at its N-terminus [22–24]. Schuierer M, et al [24] reported that the interaction of the repressor domain of AP-2rep with co-repressor, C-terminal binding protein 1 (CtBP1), could result in the repression of the AP-2α gene that is involved in embryonic development, and malignant transformation. AP-2rep is thought to bind to the DNA sequence, CAGTGGG [25]. The MATE1 promoter contains the sequence, CTCACTG, and the complementary sequence, CAGTGAG, is very similar to the consensus sequence of AP-2rep. The variant, g.−66T>C results in CAGTGGG, an exact match to the consensus sequence of AP-2rep. We also found that over-expression of AP-2rep inhibited AP-1 binding to DNA. In particular, AP-2rep had a stronger inhibitory effect on the binding of AP-1 to the g.−66C allele than on the binding of AP-1 to the g.−66T allele (Fig. 4 (c)). We performed competition assays using unlabelled AP-2rep oligonucleotides to confirm that AP-1 and AP-2rep both bind competitively within the g.−66T>C region (Fig. 4 (c)). We also observed that AP-2rep could act as a repressor of MATE1 transcription (Fig. 5 (b)). In the presence of AP-2rep, the luciferase activity was reduced dramatically in the presence of the reference g.−66T (Fig. 5 (b)). However, in the presence of the variant g.−66C, the luciferase activity was reduced significantly only by large amounts of AP-2rep (at 50 ng) (Fig. 5 (b)). This may suggest that AP-2rep binds more avidly to the variant g.−66C.
Finally we examined the effect of g.−66T>C on MATE1 expression in human kidney. MATE1 expression levels in T/C or C/C genotype groups were significantly lower than in the T/T group, which is consistent with our findings in the luciferase in vitro and in vivo assays. T hese data suggest that g.−66T>C can affect the pharmacokinetics or pharmacodynamics of drugs that are transported by MATE1 in the body. However, the g.−66T>C was not associated with lower expression levels in liver samples (data not shown). Although AP-1 is expressed in both kidney and liver, the g.−66T>C associates with lower MATE1 expression in the kidney only [26, 27]. In contrast, no significant association between the g.−66T>C and MATE1 expression levels in the liver was observed. This may be explained by differences in samples and sample collection between liver and kidney. In particular, the liver samples, unlike the kidney samples, consisted of normal, post-mortem tissue. Variation in sample collection times, in particular time from death to sample collection, could have confounded the observed MATE1 expression levels in the liver. Kidney samples, on the other hand, were from surgical resection and collection times and conditions were more standardized. It is also possible that additional hepatic transcription factors (not present in HepG2 cells or in mouse liver under the experimental conditions), e.g., inducible nuclear receptors such as Pregnane X receptor (PXR), which bind to response elements elsewhere in the MATE1 gene may play a role in regulating the transcription rate of MATE1 in the liver, but not in the kidney.
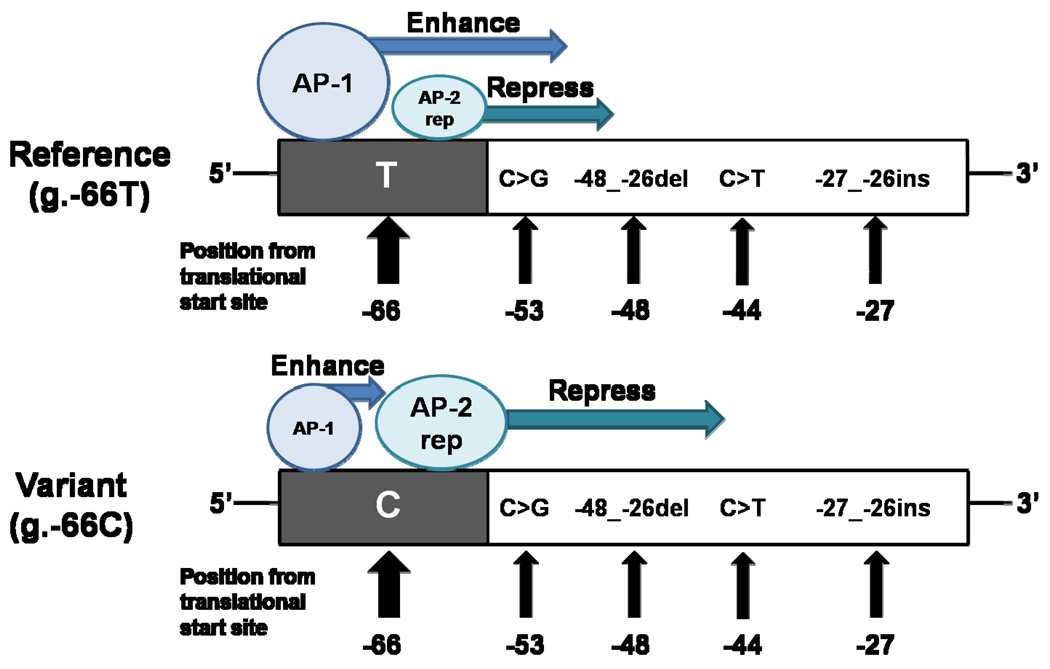
In conclusion, we identified novel polymorphisms in the promoter region of MATE1. One of the polymorphisms, g.−66T>C, present at a very high allele frequency in all major ethnic groups, is associated with a significant reduction in the promoter activity of MATE1 and with lower expression levels of the transporter in the kidney. The mechanism appeared to be related to reduced binding of the transcription factor, AP-1, which acts as an activator of transcription, and to an increased binding of the repressor, AP-2rep, to the region containing the variant g.−66T>C (Fig. 7). Our data suggest that both transcription factors, AP-1 and AP-2rep, are involved in the transcriptional regulation of MATE1. Clinical studies examining the effect of the promoter variant, g.−66T>C, on the pharmacokinetics and pharmacodynamics of MATE1 substrates are ongoing.
Figure 7.
Schematic of the interaction of the MATE1 promoter with two transcription factors that are proposed to be involved in the regulation of MATE1. The upper drawing shows the binding of the transcription factors to the reference allele and the lower drawing shows the binding to g.−66T>C. The transcription factors, AP-1 and AP-2rep, can bind with the site containing the variant, g.−66T>C, competitively. AP-1 works as an enhancer of MATE1 transcription while AP-2rep represses MATE1 transcription. The relative sizes of AP-1 and AP-2rep depict the relative binding affinity with this region; AP-1 has a higher binding affinity for the g.−66T allele whereas AP-2rep has a higher binding affinity for the g.−66C allele. Promoter polymorphisms of MATE1 identified in this study are included in the schematic. Their positions are based upon the translational start site.
Acknowledgements
Data have been deposited in www.pharmgkb.org.
Sponsorship: This work was supported by the National Institute of Health (NIH) grant GM61390 and in part by the Sandler Family Supporting Foundation (N.A.). J. H. Choi was supported by the Korea Research Foundation Grant KRF-2007-357-E00006 funded by the Korean Government (MOEHRD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shitara Y, Horie T, Sugiyama Y. Transporters as a determinant of drug clearance and tissue distribution. Eur J Pharm Sci. 2006;27:425–446. doi: 10.1016/j.ejps.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Koepsell H. Organic cation transporters in intestine, kidney, liver, and brain. Annu Rev Physiol. 1998;60:243–266. doi: 10.1146/annurev.physiol.60.1.243. [DOI] [PubMed] [Google Scholar]
- 3.Otsuka M, Matsumoto T, Morimoto R, Arioka S, Omote H, Moriyama Y. A human transporter protein that mediates the final excretion step for toxic organic cations. Proc Natl Acad Sci U S A. 2005;102:17923–17928. doi: 10.1073/pnas.0506483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanihara Y, Masuda S, Sato T, Katsura T, Ogawa O, Inui K. Substrate specificity of MATE1 and MATE2-K, human multidrug and toxin extrusion/H+-organic cation antiporters. Biochem Pharmacol. 2007;74:359–371. doi: 10.1016/j.bcp.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Zhang S, Sorani M, Giacomini KM. Transport of paraquat by human organic cation transporters and multidrug and toxic compound extrusion family. J Pharmacol Exp Ther. 2007;322:695–700. doi: 10.1124/jpet.107.123554. [DOI] [PubMed] [Google Scholar]
- 6.Dressor MJ, Leabman MK, Giacomini KM. Transporters involved in the elimination of drugs in the kidney: organic anion transporters and organic cation transporters. J Pharm Sci. 2001;90:397–421. doi: 10.1002/1520-6017(200104)90:4<397::aid-jps1000>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 7.Masuda S, Terada T, Yonezawa A, Tanihara Y, Kishimoto K, Katsura T, et al. Identification and functional characterization of a new human kidney-specific H+/organic cation antiporter, kidney-specific multidrug and toxin extrusion 2. J Am Soc Nephrol. 2006;17:2127–2135. doi: 10.1681/ASN.2006030205. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Teranishi K, Li S, Yee SW, Hesselson S, Stryke D, et al. Genetic variants in multidrug and toxic compound extrusion-1, hMATE1, alter transport function. Pharmacogenomics J. 2009;9:127–136. doi: 10.1038/tpj.2008.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajiwara M, Terada T, Asaka J, Ogasawara K, Katsura T, Ogawa O, et al. Critical roles of Sp1 in gene expression of human and rat H+/organic cation antiporter MATE1. Am J Physiol Renal Physiol. 2007;293:F1564–F1570. doi: 10.1152/ajprenal.00322.2007. [DOI] [PubMed] [Google Scholar]
- 10.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 11.Antonarakis SE the nomenclature working group. Recommendations for a nomenclature system for human gene mutations. Hum Mut. 1998;11:1–3. doi: 10.1002/(SICI)1098-1004(1998)11:1<1::AID-HUMU1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 12.den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109:121–124. doi: 10.1007/s004390100505. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Wu Y, He L, Yang Y, Moore SA, Hsu CY. Regulation of cytokine-induced iNOS expression by a hairpin oligonucleotide in murine cerebral endothelial cells. Biochem Biophys Res Commun. 1997;235:394–397. doi: 10.1006/bbrc.1997.6800. [DOI] [PubMed] [Google Scholar]
- 14.Zhang G, Vargo D, Budker V, Armstrong N, Knechtle S, Wolff JA. Expression of naked plasmid DNA injected into the afferent and efferent vessels of rodent and dog livers. Hum Gene Ther. 1997;8:1763–1772. doi: 10.1089/hum.1997.8.15-1763. [DOI] [PubMed] [Google Scholar]
- 15.Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007;26:1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- 16.Imhof A, Schuierer M, Werner O, Moser M, Roth C, Bauer R, et al. Transcriptional regulation of the AP-2α promoter by BTEB-1 and AP-2rep, a novel wt-1/egr-related zinc finger repressor. Mol Cell Biol. 1999;19:194–204. doi: 10.1128/mcb.19.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manna SK, Mukhopadhyay A, Aggarwal BB. INF-α suppresses activation of nuclear transcription factors NF-κB and activator protein 1 and potentiates TNF-induced apoptosis. J Immunol. 2000;165:4927–4934. doi: 10.4049/jimmunol.165.9.4927. [DOI] [PubMed] [Google Scholar]
- 18.Nishihara K, Masuda S, Ji L, Katsura T, Inui K. Pharmacokinetic significance of luminal multidrug and toxin extrusion 1 on chronic renal failure rats. Biochem Pharmacol. 2007;73:1482–1490. doi: 10.1016/j.bcp.2006.12.034. [DOI] [PubMed] [Google Scholar]
- 19.Pfäfflin A, Brodbeck K, Heilig CW, Häring HU, Schleicher ED, Weigert C. Increased glucose uptake and metabolism in mesangial cells overexpressing glucose transporter 1 increases interleukin-6 and vascular endothelial growth factor production: role of AP-1 and HIF-1α. Cell Physiol Biochem. 2006;18:199–210. doi: 10.1159/000097667. [DOI] [PubMed] [Google Scholar]
- 20.Duane WC, Xiong W, Lofgren J. Transactivation of the human apical sodium-dependent bile acid transporter gene by human serum. J Steroid Biochem Mol Biol. 2008;108:137–148. doi: 10.1016/j.jsbmb.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 21.Ramírez-Sotelo G, López-Bayghen E, Hernández-Kelly LC, Arias-Montaño JA, Bernabé A, Ortega A. Regulation of the mouse Na+-dependent glutamate/aspartate transporter GLAST: putative role of an AP-1 DNA binding site. Neurochem Res. 2007;32:73–80. doi: 10.1007/s11064-006-9227-3. [DOI] [PubMed] [Google Scholar]
- 22.Pearson R, Fleetwood J, Eaton S, Crossley M, Bao S. Krüppel-like transcription factors: a functional family. Int J Biochem Cell Biol. 2008;40:1996–2001. doi: 10.1016/j.biocel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 23.Kaczynski J, Cook T, Urrutia R. Sp1- and Krüppel-like transcription factors. Genome Biol. 2003;4:206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuierer M, Hilger-Eversheim K, Dobner T, Bosserhoff AK, Moser M, Turner J, et al. Induction of AP-2α expression by adenoviral infection involves inactivation of the AP-2rep transcriptional corepressor CtBP1. J Biol Chem. 2001;276:27944–27949. doi: 10.1074/jbc.M100070200. [DOI] [PubMed] [Google Scholar]
- 25.Roth C, Schuierer M, Günther K, Buettner R. Genomic structure and DNA binding properties of the human zinc finger transcriptional repressor AP-2rep (KLF12) Genomics. 2000;63:384–390. doi: 10.1006/geno.1999.6084. [DOI] [PubMed] [Google Scholar]
- 26.Woo CW, Siow YL, O K. Homocysteine induces monocyte chemoattractant protein-1 expression in hepatocytes mediated via activator protein-1 activation. J Biol Chem. 2008;283:1282–1292. doi: 10.1074/jbc.M707886200. [DOI] [PubMed] [Google Scholar]
- 27.Le NH, van der Wal A, van der Bent P, Lantinga-van Leeuwen IS, Breuning MH, van Dam H, et al. Increased activity of activator protein-1 transcription factor components ATF2, c-Jun, and c-Fos in human and mouse autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2005;16:2724–2731. doi: 10.1681/ASN.2004110913. [DOI] [PubMed] [Google Scholar]